Abstract
Background and Aims
Phosphate (Pi) deficiency in soils is a major limiting factor for crop growth worldwide. Plant growth under low Pi conditions correlates with root architectural traits and it may therefore be possible to select these traits for crop improvement. The aim of this study was to characterize root architectural traits, and to test quantitative trait loci (QTL) associated with these traits, under low Pi (LP) and high Pi (HP) availability in Brassica napus.
Methods
Root architectural traits were characterized in seedlings of a double haploid (DH) mapping population (n = 190) of B. napus [‘Tapidor’ × ‘Ningyou 7’ (TNDH)] using high-throughput phenotyping methods. Primary root length (PRL), lateral root length (LRL), lateral root number (LRN), lateral root density (LRD) and biomass traits were measured 12 d post-germination in agar at LP and HP.
Key Results
In general, root and biomass traits were highly correlated under LP and HP conditions. ‘Ningyou 7’ had greater LRL, LRN and LRD than ‘Tapidor’, at both LP and HP availability, but smaller PRL. A cluster of highly significant QTL for LRN, LRD and biomass traits at LP availability were identified on chromosome A03; QTL for PRL were identified on chromosomes A07 and C06.
Conclusions
High-throughput phenotyping of Brassica can be used to identify root architectural traits which correlate with shoot biomass. It is feasible that these traits could be used in crop improvement strategies. The identification of QTL linked to root traits under LP and HP conditions provides further insights on the genetic basis of plant tolerance to P deficiency, and these QTL warrant further dissection.
Keywords: Phosphate, phosphorus, root, Brassica napus, oilseed rape, QTL, biomass, genetic, heritability
INTRODUCTION
Phosphorus (P) is essential to plants. Their roots acquire P from the rhizosphere solution as phosphate (Pi), primarily in the form of H2PO4– (Vance et al., 2003; Hammond et al., 2004; White and Hammond, 2008). The concentration of Pi in the soil solution is often low (2–10 µm) and, consequently, the supply of Pi to the root surface by diffusion and mass flow is slow (Bieleski, 1973; Barber, 1995). Hence, P is one of the least-available mineral elements in the soil and frequently limits plant growth (Vance et al., 2003; Tiessen, 2008).
Over 85 % of mined P is used in food production (Heffer et al., 2006) and consumption of this non-renewable resource could lead to a peak P scenario (akin to peak oil; Raven, 2008; Cordell et al., 2009). It is therefore likely that there will be increasing pressures on Pi fertilizer availability and, consequently, cost in the future. These pressures will be exacerbated by increasing demand on food production systems as the human population increases and by fluctuation in oil prices (Cordell et al., 2009). Inappropriate use of inorganic Pi fertilizers can also perturb the nutrient balance of natural ecosystems and reduce biodiversity (White and Hammond, 2008, 2009).
Breeding crops that acquire Pi and/or use P more efficiently is one strategy to reduce the use of Pi fertilizers (White et al., 2005; Veneklass et al., 2012). Root traits represent a potential source of genetic variation to improve P acquisition for breeding such crops (Lynch, 2007; Römheld and Kirkby, 2010; Powlson et al., 2011; De Smet et al., 2012; Lynch and Brown, 2012). Assessment of root traits in crop breeding material can be slow and expensive, involving a combination of field-, glasshouse- and laboratory-based screens (Clark et al., 2011). The latter of these is amenable to high-throughput screens to identify germplasm with altered root growth and morphology. Genetic loci associated with these traits have the potential for use in breeding new crop varieties with improved root phenotypes.
In arabidopsis and other plant species, root responses to low Pi availability have been well characterized (White et al., 2005). Typically, a reduction in the development of the primary root (Ticconi et al., 2004; Sánchez-Calderón et al., 2005; Svistoonoff et al., 2007; Jain et al., 2007; Fang et al., 2009; Hammond et al., 2009) and increases in the number and length of lateral roots are observed under low Pi availability (Williamson et al., 2001; Linkohr et al., 2002; López-Bucio et al., 2002, 2003, 2005; Al-Ghazi et al., 2003; Nacry et al., 2005; Reymond et al., 2006). In some species, root agravitropism or topsoil foraging, is observed (Zhu et al., 2005). Both density and length of root hairs are also increased when plants are grown on a low Pi supply (Bates and Lynch, 1996; Gahoonia and Nielsen, 1997; Brown et al., 2012), thus increasing the capacity for Pi acquisition. Biochemical adaptations, including the release of organic anions to release Pi bound to clay particles (Shane and Lambers, 2005) and enzymes to release Pi from organic compounds are also observed (Li et al., 2002; George et al., 2011; Plaxton and Tran, 2011; Richardson et al., 2011).
Root traits affecting Pi acquisition are complex and regulated by multiple genetic loci. In rice, a major quantitative trait locus (QTL) for P-deficiency tolerance, P uptake 1 (Pup1) was mapped to a 150-kb region of chromosome 12 containing 60 predicted genes (Wissuwa et al., 2002; Chin et al., 2011). Subsequently the gene conferring the phenotype (PSTOL1) was identified as a protein kinase (Gamuyao et al., 2012). Among Brassicaceae species, QTL have been associated with leaf and seed P and phytate concentrations, and primary root growth responses to low Pi availability (Bentsink et al., 2003; Vreugdenhil et al., 2004; Reymond et al., 2006; Svistoonoff et al., 2007; Zhao et al., 2007, 2008; Hammond et al., 2009, 2011; Ding et al., 2010, 2012; Yang et al., 2010). Significant QTL associated with shoot-P and measures of P-use efficiency (PUE) on chromosome C03 and C07 of B. oleracea have previously been identified, and a significant correlation between these shoot traits and root morphological traits was observed (Hammond et al., 2009). The locations of these QTL have been confirmed using substitution lines. Recently, a new mapping population of B. napus has been developed, utilizing parents with contrasting PUE characteristics (Duan et al., 2009) and used to identify QTL associated with seed elemental concentrations (Ding et al., 2010), seed yield (Ding et al., 2012) and root morphological traits (Yang et al., 2010) under contrasting Pi supplies. The latter of these studies identified significant QTL associated with root length, volume and surface area on chromosomes A01, A03, C03 and C02.
