Abstract
Sensitive and reliable methods for simultaneous determination of polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) in limited volumes of human serum or plasma need to be further documented especially when they accumulate at low levels that are still capable of disrupting endocrine and immune functions, and affecting neurodevelopment and reproduction. The objective of this study was to develop and validate a sensitive and quantitative method that simultaneously quantifies PBDEs and PCBs in 0.5 ml of human serum or plasma. We optimized a solid-phase extraction (SPE) method and used silica particle purification for the extraction of PBDEs and PCBs. Two multiple reactions monitoring (MRM) transitions were optimized for each congeners. The sum of the transitions was used for quantification, and their abundance ratios were used for identification. The combined method optimization techniques resulted in limits of detection from 3–145 pg/ml for 10 PBDEs and 1–12 pg/ml for 15 PCBs. Method was solidly validated by analyzing serum fortified with a certified PBDE and PCB standard mixture from the National Institute of Standards and Technology (NIST). The accuracy was 88–118% and day-to-day precision was within 19%. The method was successfully applied to quantify native concentrations of PBDE and PCB in commercially available human serum. The sensitivity and selectivity of the GC/EI-MS/MS analysis enables it to be the method of choice for investigations of exposures to PBDE and PCB congeners, especially when sample volume is limited.
Keywords: gas chromatography-tandem mass spectrometry, polybrominated diphenyl ethers, polychlorinated biphenyls, human plasma, human serum
1. Introduction
Epidemiological studies show that exposure to specific persistent organic pollutants (POPs) such as polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) are strongly associated with disrupted endocrine [1–5] and immune system functions[6–9], adverse effects on neurodevelopment[10–13], and alterations of reproductive physiology[14–16]. In addition, experiments conducted in animal models and cell cultures indicate that PBDE and PCB burden are associated with obesity [17, 18] neurodevelopment disorders [19, 20] reproductive dysfunction [21, 22] and cardiovascular diseases [23, 24]. However, the potential for adverse effects in infants and children remains unresolved and thus accurate methods of exposure assessment are urgently needed. PBDEs continue to be used as flame retardants in the manufacture of household furnishings, appliances, textiles, paints, and electronics including televisions, cell phones, and computers [25]. PCBs were widely used in heat exchangers and dielectric fluid, as stabilizers in paints, polymers, and adhesives, and as lubricants in various industrial processes until banned in 1977 [26]. There are 209 PBDE and 209 PCB congeners. The degrees of bromination/chlorination and substitution patterns determine chemical stability, planarity and, lipophilicity and biological activity. PCBs and PBDEs degrade very slowly in the environment, biomagnifying up through the food chain [27–29], and accumulate in adipose tissue compartments in the body. Cord blood [30–33], serum or plasma [34–41], breast milk [42–46], placenta [47, 48] and adipose tissues [49–51] have been used as substances to assess the extent of human exposure to PBDEs and PCBs.
Accurate quantization of PBDEs and PCBs in complex biological matrices presents a challenge because of their expected concentrations in the low ng/ml to sub-ng/ml range. Furthermore, lipids, proteins, and inorganic salts present in serum and plasma result in a very complex matrix requiring extensive cleanup procedures prior to analysis [52]. Gas chromatography (GC) coupled to mass spectrometry (MS) with electron impact (EI) or electron capture negative (ECNI) ionization are the most commonly used techniques for PBDE and PCB congener analysis [53]. Utilizing triple quadruple mass spectrometry under positive EI with multiple reaction monitoring (MRM) mode greatly enhances the sensitivity and selectivity of detection compared to selective ion monitoring (SIM) mode. Detection of PCBs in sediments at concentrations of 0.02 to 0.07 ng/g dry weight was reported in Hebei and Hubei Province, China, using a GC-MS/MS triple-quadruple method [54]. PCBs have been detected in human adipose tissue at levels of 0.6–1.8 ng/g under EI MRM positive mode using triple-quadruple GC-MS/MS [50]. Recently, select PBDE congeners were detected in human serum between 0.10 and 0.20 ng/ml utilizing triple-quadruple GC-MS/MS [55]. Although the described GC-MS/MS methods are sensitive and selective, there is increasing demand to accurately measure PBDE and PCB concentrations in limited volumes of serum or plasma, as would be the case for blood samples collected from neonates and infants.
Of the 209 possible PCB congeners, most of the scientific and regulatory attention has been directed toward the so-called dioxin-like PCBs that lack at least two chlorines in the ortho-positions [56, 57]. Recent studies indicate that non-dixon-like PCBs currently predominate in biological and environmental samples, especially the penta- to hepta-chlorinated biphenyls [58]. For example, PCB-153 has accounted for nearly 15% of the total PCB burden determined in human serum, while PCB-138 and PCB-180 contributed 14% and 11%, respectively[58]. PCB-84, -95, -136, -149 and -176 were determined to be the most potent congeners in altering local and global Ca2+ signaling properties with possible subsequent short and long-term consequences on neurodevelopment and neurodegeneration [59, 60]. Amongst PBDEs, tetra- to hexa-brominated diphenyl ethers are the dominant contributors to total human burden [55, 61, 62]. BDE-47, -99, -100, -153, and -154 are reported to account for 90% of the total body burden [63]. Recently, PBDEs with bromine at the 5 and/or 5′ position (such as BDE-49) appear to be present in disproportionally high concentrations in human gestational tissues and blood [64, 65]. The objective of the present study was to develop a sensitive and selective method to simultaneously quantitate multiple PCB and PBDE congeners in very limited volume of samples. Based on current biological research results, 15 PCB congeners (PCB-84, -91, -95, -131, -132, -135, -136, -138, -149, -153, -174, -175, -176, -180, -196) and 10 PBDE congeners (BDE-28, -47, -49, -52, -95, -99, -100, -136, -153, -183) were targeted in this study due to extensive concerns about their high potentials in bioaccumulation and toxicity. After development and validation, the method was applied to analyze native concentrations of human serum purchased from Sigma-Aldrich. To our knowledge, this is the first paper describing a GC/EI-MS/MS method based on triple quadruple mass spectrometry with high sensitivity and selectivity for simultaneous detection of multiple PBDEs and PCBs in low-volume (0.5 ml) of serum samples.
2. Experimental
2.1. Materials
Individual PBDE and PCB analytical reference standards (listed in Table 1) were purchased from AccuStandard Inc. (New Haven, CT, USA). The 13C12-labelled surrogate internal standards, 13C12-BDE-118 and 13C12-PCB-97, were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). Mirex (Sigma-Aldrich Corp, St. Louis, MO, USA) was used as a secondary internal standard to evaluate instrument performance during analysis. Filtered serum spiked with known concentrations of PCB and PBDE congeners (standard serum matrix powder, SRM®1958) was purchased from the National Institute of Standards and Technology (NIST) (Gaithersburg, MD, USA). Human control serum (Lot No.051M0917) used for method development was purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). This serum represents a composite sample of sera obtained from healthy male donors.
Table 1.
