Abstract
Extracting and concentrating mitochondrial protein complexes from gel strips after blue native PAGE (BN-PAGE) can be daunting tasks using the traditional methods, such as electroelution, passive diffusion and centrifugal concentration. We present a simplified gel electrophoresis method to concentrate mitochondrial protein complexes with excellent recovery rate. Mitochondrial complex I contained on a long gel strip from BN-PAGE can be easily concentrated into a 0.8 cm gel strip when a second BN-PAGE is performed with a Y-shaped gel and the addition of 0.01% n-dodecyl β-D-maltoside and 0.001% SDS in the cathode buffer. Once completed, the concentrated protein complex in the gel strip is ready for SDS-PAGE or proteomic studies.
Keywords: Blue native PAGE, Complex I, Electrophoretic concentration, Mitochondria
Blue native PAGE (BN-PAGE), first developed by Schägger and von Jagow [1], has been successfully applied not only to the separation of mitochondrial respiratory protein complexes but also to the study of mitochondrial proteomics and disorders [2–4]. After BN-PAGE, the protein complex of interest is excised from the gel and further analyzed by SDS-PAGE for immunoblotting [5]. Nevertheless, due to the inherent hydrophobicity of mitochondrial membrane proteins, no more than 400 μg of sample can be loaded into a 1 cm wide, 1.6 mm thick well in the gel for BN-PAGE [5]. Otherwise, the proteins would aggregate and cannot enter the stacking gel. The limited protein load in BN-PAGE has made the subsequent 2-D SDS-PAGE on a mini-gel system difficult. In order to obtain a sufficient amount of protein for SDS-PAGE, a wide blue native gel strip is required and must be rotated 90 degrees to be embedded into the SDS stacking gel. However, this long stacking gel occupies too much space and hampers the resolution of the resolving gel. Alternatively, electroelution or passive diffusion of protein from the gel strips with subsequent centrifugal concentration can be done but it is time-consuming and not applicable when the amount of sample is limited. In our experience, only about 150 μg of complex I could be obtained from 4 mg of rat heart mitochondrial protein. Furthermore, a significant dissociation of complex I after the long isolation process was noted. Here we present an electrophoretic concentration method for mitochondrial membrane proteins using a mini-gel system. The protein complex of interest can be concentrated in a gel strip less than 1 cm wide, which can be easily sealed for SDS-PAGE. As a result, more protein is transferred to the 2-D SDS-PAGE and the sensitivity and accuracy of western blotting is improved.
Rat heart mitochondria were prepared as described previously [6] and frozen at −80 °C until use. The electrode, gel and solubilization buffers for BN-PAGE were made according to Wittig et al. [5]. All gels were hand-cast and run using a mini-gel system (Bio-Rad Mini-Protean, Hercules, CA). For BN-PAGE and electrophoretic concentration, all procedures were performed at 0–4 °C. One milligram of mitochondria was first resuspended in 300 μl buffer (250mM sucrose, 10 mM HEPES, 1 mM EGTA and 1 mM PMSF, pH 7.2), followed by three cycles of freezing and thawing, and then pelleted at 16100 g for 20 min. The mitochondrial pellet was solubilized in 77.5 μl solubilization buffer (50 mM imidazole, 500 mM 6-aminohexanoic acid and 1 mM EDTA, pH 7.0 at 4°C), added with 22.5 μl 10% n-dodecyl β-D-maltoside (DDM, detergent/protein ratio (w/w): 2.25:1) and incubated on ice for 30 minutes. The solution was centrifuged at 20000 g for another 30 min to remove insolubles and the supernatant was added with 10 μl loading buffer (solubilization buffer with 50% glycerol and 5% Coomassie blue G-250).
