Abstract
The neural crest is an excellent model system for the study of cell type diversification during embryonic development due to its multipotency, motility, and ability to form a broad array of derivatives ranging from neurons and glia, to cartilage, bone, and melanocytes. As a uniquely vertebrate cell population, it also offers important clues regarding vertebrate origins. In the past 30 yr, introduction of recombinant DNA technology has facilitated the dissection of the genetic program controlling neural crest development and has provided important insights into gene regulatory mechanisms underlying cell migration and differentiation. More recently, new genomic approaches have provided a platform and tools that are changing the depth and breadth of our understanding of neural crest development at a “systems” level. Such advances provide an insightful view of the regulatory landscape of neural crest cells and offer a new perspective on developmental as well as stem cell and cancer biology.
The neural crest is an embryonic cell population with stem cell-like properties, including multipotency and the ability to self-renew. Unique to vertebrates, neural crest cells contribute to a wide variety of derivatives, including sensory and autonomic ganglia of the peripheral nervous system, adrenomedullary cells, cartilage and bone of the face, and pigmentation of the skin. Although similar cell types, such as pigment cells and sensory neurons, already exist in nonvertebrate chordates and other multicellular organisms, these derivatives arise de novo under the umbrella of the neural crest in the vertebrate lineage.
Since its discovery by His (1868), the neural crest has occupied a prominent place in developmental biology due to its extensive migratory properties and remarkable developmental potential. Interest in this cell population has been further fueled by its medical and evolutionary importance. For example, numerous congenital birth defects and neoplastic diseases are linked to abnormal development of the neural crest development and its derivatives (Hall 1999). Due to its inherent stem cell properties, there is great interest in using these cells in regenerative medicine to treat disorders like familial dysautonomia, cleft palate, and some heart conditions (Jones and Trainor 2004; Lee et al. 2009). Furthermore, as the neural crest gives rise to a number of vertebrate-specific traits, it is thought to have played an important role in chordate evolution (Gans and Northcutt 1983; Northcutt 2005).
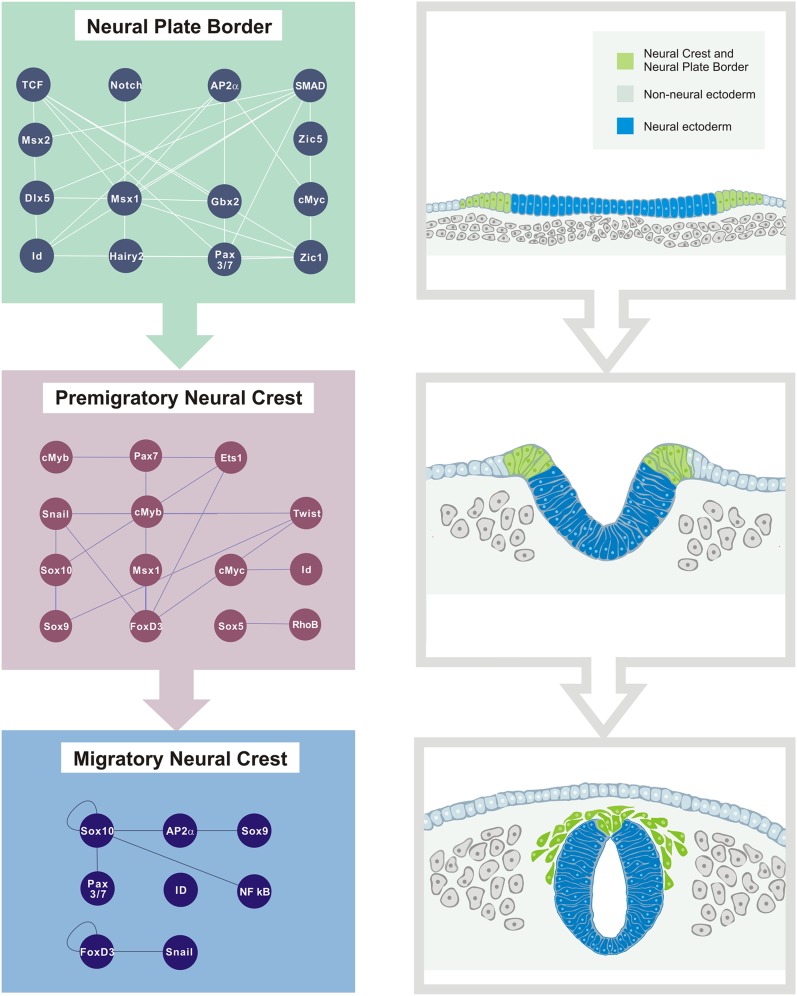
The initial phases of neural crest formation include some of the most extensive morphogenetic movements observed during vertebrate embryonic development (Fig. 1). Initially the prospective neural crest cells reside in a territory known as the neural plate border, which is located at the edges of the neural plate, the embryonic region destined to form the central nervous system. Through a process called neurulation, the neural plate invaginates by elevation of the edges, or neural folds. The end result is the conversion of the flat neural plate into a cylindrical structure called the neural tube, which will later form the brain and spinal cord. During the process of neural tube closure, premigratory neural crest cells reside first within the neural folds as they converge toward the midline and then in the dorsal aspect of the neural tube. Shortly thereafter, they lose their intercellular connections, undergo an epithelial to mesenchymal transition (EMT), and acquire mesenchymal, migratory characteristics that endow these cells with the ability to leave the neural tube (Gammill and Bronner-Fraser 2003; Sauka-Spengler and Bronner-Fraser 2008b).
Figure 1.
Morphogenetic movements during early neural crest development. (A) Schematic diagram of transverse sections through chick embryo during neurulation. Prospective neural crest cells reside in the neural plate border (green), a territory between the neural plate and the non-neural ectoderm. (B) As neurulation proceeds, the neural plate invaginates, resulting in the elevation of the neural folds, which contain neural crest precursors. (C) After neural tube closure, neural crest cells lose intercellular connections and undergo an epithelial to mesenchymal transition. Once they have delaminated from the neural tube, they migrate extensively to populate different niches throughout the embryo.
Once they have emigrated from the neural tube, neural crest cells migrate in organized streams to populate different niches throughout the embryo. Depending upon their starting position along the body axis and the subsequent path taken, they give rise to different cell types and contribute to the formation of a variety of tissues and organs (Fig. 2). The cranial neural crest forms a large portion of the facial skeleton as well as cranial ganglia, smooth muscle, and pigment cells. The vagal neural crest has an important role in cardiac development since it contributes to the valves and septa of the heart and also forms the enteric nervous system that innervates the entire length of the gut. The trunk neural crest gives rise to dorsal root and sympathetic ganglia of the peripheral nervous system, as well as secretory cells and melanocytes. Finally, the neural crest formed at the sacral region cooperates with the vagal crest to form a small portion of the enteric nervous system (Le Douarin 1986; Le Douarin and Kalcheim 1999). Although neural crest cells appear to become progressively restricted as they reach particular targets, many early migrating cells are multipotent, with the ability to form several derivatives (Bronner-Fraser and Fraser 1988, 1989). Moreover, many neural crest-derived tissues appear to retain neural crest stem cells that are multipotent (Crane and Trainor 2006).
Figure 2.
Contributions of different neural crest cell populations to adult tissues and organs. Depending on their axial level of origin and migratory pathway followed, neural crest cells adopt different fates and contribute to distinct tissues and organs. Cranial neural crest forms a large portion of the facial skeleton as well as cranial ganglia, most of the dental tissues, and the cornea. Vagal neural crest contributes to the valves and septa of the heart, the smooth muscle of the great vessels, and the enteric nervous system. Trunk neural crest gives rise to dorsal root and sympathetic ganglia of the peripheral nervous system and the chromaffin cells of the adrenal gland. Most caudally, the neural crest formed at the sacral region contributes to a small portion of the enteric nervous system. Melanocytes of the skin and integuments are derived from neural crest at all axial levels.
