Abstract
Dysregulation of eating behavior can lead to obesity, which affects 10% of the adult population worldwide and accounts for nearly 3 million deaths every year. Despite this burden on society, we currently lack effective pharmacological treatment options to regulate appetite. We used Drosophila melanogaster larvae to develop a high-throughput whole organism screen for drugs that modulate food intake. In a screen of 3630 small molecules, we identified the serotonin (5-hydroxytryptamine or 5-HT) receptor antagonist metitepine as a potent anorectic drug. Using cell-based assays we show that metitepine is an antagonist of all five Drosophila 5-HT receptors. We screened fly mutants for each of these receptors and found that serotonin receptor 5-HT2A is the sole molecular target for feeding inhibition by metitepine. These results highlight the conservation of molecular mechanisms controlling appetite and provide a method for unbiased whole-organism drug screens to identify novel drugs and molecular pathways modulating food intake.
About 1 in every 10 adults worldwide is overweight or obese1 and obesity is a risk factor for morbidity and mortality from cardiovascular diseases, diabetes, certain cancers, and musculoskeletal disorders. Despite strong interest in research addressing obesity, safe and effective pharmacological options for the prevention and treatment of this condition remain elusive2,3. Currently, most drug-discovery efforts are based on in vitro assays with candidate targets, but in vitro assays do not reconstitute the complexity of whole organisms. This is particularly relevant for drugs modulating feeding behavior and metabolic homeostasis, since both arise from complex interactions within and between the central nervous system, the digestive tract, and fat-storage organs4,5,6, which cannot be modeled in vitro.
An alternative to in vitro-based screens is phenotype-based whole organism screens7,8,9,10. Whole organism drug screens provide several advantages over in vitro assays. Active drugs are by definition bio-available and potential toxicity can be evaluated at early project stages. Further, an effective drug need not act through a well-validated target, but can have novel or complex mechanisms of action. However, whole organism drugs screens can be costly and time-consuming. These disadvantages can be partially overcome by using model organisms that can be raised cost-effectively in large quantities, like the vinegar fly Drosophila melanogaster. Flies and vertebrates share many metabolic functions, molecular machinery, and analogous organ systems that control nutrient uptake, storage, and metabolism11,12,13,14,15. Like humans, flies regulate circulating sugar levels according to food availability, and store excess energy in the form of glycogen and lipids. These reserves are mobilized during periods of energy consumption12,16,17. As seen in many animals, fasted flies increase food foraging and intake18. Two master metabolic regulators in vertebrates, insulin and leptin, have functional homologues in the fly13,14. In addition, an unbalanced diet can trigger a type-2 diabetes-like insulin resistance and obesity phenotypes in the fly19.
Here we report the development of a high-throughput drug-screen for Drosophila larval feeding. We identify the serotonin (5-hydroxytryptamine or 5-HT) receptor antagonist metitepine as a potent anorectic drug and show that all five fly 5-HT receptors are inhibited by this drug. Despite its broad spectrum antagonism of Drosophila 5-HT receptors, metitepine requires only receptor 5-HT2A for its in vivo anti-feeding activity. Our results highlight the potential of Drosophila as a tool for pharmacological study of feeding behavior and provide a powerful method for drug discovery and target identification.
Results
High-throughput feeding assay for Drosophila larvae
To screen for drugs that modify food intake in whole animals, we developed a high-throughput assay that allowed us to monitor ingestion of fluorescent liquid food by Drosophila first instar larvae in 96-well plates read by a plate reader (Fig. 1a). Larvae were dispensed into plates and fed liquid food consisting of sugar and yeast extract and supplemented with fluorescein for visualization. After washing uningested fluorescent food from the wells, we quantified the fluorescein ingested by the larvae that was visible in the digestive tract (Fig. 1b).
Figure 1. A high-throughput assay to monitor Drosophila larval feeding.
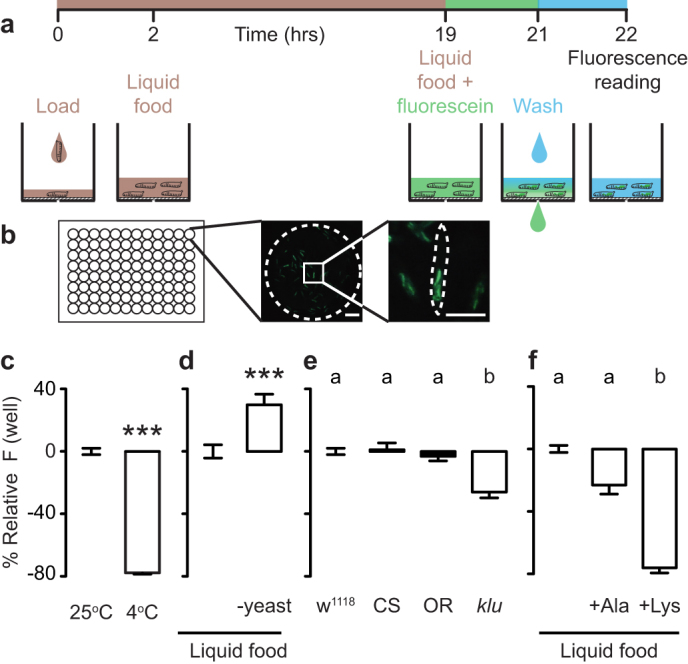
(a) Assay schematic. (b) Representative picture of the bottom of a single well of a 96-well plate with larvae treated as in (a). Scale bars: 250 μm. (c) Relative fluorescence of larvae incubated at either 25°C or 4°C during the fluorescein feeding stage (n = 32). Fluorescence normalized to 25°C. Data were compared using Mann Whitney test. (d) Relative fluorescence of larvae that were pre-fed either complete liquid food or liquid food lacking yeast extract overnight (n = 16). Fluorescence plotted relative to animals fed liquid food. Data were compared using t test. (e) Relative fluorescence of larvae of different genotypes: w1118, Canton-S (CS), Oregon-R (OR), klumpfuss09036 (klu) (n = 22–24). Fluorescence plotted relative to w1118. (f) Relative fluorescence of larvae that were fed liquid food or liquid food supplemented with 400 mM alanine or lysine. Fluorescence plotted relative to liquid food (n = 12). In (e–f), data was compared with Kruskal-Wallis test, followed by Dunn's test. In (c–f), error bars indicate s.e.m. In (c–d) *** p < 0.001. Significant differences are labeled with different letters in (e–f).