Parameters of GC/EI-MS/MS analysis of PBDE and PCB congeners and internal standards
| Initial ID | Rt (min) | RRT* | MW (Da) | MRM 1 (Q1-Q3, m/z) | CE# | MRM 2 (Q1-Q3, m/z) | CE | MRM ½ ratio$ | ||
|---|---|---|---|---|---|---|---|---|---|---|
| PCB-95 | 13.17 | 326.4 | 326 | 291 | 10 | 326 | 256 | 10 | 2.33±0.04 | |
| PCB-91 | 13.32 | 3.02 | 326.4 | 326 | 291 | 10 | 326 | 256 | 10 | 2.08±0.04 |
| PCB-84 | 13.67 | 7.08 | 326.4 | 326 | 291 | 10 | 326 | 256 | 10 | 1.75±0.11 |
| 13C-PCB-97 | 14.19 | 10.38 | 338.4 | 338 | 268 | 10 | 338 | 303 | 10 | 1.34±0.04 |
| PCB-136 | 14.49 | 5.92 | 360.9 | 360 | 325 | 10 | 325 | 290 | 15 | 1.22±0.04 |
| PCB-135 | 14.92 | 8.62 | 360.9 | 360 | 325 | 10 | 325 | 290 | 15 | 1.02±0.03 |
| PCB-149 | 15.09 | 3.46 | 360.9 | 360 | 325 | 10 | 325 | 290 | 15 | 1.06±0.12 |
| BDE-28 | 15.18 | 1.80 | 406.9 | 406 | 246 | 20 | 406 | 248 | 20 | 2.01±0.39 |
| PCB-131 | 15.44 | 5.18 | 360.9 | 360 | 325 | 10 | 325 | 290 | 15 | 1.11±0.37 |
| PCB-153 | 15.63 | 3.78 | 360.9 | 360 | 325 | 15 | 360 | 290 | 20 | 2.86±0.02 |
| PCB-132 | 15.71 | 1.66 | 360.9 | 360 | 325 | 10 | 325 | 290 | 15 | 1.07±0.02 |
| PCB-176 | 16.14 | 8.46 | 395.3 | 394 | 324 | 20 | 394 | 359 | 10 | 5.31±0.25 |
| PCB-138 | 16.27 | 2.55 | 360.9 | 360 | 290 | 28 | 360 | 325 | 12 | 2.00±0.43 |
| PCB-175 | 16.57 | 5.88 | 395.3 | 394 | 324 | 20 | 394 | 359 | 10 | 1.19±0.24 |
| BDE-52 | 16.97 | 8.02 | 485.8 | 326 | 138 | 40 | 486 | 326 | 25 | 1.91±0.38 |
| PCB-174 | 17.16 | 3.74 | 395.3 | 394 | 324 | 20 | 394 | 359 | 10 | 1.23±0.05 |
| BDE-49 | 17.39 | 4.56 | 485.8 | 326 | 138 | 40 | 486 | 326 | 25 | 1.17±0.10 |
| PCB-180 | 17.70 | 6.08 | 395.3 | 394 | 324 | 35 | 394 | 359 | 12 | 2.08±0.61 |
| BDE-47 | 17.84 | 2.75 | 485.8 | 486 | 326 | 25 | 326 | 138 | 40 | 1.21±0.11 |
| Mirex | 18.43 | 11.82 | 545.5 | 272 | 237 | 15 | 270 | 235 | 15 | 1.59±0.01 |
| PCB-196 | 18.71 | 5.68 | 429.8 | 430 | 360 | 20 | 430 | 395 | 15 | 1.27±0.10 |
| BDE-95 | 19.51 | 16.00 | 564.7 | 564 | 404 | 20 | 404 | 137 | 40 | 1.02±0.27 |
| BDE-100 | 19.76 | 5.10 | 564.7 | 564 | 404 | 20 | 404 | 137 | 40 | 3.71±0.11 |
| BDE-99 | 20.28 | 10.38 | 564.7 | 564 | 404 | 20 | 404 | 137 | 40 | 3.02±0.08 |
| 13C-BDE-118 | 20.71 | 8.54 | 576.7 | 576 | 416 | 20 | 576 | 418 | 20 | 2.10±0.07 |
| BDE-136 | 22.39 | 33.66 | 643.6 | 644 | 484 | 20 | 484 | 377 | 40 | 1.72±0.33 |
| BDE-153 | 22.50 | 2.04 | 643.6 | 644 | 484 | 20 | 484 | 377 | 40 | 3.33±0.06 |
| BDE-183 | 24.88 | 47.58 | 801.3 | 562 | 455 | 40 | 722 | 562 | 10 | 1.10±1.13 |
RRt: Retention time resolution, RRt = [Rt(A)-Rt(B)]/[W(A)/2 + W(B)/2], where Rt(A) is the retention time of analyte A, Rt(B) is the retention time of analyte B, W(A)/2 is the peak width of analyte A at its half height, W(B)/2 is the peak width of analyte B at its half height. Baseline separation achieves, when RRT > 1.6.
CE: collision energy to generate product ions.
Data shown is the mean value with standard deviation over the linearity range of each congener.
Analytical grade formic acid, methanol, hexane, isopropanol, and dichloromethane were purchased from Fisher Scientific (Pittsburgh, PA, USA). Ultrapure water (> 18 Ω) was supplied by a Milli-Q system (Millipore, Billerica, MA, USA). Waters Oasis HLB (Oasis, polydivinylbenzene-co-N-vinylpyrrolidone, 200 mg/3 cc) cartridges (Milford, MA, USA) and Phenomenex Strata-X (STX, polymeric-based sorbent, 200 mg/6ml) (Torrance, CA, USA) were used for solid phase extraction (SPE). Sep-pak® Light Silica cartridges (55–105 m) and Sep-Pak® Plus Florisil cartridge (50 -200 m) (Waters, Milford, MA, USA) were serially combined with SPE cartridges for further clean-up.
2.2. Extraction of analytes from serum
Serum samples were stored at -80°C and were thawed on ice overnight before preparation. An aliquot of 0.5 ml serum was removed and placed into disposable glass tubes. Serum samples were spiked with 10 μL of a solution containing 100 ng/ml 13C12-BDE-118 and 13C12-PCB-97. Serum was then mixed with 0.5 ml of pure formic acid and ultrasonicated for 10 minutes. SPE cartridges were gravimetrically conditioned with two aliquots of 3 ml pure methanol and two aliquots of 3 ml ultrapure water with formic acid and methanol (v/v/v, 94.5/0.5/5). Serum and formic acid matrix was applied to the SPE cartridges and filtered gravimetrically. The cartridges were washed with two aliquots of 3 ml water containing formic acid and methanol (v/v/v, 94.5/0.5/5) before being dried under vacuum (-5 mmHg) for five minutes. Disposable Sep-Pak® cartridges were placed underneath the SPE cartridges, and analytes were eluted with three aliquots of 3 ml dichloromethane under vacuum (-10 mmHg) into disposable glass tubes fortified with 100 μL of 1 ng/ml Mirex. Extracts were evaporated to dryness under a gentle nitrogen stream in a water bath (40°C). The residue was reconstituted in 100 μL of isooctane, vibrated for two minutes and then transferred into an auto-sampler vial for GC/EI-MS/MS analysis.
NIST serum powder was reconditioned according to manufacturer’s instructions. Briefly, an aliquot of 10.7 ml of ultrapure water was carefully added to the vial at room temperature (~22°C). Contents were mixed by gentle swirling and then allowed to stand for approximately 30 minutes. The vial was mixed again and allowed to stand for an additional 10 minutes. The vial was never shaken vigorously in order to avoid frothing. Total time for reconstitution was approximately one hour. Reconstituted NIST serum was prepared following the same procedures as described for serum samples above.