A number of classical experiments were fundamental for understanding the main characteristics of the neural crest. For example, early studies with amphibian embryos suggested that there was regionalization of neural crest populations along the body axis and broad developmental potential (Horstadius 1950). Importantly, the elegant quail-chick chimera experiments pioneered by Nicole Le Douarin in the 1960s and 1970s were central in defining the precise contributions of different populations of the neural crest to distinct derivatives in higher vertebrates (Le Douarin and Kalcheim 1999). Bronner-Fraser and Fraser (1988) tackled the question of the developmental potential of individual neural crest by labeling single premigratory cells with a vital dye. They showed that the progeny of a single neural crest cell included cell types as diverse as sensory neurons, presumptive pigment cells, ganglionic supportive cells, and neural tube cells (Bronner-Fraser and Fraser 1988). Clonal analysis of the neural crest in tissue culture further demonstrated multipotency of single neural crest cells to form multiple derivatives (Sieber-Blum and Cohen 1980; Trentin et al. 2004; Dupin et al. 2010) as well as their ability to self-renew (Stemple and Anderson 1992). However, the presence of some clones that formed single derivatives has been interpreted as suggesting a possible fate restriction in some subpopulations (Stemple and Anderson 1992). These and innumerous other experiments have characterized cellular properties of the neural crest that can now be tackled in molecular terms due to advances in molecular biology and genomics.
In the last three decades, advances in recombinant DNA technology and molecular genetics techniques have made it possible to start examining the genetic program controlling neural crest development. Identification of a number of tissue-specific transcription factors allowed for the description of the first regulatory interactions necessary for establishment of neural crest identity. Studies in several model organisms have provided a wealth of molecular data that has helped to formulate our views on how complex processes such as induction, specification, differentiation, and migration are regulated. This extensive body of work has established the neural crest as a prime model for the exploration of questions central to developmental biology—such as how complex and diverse tissues arise from a seemingly homogeneous population of cells.
Currently, genomic approaches are being used to address essential aspects of neural crest development. The extensive work done on the signaling and transcriptional control of neural crest development culminated with the assembly of a gene regulatory network that attempts to explain, in molecular terms, the process of neural crest formation. Sequencing of chordate genomes has allowed for identification of novel genes involved in neural crest formation and provided numerous tools to scrutinize the regulatory mechanisms underlying this process. Furthermore, comparative genomic analyses are providing important insights about the evolution of the neural crest and the origins of the vertebrate clade. Here, we will present a synopsis of the current state of the field of neural crest biology from a gene regulatory perspective and will also discuss how recent technological advances can help shape future research.
Overview of the current state of the neural crest gene regulatory network (GRN)
The regulatory machinery controlling neural crest formation and diversification is comprised of an intricate array of transcription factors and signaling molecules that act in concert to provide this cell population with its defining features (Sauka-Spengler and Bronner-Fraser 2008b). In order to interrogate such a complex regulatory program, we and others have assembled a multistep neural crest GRN that integrates transcriptional inputs and diverse environmental signals (Meulemans and Bronner-Fraser 2004; Betancur et al. 2010a). A simplified version of the cranial neural crest GRN is represented in Figure 3. It is composed of a series of regulatory steps arranged hierarchically, which include induction of the prospective neural crest, specification of the neural plate border, specification of the bona fide neural crest cells, and the diversification of the neural crest cells through the action of neural crest effector genes (Sauka-Spengler and Bronner-Fraser 2008b). Remarkably, these regulatory stages not only define cell identity and behavior at a given time point, but also drive seamless transitions to the next regulatory state.
Figure 3.
Cranial neural crest gene regulatory network (GRN). The neural crest gene GRN is composed of different regulatory modules arranged hierarchically. Each regulatory state defines cell identity and behavior at a given time and also drives the transition to the next level of the network. This is a simplified representation of the cranial neural crest GRN (Betancur et al. 2010a).
The first level of the neural crest GRN is comprised of induction events that lead to the formation of the neural plate border. This process is dependent upon the interplay of different signaling pathways such as Wnt, Fgf, BMP, and Notch/Delta. Secreted signaling molecules are produced by adjacent tissues and result in the activation of a particular set of genes in the edges of the neural plate. These neural plate border specifier genes (e.g., Msx, Pax3/7, Zic1, Dlx3/5) initiate expression prior to the traditional neural crest marker genes and often are down-regulated once neural crest identity is established. Their region of overlap at the neural plate border defines a broad territory of cells competent to respond to neural crest specifying signals and later form migrating neural crest cells (Meulemans and Bronner-Fraser 2004).
The next step in the neural crest GRN is initiated when neural plate border specifiers in combination with signaling molecules activate transcription of neural crest specifier genes. These include transcription factors like Snai1/2, Tfap2a, Foxd3, Twist, Id, Myc, and Sox9/10. Expression of these factors reflects the specification state of the neural crest population and endows them with its defining features, like the ability to undergo EMT and become migratory. Activation of the neural crest specifiers can be quite complex and depends upon various inputs from all levels of the neural crest GRN. For example, Snai2 activation requires cooperation between elements of the Wnt pathway, Zic1, and Pax3/7 (Sato et al. 2005). In addition, SoxE transcription factors are necessary for the maintenance of snai2 expression (Honoré et al. 2003). Thus, factors from different hierarchical levels of the neural crest gene regulatory network operate in concert to establish and maintain the neural crest transcriptional state (Sauka-Spengler and Bronner-Fraser 2008b).
The neural crest specifier genes, in turn, regulate expression of effector genes involved in cell cycle control, epithelial to mesenchymal transition, and migration. Neural crest effector genes jumpstart a number of gene batteries that instruct the behavior of the newly formed neural crest cells, allowing them to delaminate from the neural tube, proliferate and maintain population size, migrate along different pathways, and finally differentiate into a wide variety of derivatives (Meulemans and Bronner-Fraser 2004; Sauka-Spengler and Bronner-Fraser 2008b). At the same time, the effector genes activate the expression of receptors and signaling molecules that equip the cells with the capacity to respond to environmental cues. This molecular toolkit also allows cell–cell interactions that influence not only other neural crest cells, but also numerous embryonic tissues with which they interact during migration. For example, neural crest cells instruct somite cells to differentiate into muscle precursors (Rios et al. 2011).
The neural crest GRN integrates >20 yr of work from many research groups performed in different model organisms, and provides a conceptual framework for the study of the neural crest genetic program. It is, however, not yet complete and will greatly benefit from the high-throughput genomic approaches that are currently part of the scientific repertoire. For instance, macroarray screens have uncovered numerous new molecules that are up-regulated in the neural crest (Gammill and Bronner-Fraser 2002; Adams et al. 2008). Transcriptome analysis of pure neural crest populations has uncovered a variety of new specific transcription factors, nuclear receptors and signaling molecules that are expressed during different stages of development (M Simões-Costa and ME Bronner, unpubl.). Furthermore, novel techniques for genome-wide chromatin profiling allow genome-wide identification of active and poised cis-regulatory modules (Rada-Iglesias et al. 2012) and facilitate functional analysis for rapid characterization of epistatic interactions. Such approaches will increase the complexity of the neural crest GRN by providing a more accurate and complete representation of the different regulatory modules and their interactions.