To evaluate the dynamic range of our assay, we carried out control experiments to either decrease or increase food intake in the larvae. When the animals were cold-paralyzed while feeding on fluorescent food, ingestion was reduced (Fig. 1c). When larvae were selectively fasted for protein by removing yeast extract from the liquid food before being exposed to fluorescent food, they showed a post-fasting rebound in which they ingested more standard liquid food than control animals continuously fed standard liquid food (Fig. 1d). The dynamic range of feeding suppression was more than two times greater than feeding enhancement in these experiments, perhaps because larvae are continual feeders and may be ingesting at a near maximal rate during basal conditions20.
We next tested a known feeding mutant in our assay by comparing the fluorescent signal accumulated in three different wild-type strains (w1118, Canton-S, and Oregon-R) and a klumpfuss09036 mutant (klu, ref. 20). klu encodes a transcription factor necessary for proper expression of the neuropeptide hugin, whose activity is required for normal feeding behavior in Drosophila20. All three wild-type strains ingested significantly more than the feeding mutant (Fig. 1e). In addition, we confirmed previous reports21 that high concentrations of dietary amino acids suppressed food intake (Fig. 1f).
Drug screen for modulators of food intake
After validating our feeding assay, we screened for small molecules that modulated food intake. In a pilot screen of 415 compounds tested individually at 20 μg/ml, we identified one compound, cycloheximide, which inhibited feeding (data not shown). This established a hit rate of 0.24%. To improve the throughput of the subsequent primary screen, we used pools of 3 to 4 compounds per well (see Methods for details).
3630 small molecules were tested in the primary screen (Fig. 2a). The compounds were obtained primarily from annotated chemical libraries, such that each compound had at least one known cellular target (see Methods for details). The average signal of all the drug-treated wells was less than 3% different from the average of solvent-treated wells, indicating that the drugs did not cause generalized toxicity (Fig. 2b). 279 and 114 compounds were identified as candidate anorectic (feeding suppressant) and orexigenic (feeding stimulant) drugs, respectively, by the criterion that they differed from solvent controls by more than one standard deviation (Fig. 2a). The anorectic compounds caused an average decrease in fluorescent signal of 27%, while the orexigenic compounds caused an average increase of 18% (Fig. 2b).
Figure 2. A small molecule screen identifies metitepine as a feeding suppressant.
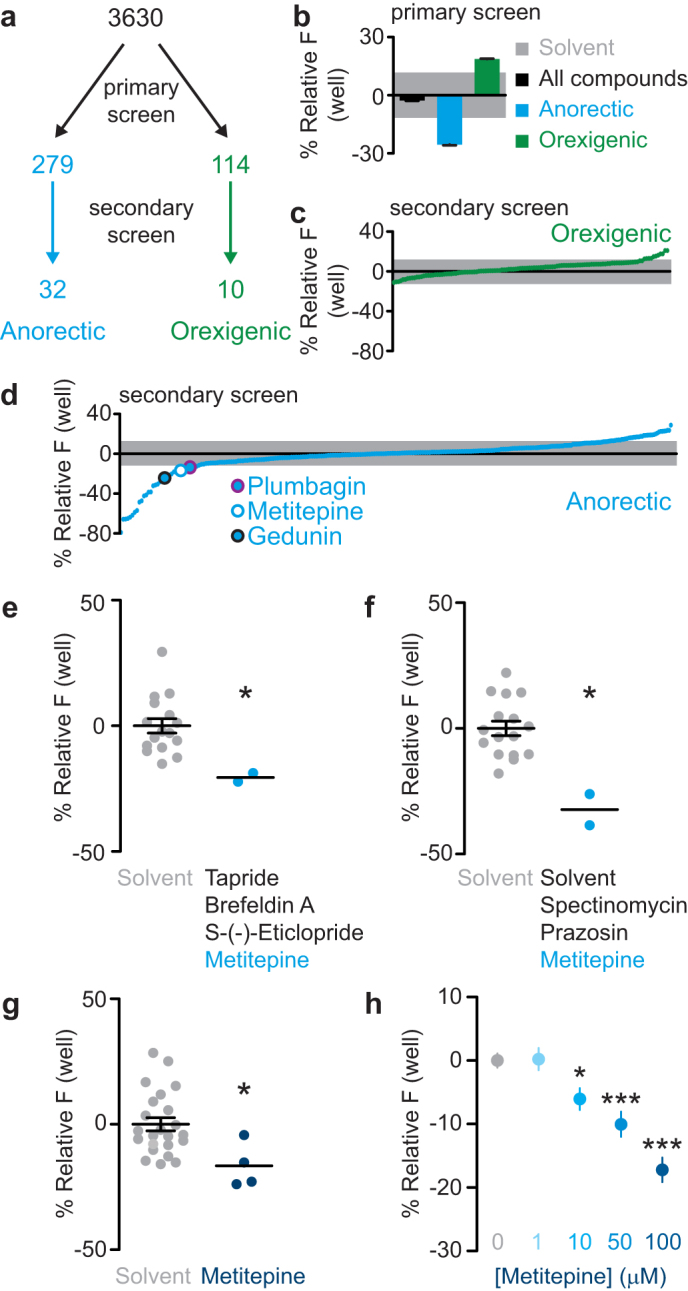
(a) Diagram of the drug screen. (b) Fluorescence, plotted relative to solvent-treated wells, of all compounds tested in the primary screen (black), anorectic compounds (cyan), and orexigenic compounds (green). The gray shaded area indicates the standard deviation of all wells treated only with solvent. (c, d) Average fluorescence of primary screen orexigenic (c) or anorectic (d) compounds tested in the secondary screen plotted relative to the solvent-treated wells. Individual compounds are indicated as green (c) or cyan (d) dots and the gray shaded area indicates the standard deviation of all wells treated only with solvent. In (d) three anorectic compounds are highlighted by circles. (e, f) Relative fluorescence accumulation in wells treated with the mixtures of four (e) or three (f) compounds including metitepine during the primary screen, plotted relative to their solvent control wells. Each dot is the signal from a single well, horizontal lines are mean ± s.e.m. (e–g). (g) Fluorescence accumulation in solvent or metitepine treated wells during the secondary screening. (h) Dose-response effects of metitepine. Y-axis shows relative accumulated fluorescence (n = 31 for solvent; n = 15–16 for all concentrations of metitepine); mean ± s.e.m. is plotted. In (e–g) data were compared with Mann Whitney test. In (h) ANOVA followed by Dunnett's test was used. * p < 0.5, *** p < 0.001 compared to solvent-treated controls.