2.3. Instrumentation
PBDE and PCB congeners were analyzed using a Bruker Scion TQ triple quadruple mass spectrometer (Bruker, Fremont, CA, USA) equipped with a Bruker 451 GC and CP 8400 autosampler and series split/splitless injector set at 250°C. GC separation was performed by a 30 m BR-5MS, 0.25 mm i.d. column with 0.25 μm film thickness (Bruker, Fremont, CA, USA). The GC oven temperature was started at 90°C and held at that temperature for one minute. Temperature was then increased to 150°C at a rate of 50°C/min and held for one minute. Finally, oven temperature was increased to 310°C at a rate of 8°C/min and held for three minutes. The flow rate of the carrier gas, helium, was set at 1.0 ml/min. Source temperature was set at 250°C and transfer line temperature at 280°C. PBDE/PCB concentrations were determined in multiple reactions monitoring (MRM) mode. MS/MS was operated in EI positive mode at 70 eV. An aliquot of 2 μL sample was injected by pulsed splitless method (40 psi, 0.2 min).
A unique new compound based scanning (CBS) workflow for setting up the multi-residue MRM method was employed with the Bruker MS workstation software. After several initial runs to identify the retention time window and MS/MS transitions for each compound, the optimal scan time and dwell time for each MRM was calculated based on the average peak width and automatically set by the software based on consideration of all overlapping retention time windows. Thus, there was no time segment in the method. Two MRM transitions were set up for each analyte. The CBS workflow allows MRM information for each compound from the acquisition mode to pass directly into the data processing method. Therefore, a separate data processing method was not needed. However, the area under each chromatographic peak was integrated for further quantization.
2.4. Method validation
Lower limit of detection and quantification
The lower limit of detection (LOD) was determined according to the EPA Regulation 40 CFR part 136 method [66]. Seven replicates of human serum spiked with a mixture of target analytes at final concentrations of 0.02 ng/ml each were processed through the entire extraction procedure and analyzed. Standard deviation of 7 replicates multiplied by the Student’s t-value of 3.143 (99% confidence level for 7 replicates) was used to estimate the method detection limit. The lower limit of quantification (LOQ) was calculated as 10 times the standard deviation of the result for seven replicates of an individual compound.
Linearity
The linearity of the method was examined by plotting the peak area ratio (analytical/surrogate internal standard) of each PBDE and PCB congener against the concentration added to the control serum. Calibration samples were prepared by adding a specific amount of analytical standard in solvent to 0.5 ml of quality control serum to obtain ten calibration samples with known concentrations of selected PBDEs and PCBs (0.04, 0.1, 0.2, 0.8, 2, 4, 10, 20, 50, 100 ng/ml, based on serum volume). The correlation between concentration and peak area ratio (analytical/surrogate) were examined using least-squared linear regression. Weighting factors of calibration curve was selected from 1, 1/x1/2, 1/x, 1/x2, 1/y1/2, 1/y, and 1/y2 based on the respective sums of the relative errors, which represented the differences between calculated concentrations and theoretical amount. Weighting factor was chosen to be the one produced the least sum of relative errors. Correlation coefficient was used to assess linearity. All PBDE and PCB concentrations are expressed in ng/ml serum.
Accuracy and precision
Accuracy and precision were assessed using the certified NIST reference standard serum (SRM®1958). An aliquot of 0.5 ml NIST reconstituted serum was spiked with surrogate internal standards (1 ng, 13C12-PCB-97 and 13C12-BDE-118) and prepared as described in 2.2. The ratios of determined PCB and PBDE levels to the theoretical concentration listed in the NIST serum certificate were reported to assess the accuracy. The residual standard deviations (RSD) of six replicate analyses of the same sample were used to determine precision. In order to determine run-to-run and day-to-day precisions, one NIST human serum sample was independently prepared and analyzed in six replicates each day for three days. 2.5. Method application The native concentrations of targeted PBDE and PCB congeners in the unspiked control human serum were evaluated by proposed method. Control serum (purchased from Sigma-Aldrich) was prepared in six replicates to determine the native concentrations of PBDE and PCB congeners.
3. Results and discussion
3.1. GC separation of analytes
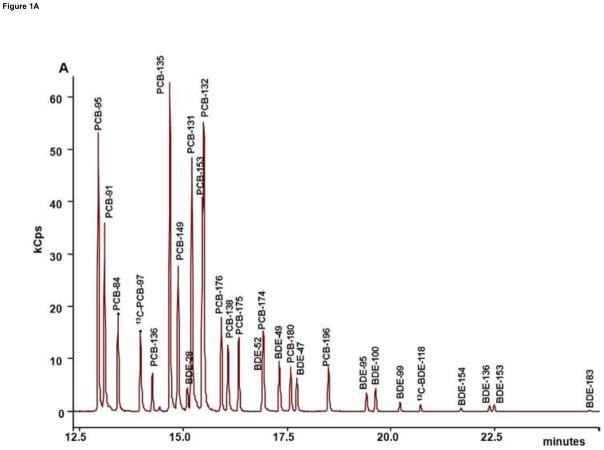
The total ion current (TIC) chromatograms of control serum spiked by 10 μL of 20 ng/ml of 10 PBDE and 15 PCB congeners were shown in Figure 1A. Congeners were eluted between 13 and 25 minutes. With the high selectivity of optimized transitions, co-elution was eliminated for all congeners. Retention time resolution (RRT) is defined as the ratio of differences between two adjacent chromatographic peaks and the sum of their half-height peak width. Baseline separation is achieved when RRT > 1.6. The analytes were all baseline separated, PCB-132 following PCB-153, and BDE-28 following PCB-149 showing the smallest RRTs of 1.66 and 1.80, respectively (Table 1). In general, PCBs were eluted before PBDEs due to their higher volatility. Signal abundances of PCBs were generally higher than that of PBDEs at the same concentration.
Figure 1.
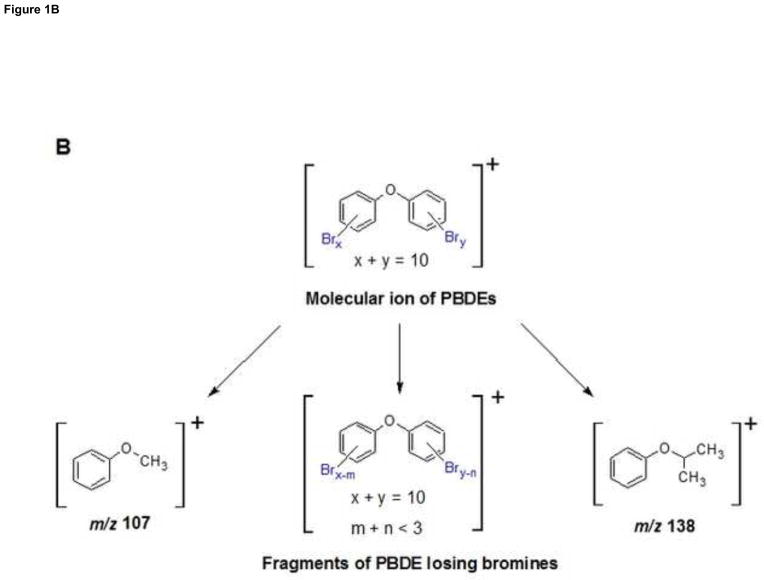
GC/EI-MS/MS analysis of PBDEs and PCBs. (A) TIC Chromatogram of PBDE and PCB congeners spiked of 10 ng into 0.5 ml of control serum followed by SPE and silica clean-up. (B) Proposed fragmentation patterns of PBDE congeners.
