Abstract
When the human genome project started, the major challenge was how to sequence a 3 billion letter code in an organized and cost-effective manner. When completed, the project had laid the foundation for a huge variety of biomedical fields through the production of a complete human genome sequence, but also had driven the development of laboratory and analytical methods that could produce large amounts of sequencing data cheaply. These technological developments made possible the sequencing of many more vertebrate genomes, which have been necessary for the interpretation of the human genome. They have also enabled large-scale studies of vertebrate genome evolution, as well as comparative and human medicine. In this review, we give examples of evolutionary analysis using a wide variety of time frames—from the comparison of populations within a species to the comparison of species separated by at least 300 million years. Furthermore, we anticipate discoveries related to evolutionary mechanisms, adaptation, and disease to quickly accelerate in the coming years.
The human genome project pioneered not only the bacterial artificial chromosome (BAC)-based sequencing of a mammalian-sized genome (International Human Genome Sequencing Consortium 2001), but also the methodology of whole-genome shotgun (WGS) sequencing (Venter et al. 2001). WGS sequencing was further improved and applied to the mouse genome (Mouse Genome Sequencing Consortium 2002) and then became the technique of choice for many vertebrate genomes (International Chicken Genome Sequencing Consortium 2004; Lindblad-Toh et al. 2005; Mikkelsen et al. 2007; Warren et al. 2008). This methodology has two advantages: It allows a relatively unbiased approach to sequencing a genome and it has the ability to be automated and hence cost effective. Thus, it revolutionized the study of comparative genomics of vertebrate genomes. New sequencing technologies have further reduced the cost of WGS sequencing, making vertebrate genome sequencing even more popular (Li et al. 2010).
Prior to whole-genome sequencing of many vertebrates, the ENCODE project had selected a representative ∼1% on the human genome to be systematically sequenced in a BAC-by-BAC approach across mammals and some vertebrates. The comparative ENCODE project demonstrated the presence of widespread orthology between species, high levels of conservation within genes, as well as extensive signals of conservation outside genes. Noncoding features lacking experimental validation, however, were harder to interpret than protein-coding genes (Margulies et al. 2007).
The human genome sequence described many of the features of the human genome such as transposable elements (TEs), segmental duplications, genes, and their promoters. The human gene count predicted at approximately 40,000 (International Human Genome Sequencing Consortium 2001) was a huge refinement from the previously cited estimate of 100,000 genes. Nonetheless, it was far above the current tally of somewhere close to 22,000 human genes (Clamp et al. 2007).
For many scientists, the comparison of the mouse and human genomes came as a strong confirmation that large-scale comparative genomics is essential for understanding the human genome. Comparison of these two mammals refined the mammalian gene count to ∼30,000 and allowed the first genome-wide estimate of the minimum fraction of the human genome that is conserved across placental mammals and is hence functional: a full 5%, much more than the ∼1.2% occupied by protein-coding sequence (Mouse Genome Sequencing Consortium 2002).
The principles of comparative genomics
Since then, comparative genomics has proven itself invaluable, not only for illuminating evolutionary mechanisms and forces, but for informing the understanding of the human genome. The essence of the field of comparative genomics is that sequence that stays conserved (similar) across multiple and/or distant species is likely to be constrained (similar due to evolutionary pressures), which implies a biological function (Fig. 1). However, the converse is not necessarily true: A DNA sequence can, of course, have a biological function without being conserved with any other species' genome. This is especially true for novel lineage-specific changes where time has not yet afforded the sequence a signature of conservation. Conservation does not necessarily imply identity: Sequence can be constrained to prefer two or three out of the four bases.
Figure 1.
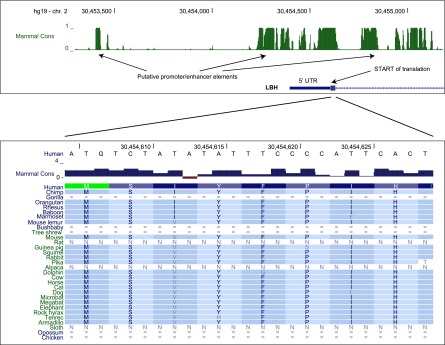
A comparative genomics display derived from the UCSC Genome Browser (Meyer et al. 2013). The top panel depicts the genomic region surrounding the 5′ end of the gene LBH (limb bud and heart development homolog) in the human genome. The top track indicates mammalian conservation as determined by phastCons (Pollard et al. 2010). Putative promoter and enhancer elements are indicated. The second track shows the intron/exon structure of the 5′ end of LBH. The 5′ untranslated region (UTR) and start site are indicated. The bottom panel shows a close up on the protein-coding portion of the first exon of LBH. Here, the top track shows the human DNA sequence, and the second track shows the degree of mammalian conservation as determined by PhyloP (Pollard et al. 2010). The bottom series of tracks shows the homologous protein sequence in selected vertebrate genomes. (N) Gaps in sequence; (=) unalignable sequence.