Here we used a mapping population, derived from a cross between a winter and a semi-winter cultivar of oilseed rape (OSR), to characterize the component traits of root architectural adaptations to low Pi availability, and identify QTL associated with them. Root architectural traits were scored using an agar-based high-throughput root phenotyping system to assess the roots of over 6600 individual seedlings.
MATERIALS AND METHODS
Plant material and growth conditions
Plant material consisted initially of 190 double haploid (DH) lines representing the ‘Tapidor’ × ‘Ningyou 7’ (TNDH) mapping population (Qiu et al., 2006), plus the parents. The TNDH mapping population was generated through anther culture of the F1 generation of a cross between Brassica napus cultivar ‘Tapidor’ (a European winter OSR) and B. napus cultivar ‘Ningyou 7’ (a Chinese semi-winter OSR). A new genetic linkage map was developed combining 53 gene-based markers (Ding et al., 2011) with an existing well-defined genetic map (Long et al., 2011). The new genetic linkage map had a total of 798 molecular markers and an average distance between two adjacent markers of 2·6 cM. The linkage map was constructed using JoinMap 4·0 (Van Ooijen, 2006) and the mapping procedure followed the method of Long et al. (2011) using RFLP, SSR and STS with default parameters and linkage groups distinguished at LOD (logarithm of the ratio of likelihoods) values between 8 and 19. The order of the markers on the new linkage map agreed well with our published maps (Shi et al., 2009; Long et al., 2011).
Seeds were first surface sterilized in 70 % (v/v) ethanol, rinsed in deionized water and then surface sterilized for 1 min using NaOCl (2·5 % active chlorine). Seeds were rinsed three times in sterile deionized water, before being placed in sterile deionized water at 4 °C for 24 h to imbibe. Imbibed seeds were sown into vented polystyrene trays (QTray; 240 × 240 × 20 mm; Molecular Devices, Hampshire, UK) containing 300 mL 0·8 % (w/v) agar and a modified basal salt mix (Murashige and Skoog, 1962) with either 0·625 mm P (HP) added as KH2PO4 or 0 mm P (LP), with 0·625 mm KCl added to provide K. The mean (± s.e.m., n = 3) total P concentration was 0·652 ± 0·011 mm P for HP agar and 0·082 ± 0·002 mm P for LP agar. Root responses of parental genotypes to external Pi concentrations ([P]ext) were also assessed at 0, 0·006, 0·312, 0·625 and 1·25 mm P, with KCl added to balance K. Seeds were sown 3 cm from the top edge of the tray, with four seeds per line and two lines per tray. Trays were sealed with Nescofilm and placed 10 ° from vertical in a growth room under a 16-h photoperiod at a constant temperature of 24 °C. Illumination was provided by a bank of 84 100-W cool fluorescent tubes (Philips, Eindhoven, Netherlands), giving a photon flux density between 400 and 700 nm of 80–100 µmol photons m−2 s−1 at plant height.
For each line, 16 seeds were sown across four independent replicates, at both LP and HP. Trays were placed randomly within the growth room. However, due to variation in germination rate, the total number of observations for each line varied between 4 and 16, with the average number of observations per line per treatment being 11.
Images of the root systems were captured using a flatbed scanner (Scanjet 3670; Hewlett-Packard, Palo Alto, CA, USA) 12 d after sowing. At harvest, shoot and root fresh weight (SFW and RFW, respectively) were determined. Tissue samples were dried at 80 °C and dry weights (shoot, SDW; root, RDW) determined. Tissue samples were digested by the addition of 2 mL nitric acid to 0·3 g dried ground material and processed in a closed vessel acid digestion microwave (MARSXpress; CEM Corporation, Matthews, NC, USA). Digested samples were diluted with 23 mL of deionized water and analysed using inductively coupled plasma emission spectrometry (JY Ultima 2; Jobin Yvon Ltd, Stanmore, Middlesex, UK) to determine tissue P concentrations.
Image analysis
Images were loaded into ImageJ (Abramoff et al., 2004). Primary root length (PRL, cm) and lateral root length (LRL, cm) were measured. Lateral root numbers were counted and used to calculate lateral root density (LRD, cm−1). Total root length (TRL, cm) was calculated as the sum of PRL and LRL.
Data analysis
Raw data were entered into GenStat (Release 13·1·0·4470; VSN International, Oxford, UK). Data for parent lines grown at different [P]ext were analysed using a two-way ANOVA. Due to variability in germination of lines within the mapping population, data for the mapping population grown at two [P]ext were analysed using REML (residual maximum likelihood) procedures to allocate sources of variation and estimate individual line means (Patterson and Thompson, 1971; Robinson, 1987). Prior to analysis SDW and RDW were ln transformed and LRN, LRD and TRL were square root transformed to improve the normality and variance of the data. PRL and LRL did not require transformation. A random term [(Replicate/Run/Plate/Position) + ([P]ext × Line)] and no defined fixed factors was used to allocate sources of variation for individual traits. Subsequently, line means were estimated using the [([P]ext × Line)] term as a fixed factor, retaining [(Replicate/Run/Plate/Position)] as a random factor.
QTL positions were estimated using the zmapQTL model 6 CIM (composite interval mapping) option in WinQTL cartographer 2·5 software (Wang et al., 2011a) and the estimated line means obtained from the REML procedures with the [([P]ext × Line)] as a fixed factor. For each trait, the threshold LOD value for the detection of a significant QTL (P < 0·05) was estimated from 1000 permutations (Churchill and Doerge, 1994). Thresholds for the LOD ranged between 3·04 and 3·43.