The 393 candidate small molecules were re-tested individually in the secondary screen (Fig. 2a). Of the anorectic and orexigenic candidates, 32 and 10 compounds, respectively, were reconfirmed as hits, defined as differing from solvent controls by more than one standard deviation (Fig. 2a, c, d, Supplementary Table 1). We searched for reported molecular targets for each of the 42 hits of the secondary screen using a drug-discovery database (https://www.collaborativedrug.com/). Two known insect anti-feedants, gedunin and plumbagin22,23, were among these compounds (Fig. 2d), confirming the efficacy of our screen. We chose 14 compounds for verification of a dose-response curve (Supplementary Table 1), based on their annotation as drugs that target neuromodulators, cell signaling, and/or neuronal activity. From these, only metitepine, a non-selective antagonist of 5-HT receptors, and reserpine, an inhibitor of the vesicular mono-amine transporter (VMAT), showed reliable dose-dependent responses and were selected for further characterization. Reserpine was subsequently discarded because it dramatically reduced larval locomotion at concentrations as low as 10 μM (data not shown). In light of these results, we concluded that the effects of reserpine on feeding were secondary to a general effect on locomotion and muscular tone, as confirmed by the sluggish phenotype of the dVMAT mutant larvae24.
Metitepine decreased food accumulation by more than one standard deviation when tested in combination with other drugs, during the primary screen (Fig. 2e, f), or alone, in the secondary screen (Fig. 2g). Dose-response experiments indicated that the threshold concentration for metitepine efficacy was 10 μM, with increasing effects at 50 and 100 μM (Fig. 2h).
Metitepine decreases feeding persistently, reversibly, and specifically
To ask if metitepine decreases larval feeding on conventional fly food, we tested the effect of the drug on larvae fed a standard laboratory cornmeal-agar-molasses diet supplemented with the dye bromophenol blue. We quantified the amount of food ingested by measuring the optical density (O.D.) of the gut in individual larvae. Larvae treated with the drug accumulated less solid food in their digestive tract as reflected by a lower O.D. (Fig. 3a). The effect of metitepine on solid food accumulation was dose-dependent (Fig. 3b) with a threshold efficacy dose of 50 μM.
Figure 3. Metitepine decreases food ingestion persistently but reversibly.
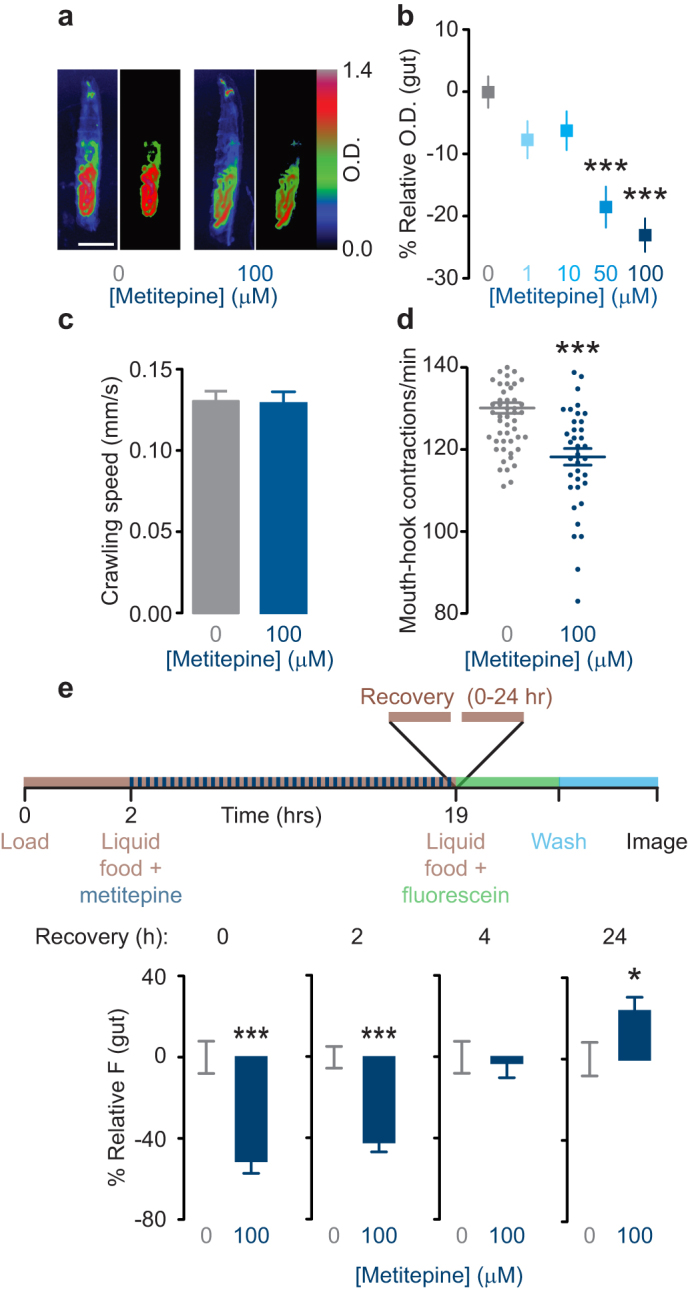
(a) Representative pseudo-color pictures of larvae fed either solvent or 100 μM metitepine in solid food with bromophenol blue. In each pair of images, the left is a whole larva and the right is the gut region quantified in MetaMorph. Scale bar: 250 μm. (b) Dose-response effects of metitepine in solid food with bromophenol blue. Y-axis shows relative optical density of gut-region (n = 60 for solvent; n = 28–38 for metitepine). Data were compared using ANOVA followed by Dunnett's test. (c) Crawling speed on an agar surface of larvae treated with either solvent or 100 μM metitepine (n = 18–20). (d) Mouth-hook contraction rate in yeast-suspension of larvae treated with either solvent or 100 μM metitepine (n = 58 and 38, respectively). t test was used for comparison. (e) Time course of recovery after metitepine treatment. The upper diagram shows a schematic of the experiment. Y-axis shows relative fluorescence (0 h: n = 42 and n = 30; 2 h: n = 47 and n = 42; 4 h: n = 43 and n = 45; 24 h: n = 49 and n = 56; for solvent and metitepine, respectively). Mann-Whitney test was used for comparisons. * p < 0.5, *** p < 0.001 compared to solvent-treated controls. In all graphs, error bars are s.e.m.
To rule out the possibility that metitepine was causing a general locomotor defect in larvae, we measured their crawling speed when fed solvent or metitepine. Metitepine-treated animals crawled at the same speed as solvent treated controls (Fig. 3c).