Exons generally are conserved across species in a very specific pattern (3-bp code with a third degenerate base), and sequence conservation has proven to be an important asset for the creation of gene models (as well as gene structure algorithms and species-specific RNA sequencing) (Flicek et al. 2013). However, regulatory elements are much harder to model: This is where comparative genomics shines. Early studies identified hundreds of so-called ultra-conserved elements—elements several hundred bases long and almost identical across mammals (Bejerano et al. 2004)—and functional studies demonstrated that some of these had a role as enhancers (Woolfe et al. 2005). Comparison of human, mouse, rat, and dog identified several hundreds of thousands of conserved noncoding elements (CNEs) that cluster near developmental genes (Lindblad-Toh et al. 2005), driving home the importance of regulation for determining body plan and neurological development.
Detecting the features and functions in the human genome
To more fully elucidate the constrained elements in the human genome, 29 placental mammals were sequenced. This number was chosen to gain enough power to find constrained elements >12 bp. This approach identified 3.6 million constrained elements encompassing 4.2% of the human genome (Lindblad-Toh et al. 2011). While 6%–15% of the human genome has been estimated to be constrained among placental mammals or under more recent lineage-specific selection (Pollard et al. 2010; Lindblad-Toh et al. 2011; Ward and Kellis 2012), only 4.2% of the human genome was annotated with constraint in the 29 placental mammals study, showing that many more constrained elements remain to be pinpointed.
While constraint shows that a base has a function, it does not necessarily reveal what that function is. To help assign function, one can use the constraint pattern for some types of features, for example, exons, splice site motifs (Wang and Burge 2008), and RNA-folding structures (Washietl et al. 2007). But for most elements, one must instead rely on combining constraint with other types of annotation data including RNA-seq, ChiP-seq data for transcription factors, methylation or histone marks, and DNase hypersensitivity. These techniques are very useful and are often complementary to sequence conservation analysis. However, sequence conservation has more specificity than assays such as transcription factor binding, or even transcription, unless very stringent binding conditions are used. As an example of this, the ENCODE Consortium annotated 80% of the human genome using a variety of overlapping experimental markers including transcription and histone modification (The ENCODE Project Consortium et al. 2012); however, this catalog does not uniquely assign the function to specific bases but to an overall region of the genome. Thus, one should combine the functional signature of the cell type of interest with constraint patterns to delineate the specific function of constrained elements (The ENCODE Project Consortium et al. 2007; Lindblad-Toh et al. 2011; Wenger et al. 2013).
Innovation in vertebrate genomes
An important indication of innovation in a genome is a recent change in an otherwise well-conserved element. Elements that are highly conserved in vertebrates sometimes show accelerated evolution in humans (almost all have proved to be regulatory elements) (Pollard et al. 2006) and are called human accelerated regions (HARs). Alternatively, noncoding regions that are highly conserved in mammals may be deleted in humans, chimps, or other species (McLean et al. 2011). This approach is particularly valuable to understand species-specific biology as well as recent evolutionary history.
To learn about mechanisms of innovation, placental mammalian genomes were compared with the first sequenced marsupial genome—the opossum genome. More than 20% of placental mammalian CNEs were not found in opossum, suggesting that they are novel innovations. In contrast, only 1% of exons conserved within placental mammals were not conserved with the opossum (Mikkelsen et al. 2007). This shows that the past 100 million yr of functional innovation in mammalian genomes derives largely from regulatory change (Mikkelsen et al. 2007), and also highlights the insights into evolution that can only come from comparative genomics.