RESULTS AND DISCUSSION
Parents of the TNDH mapping population show differences in root architecture
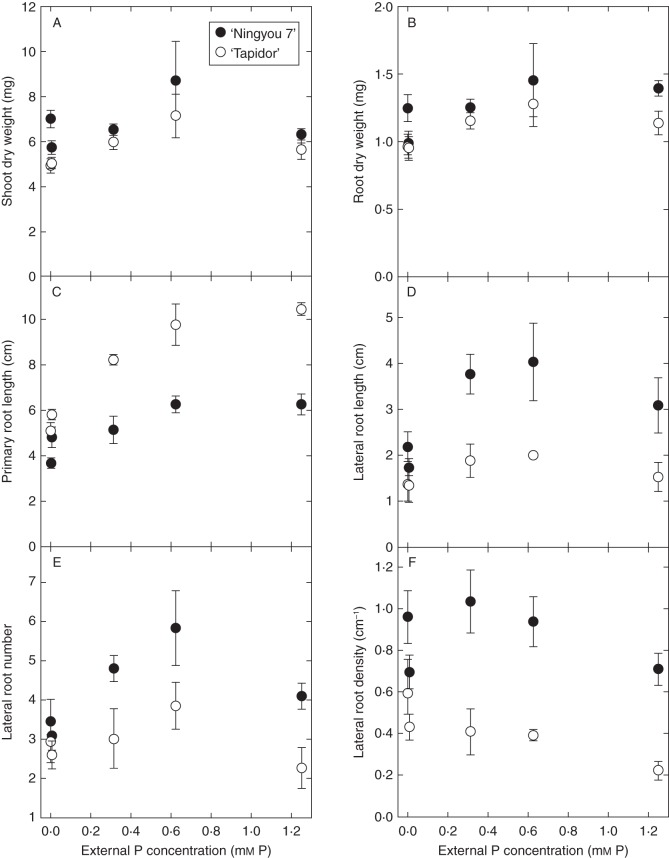
To determine optimal assay conditions for screening the large TNDH mapping population, root responses to different [P]ext were quantified in the parents of the mapping population (Fig. 1). The effect of [P]ext was significant (P < 0·01) for both RDW and SDW. Root and shoot DW was greatest in both parents at 0·625 mm [P]ext, with SDW declining with declining [P]ext and also being lower at 1·25 mm [P]ext compared with 0·625 mm [P]ext, and RDW declining with declining [P]ext and remaining similar at 1·25 mm [P]ext compared with 0·625 mm [P]ext (Fig. 1A, B). There was a significant difference between lines for both RDW (P = 0·012) and SDW (P < 0·001), with ‘Ningyou 7’ accumulating a greater biomass than ‘Tapidor’ at all [P]ext (Fig. 1A, B).
Fig. 1.
Changes in shoot dry weight (A), root dry weight (B), primary root length (C), lateral root length (D), lateral root number (E) and lateral root density (F) in ‘Tapidor’ and ‘Ningyou 7’ at different external Pi concentrations. Seedlings were grown on trays containing 300 mL 0·8 % (w/v) agar and a modified basal salt mix with 0, 0·006, 0·312, 0·625 or 1·25 mm P for 8 d and images of root systems analysed for root traits. Symbols represent means ± s.e.m. (n = 4).
Root traits differed significantly between the parents of the TNDH mapping population and showed characteristic changes in root architectural traits with declining [P]ext (Fig. 1). There was a significant decrease in PRL with declining [P]ext, with the PRL of ‘Tapidor’ seedlings declining from 10·45 cm at 1·25 mm [P]ext to 5·11 cm at 0 mm [P]ext, and ‘Ningyou 7’ seedlings declining from 6·27 cm at 1·25 mm [P]ext to 3·68 cm at 0 mm [P]ext (Fig. 1C). This is consistent with primary root responses of other B. napus (Akhtar et al., 2008; Yang et al., 2010) and B. oleracea (Hammond et al., 2009) cultivars observed previously. At all [P]ext, the PRL of ‘Tapidor’ was greater than the PRL of ‘Ningyou 7’ seedlings. In contrast, the LRL of ‘Ningyou 7’ seedlings was consistently greater than the LRL of ‘Tapidor’ seedlings at all [P]ext (Fig. 1D). Whilst the LRL of ‘Ningyou 7’ seedlings increased up to 0·625 mm [P]ext, before declining again at 1·25 mm [P]ext, the LRL of ‘Tapidor’ seedlings did not change significantly with [P]ext (Fig. 1D). The increase in LRL with [P]ext for ‘Ningyou 7’ seedlings is consistent with the root responses of other B. napus (Solaiman et al., 2007; Hu et al., 2010; Yang et al., 2010) and B. oleracea (Hammond et al., 2009) cultivars observed previously, but the LRL response of ‘Tapidor’ seedlings is atypical. The reduction in LRL of ‘Ningyou 7’ seedlings when [P]ext is reduced contrasts with the increase in LRL observed for arabidopsis when [P]ext is reduced (Williamson et al., 2001; Linkohr et al., 2002; López-Bucio et al., 2002, 2003, 2005; Al-Ghazi et al., 2003; Nacry et al., 2005; Reymond et al., 2006). Also, in a comparison between two B. napus cultivars with either high or low physiological PUE, Akhtar et al. (2008) showed a significant increase in LRL in both cultivars when supplied with low [P]ext or rock phosphate. Consequently, this phenotype may be specific to the parental lines used in this study, or may represent a more general phenotype within B. napus species. The number of lateral roots was greatest in both parents at 0·312 and 0·625 mm [P]ext, but decreased significantly with both increasing and decreasing [P]ext (Fig. 1E). LRD increased with declining [P]ext for ‘Tapidor’, and increased for ‘Ningyou 7’ up to 0·312 mm [P]ext before declining slightly at lower [P]ext (Fig. 1F).
Given the significant differences observed between the two parental lines for key root traits under low [P]ext, the TNDH mapping population was screened for root architectural traits at low (0 mm added P, LP) and high (0·625 mm added P, HP) [P]ext to identify genomic regions (QTL) associated with these traits.