The current version of the neural crest GRN is grounded in interactions between transcription factors and signaling molecules, and does not include extra levels of regulation such as those imparted by epigenetic, post-transcriptional and post-translational control. Nevertheless, there is increasing evidence pointing to an important role for chromatin modification in the development of the neural crest. For example, the histone demethylase JMJD2A directly regulates a number of neural crest specifier genes, thus controlling the time of onset of their expression in the neural folds (Strobl-Mazzulla et al. 2010). Furthermore, the neural crest specifier, SNAI2, forms a complex with histone deacetylases (HDACS) to silence genes such as CAD6B, thus allowing neural crest delamination (Strobl-Mazzulla and Bronner 2012). Epigenetic silencing is also crucial for the initial demarcation of the neural crest territory. Hu and colleagues have shown that DNA methyltransferase3A promotes neural crest specification by repressing neural genes SOX2 and SOX3, thus acting as a fate switch in the cells of the neuroectoderm (Hu et al. 2012). Recent results also emphasize the role of post-translational modifications in neural crest regulation. Lee and colleagues have shown that SUMOylation of SoxE factors causes recruitment of the repressor Grg4 (also known as Tle4) instead of its usual coactivator partners. This study highlights how post-translational regulation can alter gene function and drive the same factor to play opposing roles in a context-dependent manner (Lee et al. 2012).
At the present time, the neural crest GRN (Meulemans and Bronner-Fraser 2004; Betancur et al. 2010a) primarily focuses on the regulatory interactions of the cranial neural crest. Since important differences exist between distinct subpopulations of the neural crest along the body axis (Le Douarin et al. 2004), it would be naïve to assume that such interactions are common to all neural crest populations. Indeed, there are likely to be important modifications that help explain regional differences in neural crest cell migratory properties and cell fates (Simões-Costa et al. 2012). Thus a more inclusive version of the GRN should include the molecular circuits that are particular to each of the neural crest subpopulations. Studies scrutinizing the regulatory states of these distinct populations will reshape the GRN and provide important clues about the plasticity of the neural crest. Ultimately and optimally, the GRN will have predictive value, which will help anticipate the molecular outcome of particular perturbations. This will be useful for etiological studies, of particular importance due to the large number of neural crest-related birth defects. A complete neural crest GRN will also inform upon genetic reprogramming of induced pluripotent stem (iPS) cells or embryonic stem (ES) cells into neural crest cells, which will have significant implications for regenerative medicine and cancer.
Cis-regulatory analysis of neural crest enhancers
The first version of the neural crest gene regulatory network was based on data assembled from a combination of gene expression patterns and functional studies obtained from several model organisms (Meulemans and Bronner-Fraser 2004). Although this approach provided a conceptual framework for investigating neural crest identity, further resolution of the GRN requires the identification of direct epistatic interactions among its players. This can only be accomplished through systematic identification and characterization of the cis-regulatory apparatus that controls expression of specific components of the neural crest GRN (Fig. 4).
Figure 4.
Strategies for cis-regulatory analysis of the neural crest. (A) Early approaches for identifying enhancers of neural crest genes included screening long stretches of noncoding DNA. Fragments from the locus of the gene of interest were cloned upstream of a minimal promoter and a reporter gene and tested in vivo by cell transfection or transgenesis. (B) Sequencing of vertebrate genomes facilitated the search for cis-regulatory modules. Computational comparison between different species reveals evolutionary conserved regions (ECRs) that are putative regulators of nearby genes. The ECRs are subsequently tested for activity by stable or transient transgenesis in different model organisms. (C) Novel approaches allow for genome-wide analysis of the cis-regulatory modules. Profiling of histone modification through ChIP-seq allows mapping of chromatin mark patterns and identification of active and poised enhancers. This method was used to annotate enhancers that are active in human neural crest cells induced from human embryonic stem cells (Rada-Iglesias et al. 2012). Once enhancers are validated in vivo, they become valuable tools that can be exploited in different contexts. (MP) Minimal promoter; (REP) reporter gene; (TF) transcription factor.
Tissue-specific enhancers have been invaluable for investigation of the genetic program underlying neural crest formation and can be exploited in several different contexts. This is exemplified by numerous studies that have taken advantage of the mouse Wnt1 enhancer to address the contribution, potential, and genetic regulation of the neural crest. This enhancer region, which is a 5.5-kb element at the 3′ region of the Wnt1 locus, activates transcription in the dorsal neural tube and migrating neural crest cells of murine embryos (Echelard et al. 1994). It has been employed extensively in mapping the contribution of different neural crest populations (Jiang et al. 2000; Nakamura et al. 2006; Barraud et al. 2010) through the use of the Rosa26 system (Soriano 1999). It has also been employed for targeted genetic manipulation through conditional knockout animals (Ito et al. 2003; Akiyama et al. 2004; Brewer et al. 2004; Dudas et al. 2004; Nakamura et al. 2006) that have genes inactivated specifically in the neural crest. This has allowed for the functional characterization of a number of genes in a neural crest cell-autonomous manner.
The initial approach for the identification of enhancers of neural crest genes was done by screening noncoding DNA isolated from the locus of the gene of interest (Fig. 4; Echelard et al. 1994). Fragments from the noncoding DNA were cloned upstream of a minimal promoter plus reporter sequence and tested in vivo or in vitro. The sheer size of the genetic loci and number of subregions that had to be tested made this approach laborious and time consuming, particularly given the time and expense of using mice. New genomic tools have since emerged and revolutionized the way cis-regulatory modules are identified. Importantly, the sequencing of complete genomes has greatly benefited the search for conserved cis-regulatory elements (Cooper and Sidow 2003; Uchikawa et al. 2004). Comparison of the noncoding regions between multiple species has proved an efficient approach for uncovering evolutionarily conserved regions (ECRs) that may regulate transcription of neighboring genes (Uchikawa et al. 2003; Betancur et al. 2010b; Simões-Costa et al. 2012). The activity of the ECRs subsequently can be investigated by stable or transient transgenesis. In this context, the chicken embryo has proved to be an excellent amniote model for cis-regulatory analysis in higher vertebrates, due to highly efficient transient transgenesis accomplished through electroporation techniques (Sauka-Spengler and Barembaum 2008; Takemoto et al. 2011). Similarly, tol2-mediated transgenesis in zebrafish is an efficient way to test enhancers, including those that work across distant species (Fisher et al. 2006; Rada-Iglesias et al. 2012).
A number of neural crest-specific enhancers have been identified in the past few years in different model organisms. Comparative genomics was used to identify neural crest-specific enhancers for Zic3, Sox9, Foxd3, Sox10, Snai2, and Twist, among others (Smith et al. 1997; Bagheri-Fam et al. 2006; Betancur et al. 2010b; Garnett et al. 2012; Simões-Costa et al. 2012). For example, cis-regulatory analysis of murine Pax3 revealed intriguing aspects of how neural crest genes are regulated. This analysis uncovered two distinct enhancers that are capable of driving very similar neural crest-specific expression. Mutation of either region by itself does not disrupt gene expression, and thus these enhancers constitute an interesting case of redundancy at the cis-regulatory level (Milewski et al. 2004; Degenhardt et al. 2010). The lack of similarity between the two regions also suggests that they require different inputs, highlighting not only the complexity of the regulatory code but also the necessity for thorough analysis of the cis-regulatory apparatus (Degenhardt et al. 2010). Similar to the case in mice, a study in zebrafish by Garnett and colleagues shows that the zebrafish pax3a gene expression is also controlled by two distinct enhancers. Although manipulations in BMP, Wnt, and FGF signaling disrupt the activity of the individual enhancers, when combined in the same construct, they are less susceptible to environmental perturbations. Thus, synergistic activity of different regulatory modules can bring a level of robustness to transcriptional control, particularly in the case of genes downstream from signaling pathways (Garnett et al. 2012).
Tissue-specific enhancers for neural crest derivatives have also been identified. For example, evolutionarily conserved regions of the gene Mef2c (Agarwal et al. 2011), and tyrosinase-related family member Tyrp1 (Murisier et al. 2006), can drive specific reporter expression in melanocytes. Interestingly, both enhancers seem to be regulated by the transcription factor Sox10, a key regulator of the melanocytic lineage. Similarly, a Hoxb3 enhancer that is active in the vagal neural crest drives reporter expression in the enteric nervous system (Chan et al. 2005). Such regulatory modules are useful tools for targeted functional studies, particularly in chicken and zebrafish embryos, given the new approaches for efficient and rapid integration of transgenes in these embryos (Fisher et al. 2006; Yokota et al. 2011).