More recent analysis has demonstrated three different waves of genes that showed regulatory innovation in different eras of vertebrate evolutionary history. The investigators first identified CNEs on the genomes of five disparate vertebrates, inferred the time when the CNE first came under selective constraint and then associated those CNEs with the closest gene. They found that novel CNEs first arose for transcription factors and developmental genes, then extracellular signaling genes, and then finally, genes involved in post-translation protein modification (Lowe et al. 2011).
While the above examples show that novel regulatory elements play a role in evolution over long periods of time, other studies of natural selection in stickleback (Jones et al. 2012) and human populations (Grossman et al. 2013), as well as of artificial selection in domestic animals (Rubin et al. 2010, 2012; Olsson et al. 2011), also demonstrate the importance and preponderance of regulatory innovation in a shorter time perspective. In contrast, two regulatory elements were shown to have remained conserved between humans and very distant invertebrate species, showing no innovation over a billion years (Clarke et al. 2012).
Shedding light on vertebrate evolution
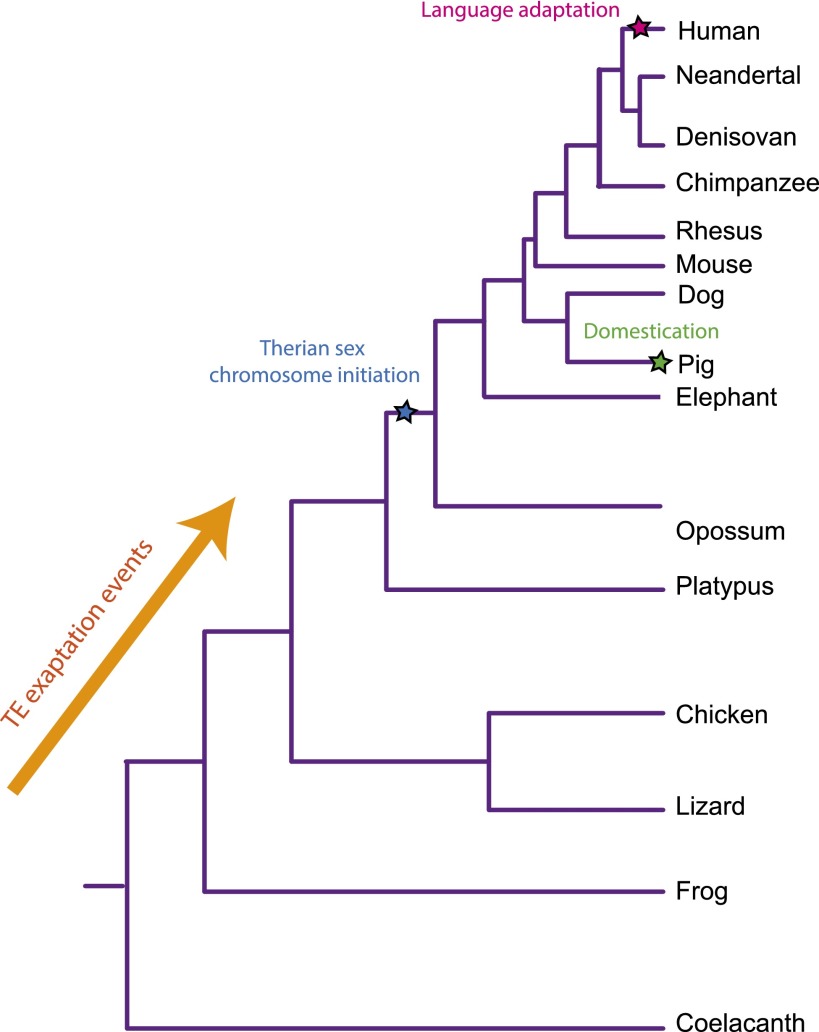
When specifically studying vertebrate genome evolution, scientists today can take advantage of more than 60 mammalian deep coverage genome assemblies, as well as a few dozen nonmammalian vertebrate deep coverage genome assemblies. These, combined with the 29 mammals (Lindblad-Toh et al. 2011) and 46 vertebrates alignments and conservation data sets (Meyer et al. 2013), supplemented by the within-species 1000 Genomes Project (Abecasis et al. 2012) displayed in viewable databases (UCSC, Ensembl, NCBI, and IGV), provide impressive resources for comparative genomics research. However, we must note that it is very important to choose the appropriate time-scale of conservation for the question being addressed. If one's phylogenetic scope (the time-scale of the relationship of species being studied) is too narrow, then the specificity of constraint is reduced. In contrast, lineage-specific biology is inevitably lost as phylogenetic scope is widened (Cooper and Shendure 2011). Here we give some examples that show how scientists have used a wide variety of genomes to their advantage to explain both general evolutionary principles and species-specific biology (Fig. 2):
Figure 2.