3.2. Mass spectra of analytes
Mass spectra give characteristics of isotope patterns according to mass and natural abundance. Bromine has two isotopes, 79 Br at 78.9183 and 81Br at 80.9163; the natural abundance of these two isotopes is 50.5 and 49.5%, respectively. Thus, the mass spectrum will show a cluster of ions differing by 2 atomic mass units with one or two most abundant peaks depending on the even or odd number of bromine atoms. For example, tetra-bromodiphenyl ether has an exact mass of 481.71508 with an isotope cluster of five peaks separated by 2 mass units and a single most abundant peak at 486 amu, which represents the molecular ion ([M]+) under EI source. On the other hand, chlorine also has two isotopes, 35Cl at 34.9689 with a natural abundance of 75.77%, and 37Clat 36.9659 with a natural abundance of 24.23%. Since the natural abundance ratio of the two chlorine isotopes is approximately 3:1, the mass spectrum of PCBs will be an isotope cluster of ions separated by 2 mass units centered by a single most abundant peak independent of the number of chlorine atoms. By convention, we will refer to ions by the nominal mass of the highest intensity peak; the calculation of nominal masses is based on the masses of all atoms rounded to the nearest integer.
The fragmentation patterns of PBDEs and PCBs under EI source with positive detection were characterized by the presence of intense molecular cluster ions, and fragment ions corresponding to successive loses of bromine or chlorine atoms from the molecular ions, respectively (Fig. 1B). Although two transitions were optimized for each analyte, the signal intensities of these two transitions were not equally abundant (refer to the MRM 1/2 ratios in Table 1). For all analyzed PBDE congeners except BDE-183, the EI spectra of precursor ions were dominated by the molecular ions. The most abundant fragment ions were [M-2Br]+ ions. For BDE-183, the most abundant transition was from [M-3Br]+ to ion [M-3Br-107]+. Congeners BDE-49 and BDE-52 generated [M-2Br]+ to ion m/z 138 as the most abundant transitions, as shown in Figure 1B. It is not possible to identify the substitution patterns of bromines on aromatic rings. For PCB congeners, the EI spectra of precursor ions were dominated by the molecular ions. The most abundant transitions of PCB-84, -91, -95, -131, -132, -135, -136, -149, and -153 resulted in fragment ion [M-Cl]+. For PCB-138, -174, -175, -176, -180 and -196, the most abundant fragment ion was [M-2Cl]+ generated from the molecular ions. Therefore, the most intensive ion peak of the molecular cluster ions of each homologue group were selected as the precursor ion, while the most dominant fragment ion resulting from the various losses of bromine or chlorine, respectively, was selected for the most abundant transition. For most PBDE and PCB congeners, a second transition from an additional loss of bromine or chlorine, was observed.
The fragmentation patterns of PBDE and PCB congeners are directly influenced by the ionization of the EI source. In the EI process, the analyte of interest is vaporized into the mass spectrometer ion source, where it is impacted by a beam of electrons with sufficient energy to ionize the molecule. Every electron in an organic molecule is paired with an electron of opposite spin. Therefore, when a molecular ion is generated by removing one electron, an unpaired electron will de-stabilize the molecule, which subsequent fragmentation. Since the bromine and chloride are electrophilic atoms, their existence on biphenyl rings is more likely to generate positive fragment ions by the successive losses of bromine or chlorine along with electron. Our analysis showed that the major fragment ions of PBDE and PCB congeners resulted from losses of various amounts of bromine and chlorine atoms under the positive detection of EI source.
Since we optimized two transitions for each congener, the ratios of the transitions were obtained by dividing the integrated peak area of the most dominant transition with that of the second major transition. Transition ratios over the entire calibration range from 0.04 to 100 ng/ml are listed in Table 1. The relative standard deviation (RSD) of each transition ratio is less than 20%, which suggests that the ionization efficiency of the EI source was relatively consistent at various concentrations of each congener. Our data shows that the transition ratios of each congener can be used to identify a specific congener. In this paper, to further enrich the signal abundance, two transitions were added up for quantification.
3.3. Sample preparation
The selection of appropriate SPE cartridges with different sorbent materials played a key role in the achievement of high and reproducible recovery for our congeners. The most commonly used sorbents are porous silica particles surface-bonded with C18 or other hydrophobic alkyl groups, and polymeric sorbents, such as styrene-divinylbenzene, and activated carbon to further remove water [52]. The purification of PBDE and PCB congeners from human serum was accomplished by denaturation, solid phase extraction and clean-up procedures. During the method development stage, the purification efficiency of each step was optimized one by one (Table 2). Mixed PBDE and PCB standards, containing 5 ng of each compound, were spiked into 0.5 ml of control serum and allowed to sit for 15 minutes. Formic acid and isopropanol were tested as denaturation solvents before SPE. The results showed that recoveries of 82.4–97.5% and 72.5–89.4% were obtained for formic acid and isopropanol, respectively. Two types of SEP cartridges, Oasis HLB and Strata-X, were evaluated and gave recoveries of 74.3–88.5% and 50.0–63.6%, respectively. Subsequently, dichloromethane (DCM) with an eluting efficiency of 85.6–99.7% was chosen over iso-hexane with an efficiency of 56.6–87.8%. Finally, silica and Florisil particles were examined separately for their ability to further clean up the human serum matrix, and resulted in recoveries of 76.9–93.4% and 67.6–78.6%, respectively. In order to maximize PBDE/PCB congener recovery from serum, formic acid for denaturation, Oasis HLB as SPE cartridges hyphenated with Sep-pak® Silica cartridges, and DCM as the eluting solvent were selected to extract and purify low-volume (only 0.5 ml) human serum samples.
Table 2.