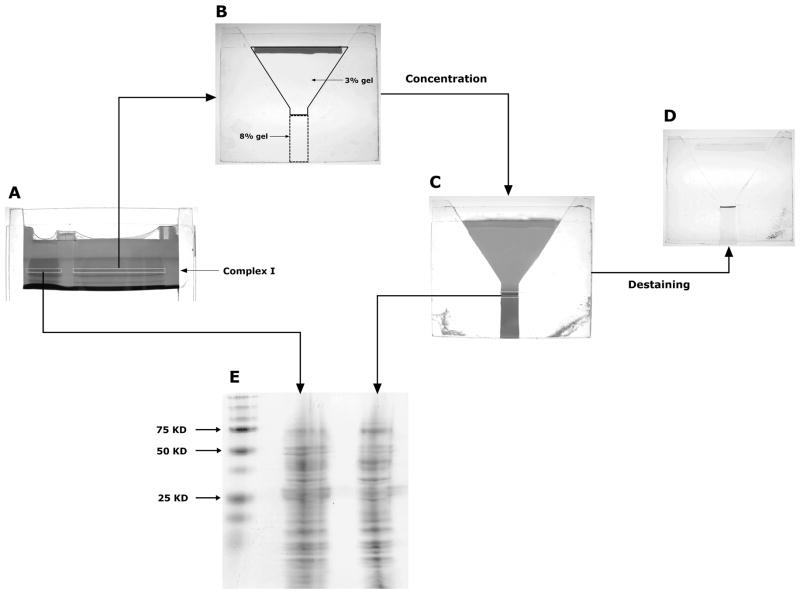
On a 1.5 mm-thick blue native gel composed of 4–6% acrylamide resolving gel and 3% acrylamide stacking gel, half of the sample (0.5 mg mitochondrial protein) was loaded onto each well (Fig. 1A). A lower concentration of gradient gel was selected because complex I was the desired protein complex in our study. For other mitochondrial membrane complexes of interest, a different acrylamide concentration may be chosen for better resolution. The BN-PAGE was run at 70 V for 80 min until the complex I band was distinctively separated. To verify the position of the complex I band, a duplicate gel was stained for the in-gel complex I activity [7]. Both short and long complex I gel strips were cut by a razor blade and only the long gel strip underwent the electrophoretic concentration. Finally, the short gel strip from the initial BN-PAGE and the concentrated gel strip were analyzed by SDS-PAGE to estimate the efficiency of electrophoretic concentration (Fig. 1).
Figure 1.
The procedures of electrophoretic concentration and evaluation of concentration efficiency. A) Mitochondrial protein complexes separated by BN-PAGE. Five hundred micrograms of mitochondrial protein were loaded onto each well. The first band of BN-PAGE was identified as complex I (marked in white dashed rectangle). B) The concentration gel. The excised complex I gel strip was sealed in a Y-shaped gel composed of 3% stacking gel and 8% resolving gel. C) The concentration gel after second BN-PAGE. A visible dark band of concentrated complex I was located at the top of the resolving gel. D) Destaining of the concentration gel. There was no residual protein identified in the gel strip and stacking gel and only one band (complex I) was noted in the resolving gel. E) Second SDS-PAGE. The concentrated gel strip has the identical protein band pattern as the non-concentrated gel strip. Gel photos A–D were taken by a Canon A570 IS digital camera and gel photo E was scanned by an Epson Perfection V30 scanner. Original color images were transformed into grayscale images and resized by FastStone Image Viewer.
The Y-shaped blue native gel (Fig. 1B) for electrophoretic concentration was cast by inserting two 1.5 mm-thick trapezoidal glass spacers between the plates. The neck was filled with 8% acrylamide resolving gel (2.0 cm x 0.8 cm) topped with a layer of 3% acrylamide stacking gel (4.5 cm in height). The excised gel strip was immobilized by 1% agarose gel. Then the gel underwent a 3-hour BN-PAGE in which the voltage was set at 100 V and the cathode buffer was added with 0.01% DDM and 0.001% SDS. After electrophoresis, all complex I in the gel strip was concentrated at the top of the resolving gel because of the small pore size (Fig. 1C). A duplicate concentration gel was destained overnight with 5% methanol and 7.5% acetic acid to demonstrate excellent migration of complex I and indiscernible dissociation (Fig. 1D). We noticed that the addition of minimal SDS was needed to facilitate the complete migration of complex I from the gel strip while a concentration greater than 0.001% caused protein dissociation.