A tree schematic depicting the relationships between the vertebrate species discussed. Important events such as the origin of therian sex chromosomes, pig domestication, human language evolution, and transposable element exaptation are indicated. Note the very different time spans used depending on the evolutionary question at hand.
Exaptation—recycling the genome
Many have been intrigued by the large size of the human genome and the fact that the majority of it contains no protein-coding genes. More precisely, a large fraction of the genome is made up of TEs that hop around the genome for their own purposes. Only recently did we learn why vertebrate genomes may have taken on the burden of all these TEs: It allows genomes to evolve by way of exaptation. Exaptation is the repurposing of a sequence element for an entirely different function, and the “random” insertion of TEs provides excellent templates for exaptation. The exaptation of TEs was first described by Bejerano et al. (2006), who aligned human ultraconserved elements to coelacanth BAC sequence. They found that two different SINE classes still active in the Indonesian coelacanth genome are homologous to an enhancer of the gene ISL1 and an alternatively spliced exon of PCBP2 in humans, respectively. In the past ∼400 million yr, vertebrate genomes repurposed these TE sequences to serve entirely new functions. Since then, many more examples of exaptation have been detailed (Mikkelsen et al. 2007), including 96 human noncoding elements and one human exon discovered by comparison to the green anole lizard genome (Alfoldi et al. 2011). Significantly, approximately 280,000 human regulatory elements (7 Mb of sequence) were shown to have been exapted from TEs using the 29 placental mammals data set; this constitutes 20% of the conserved regulatory sequence annotated (Lowe and Haussler 2012). In this case, comparative genomics has taught us about the incredible ingenuity of evolution, and given us a window into the origins of new regulatory elements.
Sex-chromosomes—will there still be a Y chromosome in the future?
The process of sex chromosome evolution has been studied for several decades—but a true understanding of it was only made possible through comparative genomics. The X and Y chromosomes originate from a homologous pair of chromosomes, and so many have sought to understand sex chromosome evolution by comparing the X and Y chromosomes within a species. Observing that the current human Y chromosome has lost many genes in comparison to the formerly identical X chromosome have led some to believe that the human Y chromosome was destined to disappear from the genome forever (Aitken and Marshall Graves 2002). However, comparisons with other primate Y chromosomes have shown an entirely different picture of sex chromosome evolution. First, the iterative mapping and sequencing of the chimpanzee Y chromosome showed rapid evolution—not just the loss of genes from the Y, but the addition and duplication of novel sequence (Hughes et al. 2010). Then, the sequencing of the rhesus macaque Y chromosome showed that neither the human nor the rhesus had lost any of the Y chromosome genes that had been on the homologous chromosome predecessors of the X and Y. This led to a new theory of sex chromosomes, where Y chromosomes lose significant gene content soon after they lose the ability to cross-over with a homologous partner, but the original genes that remain exhibit strict purifying selection, giving us more confidence in the continuing existence of the human Y (Hughes et al. 2012).
Language adaptation by altering a key protein
Much has been learned about recent human evolution by the comparison of the modern human genome with those of extinct human species and the chimpanzee. Comparison of the Neanderthal genome to our own revealed several examples of recent fixation in the human genome, and showed that 91% of HARs lost their mammalian conservation earlier than the Neanderthal/modern human split (Green et al. 2010). The FOXP2 gene is known for having mutations that cause linguistic and grammatical impairments as part of a Mendelian disorder in humans. Comparison of the sequence of this gene in several humans, two chimpanzees and an orangutan showed a recent selective sweep of this gene in modern humans, making it likely that the two nonsynonymous changes in human FOXP2 are at least partially responsible for the orofacial movement control that allows us, unlike our ape cousins, to speak (Enard et al. 2002). Another comparison—this time with the Denisovan genome (another genome of an extinct human species)—demonstrated that the Denisovans possessed the ancestral FOXP2 allele, showing that this crucial change in the human lineage happened only in the last 800,000 yr, after the divergence from Denisovans and Neanderthals (Meyer et al. 2012).