Effects of extraction procedures on the recoveries (%) of PCB and PBDE congeners in human serum
| Recovery (%) | Denaturation | SPE cartridge | Eluting solution | Clean-up | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Formic acid | Iso-proponal | Oasis HLB | Strata-X | hexane | DCM | Silica | Florisil | |
| PCB-84 | 82.4 | 78.3 | 80.7 | 53.3 | 56.6 | 96.9 | 86.5 | 76.1 |
| PCB-91 | 83.2 | 77.1 | 76.7 | 53.6 | 71.9 | 98.7 | 83.4 | 74.4 |
| PCB-95 | 85.6 | 76.3 | 84.4 | 53.9 | 82.2 | 97.6 | 80.3 | 72.3 |
| 13C-PCB-97 | 86.1 | 75.9 | 84.2 | 54.2 | 75.3 | 93.1 | 81.2 | 73.1 |
| PCB-131 | 83.7 | 75.1 | 77.2 | 53.2 | 77.8 | 96.5 | 79.9 | 67.6 |
| PCB-132 | 90.3 | 72.5 | 80.5 | 52.5 | 61.6 | 95.1 | 78.6 | 69.6 |
| PCB-135 | 88.4 | 76.8 | 74.3 | 50.0 | 84.8 | 99.7 | 82.1 | 75.1 |
| PCB-136 | 93.6 | 79.8 | 82.5 | 51.3 | 82.1 | 90.9 | 84.7 | 71.7 |
| PCB-149 | 86.7 | 77.9 | 78.2 | 52.6 | 76.4 | 92.9 | 85.2 | 74.3 |
| PCB-153 | 84.2 | 78.7 | 82.3 | 53.1 | 82.8 | 96.1 | 79.1 | 68.1 |
| PCB-174 | 90.4 | 81.9 | 78.2 | 52.3 | 83.1 | 97.7 | 90.6 | 78.6 |
| PCB-175 | 95.3 | 85.4 | 80.9 | 50.5 | 78.4 | 95.6 | 89.9 | 77.9 |
| PCB-176 | 92.2 | 87.2 | 79.3 | 52.3 | 80.8 | 97.5 | 93.4 | 73.4 |
| PCB-196 | 97.5 | 86.4 | 78.6 | 51.7 | 84.1 | 98.1 | 91.7 | 71.6 |
| BDE-28 | 96.5 | 78.1 | 81.7 | 63.6 | 75.9 | 96.7 | 80.4 | 73.4 |
| BDE-47 | 88.1 | 74.5 | 77.7 | 59.9 | 78.2 | 94.6 | 86.3 | 70.3 |
| BDE-49 | 85.7 | 75.8 | 82.4 | 56.2 | 81.3 | 95.1 | 87.2 | 70.1 |
| BDE-52 | 91.3 | 73.8 | 86.2 | 58.2 | 87.8 | 96.0 | 76.9 | 77.6 |
| BDE-95 | 85.4 | 75.9 | 75.2 | 55.5 | 71.6 | 95.7 | 79.6 | 75.6 |
| BDE-99 | 90.6 | 79.7 | 88.5 | 56.0 | 81.8 | 92.7 | 80.1 | 72.1 |
| BDE-100 | 87.7 | 80.9 | 84.3 | 54.3 | 78.1 | 93.9 | 78.7 | 76.7 |
| 13C-BDE118 | 83.2 | 87.4 | 86.5 | 57.6 | 86.4 | 90.6 | 80.2 | 70.3 |
| BDE-136 | 91.4 | 84.2 | 75.2 | 55.1 | 77.8 | 86.1 | 79.8 | 68.3 |
| BDE-153 | 96.3 | 89.4 | 83.3 | 51.3 | 80.1 | 90.7 | 85.6 | 72.6 |
| BDE-183 | 90.5 | 80.4 | 76.2 | 53.9 | 78.2 | 85.6 | 81.9 | 75.9 |
3.4. Validation of the GC/EI-MS/MS method
Lower limits of quantification and detection
As shown in Table 3, LOD varied from 0.003 to 0.036 ng/ml for PBDEs, except BDE-118 which is 0.145 ng/ml, and from 0.001 to 0.012 ng/ml for PCBs. When comparing our LODs to methods using ion trap mass spectrometry for analysis of human serum or tissues, for PBDEs and PCBs, our LODs conducted by triple quadruple tandem mass spectrometry were up to 10 times lower than previously reported LODs values ranging from 0.07–1.3 ng/ml for PBDEs [67], and 0.05–0.13 ng/ml for PCBs [68, 69].
Table 3.
Lower limits of detection (LOD) and quantification (LOQ), and linearity of GC-MS/MS method for the analysis of PBDE and PCB congeners.
| Initial ID | LOD (ng/ml) | LOQ (ng/ml) | LOQ (ng/ml) published methods | Linearity
|
||
|---|---|---|---|---|---|---|
| Range (ng/ml) | Regression Weight | Correlation coefficient (r) | ||||
| PCB-95 | 0.001 | 0.002 | N.A. | 0.04–100 | 1/x2 | 0.7865 |
| PCB-91 | 0.001 | 0.005 | N.A. | 0.04–100 | 1/x2 | 0.7871 |
| PCB-84 | 0.002 | 0.007 | N.A. | 0.04–100 | 1/x2 | 0.5940 |
| 13C-PCB-97 | 0.002 | 0.006 | N.A. | 2 | - | - |
| PCB-136 | 0.007 | 0.022 | 0.25b | 0.04–100 | 1/x2 | 0.9874 |
| PCB-135 | 0.004 | 0.014 | N.A. | 0.04–100 | 1/x2 | 0.9866 |
| PCB-149 | 0.006 | 0.019 | N.A. | 0.04–100 | 1/x2 | 0.9858 |
| BDE-28 | 0.016 | 0.050 | 0.10a | 0.04–100 | 1/x2 | 0.9511 |
| PCB-131 | 0.005 | 0.015 | N.A. | 0.04–100 | 1/x2 | 0.9941 |
| PCB-153 | 0.005 | 0.017 | N.A. | 0.04–100 | 1/x2 | 0.8556 |
| PCB-132 | 0.010 | 0.033 | N.A. | 0.04–100 | 1/x2 | 0.9399 |
| PCB-176 | 0.002 | 0.007 | N.A. | 0.04–100 | 1/x | 0.9906 |
| PCB-138 | 0.014 | 0.044 | N.A. | 0.04–100 | 1/x2 | 0.9479 |
| PCB-175 | 0.003 | 0.010 | N.A. | 0.04–100 | 1/x | 0.9939 |
| BDE-52 | 0.011 | 0.036 | N.A.a | 0.04–100 | 1/x2 | 0.9251 |
| PCB-174 | 0.007 | 0.021 | N.A. | 0.04–100 | 1/x2 | 0.9942 |
| BDE-49 | 0.003 | 0.010 | N.A.a | 0.04–100 | 1/x2 | 0.9449 |
| PCB-180 | 0.012 | 0.040 | N.A. | 0.04–100 | 1/x | 0.9342 |
| BDE-47 | 0.003 | 0.011 | 0.20a | 0.04–100 | 1/x2 | 0.9942 |
| Mirex | 0.003 | 0.010 | N.A. | 1 | - | - |
| PCB-196 | 0.003 | 0.011 | N.A. | 0.04–100 | 1/x | 0.9929 |
| BDE-95 | 0.023 | 0.073 | N.A.a | 0.04–100 | 1/x | 0.9932 |
| BDE-100 | 0.036 | 0.115 | 0.14a | 0.04–100 | 1/x | 0.9913 |
| BDE-99 | 0.032 | 0.101 | 0.16a | 0.04–100 | 1/x2 | 0.9940 |
| 13C-PBDE-118 | 0.019 | 0.062 | N.A.a | 2 | - | - |
| BDE-136 | 0.026 | 0.083 | N.A.a | 0.04–100 | 1/x | 0.9817 |
| BDE-153 | 0.029 | 0.092 | 0.14a | 0.04–100 | 1/x | 0.9818 |
| BDE-183 | 0.145 | 0.461 | 0.17a | 0.04–100 | 1/x | 0.8556 |
Linearity
Good linearity was obtained in a concentration range of 0.04–100 ppb (Table 3), and the correlation coefficients for each compound presented. The concentration range in biological methods is required to be dynamic and broad, in order to monitor concentrations effectively. When the range of data values is large, it might be expected that the variance of each data point might be quite different. A proper weighting factor is needed for data to adequately fit the linear model [70]. In this study, 1/x2 and 1/x were the best weighting factors producing the smallest relative error after regression. That means the most random distribution of data happens at the lower end of calibration curve. The broad range of linearity demonstrated that the method is suitable for simultaneous determination of PBDEs and PCBs in low volume of human specimens.