Root traits show transgressive segregation in the TNDH mapping population
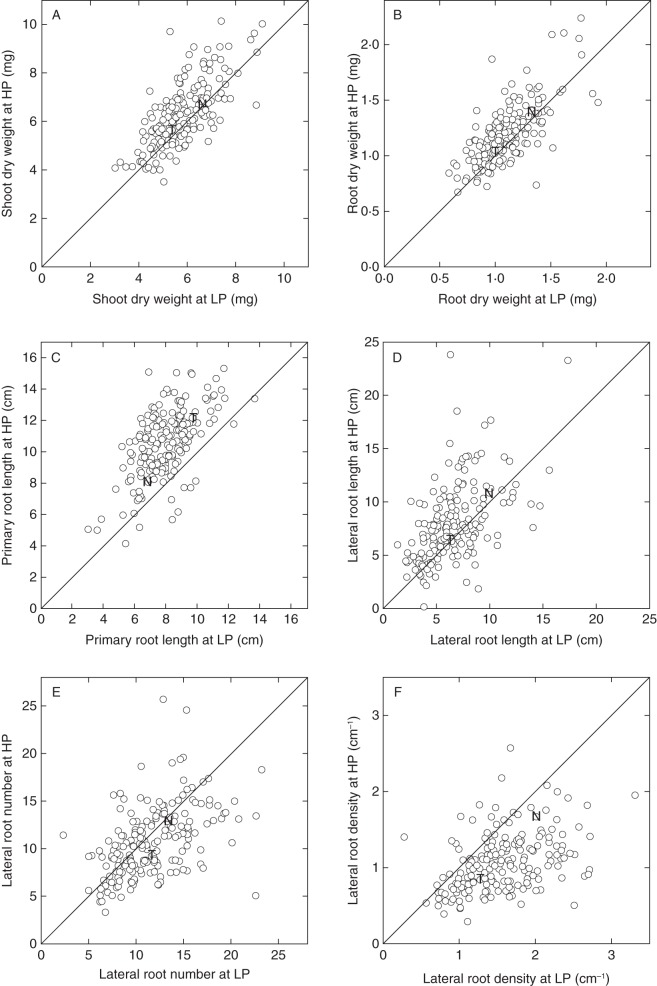
Seeds from 190 lines of the TNDH mapping population were sown on plates containing either LP or HP agar, to facilitate high-throughput phenotyping of root traits. Root architectural traits were scored for all lines at 12 d (Fig. 2 and Table 1). Transgressive segregation was observed for all traits. At the population level, variance attributed to line, [P]ext, and their interaction was highly significant (P < 0·001) for all traits (Table 1), and correlations between LP and HP treatments for individual traits were all significant (P < 0·001; Fig. 2). Mean SDW and RDW for the population did not change significantly when plants were grown on HP agar compared with LP agar, although the individuals within the population showed large responses to [P]ext with the difference in SDW between LP and HP varying from –2·2 to 4·4 mg, and the difference in RDW between LP and HP varying from –0·6 to 0·9 mg (Fig. 2A, B).
Fig. 2.
Variation in shoot dry weight (A), root dry weight (B) primary root length (C), lateral root length (D), lateral root number (E) and lateral root density (F) in the ‘Tapidor’ × ‘Ningyou 7’ double haploid (TNDH) mapping population. Seedlings were grown on trays containing 300 mL 0·8 % (w/v) agar and a modified basal salt mix with 0 (LP) or 0·625 (HP) mm P for 12 d. Data are REML-estimated means (n varies between 4 and 16, average 11 observations per line per treatment). Symbols ‘T’ and ‘N’ represent the mapping population parental values for ‘Tapidor’ and ‘Ningyou 7’, respectively. The continuous line represents the 1 : 1 line.
Table 1.
Percentage contributions for individual variance components derived from the REML analyses of biomass and root architectural data measured in the Brassica napus TNDH mapping population
| Trait | Replicate | Run | Plate | Position | Line† | [P]ext† | Line × [P]ext† | Residual |
|---|---|---|---|---|---|---|---|---|
| SDW | 0·32 | 2·97 | 10·62 | 2·74 | 30·70*** | 3·98*** | 4·72*** | 43·94 |
| RDW | 1·64 | 3·53 | 13·19 | 0·84 | 28·01*** | 2·67*** | 3·47*** | 46·64 |
| PRL | 2·29 | 9·95 | 9·64 | 3·27 | 13·08*** | 23·52*** | 4·15*** | 34·10 |
| LRL | 0·53 | 6·11 | 13·91 | 1·56 | 16·06*** | 3·12*** | 5·08*** | 53·63 |
| LRN | 0·67 | 9·98 | 10·93 | 1·17 | 15·92*** | 1·30*** | 4·54*** | 55·48 |
| LRD | 1·11 | 6·43 | 8·41 | 1·28 | 13·47*** | 17·77*** | 4·88*** | 46·65 |
| TRL | 0·79 | 12·14 | 9·88 | 1·46 | 14·81*** | 14·78*** | 4·65*** | 41·48 |
Data were analysed in GenStat using the REML procedures with a random term [(Replicate/Run/Plate/Position) + ([P]ext × Line)] and no defined fixed factors.
† A Wald test statistic was calculated so that significant sources of variation could be identified using a χ2 function (degrees of freedom for Line = 191, [P]ext = 1, Line × [P]ext = 191; *** = P < 0·001).
Across the TNDH population, mean PRL and LRL were both significantly lower at LP compared with HP; however, there were a range of responses within the population with some individuals increasing their LRL or PRL by 1·0 and 0·4 cm, respectively, at LP compared with HP. ‘Tapidor’ had a longer primary root, and shorter lateral roots, compared with ‘Ningyou 7’ (Fig. 2C, D). The mean LRN and LRD were both higher at LP compared with HP, with a greater range in values for LRD when plants were grown at LP compared with plants grown at HP (Fig. 2E, F). Overall, TRL (data not shown) was greater at HP (mean 17·60 cm, n = 4536) compared with LP (mean 13·76 cm), with a greater range in values observed at HP (5·13–34·22 cm) than at LP (5·35–25·12 cm).