Signals of selection enhanced by domestication
Comparisons of the genomes of domesticated pigs and wild boars demonstrate multiple points about selection mechanisms and biological traits. This study found an excess of derived nonsynonymous substitutions in domestic pigs, most likely reflecting both positive selection and relaxed purifying selection after domestication. Analysis of the KIT locus in white or white-spotted pigs identified four different structural variants, emphasizing how structural changes have contributed to rapid phenotypic evolution in domestic animals and how alleles in domestic animals may evolve by the accumulation of multiple causative mutations as a response to strong directional selection. Selective sweep analyses were performed by searching the genome for regions with elevated homozygosity, and revealed strong signatures of selection at loci likely involved in behavior, morphology, and production traits. Three genes (NR6A1, PLAG1, and LCORL) at different loci together explain the majority of the genetics underlying the elongation of the back and an increased number of vertebrae in the domestic pig. PLAG1 and LCORL also control stature in other domestic animals and in humans (Rubin et al. 2012).
Great things to come—fully applying comparative genomics to disease
We have demonstrated the benefits that comparative genomics has provided to the field of evolutionary biology. With the currently available information, we believe that the time is now ripe for comparative genomics to be more fully embraced in the study of human disease. A very large fraction, 88%, of trait- or disease-associated loci identified in genome-wide association studies (GWAS) are intronic or intergenic in nature (Hindorff et al. 2009). A major hurdle to finding the actual mutations responsible for a given human trait or disease is the lack of understanding of the function of the noncoding portion of the genome. Many studies therefore stop at the general locus and, understandably, assume that the most nearby gene is affected. While genomic distance plays a role, more and more examples are now surfacing where regulatory elements or lincRNAs affect genes at a distance or even genes on other chromosomes (Sanyal et al. 2012), thus making such assumptions overly simplistic. Comparative genomics, together with other genomic resources, is now on the verge of offering the tools for developing testable hypotheses for candidate regulatory mutations. In fact, SNPs associated with human disease in GWAS are 1.37-fold enriched in placental mammalian conserved elements relative to total HapMap SNPs (Lindblad-Toh et al. 2011), suggesting that a portion of the tagged SNPs are indeed functional. In a disease-associated haplotype, 10–100 SNPs might be present. Of these ∼5% will be constrained, allowing the selection of only one or a few SNPs for first-tier functional characterization. Thus, sequence conservation is extremely useful for prioritizing GWAS SNPs or resequencing variants for functional studies (Fig. 3).
Figure 3.
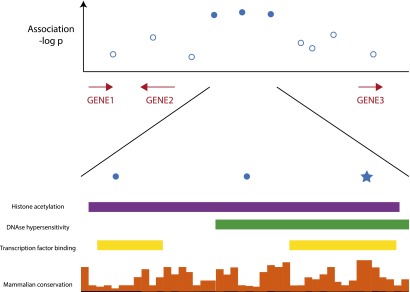
Hypothetical data showing how conservation can be used to identify the most likely functional variants in a disease locus. GWAS or sequencing data can be combined with constraint information and other genome annotations in the appropriate cell types to assess SNPs and other variants present on human disease-associated haplotypes. From the multitude of associated variants, one or a few candidate mutations often stand out based on the overlap of a candidate SNP (best variant indicated by star in lower panel) with constraint and other genome annotation. In a best-case scenario, careful analysis of sequence motifs overlapping the candidate variant will also identify a transcription factor binding site, splice site, or RNA structure that is altered by the candidate variant.
Despite the discovery of tens or hundreds of GWAS loci each for many of our most common diseases, only a fraction of the genetic risk has been accounted for. Multiple hypotheses to explain this have been proposed (Visscher et al. 2012), but no ultimate conclusion has been reached. One potential scenario is that rare variants of strong effect, which theoretically should not become common due to purifying selection, have indeed become more common in certain populations due to drift or selection. Furthermore, it seems reasonable that deleterious noncoding variants affecting the expression of a gene in a specific tissue would be more tolerated than deleterious mutations destroying the protein in all tissues, and therefore would be more conducive to a viable and reproductively fit individual. The observation that a large fraction of evolutionary innovation occurs in noncoding sequence, where it can fine-tune the regulation of specific genes under certain conditions without overall changing the function of the protein, also supports this notion. Many of these rare variants could potentially regulate pathways involved both in normal physiology and in disease pathology. We therefore hypothesize that rare variants in noncoding functional elements will play a relatively large role in common disease, and advocate for more whole-genome sequencing or targeted efforts that examine both coding and noncoding constrained sequence.