Cis-regulatory analysis can also provide insights into how differences between distinct neural crest populations arise. The cis-regulatory analyses of both SOX10 (Betancur et al. 2010b) and FOXD3 (Simões-Costa et al. 2012) point to distinct genetic programs controlling gene expression at different axial levels of the chick embryo. Although both SOX10 and FOXD3 are pan-neural crest markers, different enhancers are responsible for driving transcription in the cranial versus trunk neural crest populations. Furthermore, cranial and trunk enhancers are activated by different inputs (Simões-Costa et al. 2012), which suggests an intrinsic heterogeneity in the neural crest population that precedes specification. For example, two enhancers, NC1 and NC2, were uncovered, which collectively drive reporter expression that recapitulates the endogenous pattern of FOXD3 in the neural crest but separately control expression in nonoverlapping neural crest populations. NC1 drives initial expression in the cranial neural crest but is not active in the trunk region. In contrast, NC2 mediates initial expression in trunk neural crest and only drives reporter expression later in migrating cranial crest cells that no longer have NC1 activity. Whereas PAX7 and MSX1/2 are common inputs to both NC1 and NC2, ETS1 is a direct input into NC1 and ZIC1 into NC2. Consistent with axial level specific inputs, ETS1 is a cranial-specific transcription factor, whereas ZIC1 is expressed in a graded fashion, from posterior to anterior along the body axis (Simões-Costa et al. 2012). This differential regulation is consistent with the differences observed in the migratory patterns, potential, and behavior of the trunk versus cranial neural crest cells (Le Douarin and Kalcheim 1999). Further characterization of unique regulatory modules in distinct neural crest subpopulations hold the promise of providing important insights into neural crest heterogeneity and developmental potential.
Currently, an array of new technologies allows high-throughput identification of cis-regulatory modules. In the future, these will allow a systems biology-level analysis of gene regulation during neural crest formation. Importantly, novel approaches such as ChIP-seq allow for genome-wide analysis of the cis-regulatory apparatus at specific times and axial levels, promising to yield new insight into the regulatory events underlying neural crest development. This technique can be used to map the genome-wide occurrence of histone modifications as well as active binding sites for transcription factors and coactivators (Valouev et al. 2008; Creyghton et al. 2010). In a remarkable effort to describe the epigenomic landscape of neural crest cells, Rada-Iglesias et al. (2012) mapped the chromatin mark patterns and transcription factor occupancy in induced neural crest cells produced from human embryonic stem cells. This study identified TFAP2A and nuclear receptors NR2F1 and NR2F2 as key players in neural crest development, as the regulators occupy active enhancers characterized by high levels of EP300 and H3K27ac occupancy (Rada-Iglesias et al. 2012). It also revealed thousands of active enhancers functioning during neural crest development. Moreover, the resulting epigenomic annotation of these cells constitutes an invaluable resource for further investigation of the neural crest genetic program.
Such studies provide a wealth of information and highlight the extraordinary complexity of the regulatory events controlling neural crest formation. Not surprisingly, new paradigms are currently emerging in the field of gene regulation that may reshape our views of the neural crest gene regulatory network and bring an extra layer of complexity to the proposed regulation of this cell population. Increasing evidence indicates that a complete understanding of transcriptional events will require taking into consideration nuclear architecture and dynamics (Cremer and Cremer 2001; Mateos-Langerak et al. 2007). In the context of the neural crest, a recent study has shown that mutations of histone H3.3 cause down-regulation of neural crest specifiers and loss of crest-derived ectomesenchyme (Cox et al. 2012). This is particularly interesting since the H3.3 variant histones tend to be associated with permissive chromatin structures and have been described as regulators of transcription and specification of germ cells. The results highlight how chromatin rearrangements can impact the transcriptional state of the neural crest and their importance for its developmental potential (Cox et al. 2012). Interestingly, transcription factors also have been shown to interact with repressive molecules involved in epigenetic silencing (Wang et al. 2011; Strobl-Mazzulla and Bronner 2012). Thus, increasing evidence points to a new paradigm in which transcription factor complexes are constantly rearranging chromatin structure, resulting in dynamic shifts of the regulatory state.
The development of novel techniques such as ChIA-PET, 5C, and Hi-C have allowed the identification of transcription factor modules that are comprised of trans-regulators complexed with different cis-regulatory regions and transcription start sites (Sanyal et al. 2011; Lan et al. 2012). These innovative approaches allow for the characterization of chromosome looping, interaction between different enhancers and promoter regions, and provide a three-dimensional assessment of the state of transcriptional regulation. Recent studies using chromosome conformation capture carbon copy (5C) in different human cell lines have pointed to a large number of long-range interactions between enhancers and promoters and indicate that only a minority of such interactions occur with the nearest gene (Sanyal et al. 2012). Furthermore, ChIA-PET analysis highlights a large number of promoter–promoter interactions between often-distant genes, suggesting the formation of multigene complexes that are coregulated by the same transcriptional machinery (Li et al. 2012). These findings infer a different model of gene regulation in which promoters and enhancers coalesce in transcription “factories” assembled at particular compartments in the nuclei (Sandhu et al. 2012). Such emerging concepts in eukaryotic gene regulation will surely impact how the neural crest genetic program is interpreted and scrutinized in the years to come.
Evolutionary conservation of the neural crest GRN
The neural crest gives rise to a number of vertebrate-specific features such as the craniofacial skeleton and peripheral nervous system. As a vertebrate synapomorphy, neural crest cells are thought to have been crucial for the early evolution of vertebrate body plan and predatory lifestyle. This idea was put forward in the “New Head” hypothesis by Gans and Northcutt, which postulated that the emergence of neural crest derivatives allowed for remodeling of the chordate head and resulted in a shift from filter feeding to active predation (Gans and Northcutt 1983). Although the main premise of the New Head hypothesis is still supported by evidence from the fossil record and developmental studies, tracing the origin of the neural crest to an invertebrate precursor cell type has proven to be a challenge, as invertebrate chordates lack migratory cells that are formed at the neural plate border (Northcutt 2005; Hall and Gillis 2013; Medeiros 2013). Assessment of the regulatory identity of the neural crest and its putative evolutionary precursors holds the promise of shedding light on the evolutionary origins of the vertebrate neural crest.
Genomic studies have been key in informing scenarios that attempt to explain the origins of neural crest and its role in vertebrate evolution. Sequencing of chordate genomes allowed for the systematic characterization of expression and function of the neural crest GRN homologs in the chordate lineage (Yu et al. 2008). Studies from basal vertebrates such as the lamprey have offered interesting insights pertaining to the origin and evolution of the neural crest (Sauka-Spengler et al. 2007). Finally, whole genome phylogenetic analysis also has had considerable impact on how vertebrate evolution and neural crest evolution are surveyed. Molecular phylogenetic studies have placed tunicates as sister groups of vertebrates (Philippe et al. 2005; Delsuc et al. 2006) and also pointed to a potential role for genome-wide duplications in facilitating the origins of the neural crest (Green and Bronner 2013).