Food accumulation in the digestive tract is a function of both ingestion and excretion. To establish a direct link between metitepine treatment and food intake, we measured mouth-hook contraction rates of larvae that were treated with either solvent or 100 μM metitepine. Mouth-hook contraction is the motor behavior associated with food intake in larvae. Animals treated with the drug displayed decreased mouth-hook contraction rates, confirming that metitepine had a direct effect on food intake (Fig. 3d). Notably, since mouth-hook contractions were measured after drug treatment (see Methods), this result also suggests that metitepine is not required to be present in food to induce a decrease in ingestion.
To further explore the time course of metitepine action on feeding behavior, we measured food intake at various time points after metitepine treatment. When tested immediately after exposure, drug-treated larvae showed reduced food accumulation (Fig. 3e, 0-h recovery). The anorectic effect of metitepine lasted for 2 hours but was not evident 4 hours after treatment (Fig. 3e). At 24 hours after treatment, metitepine-treated larvae ate significantly more, suggesting that larvae were rebounding from drug-induced fasting (Fig. 3e).
Metitepine is a non-selective antagonist of Drosophila 5-HT receptors
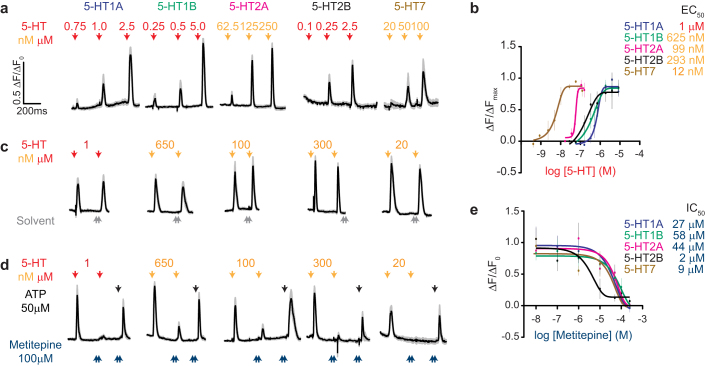
Metitepine is a broad spectrum antagonist of vertebrate 5-HT receptors. To investigate the pharmacology of this drug on Drosophila 5-HT receptors, we expressed each in mammalian tissue culture cells and carried out calcium imaging experiments. Four 5-HT receptors have been previously identified and cloned: 5-HT1A and 5-HT1B (ref. 25), 5-HT2 (ref. 26), and 5-HT7 (ref. 27). Of these, 5-HT1A, 5-HT1B, and 5-HT7 were previously expressed in heterologous systems, shown to respond to 5-HT, and to activate different intracellular effector systems25,27,28. In binding-competition assays 5-HT2 was shown to bind to both 5-HT and metitepine26. A fifth putative receptor (CG42796) was annotated as a 5-HT receptor based on homology29. We cloned CG42796 and propose that it be named 5-HT2B, since its closest homologue is 5-HT2 (ref. 29). We suggest that the gene formerly known as 5-HT2 be denoted as 5-HT2A. This revised nomenclature for 5-HT2A and 5-HT2B is used throughout the manuscript.
We expressed all five 5-HT receptors in HEK-293T cells and monitored their activity by measuring intracellular calcium concentrations. All of the receptors induced a dose-dependent response to 5-HT (Fig. 4a). From the dose-responses curves of each receptor we calculated a half effective concentration (EC50, Fig. 4b). The most sensitive receptor was 5-HT7 (EC50 = 12 nM) and the least sensitive receptor was 5-HT1A (EC50 = 1 μM). We next established stimulus conditions in which we applied two pulses of 5-HT without desensitizing any of the receptors (Fig. 4c). Under these conditions, 100 μM metitepine dramatically suppressed or completely abolished responses to 5-HT in all 5-HT receptors (Fig. 4d). Control experiments confirmed that metitepine did not kill the cells because ATP, a ligand for endogenous purinergic receptors, activated all cells after metitepine treatment (Fig. 4d). Metitepine had an effective inhibitory concentration (IC50) in the μM range, ranging from 2 μM for 5-HT2B to 58 μM for 5-HT1B (Fig. 4e). These pharmacological experiments confirmed that all five candidate 5-HT receptors in the Drosophila genome respond to serotonin. However, since they all showed sensitivity to metitepine, further genetic experiments were required to identify the molecular target of the anorectic drug in vivo.
Figure 4. Metitepine is an antagonist of all known Drosophila 5-HT receptors.
(a) Traces show calcium responses for each receptor to increasing concentration of 5-HT (red arrow: μM; orange arrow: nM). All traces are average responses in black (± s.e.m. in gray) of 10–12 simultaneously recorded cells. (b) Dose-response curves were obtained normalizing the peak response at each concentration of 5-HT to the maximal response in that cell (ΔF/ΔFMAX). n = 3–6 plates, 10–12 cells each. (c) Traces of cells after treatment with serotonin and then solvent. (d) Traces of cells after treatment with serotonin and then 100 μM metitepine. (e) Inhibitory dose-response curves of metitepine. In b and e, error bars are s.e.m.
5-HT2A is required for the anorectic actions of metitepine and for normal feeding behavior
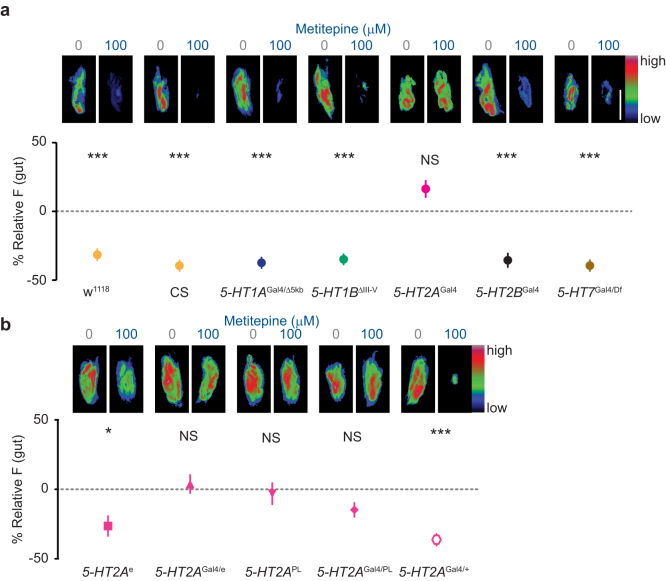
We reasoned that if metitepine were acting through a specific 5-HT receptor, mutating that gene would render larvae resistant to the drug. We generated or obtained null mutants for each one of the five Drosophila 5-HT receptors and asked if they were sensitive to the anorectic effects of 100 μM metitepine. All mutants except 5-HT2AGal4 were sensitive to metitepine and showed a decrease in feeding similar to that seen in wild-type strains (Fig. 5a). In contrast, 5-HT2A receptor mutants were resistant to the effects of this drug.