Root architectural traits are heritable in Brassica napus
Breeding new cultivars with improved acquisition and/or utilization of Pi through selection for root architectural traits requires them to be heritable. Within the variance components in the REML analyses, the Line component represents variance attributed to genetic factors. This variance component approximates the population wide additive genetic variation, or narrow-sense heritability. Shoot dry weight and RDW had the highest heritabilities, of 30·70 and 28·01, respectively (Table 1). The root architectural traits had heritabilities ranging from 13·08 to 16·06, suggesting the underlying genetic control of these traits can be selected for, and used in breeding new cultivars with improved root traits. Interestingly, the variance attributed to the [P]ext treatment varied between the root traits. For LRL and LRN, the trait variance attributed to [P]ext was relatively small, compared with the trait variance attributed to [P]ext for PRL, LRD and TRL (Table 1). This implies that PRL, LRD and TRL traits are more responsive to [P]ext in this mapping population, and in B. napus may represent key adaptive root architectural traits to low Pi availability.
QTL associated with biomass and root architectural traits are conserved within the Brassicaceae
A total of 38 QTL, associated with root architectural and biomass traits, were identified across nine of the 19 chromosomes (Table 2). Significant QTL associated with SDW at both LP and HP co-localized to 44·2 cM on chromosome A03, explaining 14·9 and 8·5 % of the genetic variation for SDW at LP and HP, respectively. With the exception of one QTL on chromosome A02 (SDW_HP_C02a), all shoot biomass QTL had a negative additive effect, indicating that the allele from ‘Ningyou 7’ increased the trait value. QTL associated with LRN at LP and RDW at LP also co-localized at 44·2 cM on A03, again with alleles from ‘Ningyou 7’ increasing the trait value. Also on chromosome A03, three markers between 60·3 and 76·8 cM were significantly associated with LRD at LP, accounting for 23·9 % of the genetic variation associated with this trait.
Table 2.
Significant QTL associated with biomass and root architectural traits in the Brassica napus TNDH mapping population
| Trait | QTL name | Chromosome | Marker | Position (cM) | LOD score | 2 LOD support interval (cM) | Additive effect | R2 (%) |
|---|---|---|---|---|---|---|---|---|
| SDW at LP | SDW_LP_A02a | A02 | znS16M07-1-230 | 80·3 | 3·46 | 77·8–83 | –0·30 | 6·2 |
| SDW_LP_A03a | A03 | BRMS-043 | 44·2 | 7·68 | 42–44·6 | –0·47 | 14·9 | |
| SDW_LP_A04a | A04 | JICB0283 | 7·2 | 5·15 | 5·3–17·2 | –0·36 | 9·9 | |
| SDW at HP | SDW_HP_A03a | A03 | BRMS-043 | 44·2 | 4·28 | 43·2–46 | –0·43 | 8·5 |
| SDW_HP_C02a | C02 | sN3761b | 11·9 | 3·33 | 8·5–15 | 0·36 | 7·2 | |
| RDW at LP | RDW_LP_A03a | A03 | BRMS-043 | 44·2 | 3·89 | 43·2–46 | –0·07 | 7·8 |
| RDW at HP | RDW_HP_A03a | A03 | CNU098 | 61·3 | 3·13 | 60·3–62·4 | –0·07 | 6·4 |
| LRN at LP | LRN_LP_A03a | A03 | HBr082 | 37·5 | 4·11 | 36·8–38 | –1·19 | 8·9 |
| LRN_LP_A03b | A03 | BRMS-043 | 44·2 | 4·81 | 43·2–46 | –1·30 | 10·4 | |
| LRN_LP_A03c | A03 | B068E07-2 | 51·9 | 3·71 | 50·8–52·5 | –1·13 | 8·4 | |
| LRN at HP | LRN_HP_C09a | C09 | CB10064 | 36·0 | 3·51 | 34·6–43·6 | 1·06 | 8·2 |
| LRD at LP | LRD_LP_A02a | A02 | em12me31-320 | 73·3 | 3·92 | 66·6–75·5 | –0·14 | 7·5 |
| LRD_LP_A03a | A03 | CNU098 | 61·5 | 4·55 | 60·3–62·4 | –0·16 | 8·7 | |
| LRD_LP_A03b | A03 | H034E17-1 | 69·3 | 3·83 | 67·3–70·7 | –0·14 | 7·4 | |
| LRD_LP_A03c | A03 | BnPYK10-A3b | 76·7 | 3·89 | 70·7–76·8 | –0·14 | 7·8 | |
| LRD_LP_A09a | A09 | B019F12-3 | 37·9 | 5·37 | 34·9–40·6 | –0·23 | 11·5 | |
| LRD_LP_C06a | C06 | JBnB061J08 | 29·0 | 3·74 | 25·7–35·2 | –0·13 | 7·1 | |
| LRD at HP | LRD_HP_A04a | A04 | JICB0283 | 16·2 | 5·98 | 7·6–21 | –0·15 | 13·2 |
| LRD_HP_C04a | C04 | sN12353c | 50·6 | 3·36 | 49·9–52·3 | –0·10 | 6·8 | |
| LRD_HP_C04b | C04 | Na10C01a | 62·2 | 3·64 | 59·6–62·7 | –0·10 | 7·5 | |
| TRL at HP | TRL_HP_A03a | A03 | BnWRKY-A3 | 14·4 | 3·64 | 10·8–17 | –1·33 | 7·5 |
| PRL at LP | PRL_LP_A03a | A03 | BnPHT3-A3 | 15·5 | 3·36 | 13·5–18·8 | –0·40 | 5·8 |
| PRL_LP_A07a | A07 | BRAS023 | 29·8 | 4·76 | 28·8–36·3 | 0·48 | 9·5 | |
| PRL_LP_A07b | A07 | HR-Tp4-305 | 42·6 | 5·70 | 39·5–46·4 | 0·50 | 10·2 | |
| PRL_LP_A07c | A07 | sR7223 | 50·6 | 4·46 | 48–54·3 | 0·44 | 8·2 | |
| PRL_LP_C06a | C06 | em18me23-350 | 27·5 | 6·07 | 17·6–34·7 | 0·53 | 12·0 | |
| PRL at HP | PRL_HP_A03a | A03 | BnPHT3-A3 | 15·5 | 4·59 | 14·4–17 | –0·67 | 8·3 |
| PRL_HP_A03b | A03 | H003M07-4 | 21·9 | 3·88 | 20·9–27·8 | –0·64 | 8·0 | |
| PRL_HP_C06a | C06 | CNU053a | 21·4 | 6·82 | 20·6–25·7 | 0·82 | 14·8 | |
| PRL_HP_C06b | C06 | em18me23-350 | 27·5 | 8·00 | 25·7–33·6 | 0·85 | 16·3 |
REML estimated means for biomass and root architectural traits for 176 lines in the TNDH mapping population were used to estimate QTL positions associated with these traits using the zmapQTL model 6 composite interval mapping function (Wang et al., 2011a). Significant (P < 0·05) LOD thresholds for individual traits were determined using 1000 permutations. A positive additive effect indicates a positive contribution of the ‘Tapidor’ allele to the trait value.