If rare variants contribute substantially to human common diseases, then there ought to be a reason why they remain at a considerable frequency in the population. In selected traits in domestic animals, identified noncoding mutations often affect both phenotypic traits and disease traits (either through pleiotropic effects or through hitchhiking of nearby loci). It is not unreasonable to think that selection will also play a role in increasing the frequency of specific disease variants in humans. A well-studied example is sickle-cell anemia, a serious disease that remains common in Africa as it offers protection against malaria (for review, see Bunn 2013). Other examples are quickly amassing, including a connection between celiac disease and bacterial infection (Zhernakova et al. 2010). While it is easy to appreciate the importance of infectious diseases and the selective effects they may have had on us, other environmental factors may also be important. These include factors such as the amount of sunlight, diet, climate, and altitude, most of which are likely to have adaptive responses also. Local selective pressures from the environment could thus contribute to selection for certain disease alleles and contribute to the variation of disease frequencies often seen between different populations.
A great deal of vertebrate sequencing has already been completed, and invaluable data sets are already available, but more mammalian genome sequencing could help us annotate the human genome even further. The 29 placental mammals data set possesses a total branch length of 4.5 substitutions per site and yielded a 12-bp resolution for placental mammalian conservation. To obtain a tantalizing 1-bp resolution, which would allow us to assess the functional potential of every base, would require an additional 100–200 placental mammal genomes (with a total branch length of 15–25 substitutions/site) (Lindblad-Toh et al. 2011). This would also enable detailed mammalian lineage-based constraint annotation and would thereby annotate the predicted further 2%–10% of the human genome.
In conclusion, the use of comparative genomics, enabled by the human genome sequence and the technological advances catalyzed by its generation, has brought a wealth of insights into vertebrate genome evolution, increased our understanding of the human genome, and now offers the potential to decipher human evolution and disease and the inevitable link between the two.
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.157503.113.
Freely available online through the Genome Research Open Access option.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken RJ, Marshall Graves JA 2002. The future of sex. Nature 415: 963. [DOI] [PubMed] [Google Scholar]
- Alfoldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441: 87–90 [DOI] [PubMed] [Google Scholar]
- Bunn HF 2013. The triumph of good over evil: Protection by the sickle gene against malaria. Blood 121: 20–25 [DOI] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, VanderMeer JE, Wenger AM, Schaar BT, Ahituv N, Bejerano G 2012. Human developmental enhancers conserved between deuterostomes and protostomes. PLoS Genet 8: e1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Shendure J 2011. Needles in stacks of needles: Finding disease-causal variants in a wealth of genomic data. Nat Rev Genet 12: 628–640 [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869–872 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463: 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, et al. 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ III, Zody MC, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438: 803–819 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Haussler D 2012. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS ONE 7: e43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR, Kingsley DM, Lindblad-Toh K, Haussler D 2011. Three periods of regulatory innovation during vertebrate evolution. Science 333: 1019–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. 2007. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res 17: 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471: 216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2013. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Olsson M, Meadows JR, Truve K, Rosengren Pielberg G, Puppo F, Mauceli E, Quilez J, Tonomura N, Zanna G, Docampo MJ, et al. 2011. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet 7: e1001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464: 587–591 [DOI] [PubMed] [Google Scholar]
- Rubin CJ, Megens HJ, Martinez Barrio A, Maqbool K, Sayyab S, Schwochow D, Wang C, Carlborg O, Jern P, Jorgensen CB, et al. 2012. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci 109: 19529–19536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J 2012. Five years of GWAS discovery. Am J Hum Genet 90: 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M 2012. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337: 1675–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermuller J, Hertel J, Lindemeyer M, Reiche K, Tanzer A, et al. 2007. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res 17: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger AM, Clarke SL, Guturu H, Chen J, Schaar BT, McLean CY, Bejerano G 2013. PRISM offers a comprehensive genomic approach to transcription factor function prediction. Genome Res 23: 889–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, de Kovel CG, Franke L, Oosting M, Barisani D, et al. 2010. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet 86: 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]