Figure 5. 5-HT2A is the in vivo target of metitepine.
(a, b) Top pictures are representative pseudo-color images of larval digestive tracts of the indicated genotypes that were fed the specified concentration of metitepine or solvent in liquid food with fluorescein. The Y-axis in the graphs is relative fluorescence of metitepine-fed larvae to solvent-fed larvae. n = 88–99 for all genotypes, except 5-HT1BΔIII-V (76), 5-HT2AGal4 (124), and 5-HT7Gal4/Df (65). Scale bar (white): 100 μm applies to all panels. Data were compared using Kruskal-Wallis test followed by Dunn's test. * p < 0.05, *** p < 0.001.
To confirm this observation, we tested additional 5-HT2A mutant alleles. 5-HT2Ae01363 and 5-HT2APL00052 are different pBac transposon insertions in the 5-HT2A locus. We tested each as homozygous insertions and in heteroallelic combinations with the original 5-HT2AGal4 mutant.
5-HT2Ae01363 (5-HT2Ae in Fig. 5b) interferes with proper splicing of the transcript, but is not a null30. Consistent with this, 5-HT2Ae was sensitive to the effects of metitepine when tested as a homozygote (Fig. 5b). However, when 5-HT2Ae was tested in combination with the original 5-HT2AGal4, the heteroallelic mutant combination 5-HT2AGal4/e was insensitive to metitepine (Fig. 5b).
5-HT2APL00052 (5-HT2APL in Fig. 5b) has dramatically reduced levels of 5-HT2A mRNA31. Both 5-HT2APL homozygous mutants and the heteroallelic mutant combination 5-HT2AGal4/PL were insensitive to metitepine (Fig. 5b). Heterozygous 5-HT2AGal4 larvae showed normal sensitivity to the drug (Fig. 5b).
If metitepine suppresses feeding by blocking the activation of 5-HT2A, mutating the gene should result in larvae that eat less. Indeed, 5-HT2AGal4/e mutants ate less than wild-type larvae (Fig. 6a).
Figure 6. 5-HT2A is necessary for normal larval feeding.
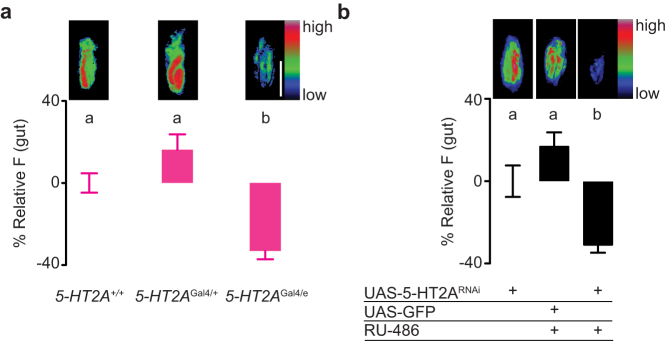
(a,b) Top pictures are representative pseudo-color images of digestive tracts of larvae of the indicated genotypes that fed in liquid food with fluorescein. Scale bar (white): 100 μm applies to all panels. The graphs show relative fluorescence to wild-type larvae (c) or to Elav-GeneSwtich > 5-HT2ARNAi larvae that were not fed RU-486 (d). n = 98–126 in (c); n = 90–104 in (d). Data were compared using Kruskal-Wallis test followed by Dunn's test. In all graphs, error bars are s.e.m. Significant differences are labeled with different letters.
To further confirm the role of 5-HT2A in larval feeding behavior, we knocked down 5-HT2A by conditional expression of a 5-HT2A RNAi with a pan-neuronal GeneSwitch system32. Larvae in which neuronal expression of 5-HT2A-RNAi was induced ate less than larvae in which the RNAi was not induced or in control larvae in which GFP was induced (Fig. 6b). Thus, genetic knock-down of 5-HT2A in a time frame similar to the action of metitepine was sufficient to phenocopy the drug-induced feeding phenotype.
Discussion
Serotonin is involved in regulating appetite, food intake, and metabolic homeostasis in organisms ranging from C. elegans to humans. In C. elegans, serotonin activates overall feeding by activating two separate neural pathways that respectively control pharyngeal pumping and isthmus peristalsis33. In Drosophila, serotonin has been shown to play a trophic role during embryonic development in the establishment of neuronal innervation to the gut34. In adult female flies, serotonin controls the postmating dietary switch to protein-rich food35. We show here that the 5-HT receptor antagonist metitepine reduces food intake in Drosophila larvae and that the drug acts selectively through the 5-HT2A receptor.
Interestingly, metitepine was previously identified in a different small molecule screen as a compound that extends lifespan in C. elegans8. There is a known connection between dietary restriction and lifespan across several organism including primates36. We have not tested the effect of metitepine on Drosophila lifespan, but this would be of interest in future studies on this drug.
In mammals, the role of serotonin in controlling appetite and body-weight is complex37. It appears that the level of brain serotonin signaling has an inverse relationship with food intake: when brain serotonin signaling is increased, food intake is reduced, and vice versa. For example, serotonin re-uptake inhibitors reduce food intake2,37. Part of the complexity of the serotonin system that controls metabolic homeostasis in mammals might arise from the fact mammals have 14 5-HT receptor subtypes. This raises the possibility of serotonin having divergent effects on food intake and body weight depending on the receptor subtype activated. Drosophila, with only five receptor subtypes, offers a simplified model in which to study the core mechanisms by which serotonin regulates food intake and coordinates metabolic homeostasis.
Here we established that Drosophila larvae can be used to screen for drugs that modulate food intake. Few model organisms allow the combination of large-scale drug screens with genetic screens to identify bioactive small molecules and their in vivo targets2. Drosophila is an appealing model to study appetite control because 60% of functional human genes have orthologues in the fly38 and Drosophila has a specialized tissue for fat storage that controls metabolic homeostasis using a leptin-like signaling mechanism13. Future work can apply these methods to larger scale screens of novel compounds to identify new pathways regulating feeding behavior.
Methods
Fly stocks
Flies were maintained on conventional cornmeal-agar-molasses medium and, unless otherwise stated, under a natural light-dark cycle, at room temperature. klumpfuss09036 (stock #11733), 5-HT1AΔ5Kb (stock #27640), 5-HT1BMB08181 (stock #24240), 5-HT2APL00052 (stock #19367), 5-HT2ARNAi (stock #31882) and Df(3R)tll-e (stock #5415) were obtained from the Bloomington Stock Center. 5-HT2Ae01363 was obtained from the Harvard Exelixis Collection. 5-HT1AGal4, 5-HT2AGal4, 5-HT2BGal4, and 5-HT7Gal4 were generated by J.H and Y.R. by replacing the first coding exon of each gene by Gal4, and will be reported elsewhere (Huang and Rao, in preparation).