Primary root length at LP and HP was associated with nine loci within the B. napus genome. At LP, PRL was associated with three loci on A07 between 28·8 and 54·6 cM, accounting for 27·9 % of the genetic variation for PRL at LP (Table 2). QTL associated with PRL at LP and HP, and also TRL at HP, were located between 10·8 and 18·8 cM on A03, and were associated with the functional markers BnWRKY-A3 and BnPHT3-A3 (Ding et al., 2011; Table 2). The remaining three loci associated with PRL at LP and HP co-located to chromosome C06 at 16 cM (support interval from 17·6 to 33·6 cM), together with a QTL associated with LRD at LP. No QTL were identified for LRL traits at either LP or HP within this population, despite the large variation between the parents in the initial experiment (Fig. 1D).
The ancestral genome segments of Brassicaceae species have been relatively well defined in terms of rearrangements and duplication events (Parkin et al., 2005; Mun et al., 2009; Wang et al., 2011b). These segments facilitate comparative genomics between Brassicaceae species, including other Brassica species and the model plant arabidopsis. QTL associated with plant responses to low Pi availability have been identified in both Brassica species and arabidopsis (Bentsink et al., 2003; Vreugdenhil et al., 2004; Reymond et al., 2006; Svistoonoff et al., 2007; Zhao et al., 2007, 2008; Hammond et al., 2009, 2011; Ding et al., 2010, 2012; Yang et al., 2010, 2011) and share common genomic regions to those identified in this study. For example, QTL identified here, associated with SDW at LP and HP, and RDW and LRN at LP, and located on chromosome A03 between 36·8 and 46 cM, co-locate with QTL associated with SDW, RDW, root volume and root surface area at LP, and plant height at LP and HP, determined in a cross between P-efficient and P-inefficient B. napus cultivars (Ding et al., 2010, 2012; Yang et al., 2010, 2011). This corresponds to a pleiotropic QTL associated with multiple measures of biomass and flowering time and seed weight (Shi et al., 2009) and overlaps with a region of chromosome C03 in B. oleracea which has previously been associated with shoot biomass and PUE traits (Hammond et al., 2009). This region is syntenous with ancestral block J on arabidopsis chromosome 2, where QTL for SDW (Loudet et al., 2003) and rosette and root weight (Prinzenberg et al., 2010) have previously been identified, further supporting the presence of a pleiotropic gene in this region.
The unique QTL associated with PRL at LP between 28·8 and 54·6 cM on chromosome A07, corresponds to two ancestral blocks, E and N, on the bottom of arabidopsis chromosomes 1 and 3, respectively. The region containing QTL associated with PRL and LRD on chromosome C06 (Table 2), is also syntenous with the ancestral block E, suggesting these regions may contain paralogues of genes involved in the regulation of PRL. Arabidopsis genes identified as regulating primary root development under low Pi availability have previously been identified in block E. Arabidopsis LPR2 (At1g71040) encodes multicopper oxidases in arabidopsis, and is a paralogue of arabidopsis LPR1 (At1g23010). These proteins are critical for the reduction in primary root growth observed when the root tips are in contact with low-Pi media (Reymond et al., 2006; Svistoonoff et al., 2007). An AINTEGUMENTA-like gene, named PRD (At1g79700), the mutant of which was identified as having reduced primary and lateral root development under low Pi availability compared with the wild-type, also co-locates to this region (Camacho-Cristóbal et al., 2008). The latter of these ancestral blocks also co-locates with a QTL for PRL in arabidopsis (Loudet et al., 2005).
Conclusions
The development of crops that can acquire and/or utilize P more efficiently is essential for the sustainability of future crop production. Plant adaptations to low Pi availability include alterations in the allocation of resources to roots and changes in the distribution of those roots in the soil. Since the assessment of root traits in plants can be slow and expensive, we employed an agar based high-throughput root phenotyping screen to characterize the root traits of a large B. napus mapping population and identify genetic loci controlling these traits under low Pi availability (Fig. 2 and Table 2). Significant QTL associated with biomass and root architectural traits were identified on A03, and co-locate with QTL for biomass traits in B. napus, B. rapa and arabidopsis. Significant QTL associated with root architectural traits were also identified, including one for PRL at LP on A07, which co-locates with several arabidopsis genes implicated in primary root development. Identification of the genetic elements associated with these traits will provide targets for the future development of crops adapted to growth in low Pi soils. The use of high-throughput root phenotyping assays has the potential to advance the breeding and selection of these cultivars, but requires cross validation with root characteristics and yield determined under field conditions.