The use of the lamprey Petromyzon marinus as a model organism has provided important information about the ancestral state of the neural crest. Lampreys are basal vertebrates that lack several neural crest derivatives, including a neural crest-derived jaw and sympathetic ganglia (Nicol 1952; Häming et al. 2011). Nevertheless they possess a SoxE expressing population of migrating neural crest cells (McCauley and Bronner-Fraser 2006) that contribute to branchial arch cartilage, as well as the formation of pigment cells, cranial, and dorsal root ganglia. In an extensive analysis of the lamprey neural crest genetic program, Sauka-Spengler et al. (2007) described the expression patterns of orthologs of numerous genes occupying different hierarchical positions within the neural crest GRN. The results from this study definitively show conservation of most of the neural crest genetic program in this basal vertebrate with genes maintained in the same spatial and temporal relationships as those observed in jawed vertebrate models (Sauka-Spengler et al. 2007). This was corroborated by functional studies demonstrating that the epistatic relationships between neural plate border genes and neural crest specifiers are largely conserved (Nikitina et al. 2008). Although these results point to a strong conservation of the neural crest GRN across vertebrates, some intriguing differences have emerged from this work. For example, the neural crest specifier Twist, which is thought to be crucial in the acquisition of the mesenchymal state in higher vertebrates, is only expressed in lamprey neural crest cells at a later time, when they populate the branchial arches rather than within the “neural crest specifier” module. Similarly, Ets1, which acts as a critical neural crest specifier gene in jawed vertebrates, functioning directly upstream of both Sox10 and Foxd3, is only employed in the effector module in the lamprey GRN (Sauka-Spengler et al. 2007; Sauka-Spengler and Bronner-Fraser 2008c). Such differences suggest that shifts in the circuitry of the specifier module within the neural crest GRN might result in species-specific traits.
Since comparative genomic analyses support the position of tunicates as the sister group of vertebrates, Ciona intestinalis or other ascidians now occupy a key phylogenetic position for examining the origins of vertebrate features (Schubert et al. 2006). Studies of the development of the ascidian, Ecteinascidia turbinate, led to the identification of pigmented cells from the A7.6 lineage that arises near the neural tube and undergoes migration (Jeffery et al. 2004). This cell lineage, which is known as the trunk lateral cells, expresses the HNK-1 epitope as well as the Twist, FoxD, AP2, and Myc orthologs (Jeffery 2006; Jeffery et al. 2008). Further studies are necessary to determine unequivocally if these cells are derived from the neural plate border, neural tube, or other tissue (Hall and Gillis 2013). In an intriguing recent study, Abitua et al. (2012) showed that a lineage a9.49 in Ciona intestinalis expresses ID, Snail, ETS, and FoxD, as well as melanogenic genes like MITF, TYR, and TYRP. This lineage normally contributes to the formation of the otolith and the ocellus and does not migrate extensively. However, misexpression of Twist causes these cells to adopt a mesenchymal fate and exhibit long-range migration (Abitua et al. 2012). Because these lineages share some common gene signatures with vertebrate neural crest cells, they may hold important clues to neural crest origins.
The expression of neural crest orthologs also has been investigated in the amphioxus (Branchiostoma floridae) (Meulemans and Bronner-Fraser 2002; Meulemans et al. 2003; Yu et al. 2008), a cephalochordate. Although the amphioxus body plan shares important morphological traits with vertebrates, molecular phylogeny has now placed cephalochordates at the base of the chordate tree (Philippe et al. 2005). A systematic analysis of neural crest-related genes indicates conservation of specification mechanisms at the neural plate border between cephalochordates and vertebrates. The transcription factor Snail is expressed specifically in the lateral portion of the neural tube in the neurula, although no other neural crest-specifier genes are coexpressed at this location (Yu et al. 2008). Thus, it is tempting to speculate that this transient expression of Snail may reflect the beginnings of the assembly of a neural crest-specifier module.
Co-option of regulatory elements is likely to have played an important role in the origins of the neural crest in the vertebrate lineage. As an example, Manzanares et al. (2000) performed transgenesis experiments in which they expressed cis-regulatory regions encoding amphioxus Hox gene expression in vertebrates. Surprisingly, despite the fact that amphioxus lacks neural crest, these enhancers drove spatially localized expression in vertebrate neural crest and placode derivatives like cranial ganglia and branchial arches (Manzanares et al. 2000). This study demonstrates that cis-regulatory elements capable of driving gene expression in neural crest cells are already present in the cephalochordate genome. Such elements might have been co-opted to control expression of neural crest-specifier genes, thereby facilitating emergence of this cell type.
Although invertebrate cell lineages described above share some regulatory and behavioral traits with the neural crest, it is difficult to ascertain their position with respect to the evolutionary precursors of neural crest cells. If one defines the neural crest by virtue of its specification kernel, that is, the network module that comprises the neural crest specifier genes, then a bona fide neural crest must have factors that control multipotency, EMT, and diverse lineage specification. As highlighted by Davidson (2009), there are numerous instances in which a given regulatory gene functions in different levels of the GRN, often within the commitment and differentiation of the same cell type. This is observed repeatedly in neural crest development with genes such as Foxd3, SoxE, and Msx1/2, being employed early for specification purposes, and again later to activate differentiation of various cell types (e.g., melanocytes, cartilage, and neuronal cell types). Further work will be necessary to define whether the regulatory similarities observed between neural crest cells and intriguing lineages in invertebrate chordates are a consequence of conservation of ancient differentiation programs or if they indeed are part of the novel regulatory module that characterizes the neural crest.
Taken together, these studies indicate that the proximal levels of the neural crest gene regulatory network, such as induction of the neural plate border, are primitive features shared amongst chordates, whereas the core neural crest specification modules of the network might have been added during early vertebrate evolution. This is expected since delimitation of the neural plate border is necessary for development of the central nervous system. The terminal differentiation programs of the neural crest GRN also are deeply conserved in both cephalochordates and tunicates, particularly the control of pigment cell differentiation (Yu et al. 2008; Davidson 2009). This is not surprising as most of the differentiation batteries are thought to be ancient (Davidson 2009). Placement of the origin of the specification kernel of the GRN still demands further studies, but it is clear that it has a high degree of conservation among vertebrates, as supported by studies in the lamprey (Sauka-Spengler et al. 2007; Sauka-Spengler and Bronner-Fraser 2008a).
Neural crest cells and disease
The above data show that scrutinizing the regulatory mechanisms controlling neural crest formation can expand our understanding of essential cellular processes such as delamination, migration, and differentiation, as well as shed light on important evolutionary questions pertaining to vertebrate origins and adaptation. Beyond its relevance for basic biological questions, the neural crest is also of remarkable medical importance for its role in birth defects and malignant diseases. Studies on the plasticity of the neural crest and its fate switches also offer clues for strategies for cell reprograming and stem cell manipulation. In addition, the persistence of neural crest stem cells in adult tissues offers potential targets for therapies aiming to utilize endogenous progenitors for repair and regeneration.
Given the sheer number of cell types and derivatives formed by the neural crest progenitor cell population and the complexity of its developmental program, it is not surprising that a large fraction of congenital birth defects can be traced back to the neural crest (Hall 1999). Neural crest developmental anomalies, or neurocristopathies, are responsible for the vast majority of craniofacial malformations, and more than 700 different syndromes have been described (Trainor 2010). In addition to the skeletal elements of the face, abnormal neural crest development can also affect the heart, adrenal medulla, pigment cells, and the peripheral nervous system. Although treatment of neurocristopathies has improved considerably over the years, dissection of the molecular mechanisms underlying many of these disorders could bring considerable improvements to management and prevention of these conditions.
In the past few decades, the molecular basis of several neural crest-related syndromes has been investigated through genetic linkage as well as the use of animal models. One example is Treacher Collins disease, an autosomal dominant disorder that is marked by severe hypoplasia of skeletal elements of the face and irregularities in otic and ophthalmic development (The Treacher Collins Syndrome Collaborative Group 1996; Trainor 2010). Genetic mapping and gene expression analysis identified mutations in the Tcof1 gene as the cause of the disease. Mapping of TCOF1 expression in mouse embryos shows that it is enriched in neural crest cells and facial mesenchyme, raising the intriguing possibility that the mutations could be interfering with neural crest development (Dixon et al. 2006). In fact, TCOF1 is important for ribosome biogenesis and disruption of this process results in TP53-mediated apoptosis, and analysis of Tcof1 null mice reveals strong reduction in the migratory neural crest population as a consequence of increased apoptosis and reduced proliferation of migrating cells (Dixon et al. 2006).