Embryo collection
For embryo collection, grape-juice 2%-agar plates were used. Flies were allowed to lay eggs for 24 hours at 25°C. Eggs were further incubated at 18°C for another 24 hours. Egg laying and embryo development were both performed at 70% humidity and in a 12 hour light: 12 hour dark cycle.
First instar larva collection
Previously hatched larvae were removed from the embryo-collection plates under a stream of water. Plates were further incubated for 2 hours at 25°C to allow new larvae to hatch.
For primary and secondary screens
Newly hatched larvae were collected in a cell strainer with a gentle stream of distilled water, rinsed and re-suspended in liquid food (100 g/l yeast extract, 100 g/l glucose, 7.5% sucrose, 0.15% nipagin, 6.25 μg/ml cholesterol).
For individual larval assays
Newly hatched larvae were collected with a brush, and transferred to a vial with conventional medium or to a 96-well plate with liquid food. To facilitate penetration of the larvae into the solid food, incisions were made on the surface of the medium. Larvae were incubated at 25°C and 70% humidity.
Liquid food feeding assay
Seventy-five larvae were dispensed into each well a filter-bottom 96-well plate (Millipore, Part. No. MSRLN04) using a COPAS Select worm sorter (Union Biometrica). Once loading was completed, the liquid content of the plates was filtered away and replaced with 100 μl of liquid food. The plates were incubated for 16–18 hours at 25°C and 70% humidity. Thereafter, the food was replaced with food that contained 0.3% fluorescein (sodium salt, Sigma Cat. No. 6377). Before the fluorescent signal was quantified the plates were washed 4× with 300 μl double-distilled water, 10× with 0.05% PBT, 6× with 400 mM lysine and 2× with 100 mM Na-citrate, 100 mM NaCl, pH2 (citrate buffer). The larvae were kept in 50 μl of citrate buffer for quantification or imaging capture. The fluorescent signal from the plate was acquired in a 5 × 5 circular grid using an EnVision Plate Reader (Perkin Elmer). The total fluorescent value of each well was calculated by adding the data points.
Small-molecule screen
Primary screen
The small molecules were obtained from the LOPAC1280, Prestwick, GreenPharma, and MicroSource Spectrum libraries, and were provided by the High-Throughput Screening Resource Center (HTSRC) of The Rockefeller University. A total of 3688 compounds, representing 3630 unique structures, were screened. Small molecules were pooled at 10 μg/ml each such that each compound was represented twice in two independent mixtures. Only those compounds that showed the required effect two times were chosen for confirmation in the secondary screen. For the primary screen, sixteen 384-well plates with a different small-molecule in each well were mixed using a 4 × 4 grid into eight 384-well destination plates. Since we screened 3688 small molecules, and the 384-well plates contain 352 usable wells each (32 wells in each plate are reserved for solvent controls), a total of 1944 wells in the source plates were empty (16 × 352 − 3688 = 1944). While the majority of mixtures contained 4 compounds, about a third contained only 3. To apply the drugs to the larvae loaded into the 96-well plates, each 384-well plate quadrant was treated as an independent 96-well plate.
The cut-off for hit-identification was arbitrarily set to one standard deviation above or below the solvent-treated wells for anorectic and orexigenic compounds, respectively. The compounds were applied to the larvae in 96-well plates in the liquid food together with fluorescein for 16–18 hours. In each 96-well plate, 80 compound-mixtures were tested and 16 wells were treated only with solvent as control (1.6% DMSO). Each plate was tested in duplicate.
Secondary screen
Compounds were screened individually at 40 μg/ml. Each hit was tested in two rounds, in duplicate. Each screening plate contained 8 solvent-treated control wells.
Solid food feeding assay
Zero-to-two hour old larvae were transferred to conventional solid food containing either solvent or 100 μM metitepine and 0.05% bromophenol blue, and allowed to feed for 17 hours at 25°C and 70% humidity. Larvae were recovered from the food with a brush, rinsed in PBS, and photograph under a SMZ1500 dissecting scope (Nikon). Images were captured with a DS-2Mv digital camera (Nikon) and a NIS-Elements F acquisition program. Larvae were immobilized in a cold plate set to 4°C. Reflective light was adjusted to make the background of each picture 12% gray. Images of individual larvae were analyzed in MetaMorph (Molecular Devices) to calculate the optical density of the digestive tract. Optical density values of drug-treated larvae were analyzed always in parallel with same day solvent-treated larvae.
Mouth-hook contractions
Zero-to-two hour old larvae were transferred to conventional solid food containing either solvent or 100 μM metitepine, and allowed to feed for 17 hours at 25°C and 70% humidity. Larvae were recovered from the food with a brush, rinsed in PBS and transfer to a 2% yeast suspension at room temperature. After a one-minute acclimation, a one-minute movie was recorded at 6.25 frames per second (fps) using a Nikon SMZ1500 dissecting scope, a Rolera-RX camera (Q Imaging), and Q Capture 6.0 Software (Q Imaging). Movies were manually scored to quantify mouth-hook contraction rate.
Larval locomotion
Zero-to-two hour old larvae were transferred to conventional solid food containing either solvent or 100 μM metitepine and 0.05% bromophenol blue, and allowed to feed for 17 hours at 25°C and 70% humidity. Bromophenol blue facilitated tracking of the animals since it enhanced contrast. Larvae were recovered from the food with a brush, rinsed in PBS, and transferred to a 3% agar plate at room temperature. After a one-minute acclimation, a one-minute movie was recorded at 6.25 fps using a Nikon SMZ1500 dissecting scope, a Rolera-RX camera (Q Imaging), and Q Capture 6.0 Software (Q Imaging). Movies were analyzed in EthoVision XT 8.0 (Noldus) to calculate linear velocity.