Accuracy and precision
Accuracy and precision of the method were determined by the use of NIST certified reference serum (Table 4). Accuracy ranged from 88 to 118% and run-to-run and day-to-day precisions were between 4% and 10%, and 12% and 19%, respectively (Table 4). The results demonstrated that PBDEs and PCBs can be extracted and simultaneously determined by the proposed method with good accuracy and precision. The combined optimized samples preparation and GC/EI-MS/MS method developed in this study can therefore be applied to plasma or serum samples for accurate quantification of the target PBDE and PCB concentrations at concentrations as low as pg/ml.
Table 4.
Accuracy and precision results of NIST certified reference standard
| Initial ID | Theoretical Value (ng/ml) | Determined concentration (ng/ml) | Accuracy (%) | Precision (RSD %)
|
|
|---|---|---|---|---|---|
| Run-to-run | Day-to-day | ||||
| PCB-138 | 0.3790 | 0.3520 | 93 | 6 | 13 |
| PCB-149 | 0.3032 | 0.3307 | 109 | 5 | 16 |
| PCB-153 | 0.3744 | 0.3305 | 88 | 7 | 16 |
| PCB-180 | 0.3760 | 0.3766 | 100 | 8 | 15 |
| BDE-28 | 0.3760 | 0.4440 | 118 | 6 | 17 |
| BDE-47 | 0.5288 | 0.5520 | 104 | 4 | 12 |
| BDE-99 | 0.3992 | 0.3900 | 98 | 4 | 13 |
| BDE-100 | 0.3856 | 0.3585 | 93 | 7 | 16 |
| BDE-153 | 0.3680 | 0.3293 | 88 | 9 | 17 |
| BDE-183 | 0.3688 | 0.3384 | 92 | 10 | 19 |
3.5. Application of optimized GC/EI-MS/MS method
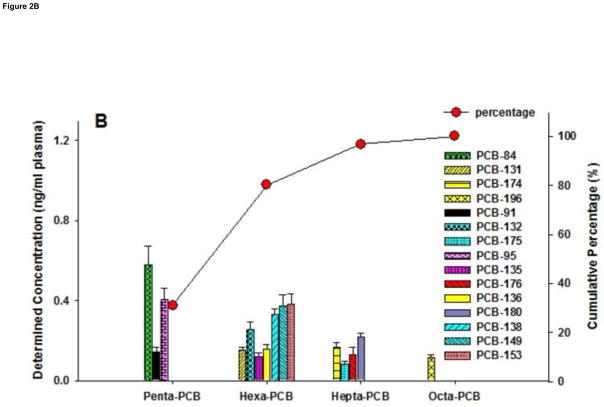
The native concentrations of commercially available control human serum, purchased from Sigma-Aldrich, are shown in Figure 2. According to the certificate of analysis of Sigma-Aldrich, the control serum is a composite sample from male donors residing in the United Stations. For PBDEs, BDE-47, -99, -28, and -100 were the dominant congeners identified in control serum. Native concentrations of BDE-136 and BDE-183 in control serum were determined to be 0.004 and 0.028 ng/ml, respectively, which were below their corresponding LODs (0.026 and 0.145 ng/ml) when extracted from 0.5 ml volume of samples. Three targeted tetra-BDEs and three targeted penta-BDEs accounted for 53.8% and 28.0% of the total burden of measured PBDEs respectively. Meanwhile, PCB-84, -95, -153, -149, and --138 were the major contributors of PCB congeners in control serum. Native concentrations of all targeted PCB congeners were above their LODs. Six hexa-PCBs and three penta-PCBs contributed 49.1% and 31.1% to the total body burden of measured 15 PCB congeners. Generally, among the analyzed 15 PCB congeners, less chlorinated congeners were the major contributors.
Figure 2.
Native concentrations (ng/ml, based on sample volume) of (A) PBDEs and (B) PCBs in commercially available human control serum determined by GC/EI-MS/MS method. Data shown as mean +/− standard deviation (n=6). Scatter and line (-○-) present the cumulative percentages of each homologue group of PBDEs/PCBs of all targeted congeners. Line (—) illustrates the lower limit of detection (LOD).
The application example demonstrated that the proposed GC/EI-MS/MS method is sensitive to simultaneously evaluate bioaccumulation of targeted PBDEs and PCBs in human serum or plasma limited to 0.5 ml sample volume.
4. Conclusion
We developed a highly sensitive and selective GC/EI-MS/MS method and combined it with specific SPE preparation to simultaneously determine PBDE and PCB congener concentrations in low-volumes of human plasma or serum. This method is a significant improvement over existing methodologies for that it provides a rapid and unequivocal determination of organic pollutants in biological fluids and it is validated to be capable of pg/ml concentrations. The major advantage of this method is the greatly increased instrument sensitivity and selectivity utilizing tandem mass spectrometry and the proven effective sample pretreatment process limited to only 0.5 ml of human serum or plasma. The accuracy and precision of the method was validated with NIST standard serum and obtained excellent recovery and repeatability. The method was successfully used to measure PBDE and PCB congeners in commercially available human serum. The sensitivity and selectivity of the GC/EI-MS/MS analysis enables it to be the method of choice for investigations of exposures to PBDE and PCB congeners.
Highlights.
Simultaneous determination of various classes of persistent organic pollutants in small volume human plasma.
Simple and efficient extraction based on solid phase extraction and silica clean-up.
Two transitions were used for quantification and qualification using GC/ESI-MS/MS.
LOQs and LODs are lower than previously published methods for PBDE and PCB analyses.
A sensitive and validated GC/ESI-MS/MS method to assess human exposure to PBDE and PCB congeners.