Genomic approaches have greatly benefited the dissection of mechanisms leading to congenital disease. Genome-wide association studies have been pivotal to the identification of a number of loci and genetic variants associated to craniofacial anomalies (Grant et al. 2009; Beaty et al. 2010; Mangold et al. 2010; Dixon et al. 2011) and also suggest genes linked to normal variation in facial structure (Liu et al. 2012). Similarly, recent studies using exome sequencing have been used to map the causes of craniofacial defects in conditions such as Miller syndrome (Ng et al. 2010) and Nager syndrome (Bernier et al. 2012). Current transcriptome and epigenomic analysis should accelerate the dissection of the genetic basis of neural crest-related syndromes through regulatory network comparisons between normal and disease states. Such studies promise to uncover more complex aspects of disease such as variable penetrance and multifactorial etiology. In this context, centralized databases of genomic information containing data sets of high-throughput studies such as Facebase and ENCODE will be key (The ENCODE Project Consortium 2004; Hochheiser et al. 2011). Such integrated databases and user-friendly bioinformatic software will become increasingly important in the neural crest field as it continues to shift toward genomic approaches.
The medical importance of neural crest cells is not restricted to congenital birth defects. This cell population has an important link to metastatic conditions due to its migratory and invasive properties. Neural crest-derived cancers, such as melanoma and neuroblastomas, tend to be particularly aggressive and prone to metastasis. This has led to the assumption that malignancy in neural crest-derived cell types involves the reactivation of regulatory circuits important for embryonic development but that are normally silenced once cell differentiation has occurred. Indeed, evidence from studies of malignant melanoma cell lines shows anomalous transcriptional reactivation of genes such as Snai, Twist, Sox10, and Myb, all of which are important neural crest specifier genes in the early GRN (Shakhova et al. 2012; Shirley et al. 2012; Weiss et al. 2012). Interestingly, transplanting melanoma cells to chicken embryo causes “regulation” of some of the cells that migrate along neural crest pathways, although some do localize in ectopic sites (Kulesa et al. 2006). This suggests that melanoma cells can respond to cues in the embryonic environment similar to neural crest cells. Thus, the embryonic environment may be able to control or inhibit the metastatic state (Hendrix et al. 2007), at least under some circumstances. Although this is an intriguing possibility, further work that compares the genetic programs controlling neural crest and neural crest-derived tumor cells is needed to inform upon the link between neural crest development and metastatic behavior in malignant cells.
The neural crest has also been an important model for the study of EMT, which is central for metastatic processes. A wealth of knowledge regarding the genetics and cell biology underlying EMT has been obtained through the use of the neural crest as a model (Barrallo-Gimeno and Nieto 2005; Lee et al. 2006; Yang and Weinberg 2008; Acloque et al. 2009). Interestingly, recent evidence suggests that cells undergoing EMT seem to acquire stem-like properties. Induction of EMT in different cell lines has been shown to up-regulate totipotency markers such as POU5F1 (also known as OCT4) and NANOG in conjunction with transition to a mesenchymal state (Mani et al. 2008). Such findings highlight similarities between the metastatic process and neural crest development. They also point to interconnectivity between cell behavior and developmental potential. In fact, it is likely that advances in the assembly of the neural crest regulatory network will highlight common nodes in the genetic circuits controlling EMT, plasticity, and migratory potential. Such events are likely to depend on the shared regulatory circuits that interconnect to define neural crest identity.
The question of plasticity has been central to neural crest biology, and interest in the topic of self-renewal and developmental potential has driven efforts to isolate neural crest stem cells. Work by several investigators has led to the identification and purification of neural crest stem cells—cells with the potential to self-renew and also to give rise to the diverse population of derivatives that are generated by the neural crest. The first neural crest progenitor cells were isolated in vitro by clonal analysis of quail cells and shown to be multipotent (Sieber-Blum and Cohen 1980). The ability of neural crest cells to self-renew was first demonstrated in elegant studies by Stemple and Anderson using clonal cultures of murine neural crest cells purified by cell sorting based on expression of the NGFR (p75) cell surface epitope (Stemple and Anderson 1992, 1993). These cells have the ability to self-renew and also to form neurons or glia, thus demonstrating true stem cell properties. More recently, clonal analysis of cranial neural crest in quail has revealed that this population has the potential to differentiate into osteoblasts (Calloni et al. 2009), and that the majority of the cranial neural crest is comprised of progenitors with both osteogenic and neural-melanocytic potential. Remarkably, this study also identified precursors that can give rise to all neural crest-derived phenotypes analyzed, which indicates that part of the migratory cranial crest remains multipotent (Calloni et al. 2009). Although isolated neural crest stem cells represent a good model for studies of plasticity and commitment, there are possible caveats. Because it is likely that in vitro culture conditions introduce changes that do not reflect in vivo behavior, embryological observations of these cells in their normal environment will be critical to uncovering the mechanism underlying maintenance and renewal of the latent neural crest stem cell population.
Neural crest-like cell populations have been derived from human embryonic stem cells by a number of different methods (Chimge and Bayarsaihan 2010). For instance, neural crest cells were isolated from neural rosettes through fluorescence activated cell sorting (FACS) using markers NGFR (p75) and B3GAT1 (previously HNK1,) and subsequently express neural crest specifiers, such as SOX10 and SNAI1 (SNAIL). These have been shown to differentiate into a wide range of neural crest derivatives, including sensory and autonomic neurons, Schwann cells, myofibroblasts, adipocytes, cartilage, and bone cells (Lee et al. 2007). Another study identified early migrating neural crest cells in neurospheres cultured on fibronectin plates. These cells express the neural crest specifier SOX10 and clonal analysis indicates they are multipotent. Importantly, when these cells are injected in chick embryos, they migrate normally and contribute to the same derivatives as endogenous neural crest cells (Curchoe et al. 2010).
Induced neural crest cells (iNCCs) from biopsied human tissue may be useful in different contexts for clinical purposes. First, iNCCs can be used as a model for the study of human neurocristopathies, as cells from patients can be reprogrammed to adopt a neural crest fate (Lee et al. 2009). This approach essentially allows the observation of an embryonic cell population from reprogrammed adult cells, which can be used for transcriptomic, epigenomic, or behavioral investigations that can uncover molecular mechanisms of disease and possibly also expand our knowledge of the neural crest gene regulatory network. There is also the promise that induced neural crest cells could be used for cell therapy for diseases that involve failure or impairment of neural crest-derived tissue (Crane and Trainor 2006; Barraud et al. 2010). For instance, transplantation of induced “neural crest-like cells” to diabetic mice resulted in improvement of the impaired nerve and vascular functions that result from diabetic neuropathy (Okawa et al. 2012). One would imagine that engineered neural crest cells could be used in such fashion to treat neurodegenerative disorders that affect peripheral nervous system, among others.
Although it is clear that iNCCs are a powerful tool that can reveal novel molecular mechanisms and hold promise in treatment of disease, it is important to bear in mind that neural crest development is intrinsically linked to the embryonic environment. In vitro conditions do not necessarily reflect the complexity and dynamism of a developing embryo and thus are unlikely to recapitulate in vivo conditions. This is especially true for the neural crest, which is continuously interacting with different cell types and embryonic environments. Therefore, in vivo approaches continue to represent the gold standard for neural crest biology, and findings obtained with isolated neural crest cells or iNCCs should be validated with model organisms when possible.
Conclusions
Over one hundred years of investigation of the neural crest has produced a remarkable body of knowledge about neural crest cell behavior, derivatives, and plasticity. The ingenuity and creativity of classical embryologists helped to frame the very questions that remain integral to the field of neural crest biology today. The introduction of recombinant DNA technology in the 1990s has finally allowed these questions to be addressed at the mechanistic level by analysis of the components of the genetic machinery that imbues the neural crest with its fascinating properties. In the last twenty years, several aspects of the genetic circuitry controlling these cells have been identified and functionally characterized, providing important insights into the logic of neural crest development.