Cloning Drosophila 5-HT receptors
5-HT1A, 5-HT1B and 5-HT2A receptors were PCR-cloned from genomic DNA extracted from transgenic flies expressing full-length cDNAs under regulation of UAS promoter (5-HT1A: Bloomington stock #27630; 5-HT1B: Bloomington stock # 27632; 5-HT2A (formerly 5-HT2): Bloomington stock #24504). The 5-HT2B receptor cDNA (GenBank accession #KC852205) was PCR-cloned from whole adult fly cDNA prepared using poly-A primers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. 5-HT7 receptor was PCR-cloned from w1118 genomic DNA. High-fidelity KOD polymerase (Novagen) was used for all cloning amplifications and the full sequences of the amplicons were verified. The following primers were used in the cloning reactions:
5-HT1A:
Forward: 5′-ATGGCGCACGAGACCAGC-3′
Reverse: 5′-CTAGAGCTTCCCGCTGCGG-3′
5-HT1B:
Forward: 5′-ATGCTGAAAACTGTGACAACAGC-3′
Reverse: 5′-TCAAATTTTCGCACTGCG-3′
5-HT2A:
Forward: 5′-ATGGAGATGCAAAGCTACTCTG-3′
Reverse: 5′-TCACCGTTTGCAGTTGCACTTG-3′
5-HT2B:
Forward: 5′-ATGGAAGAGGATGTGTATGCCT-3′
Reverse: 5′-TTATCTGCTCGGTCGCCA-3′
5-HT7:
Forward: 5′-ATGGCTTTATCTGGACAGGACT-3′
Reverse: 5′-CTAGAGAAAGCTCTCCCTCGC-3′
A 5′-GCCACC-3′ vertebrate Kozak sequence was added upstream of every forward primer. The amplicons were cloned into the XhoI-NotI sites in the pME18ST vertebrate expression vector39.
Expression of Drosophila 5-HT receptors in HEK-293T cells
HEK-293T cells were seeded on glass-bottom 35-mm Petri dishes (MatTek Corporation, Part No. P35GC-1.5-10-C) and allowed to reach ~70% confluence. Cells were transiently transfected with 2 μg of each receptor-expressing plasmid and Gα15-expressing plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Gα15 is a promiscuous G protein that couples activation of a wide variety of G-protein coupled receptor to release of calcium from intracellular stores40. Cells were kept for 24–32 hours at 37°C and 5% CO2 before imaging. To monitor intracellular Ca2+ concentrations, transfected cells were loaded for 20 minutes with 2 μM Fura2-AM. Imaging was carried out in a saline solution containing 140 mM NaCl, 5.6 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 1.25 mM KH2PO4, 2 mM Na-pyruvate, 0.17% Glucose, 5 mM HEPES (pH 7.4 with NaOH). The Petri dish was placed on an Eclipse TE 2000-U inverted microscope (Nikon) equipped with a Retiga Exi Fast Camera (Q Imaging) and excited with a Lambda DG-4 xenon lamp illumination system (Sutter Instruments). Images were acquired at 1.43 fps. Ligands and drugs were dissolved freshly every day at the indicated concentrations in saline solution and superfused directly into the Petri dish with a peristaltic pump (Miniplus 3, Gilson). Ca2+ fluctuations were recorded with MetaFluor software (version 7.1.2.0; Molecular Devices). To determine 5-HT sensitivity for each receptor, several pulses of increasing concentration of 5-HT were applied to the same cells. Pulses were delivered 2 minutes after full recovery of the previous pulse to Ca2+ resting levels. 5-HT dose-response curves were calculated normalizing the responses elicited by each concentration tested, by the maximal response elicited by 5-HT in those cells: ΔF/ΔFMAX. For metitepine inhibition, a pair pulse protocol was used. Each set of cells was stimulated twice; first they were exposed to 5-HT alone (ΔF0), then they were exposed to 5-HT and a given concentration of metitepine (ΔFMet). The concentration of 5-HT used in these experiments was closed to the calculated EC50 for each receptor and within 95% confidence intervals. The degree of metitepine inhibition was calculated by the following equation: ΔFMet/ΔF0. Data analysis and curve-fitting was done in Prism 5 (GraphPad).
Drug preparation
Primary and secondary screens
Drugs were obtained from the HTSRC of The Rockefeller University pre-dissolved in DMSO at 2.5 mg/ml.
Dose-response curves, in vitro pharmacology, and behavioral experiments
Stock solutions of metitepine (Sigma) were prepared in DMSO (100 mM or 500 mM). Stock solutions of 5-HT and ATP (both from Sigma) were prepared in water (50 mM), kept frozen at −20°C, and aliquots thawed only once.
5-HT1B mutant generation
Homozygous males carrying the Minos MB05181 transposable insertion inserted in the sixth intron of 5-HT1B were crossed to virgin females of this genotype:
SnaSco/SM6a, p(w[+mC] = hslLMiT) (Bloomington stock #24613).
The heat-shock scheme to induce expression of the Minos transposase, marker selection, and the establishment of excision lines were carried out as originally described41. Ninety-four independent excision events were analyzed by PCR with the following primers:
Forward primer: 5′-CTGCGCTCCTTCTTCAGC-3′
Reverse primer: 5′-CGTAATTGCCGCCATTATACTC-3′
One imprecise excision that removed a 1344 bp fragment from genomic DNA, thus producing a 1002 bp PCR product instead of the wild-type 2346 bp product, was selected for further characterization. The resulting allele was named 5-HT1BΔIII-V, since the deleted exons encode transmembrane segments III-V. The breakpoints of the sequence are:
AAAAAGGATGTAGAGGAATAGAATA //deletion// CTCCAAAAATAATATTTATACAATA
A precise excision was also identified in this screen by virtue of producing a 2346 bp PCR fragment, diagnostic of a clean deletion of the Minos element.
Expression of 5-HT2A-RNAi
Zero-to-two hour old transgenic larvae carrying Elav-GeneSwitch and either UAS-5-HT2A-RNAi (Bloomington stock #31882) or UAS-mCD8-GFP were transferred to fluorescent liquid food containing 160 μg/ml of RU-486 (induced) (Sigma Cat. No. M8046) or 1% ethanol (uninduced) for 17 hours.
Data presentation
Fluorescence and optical density data were always normalized to controls ran in parallel, according to the equation:
Unless otherwise noted, data are presented as mean ± s.e.m.
Statistical analysis
Statistical analysis was conducted as indicated in the figure legends using Prism (GraphPad). Every single set of data was tested for normality (Shapiro-Wilk test) and equal variance (F test or Bartlett's test). If those criteria were met, parametric comparisons were performed (t test or ANOVA). Otherwise, Mann Whitney test or Kruskal-Wallis test were used. Post hoc test (Dunnett's or Dunn's) are listed for each condition examined. Significance is as described in the figure legends with *p < 0.05; **p < 0.01; ***p < 0.001.
Author Contributions
G.G. and L.B.V. conceived the project. G.G. carried out all experiments and analyzed the data. S.C. provided technical help for the drug screen and the mouth-hook contraction experiments. J.H. and Y.R. made the Gal4 knock-in Drosophila mutant lines. G.G. and L.B.V. wrote the manuscript and prepared the figures.