ACKNOWLEDGEMENTS
This work was supported by the following: National Basic Research and Development Program (grant nos 2011CB109302 and 2011CB100301); Natural Science Funds for Distinguished Young Scholar in Hubei Province (grant no. 2011CDA090); Fundamental Research Funds for the Central Universities (grant no. 2012PY006); the Rural and Environment Science and Analytical Services Division (RESAS) of the Scottish Government through Workpackage 3·3 (2011–2016); UK Department for the Environment, Food and Rural Affairs (grant no. WQ0119); and the Australian Research Council Future Fellowship scheme (grant no. FT11010149).
LITERATURE CITED
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Akhtar MS, Oki Y, Adachi T. Genetic variability in phosphorus acquisition and utilisation efficiency from sparingly soluble P-sources by Brassica cultivars under P-stress environment. Journal of Agronomy and Crop Science. 2008;194:380–392. [Google Scholar]
- Al-Ghazi Y, Muller B, Pinloche S, et al. Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signalling. Plant, Cell & Environment. 2003;26:1053–1066. [Google Scholar]
- Barber SA. Soil nutrient bioavailability: a mechanistic approach. New York: Wiley; 1995. [Google Scholar]
- Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant, Cell & Environment. 1996;19:529–538. [Google Scholar]
- Bentsink L, Yuan K, Koornneef M, Vreugdenhil D. The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theoretical and Applied Genetics. 2003;106:1234–1243. doi: 10.1007/s00122-002-1177-9. [DOI] [PubMed] [Google Scholar]
- Bieleski RL. Phosphate pools, phosphate transport, and phosphate availability. Annual Review of Plant Physiology. 1973;24:225–252. [Google Scholar]
- Brown LK, George TS, Thompson JA, et al. What are the implications of variation in root hair length on tolerance to phosphorus deficiency in combination with water stress in barley (Hordeum vulgare)? Annals of Botany. 2012;110:319–328. doi: 10.1093/aob/mcs085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Cristóbal JJ, Rexach J, Conéjéro G, Al-Ghazi Y, Nacry P, Doumas P. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta. 2008;228:511–522. doi: 10.1007/s00425-008-0754-9. [DOI] [PubMed] [Google Scholar]
- Chin JH, Gamuyao R, Dalid C, et al. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiology. 2011;156:1202–1216. doi: 10.1104/pp.111.175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, et al. 3-Dimensional root phenotyping with a novel imaging and software platform. Plant Physiology. 2011;156:455–465. doi: 10.1104/pp.110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. The story of phosphorus: global food security and food for thought. Global Environmental Change. 2009;19:292–305. [Google Scholar]
- De Smet I, White PJ, Bengough AG, et al. Analyzing lateral root development: how to move forward. The Plant Cell. 2012;24:15–20. doi: 10.1105/tpc.111.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Yang M, Hu Y, et al. Quantitative trait loci affecting seed mineral concentrations in Brassica napus grown with contrasting phosphorus supplies. Annals of Botany. 2010;105:1221–1234. doi: 10.1093/aob/mcq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding GD, Liao Y, Yang M, Zhao ZK, Shi L, Xu FS. Development of gene-based markers from Arabidopsis thaliana functional genes involved in phosphorus homeostasis and mapping in Brassica napus. Euphytica. 2011;181:305–322. [Google Scholar]
- Ding G, Zhao Z, Liao Y, et al. Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Annals of Botany. 2012;109:747–759. doi: 10.1093/aob/mcr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan HY, Shi L, Ye XS, Wang YH, Xu FS. Identification of phosphorous efficient germplasm in oilseed rape. Journal of Plant Nutrition. 2009;32:1148–1163. [Google Scholar]
- Fang Z, Shao C, Meng Y, Wu P, Chen M. Phosphate signaling in Arabidopsis and Oryza sativa. Plant Science. 2009;176:170–180. [Google Scholar]
- Gahoonia TS, Nielsen NE. Variation in root hairs of barley cultivars doubled soil phosphorus uptake. Euphytica. 1997;98:177–182. [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature. 2012;488:535–539. doi: 10.1038/nature11346. [DOI] [PubMed] [Google Scholar]
- George TS, Frasson AM, Hammond JP, White PJ. Phosphorus nutrition: rhizosphere processes, plant response and adaptations. In: Bünemann E, Oberson A, Frossard E, editors. Phosphorus in action. Dordrecht: Springer; 2011. pp. 245–271. [Google Scholar]
- Hammond JP, Broadley MR, White PJ. Genetic responses to phosphorus deficiency. Annals of Botany. 2004;94:323–332. doi: 10.1093/aob/mch156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ, et al. Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. Journal of Experimental Botany. 2009;60:1953–1968. doi: 10.1093/jxb/erp083. [DOI] [PubMed] [Google Scholar]
- Hammond JP, Mayes S, Bowen HC, et al. Regulatory hotspots control plant gene expression under varying soil phosphorus (P) supply in Brassica rapa. Plant Physiology. 2011;156:1230–1241. doi: 10.1104/pp.111.175612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffer P, Prud'homme MPR, Muirheid B, Isherwood KF. Proceedings 586. York, UK: International Fertiliser Society; 2006. Phosphorus fertilisation: issues and outlook. [Google Scholar]
- Hu Y, Ye X, Shi L, Duan H, Xu F. Genotypic differences in root morphology and phosphorus uptake kinetics in Brassica napus under low phosphorus supply. Journal of Plant Nutrition. 2010;33:889–901. [Google Scholar]
- Jain A, Vasconcelos MJ, Raghothama KG, Sahi SV. Molecular mechanisms of plant adaptation to phosphate deficiency. Plant Breeding Reviews. 2007;29:359–419. [Google Scholar]
- Li D, Zhu H, Liu K, et al. Purple acid phosphatases of Arabidopsis thaliana. Journal of Biological Chemistry. 2002;277:27772–27781. doi: 10.1074/jbc.M204183200. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. The Plant Journal. 2002;29:751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- Long Y, Xia W, Li R, et al. Epigenetic QTL mapping in Brassica napus. Genetics. 2011;189:1093–1102. doi: 10.1534/genetics.111.131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiology. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis: identification of BIG as a mediator of auxin pericycle cell activation. Plant Physiology. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F. Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics. 2003;163:711–722. doi: 10.1093/genetics/163.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theoretical and Applied Genetics. 2005;110:742–753. doi: 10.1007/s00122-004-1900-9. [DOI] [PubMed] [Google Scholar]