Yet we are still at the “tip of the iceberg” in terms of understanding this intriguing cell population from an integrated genomic perspective. Further transcriptome and enhancesome analysis of neural crest cells at various stages and states of development will greatly increase our understanding of the neural crest GRN. Since regulatory states are dynamic and interconnected, this should be expanded to encompass more axial levels and time points of development, ranging from the neural plate border to the final differentiated state. The formulation of computational models that can predict outcomes within the GRN and account for species-specific differences across vertebrates hold the promise of greatly increasing our understanding of deuterostome evolution (Peter et al. 2012). Finally, additional levels of control have been shown to act in the control of neural crest behavior and identity, including epigenetic (Hu et al. 2012), post transcriptional (Jayasena and Bronner 2012), and post-translational regulation (Lee et al. 2012). Future research along these lines has the potential to reveal the secrets of the neural crest from its inception at the neural plate border to their acquisition of specific differentiated fates.
Another exciting feature of the neural crest is its position at the convergence of developmental, cancer, and stem cell biology. As such, neural crest cells serve as a model for the investigation of processes as diverse as the epithelial to mesenchymal transition, to cell motility and migration, and transition from stemness to the differentiated state. Genomic analyses provide tools and insights that are poised to reveal the complexities of these events with unprecedented depth and breadth. Given the inherent interconnections that exist between diverse biological processes, the neural crest represents an excellent model system for unbiased examination of the genetic basis of cellular identity and behavior. In turn, the results of these studies will undoubtedly have implications for other aspects of stem cell biology and metastatic transformation.
Acknowledgments
We would like to thank Tatiana Hochgreb, Sujata Bhattacharyya, and Stephen Green for critical reading of the manuscript and helpful suggestions. This work was supported by NIH grants HD037105 and DE16459 (M.E.B.), the Pew Fellows Program in the Biomedical Sciences (M.S-C.), and a Caltech Cell Center fellowship (M.S-C.)
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.157586.113.
References
- Abitua PB, Wagner E, Navarrete IA, Levine M 2012. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA 2009. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest 119: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MS, Gammill LS, Bronner-Fraser M 2008. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn 237: 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, McCulley DJ, Xu SM, Dodou E, Anderson JP, Wei ML, et al. 2011. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development 138: 2555–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Behringer RR, Rowitch DH, Schedl A, Epstein JA, de Crombrugghe B 2004. Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci 101: 6502–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G 2006. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol 291: 382–397 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA 2005. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Barraud P, Seferiadis AA, Tyson LD, Zwart MF, Szabo-Rogers HL, Ruhrberg C, Liu KJ, Baker CV 2010. Neural crest origin of olfactory ensheathing glia. Proc Natl Acad Sci 107: 21040–21045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. 2010. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet 42: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, Jabs EW, Innis JW, Schuette JL, Gorski JL, et al. 2012. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet 90: 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T 2010a. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol 26: 581–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T 2010b. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci 107: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T 2004. Wnt1-Cre-mediated deletion of AP-2α causes multiple neural crest-related defects. Dev Biol 267: 135–152 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser S 1988. Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature 335: 161–164 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser S 1989. Developmental potential of avian trunk neural crest cells in situ. Neuron 3: 755–766 [DOI] [PubMed] [Google Scholar]
- Calloni GW, Le Douarin NM, Dupin E 2009. High frequency of cephalic neural crest cells shows coexistence of neurogenic, melanogenic, and osteogenic differentiation capacities. Proc Natl Acad Sci 106: 8947–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KK, Chen YS, Yau TO, Fu M, Lui VC, Tam PK, Sham MH 2005. Hoxb3 vagal neural crest-specific enhancer element for controlling enteric nervous system development. Dev Dyn 233: 473–483 [DOI] [PubMed] [Google Scholar]
- Chimge NO, Bayarsaihan D 2010. Generation of neural crest progenitors from human embryonic stem cells. J Exp Zoolog B Mol Dev Evol 314: 95–103 [DOI] [PubMed] [Google Scholar]
- Cooper GM, Sidow A 2003. Genomic regulatory regions: Insights from comparative sequence analysis. Curr Opin Genet Dev 13: 604–610 [DOI] [PubMed] [Google Scholar]
- Cox SG, Kim H, Garnett AT, Medeiros DM, An W, Crump JG 2012. An essential role of variant histone H3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet 8: e1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JF, Trainor PA 2006. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol 22: 267–286 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2: 292–301 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curchoe CL, Maurer J, McKeown SJ, Cattarossi G, Cimadamore F, Nilbratt M, Snyder EY, Bronner-Fraser M, Terskikh AV 2010. Early acquisition of neural crest competence during hESCs neuralization. PLoS ONE 5: e13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. 2009. The regulatory genome: Gene regulatory networks in development and evolution. Elsevier, Burlington, MA. [Google Scholar]
- Degenhardt KR, Milewski RC, Padmanabhan A, Miller M, Singh MK, Lang D, Engleka KA, Wu M, Li J, Zhou D, et al. 2010. Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev Biol 339: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439: 965–968 [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA 2006. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci 103: 13403–13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC 2011. Cleft lip and palate: Understanding genetic and environmental influences. Nat Rev Genet 12: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V 2004. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev 121: 173–182 [DOI] [PubMed] [Google Scholar]
- Dupin E, Calloni GW, Le Douarin NM 2010. The cephalic neural crest of amniote vertebrates is composed of a large majority of precursors endowed with neural, melanocytic, chondrogenic and osteogenic potentialities. Cell Cycle 9: 238–249 [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP 1994. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development 120: 2213–2224 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640 [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS 2006. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M 2002. Genomic analysis of neural crest induction. Development 129: 5731–5741 [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M 2003. Neural crest specification: Migrating into genomics. Nat Rev Neurosci 4: 795–805 [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG 1983. Neural crest and the origin of vertebrates: A new head. Science 220: 268–273 [DOI] [PubMed] [Google Scholar]
- Garnett AT, Square TA, Medeiros DM 2012. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development 139: 4220–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, et al. 2009. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr 155: 909–913 [DOI] [PubMed] [Google Scholar]
- Green SA, Bronner ME 2013. Gene duplications and the early evolution of neural crest development. Semin Cell Dev Biol 24: 95–100 [DOI] [PubMed] [Google Scholar]
- Hall BK. 1999. The neural crest in development and evolution. Springer, Berlin. [Google Scholar]
- Hall BK, Gillis JA 2013. Incremental evolution of the neural crest, neural crest cells and neural crest-derived skeletal tissues. J Anat 222: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häming D, Simoes-Costa M, Uy B, Valencia J, Sauka-Spengler T, Bronner-Fraser M 2011. Expression of sympathetic nervous system genes in lamprey suggests their recruitment for specification of a new vertebrate feature. PLoS ONE 6: e26543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM 2007. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer 7: 246–255 [DOI] [PubMed] [Google Scholar]
- His W. 1868. Untersuchungen über die erste Anlage des Wirbeltierleibes. Die erste Entwickelung des Hühnchens im Ei. FCW Vogel, Leipzig. [Google Scholar]
- Hochheiser H, Aronow BJ, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, et al. 2011. The FaceBase Consortium: A comprehensive program to facilitate craniofacial research. Dev Biol 355: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré SM, Aybar MJ, Mayor R 2003. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol 260: 79–96 [DOI] [PubMed] [Google Scholar]
- Horstadius S. 1950. The neural crest. Oxford University Press, Oxford, UK. [Google Scholar]
- Hu N, Strobl-Mazzulla P, Sauka-Spengler T, Bronner ME 2012. DNA methyltransferase3A as a molecular switch mediating the neural tube-to-neural crest fate transition. Genes Dev 26: 2380–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr, Nakajima A, Shuler CF, Moses HL, Chai Y 2003. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130: 5269–5280 [DOI] [PubMed] [Google Scholar]