Acknowledgments
The authors would like to express their gratitude to Dr. Kefei Wang for his permission to use the facilities at the Bruker Daltonics Inc. Demonstration Laboratory in Fremont, California, USA. We also would like to thank Dr. Gwen Lim and Dr. Keyu Zhou for their technical assistance during this research. This study is supported by 1R01ES020392, P42ES04699 and P01 ES011269, US EPA STAR R833292 and R829388, and an unrestricted gift grant from the JB Johnson Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibhazehiebo K, Iwasaki T, Kimura-Kuroda J, Miyazaki W, Shimokawa N, Koibuchi N. Environ Health Perspect. 2011;119:168–175. doi: 10.1289/ehp.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang-Peronard JL, Andersen HR, Jensen TK, Heitmann BL. Obes Rev. 2011;12:622–636. doi: 10.1111/j.1467-789X.2011.00871.x. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka T, Morita A, Kato M, Hirai T, Mizoue T, Terauchi Y, Watanabe S, Noda M. Endocr J. 2011;58:589–596. doi: 10.1507/endocrj.k10e-361. [DOI] [PubMed] [Google Scholar]
- 4.Silverstone AE, Rosenbaum PF, Weinstock RS, Bartell SM, Foushee HR, Shelton C, Pavuk M. Environ Health Perspect. 2012;120:727–732. doi: 10.1289/ehp.1104247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valvi D, Mendez MA, Martinez D, Grimalt JO, Torrent M, Sunyer J, Vrijheid M. Environ Health Perspect. 2012;120:451–457. doi: 10.1289/ehp.1103862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer S, Hikel SM, Adams K, Hinds D, Moon K. Environ Health Perspect. 2012;120:1067–1075. doi: 10.1289/ehp.1104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jusko TA, Sonneborn D, Palkovicova L, Kocan A, Drobna B, Trnovec T, Hertz-Picciotto I. Environ Health Perspect. 2012;120:595–600. doi: 10.1289/ehp.1104229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MJ, Pelloux V, Guyot E, Tordjman J, Bui LC, Chevallier A, Forest C, Benelli C, Clement K, Barouki R. Environ Health Perspect. 2012;120:508–514. doi: 10.1289/ehp.1104282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett JR. Environ Health Perspect. 2010;118:A445. doi: 10.1289/ehp.118-a445a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman K, Adgent M, Goldman BD, Sjodin A, Daniels JL. Environ Health Perspect. 2012;120:1438–1442. doi: 10.1289/ehp.1205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gascon M, Fort M, Martinez D, Carsin AE, Forns J, Grimalt JO, Santa Marina L, Lertxundi N, Sunyer J, Vrijheid M. Environ Health Perspect. 2012;120:1760–1765. doi: 10.1289/ehp.1205266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskenazi B, Chevrier J, Rauch SA, Kogut K, Harley KG, Johnson C, Trujillo C, Sjodin A, Bradman A. Environ Health Perspect. 2012 [Google Scholar]
- 13.Sagiv SK, Thurston SW, Bellinger DC, Altshul LM, Korrick SA. Environ Health Perspect. 2012;120:904–909. doi: 10.1289/ehp.1104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buck Louis GM, Sundaram R, Schisterman EF, Sweeney AM, Lynch CD, Gore-Langton RE, Maisog J, Kim S, Chen Z, Barr DB. Environ Health Perspect. 2012 doi: 10.1289/ehp.1205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuliffe ME, Williams PL, Korrick SA, Altshul LM, Perry MJ. Environ Health Perspect. 2012;120:535–540. doi: 10.1289/ehp.1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelouahab N, Ainmelk Y, Takser L. Reprod Toxicol. 2011;31:546–550. doi: 10.1016/j.reprotox.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Environ Health Perspect. 2013;121:105–110. doi: 10.1289/ehp.1205421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyche JL, Nourizadeh-Lillabadi R, Almaas C, Stavik B, Berg V, Skare JU, Alestrom P, Ropstad E. J Toxicol Env Heal A. 2010;73:1032–1057. doi: 10.1080/15287394.2010.481618. [DOI] [PubMed] [Google Scholar]
- 19.Wayman GA, Bose DD, Yang D, Lesiak A, Bruun D, Impey S, Ledoux V, Pessah IN, Lein PJ. Environ Health Perspect. 2012;120:1003–1009. doi: 10.1289/ehp.1104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wayman GA, Yang D, Bose DD, Lesiak A, Ledoux V, Bruun D, Pessah IN, Lein PJ. Environ Health Perspect. 2012;120:997–1002. doi: 10.1289/ehp.1104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li ZH, Liu XY, Wang N, Chen JS, Chen YH, Huang JT, Su CH, Xie F, Yu B, Chen DJ. Environ Health Perspect. 2012;120:541–546. doi: 10.1289/ehp.1104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodavanti PRS, Coburn CG, Moser VC, MacPhail RC, Fenton SE, Stoker TE, Rayner JL, Kannan K, Birnbaum LS. Toxicol Sci. 2010;116:297–312. doi: 10.1093/toxsci/kfq105. [DOI] [PubMed] [Google Scholar]
- 23.Lind PM, Orberg J, Edlund UB, Sjoblom L, Lind L. Toxicol Lett. 2004;150:293–299. doi: 10.1016/j.toxlet.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Arsenescu V, Arsenescu RI, King V, Swanson H, Cassis LA. Environ Health Perspect. 2008;116:761–768. doi: 10.1289/ehp.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frederiksen M, Vorkamp K, Thomsen M, Knudsen LE. Int J Hyg Environ Health. 2009;212:109–134. doi: 10.1016/j.ijheh.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Brinkman UAT, DeKok A. In: Halogenated Biphenyls, Terphenyls, Naphthalenes, Dibenzodioxins and Related Products. Kimbrough RD, editor. Elsevier/North Holland; Amsterdam-New York-Oxford: 1980. pp. 1–40. [Google Scholar]
- 27.Tian SY, Zhu LY, Liu M. Environ Toxicol Chem. 2010;29:2278–2285. doi: 10.1002/etc.275. [DOI] [PubMed] [Google Scholar]
- 28.Yu YX, Huang NB, Zhang XY, Li JL, Yu ZQ, Han SY, Lu M, Van de Wiele T, Wu MH, Sheng GY, Fu JM. Chemosphere. 2011 doi: 10.1016/j.chemosphere.2010.12.049. [DOI] [PubMed] [Google Scholar]
- 29.Bruckmann P, Hiester E, Klees M, Radermacher L. Gefahrstoffe Reinhaltung Der Luft. 2011;71:151–158. [Google Scholar]
- 30.Vizcaino E, Grimalt JO, Lopez-Espinosa MJ, Llop S, Rebagliato M, Ballester F. Environ Int. 2011;37:152–157. doi: 10.1016/j.envint.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Vizcaino E, Grimalt JO, Carrizo D, Lopez-Espinosa MJ, Llop S, Rebagliato M, Ballester F, Torrent M, Sunyer J. J Environ Monit. 2011;13:422–432. doi: 10.1039/c0em00483a. [DOI] [PubMed] [Google Scholar]
- 32.Vrijheid M, Martinez D, Aguilera I, Ballester F, Basterrechea M, Esplugues A, Guxens M, Larranaga M, Lertxundi A, Mendez M, Murcia M, Marina LS, Villanueva CM, Sunyer J. J Epidemiol Community Health. 2012;66:106–113. doi: 10.1136/jech.2010.117408. [DOI] [PubMed] [Google Scholar]