- Lynch J. Roots of the second green revolution. Australian Journal of Botany. 2007;55:493–512. [Google Scholar]
- Lynch JP, Brown KM. New roots for agriculture: exploiting the root phenome. Philosophical Transactions of the Royal Society B. 2012;367:1598–1604. doi: 10.1098/rstb.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Yang TJ, et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biology. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. http://dx.doi.org/10.1186/gb-2009-10-10-r111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nacry P, Canivenc G, Muller B, et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiology. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. [Google Scholar]
- Plaxton WC, Tran HT. Metabolic adaptations of phosphate-starved plants. Plant Physiology. 2011;156:1006–1015. doi: 10.1104/pp.111.175281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlson DS, Gregory PJ, Whalley WR, et al. Soil management in relation to sustainable agriculture and ecosystem services. Food Policy. 2011;36:S72–S87. [Google Scholar]
- Prinzenberg AE, Barbier H, Salt DE, Stich B, Reymond M. Relationships between growth, growth response to nutrient supply, and ion content using a recombinant inbred line population in Arabidopsis. Plant Physiology. 2010;154:1361–1371. doi: 10.1104/pp.110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoretical and Applied Genetics. 2006;114:67–80. doi: 10.1007/s00122-006-0411-2. [DOI] [PubMed] [Google Scholar]
- Raven JA. Phosphorus and the future. In: White PJ, Hammond JP, editors. The ecophysiology of plant–phosphorus interactions. Dordrecht: Springer; 2008. pp. 271–283. [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant, Cell & Environment. 2006;29:115–125. doi: 10.1111/j.1365-3040.2005.01405.x. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Lynch JP, Ryan PR, et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant and Soil. 2011;349:121–156. [Google Scholar]
- Robinson DL. Estimation and use of variance components. The Statistician. 1987;36:3–14. [Google Scholar]
- Römheld V, Kirkby EA. Research on potassium in agriculture: needs and prospects. Plant and Soil. 2010;335:155–180. [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant and Cell Physiology. 2005;46:174–184. doi: 10.1093/pcp/pci011. [DOI] [PubMed] [Google Scholar]
- Shane MW, Lambers H. Cluster roots: a curiosity in context. Plant and Soil. 2005;274:101–125. [Google Scholar]
- Shi J, Li R, Qiu D, et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics. 2009;182:851–861. doi: 10.1534/genetics.109.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaiman Z, Marschner P, Wang DM, Rengel Z. Growth, P uptake and rhizosphere properties of wheat and canola genotypes in an alkaline soil with low P availability. Biology and Fertility of Soils. 2007;44:143–153. [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nature Genetics. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. The Plant Journal. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- Tiessen H. Phosphorus in the global environment. In: White PJ, Hammond JP, editors. The ecophysiology of plant–phosphorus interactions. Dordrecht: Springer; 2008. pp. 1–7. [Google Scholar]
- Van Ooijen. JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. 2006 Kyazma B.V, Wageningen. [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Veneklaas EJ, Lambers H, Bragg J, et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytologist. 2012;195:306–320. doi: 10.1111/j.1469-8137.2012.04190.x. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil D, Aarts MGM, Koornneef M, Nelissen H, Ernst WHO. Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant, Cell & Environment. 2004;27:828–839. [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2·5. Raleigh, NC: Department of Statistics, North Carolina State University; 2011a. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm . [Google Scholar]
- Wang X, Wang H, Wang J, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. 2011b;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR, Greenwood DJ, Hammond JP. Proceedings 568. York, UK: International Fertiliser Society; 2005. Genetic modifications to improve phosphorus acquisition by roots. [Google Scholar]
- White PJ, Hammond JP. Phosphorus nutrition of terrestrial plants. In: White PJ, Hammond JP, editors. The ecophysiology of plant–phosphorus interactions. Dordrecht: Springer; 2008. pp. 51–81. [Google Scholar]
- White PJ, Hammond JP. The sources of phosphorus in the waters of Great Britain. Journal of Environmental Quality. 2009;38:13–26. doi: 10.2134/jeq2007.0658. [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology. 2001;126:875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M, Wegner J, Ae N, Yano M. Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theoretical and Applied Genetics. 2002;105:890–897. doi: 10.1007/s00122-002-1051-9. [DOI] [PubMed] [Google Scholar]
- Yang M, Ding G, Shi L, Feng J, Xu F, Meng J. Quantitative trait loci for root morphology in response to low phosphorus stress in Brassica napus. Theoretical and Applied Genetics. 2010;121:181–193. doi: 10.1007/s00122-010-1301-1. [DOI] [PubMed] [Google Scholar]
- Yang M, Ding G, Shi L, Xu F, Meng J. Detection of QTL for phosphorus efficiency at vegetative stage in Brassica napus. Plant and Soil. 2011;339:97–111. [Google Scholar]
- Zhao J, Jamar DCL, Lou P, et al. QTL analysis of phytate and phosphate concentrations in seeds and leaves of Brassica rapa. Plant, Cell & Environment. 2008;31:887–900. doi: 10.1111/j.1365-3040.2008.01791.x. [DOI] [PubMed] [Google Scholar]
- Zhao J, Paulo MJ, Jamar D, et al. Association mapping of leaf traits, flowering time, and phytate content in Brassica rapa. Genome. 2007;50:963–973. doi: 10.1139/g07-078. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays) Functional Plant Biology. 2005;32:749–762. doi: 10.1071/FP05005. [DOI] [PubMed] [Google Scholar]