- Jayasena CS, Bronner ME 2012. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-β signaling. J Cell Biol 199: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR 2006. Ascidian neural crest-like cells: Phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J Exp Zool B Mol Dev Evol 306: 470–480 [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y 2004. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature 431: 696–699 [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Chiba T, Krajka FR, Deyts C, Satoh N, Joly JS 2008. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: Insights into the ancestry and evolution of the neural crest. Dev Biol 324: 152–160 [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM 2000. Fate of the mammalian cardiac neural crest. Development 127: 1607–1616 [DOI] [PubMed] [Google Scholar]
- Jones NC, Trainor PA 2004. The therapeutic potential of stem cells in the treatment of craniofacial abnormalities. Expert Opin Biol Ther 4: 645–657 [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, Hendrix MJ 2006. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc Natl Acad Sci 103: 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Farnham PJ, Jin VX 2012. Uncovering transcription factor modules using one- and three-dimensional analyses. J Biol Chem 287: 30914–30921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM 1986. Investigations on the neural crest. Methodological aspects and recent advances. Ann NY Acad Sci 486: 66–86 [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C 1999. The neural crest. Cambridge University Press, Cambridge, UK [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E 2004. Neural crest cell plasticity and its limits. Development 131: 4637–4650 [DOI] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW 2006. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol 172: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Kim H, Elkabetz Y, Al Shamy G, Panagiotakos G, Barberi T, Tabar V, Studer L 2007. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol 25: 1468–1475 [DOI] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461: 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C 2012. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J Cell Biol 198: 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, van der Lijn F, Schurmann C, Zhu G, Chakravarty MM, Hysi PG, Wollstein A, Lao O, de Bruijne M, Ikram MA, et al. 2012. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet 8: e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, et al. 2010. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet 42: 24–26 [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares M, Wada H, Itasaki N, Trainor PA, Krumlauf R, Holland PW 2000. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature 408: 854–857 [DOI] [PubMed] [Google Scholar]
- Mateos-Langerak J, Goetze S, Leonhardt H, Cremer T, van Driel R, Lanctot C 2007. Nuclear architecture: Is it important for genome function and can we prove it? J Cell Biochem 102: 1067–1075 [DOI] [PubMed] [Google Scholar]
- McCauley DW, Bronner-Fraser M 2006. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature 441: 750–752 [DOI] [PubMed] [Google Scholar]
- Medeiros D 2013. The evolution of the neural crest: New perspectives from lamprey and invertebrate neural crest-like cells. WIREs Dev Biol 2: 1–15 [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M 2002. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development 129: 4953–4962 [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M 2004. Gene-regulatory interactions in neural crest evolution and development. Dev Cell 7: 291–299 [DOI] [PubMed] [Google Scholar]
- Meulemans D, McCauley D, Bronner-Fraser M 2003. Id expression in amphioxus and lamprey highlights the role of gene cooption during neural crest evolution. Dev Biol 264: 430–442 [DOI] [PubMed] [Google Scholar]
- Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA 2004. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development 131: 829–837 [DOI] [PubMed] [Google Scholar]
- Murisier F, Guichard S, Beermann F 2006. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev Biol 298: 644–655 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert MC, Robbins J 2006. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res 98: 1547–1554 [DOI] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. 2010. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet 42: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JAC 1952. Autonomic nervous systems in lower chordates. Biol Rev Camb Philos Soc 27: 1–50 [Google Scholar]
- Nikitina N, Sauka-Spengler T, Bronner-Fraser M 2008. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc Natl Acad Sci 105: 20083–20088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG 2005. The new head hypothesis revisited. J Exp Zool B Mol Dev Evol 304: 274–297 [DOI] [PubMed] [Google Scholar]
- Okawa T, Kamiya H, Himeno T, Kato J, Seino Y, Fujiya A, Kondo M, Tsunekawa S, Naruse K, Hamada Y, et al. 2012. Transplantation of neural crest like cells derived from induced pluripotent stem cells improves diabetic polyneuropathy in mice. Cell Transplant doi: 10.3727/096368912X657710 [DOI] [PubMed] [Google Scholar]
- Peter IS, Faure E, Davidson EH 2012. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci 109: 16434–16442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H 2005. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol 22: 1246–1253 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Serralbo O, Salgado D, Marcelle C 2011. Neural crest regulates myogenesis through the transient activation of NOTCH. Nature 473: 532–535 [DOI] [PubMed] [Google Scholar]
- Sandhu KS, Li G, Poh HM, Quek YL, Sia YY, Peh SQ, Mulawadi FH, Lim J, Sikic M, Menghi F et al. 2012. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep 2: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Bau D, Marti-Renom MA, Dekker J 2011. Chromatin globules: A common motif of higher order chromosome structure? Curr Opin Cell Biol 23: 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y 2005. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 132: 2355–2363 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M 2008. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol 87: 237–256 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M 2008a. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis 46: 673–682 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M 2008b. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol 9: 557–568 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M 2008c. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol Bull 214: 303–314 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M 2007. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell 13: 405–420 [DOI] [PubMed] [Google Scholar]
- Schubert M, Escriva H, Xavier-Neto J, Laudet V 2006. Amphioxus and tunicates as evolutionary model systems. Trends Ecol Evol 21: 269–277 [DOI] [PubMed] [Google Scholar]
- Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, Claudinot S, Okoniewski M, Beermann F, Mihic-Probst D, et al. 2012. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol 14: 882–890 [DOI] [PubMed] [Google Scholar]
- Shirley SH, Greene VR, Duncan LM, Torres Cabala CA, Grimm EA, Kusewitt DF 2012. Slug expression during melanoma progression. Am J Pathol 180: 2479–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber-Blum M, Cohen AM 1980. Clonal analysis of quail neural crest cells: They are pluripotent and differentiate in vitro in the absence of noncrest cells. Dev Biol 80: 96–106 [DOI] [PubMed] [Google Scholar]
- Simões-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME 2012. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genet 8: e1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Robinson V, Patel K, Wilkinson DG 1997. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol 7: 561–570 [DOI] [PubMed] [Google Scholar]
- Soriano P 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ 1992. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell 71: 973–985 [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ 1993. Lineage diversification of the neural crest: In vitro investigations. Dev Biol 159: 12–23 [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Bronner ME 2012. A PHD12–Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J Cell Biol 198: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M 2010. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell 19: 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, Kondoh H 2011. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470: 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA 2010. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A 152A: 2984–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Treacher Collins Syndrome Collaborative Group 1996. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet 12: 130–136 [DOI] [PubMed] [Google Scholar]
- Trentin A, Glavieux-Pardanaud C, Le Douarin NM, Dupin E 2004. Self-renewal capacity is a widespread property of various types of neural crest precursor cells. Proc Natl Acad Sci 101: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4: 509–519 [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Takemoto T, Kamachi Y, Kondoh H 2004. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev 121: 1145–1158 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kumar RM, Biggs VJ, Lee H, Chen Y, Kagey MH, Young RA, Abate-Shen C 2011. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev Cell 21: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MB, Abel EV, Mayberry MM, Basile KJ, Berger AC, Aplin AE 2012. TWIST1 is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res 72: 6382–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA 2008. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell 14: 818–829 [DOI] [PubMed] [Google Scholar]
- Yokota Y, Saito D, Tadokoro R, Takahashi Y 2011. Genomically integrated transgenes are stably and conditionally expressed in neural crest cell-specific lineages. Dev Biol 353: 382–395 [DOI] [PubMed] [Google Scholar]
- Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M 2008. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res 18: 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]