To evaluate the recovery rate of complex I after electrophoretic concentration, the short gel strip from the initial BN-PAGE and that of the concentration gel were analyzed by standard Tris-glycine SDS-PAGE [8]. The gel strips were soaked in 1% SDS for 15 min and embedded in a 1.5 mm-thick mini-gel with 8–16% gradient gel (4.5cm) and 4% stacking gel (1cm). After SDS-PAGE, the gel was stained for one hour using a modified colloidal coomassie blue G-250 staining method [9]. Destained with water overnight, the gel was scanned and densitometrically analyzed by ImageJ software (National Institutes of Health) for comparison of total protein content in both gel strips [10]. As seen in Fig. 1E, the protein bands in both lanes were comparable. There were no protein bands missing in the lane of the concentrated gel strip. The recovery rate of complex I, determined as the ratio of the total protein content of the concentrated gel trip to the total protein content of the short gel strip from BN-PAGE, was 87.7 ± 0.8 % (mean ± SD, n = 3).
There have been a few reports applying gel electrophoresis to protein concentration [11–14]. However, none of them can be used for the concentration of high-molecular-weight protein complexes because of the SDS-PAGE system. Although conceptually similar to funnel-well SDS-PAGE [11], our system is superior for its low-neck design, which reduces both the total electrical resistance of the gel and heat production. Additionally, there is no overlay of sample gel strips which could hinder the migration of hydrophobic, large-sized protein molecules. This concentration method does not require special equipment, i.e., a funnel tube, or involve cumbersome electroelution and centrifugal concentration procedures. It is a modified 2-D BN/BN-PAGE which can be easily done with a mini-gel apparatus. Once completed, the concentrated protein complex in the gel strip is ready for SDS-PAGE or proteomic studies. Unwanted loss of protein complex is minimized, which is critical whenever the amount of mitochondrial sample is limited. In conclusion, the proposed method is a simple and effective way to concentrate mitochondrial membrane protein complexes in preparation for the subsequent experimentation.
Acknowledgments
This study was supported by grants from the American Heart Association (09PRE2170046, Yeh; 60015791, Angelos) and the National Institutes of Health (HL083237, Chen).
Abbreviations
- BN-PAGE
blue native PAGE
- DDM
n-dodecyl β-D-maltoside
Footnotes
The authors have declared no conflict of interest.
References
- 1.Schägger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 2.Schägger H. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- 3.Calvaruso MA, Smeitink J, Nijtmans L. Methods. 2008;46:281–287. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Wittig I, Schägger H. Proteomics. 2008;8:3974–3990. doi: 10.1002/pmic.200800017. [DOI] [PubMed] [Google Scholar]
- 5.Wittig I, Braun HP, Schägger H. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 6.Yeh ST, Lee HL, Aune SE, Chen CL, et al. J Mol Cell Cardiol. 2009;47:789–797. doi: 10.1016/j.yjmcc.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerbetto E, Vergani L, Dabbeni-Sala F. Electrophoresis. 1997;18:2059–2064. doi: 10.1002/elps.1150181131. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Candiano G, Bruschi M, Musante L, Santucci L, et al. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 10.Smith BJ. In: The Protein Protocols Handbook. 2. Walker JM, editor. Humana Press; Totowa, NJ: 2002. pp. 237–242. [Google Scholar]
- 11.Lombard-Platet G, Jalinot P. Biotechniques. 1993;15:668–672. [PubMed] [Google Scholar]
- 12.Terbush DR, Novick P. J Biomol Tech. 1999;10:149–152. [Google Scholar]
- 13.Sheen H, Ali-Khan Z. Anal Biochem. 2005;343:338–340. doi: 10.1016/j.ab.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 14.Rider MH, Puype M, Van DJ, Gevaert K, et al. Eur J Biochem. 1995;230:258–265. doi: 10.1111/j.1432-1033.1995.0258i.x. [DOI] [PubMed] [Google Scholar]