- 33.Govarts E, Nieuwenhuijsen M, Schoeters G, Ballester F, Bloemen K, de Boer M, Chevrier C, Eggesbo M, Guxens M, Kramer U, Legler J, Martinez D, Palkovicova L, Patelarou E, Ranft U, Rautio A, Petersen MS, Slama R, Stigum H, Toft G, Trnovec T, Vandentorren S, Weihe P, Kuperus NW, Wilhelm M, Wittsiepe J, Bonde JP. Environ Health Perspect. 2011 doi: 10.1289/ehp.1103767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stapleton HM, Eagle S, Anthopolos R, Wolkin A, Miranda ML. Environ Health Perspect. 2011;119:1454–1459. doi: 10.1289/ehp.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodruff TJ, Zota AR, Schwartz JM. Environ Health Perspect. 2011;119:878–885. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hertz-Picciotto I, Bergman A, Fangstrom B, Rose M, Krakowiak P, Pessah I, Hansen R, Bennett DH. Environ Health. 2011;10:1. doi: 10.1186/1476-069X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallagher RP, MacArthur AC, Lee TK, Weber JP, Leblanc A, Elwood JM, Borugian M, Abanto Z, Spinelli JJ. Int J Cancer. 2011;128:1872–1880. doi: 10.1002/ijc.25503. [DOI] [PubMed] [Google Scholar]
- 38.Chevrier J, Harley KG, Bradman A, Gharbi M, Sjodin A, Eskenazi B. Environ Health Perspect. 2010;118:1444–1449. doi: 10.1289/ehp.1001905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meijer L, Weiss J, Van Velzen M, Brouwer A, Bergman A, Sauerf PJJ. Environ Sci Technol. 2008;42:3428–3433. doi: 10.1021/es702446p. [DOI] [PubMed] [Google Scholar]
- 40.Linderholm L, Park JS, Kocan A, Trnovec T, Athanasiadou M, Bergman A, Hertz-Picciotto I. Chemosphere. 2007;69:403–410. doi: 10.1016/j.chemosphere.2007.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandanger TM, Dumas P, Berger U, Burkow IC. J Environ Monit. 2004;6:758–765. doi: 10.1039/b401999g. [DOI] [PubMed] [Google Scholar]
- 42.Govarts E, Nieuwenhuijsen M, Schoeters G, Ballester F, Bloemen K, de Boer M, Chevrier C, Eggesbo M, Guxens M, Kramer U, Legler J, Martinez D, Palkovicova L, Patelarou E, Ranft U, Rautio A, Petersen MS, Slama R, Stigum H, Toft G, Trnovec T, Vandentorren S, Weihe P, Kuperus NW, Wilhelm M, Wittsiepe J, Bonde JP, Eneieco O. Environ Health Perspect. 2012;120:162–170. doi: 10.1289/ehp.1103767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnerud PO, Lignell S, Glynn A, Aune M, Tornkvist A, Stridsberg M. Environ Int. 2010;36:180–187. doi: 10.1016/j.envint.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang JG, Sun XW, Ai H. J Environ Monit. 2012;14:893–900. doi: 10.1039/c2em10739b. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Li JG, Zhao YF, Li XW, Yang X, Wen S, Cai ZW, Wu YN. Chemosphere. 2011;84:625–633. doi: 10.1016/j.chemosphere.2011.03.041. [DOI] [PubMed] [Google Scholar]
- 46.Park JS, She J, Holden A, Sharp M, Gephart R, Souders-Mason G, Zhang V, Chow J, Leslie B, Hooper K. Environ Sci Technol. 2011;45:4579–4585. doi: 10.1021/es103881n. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Qiu XH, Ren AG, Jin L, Zhu T. Ecotoxicol Environ Saf. 2012;86:141–146. doi: 10.1016/j.ecoenv.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 48.Dassanayake RMAPS, Wei H, Chen RC, Li A. Anal Chem. 2009;81:9795–9801. doi: 10.1021/ac901805d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon HB, Lee DH, Lee YS, Choi M, Choi HG, Kannan K. Arch Environ Contam Toxicol. 2012;62:176–184. doi: 10.1007/s00244-011-9679-6. [DOI] [PubMed] [Google Scholar]
- 50.Wang N, Kong D, Cai D, Shi L, Cao Y, Pang G, Yu R. Environ Sci Technol. 2010;44:4334–4340. doi: 10.1021/es9038775. [DOI] [PubMed] [Google Scholar]
- 51.Wang N, Kong D, Cai D, Shi L, Cao Y, Pang G, Yu R. Environmental Science & Technology. 2010;44:4334–4340. doi: 10.1021/es9038775. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Rhind SM. Talanta. 2011;84:487–493. doi: 10.1016/j.talanta.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 53.Malavia J, Santos FJ, Galceran MT. Talanta. 2011;84:1155–1162. doi: 10.1016/j.talanta.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Zhao G, Zhou H, Zhao J, Yuan H, Gao J, Liu X, Gao B, Wan X, Lu J, Hao H. Journal of environmental science and health. Part A, Toxic/hazardous substances & environmental engineering. 2010;45:1758–1767. doi: 10.1080/10934529.2010.513272. [DOI] [PubMed] [Google Scholar]
- 55.Foster WG, Gregorovich S, Morrison KM, Atkinson SA, Kubwabo C, Stewart B, Teo K. Chemosphere. 2011;84:1301–1309. doi: 10.1016/j.chemosphere.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 56.Faroon O, Jones D, de Rosa C. Toxicol Ind Health. 2000;16:305–333. doi: 10.1177/074823370001600708. [DOI] [PubMed] [Google Scholar]
- 57.Kodavanti PR, Tilson HA. Neurotoxicology. 1997;18:425–441. [PubMed] [Google Scholar]
- 58.DeCaprio AP, Johnson GW, Tarbell AM, Carpenter DO, Chiarenzelli JR, Morse GS, Santiago-Rivera AL, Schymura MJ, Environment ATF. Environ Res. 2005;98:284–302. doi: 10.1016/j.envres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Pessah IN, Cherednichenko G, Lein PJ. Pharmacol Therapeut. 2010;125:260–285. doi: 10.1016/j.pharmthera.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pessah IN, Hansen LG, Albertson TE, Garner CE, Ta TA, Do Z, Kim KH, Wong PW. Chem Res Toxicol. 2006;19:92–101. doi: 10.1021/tx050196m. [DOI] [PubMed] [Google Scholar]
- 61.Costa LG, Giordano G. Neurotoxicology. 2007;28:1047–1067. doi: 10.1016/j.neuro.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stapleton HM. Anal Bioanal Chem. 2006;386:807–817. doi: 10.1007/s00216-006-0400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald TA. Integr Environ Assess Manag. 2005;1:343–354. [PubMed] [Google Scholar]
- 64.Qiu X, Bigsby RM, Hites RA. Environ Health Perspect. 2009;117:93–98. doi: 10.1289/ehp.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller MF, Chernyak SM, Batterman S, Loch-Caruso R. Environ Sci Technol. 2009;43:3042–3046. doi: 10.1021/es8032764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.N.A.a.R. National Archives Records Administration, Electronic Code of Federal Regulations. May 25, 2012. Administration. [Google Scholar]
- 67.Gomara B, Herrero L, Bordajandi LR, Gonzalez MJ. Rapid Commun Mass Spectrom. 2006;20:69–74. doi: 10.1002/rcm.2264. [DOI] [PubMed] [Google Scholar]
- 68.Frias MM, Torres MJ, Frenich AG, Vidal JLM, Olea-Serrano F, Olea N. Biomed Chromatogr. 2004;18:102–111. doi: 10.1002/bmc.300. [DOI] [PubMed] [Google Scholar]
- 69.Vidal JLM, Frias MM, Frenich AG, Olea-Serrano F, Olea N. Anal Bioanal Chem. 2002;372:766–775. doi: 10.1007/s00216-002-1272-4. [DOI] [PubMed] [Google Scholar]
- 70.Almeida AM, Castel-Branco MM, Falcao AC. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2002;774:215–222. doi: 10.1016/s1570-0232(02)00244-1. [DOI] [PubMed] [Google Scholar]