Supplementary Material
Supplementary Table 1
Acknowledgments
Dragana Rogulja, Vanessa Ruta, and members of the Vosshall Laboratory provided helpful comments on the manuscript. We thank Yi-Chen Hsieh for technical help in setting up the feeding assay and Fraser Glickman and members of The Rockefeller University HTSRC for advice in implementing the drug screen. G.G. was a Pew Latin American Fellow in the Biomedical Sciences. This work was funded by National Natural Science Foundation of China (Young Scientists Fund 31000547 to J.H.), Chinese Ministry of Science and Technology (973 Program 2010CB833900 to Y.R.), and The Klarman Family Foundation Grants Program in Eating Disorders Research to L.B.V. L.B.V. is an investigator of the Howard Hughes Medical Institute.
References
- Finucane M. M. et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet 377, 557–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M. O. & Horvath T. L. Limitations in anti-obesity drug development: the critical role of hunger-promoting neurons. Nat. Rev. Drug. Discov. 11, 675–691 (2012). [DOI] [PubMed] [Google Scholar]
- Huntington M. K. & Shewmake R. A. Anti-obesity drugs: are they worth it? Future. Med. Chem. 3, 267–269 (2011). [DOI] [PubMed] [Google Scholar]
- Oury F. & Karsenty G. Towards a serotonin-dependent leptin roadmap in the brain. Trends Endocrinol. Metab. 22, 382–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberger C. Brain regulation of food intake and appetite: molecules and networks. J. Intern. Med. 258, 301–327 (2005). [DOI] [PubMed] [Google Scholar]
- Yeo G. S. & Heisler L. K. Unraveling the brain regulation of appetite: lessons from genetics. Nat. Neurosc. i 15, 1343–1349 (2012). [DOI] [PubMed] [Google Scholar]
- Rihel J. et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck M., Ye X. & Buck L. B. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature 450, 553–556 (2007). [DOI] [PubMed] [Google Scholar]
- Dar A. C., Das T. K., Shokat K. M. & Cagan R. L. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S. et al. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat. Chem. Biol. 4, 256–263 (2008). [DOI] [PubMed] [Google Scholar]
- Bland M. L., Lee R. J., Magallanes J. M., Foskett J. K. & Birnbaum M. J. AMPK supports growth in Drosophila by regulating muscle activity and nutrient uptake in the gut. Dev. Biol. 344, 293–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E., Wiggins D., Fielding B. & Gould A. P. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445, 275–280 (2007). [DOI] [PubMed] [Google Scholar]
- Rajan A. & Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K. & Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118–1120 (2002). [DOI] [PubMed] [Google Scholar]
- Kim S. K. & Rulifson E. J. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431, 316–320 (2004). [DOI] [PubMed] [Google Scholar]
- Rusten T. E. et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell. 7, 179–192 (2004). [DOI] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O. & Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7, 167–178 (2004). [DOI] [PubMed] [Google Scholar]
- Carvalho G. B., Kapahi P. & Benzer S. Compensatory ingestion upon dietary restriction in Drosophila melanogaster. Nat. Methods 2, 813–815 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman L. P. et al. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 4, 842–849 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher C. & Pankratz M. J. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3, e305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Kirchner C., Chao L. C., Tetzlaff M. T. & Pankratz M. J. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126, 5275–5284 (1999). [DOI] [PubMed] [Google Scholar]
- Tokunaga T., Noburu T. & Ueda M. Mechanism of antifeedant activity of plumbagin, a compound concerning the chemical defense in carnivorous plant. Tetrahedron Lett. 45, 7115–7119 (2004). [Google Scholar]
- Kubo I. & Klocke J. A. Some terpenoid insect antifeedants from tropical sources. Adv. Pestic. Sci. 2, 284–291 (1986). [Google Scholar]
- Simon A. F. et al. Drosophila vesicular monoamine transporter mutants can adapt to reduced or eliminated vesicular stores of dopamine and serotonin. Genetics 181, 525–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F., Boschert U., Amlaiky N., Plassat J. L. & Hen R. A family of Drosophila serotonin receptors with distinct intracellular signalling properties and expression patterns. EMBO J. 11, 7–17 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas J. F., Launay J. M., Kellermann O., Rosay P. & Maroteaux L. Drosophila 5-HT2 serotonin receptor: coexpression with fushi-tarazu during segmentation. Proc. Natl. Acad. Sci. U. S. A. 92, 5441–5445 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witz P. et al. Cloning and characterization of a Drosophila serotonin receptor that activates adenylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 87, 8940–8944 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obosi L. A. et al. Functional characterisation of the Drosophila 5-HTdro1 and 5-HTdro2B serotonin receptors in insect cells: activation of a G(alpha) s-like protein by 5-HTdro1 but lack of coupling to inhibitory G-proteins by 5-HTdro2B. FEBS Lett. 381, 233–236 (1996). [DOI] [PubMed] [Google Scholar]
- Hauser F., Cazzamali G., Williamson M., Blenau W. & Grimmelikhuijzen C. J. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog. Neurobiol. 80, 1–19 (2006). [DOI] [PubMed] [Google Scholar]
- Yuan Q., Joiner W. J. & Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16, 1051–1062 (2006). [DOI] [PubMed] [Google Scholar]
- Nichols C. D. 5-HT2 receptors in Drosophila are expressed in the brain and modulate aspects of circadian behaviors. Dev. Neurobiol. 67, 752–763 (2007). [DOI] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H. & Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U. S. A. 98, 12596–12601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. M. & Avery L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J. Neurosci. 32, 1920–1931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer W. S. A trophic role for serotonin in the development of a simple feeding circuit. Dev. Neurosci. 32, 217–237 (2010). [DOI] [PubMed] [Google Scholar]
- Vargas M. A., Luo N., Yamaguchi A. & Kapahi P. A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr. Biol. 20, 1006–1011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W. & Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754 (2008). [DOI] [PubMed] [Google Scholar]
- Lam D. D., Garfield A. S., Marston O. J., Shaw J. & Heisler L. K. Brain serotonin system in the coordination of food intake and body weight. Pharmacol. Biochem. Behav. 97, 84–91 (2010). [DOI] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6, 9–23 (2005). [DOI] [PubMed] [Google Scholar]
- Chalasani S. H. et al. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat. Neurosci. 13, 615–621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns S. & Simon M. I. G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J. Biol. Chem. 270, 15175–15180 (1995). [DOI] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A. & Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171, 571–581 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1