Abstract
Cancer evolution involves cycles of genomic damage, epigenetic deregulation, and increased cellular proliferation that eventually culminate in the carcinoma phenotype. Early neoplasias, which are often found concurrently with carcinomas and are histologically distinguishable from normal breast tissue, are less advanced in phenotype than carcinomas and are thought to represent precursor stages. To elucidate their role in cancer evolution we performed comparative whole-genome sequencing of early neoplasias, matched normal tissue, and carcinomas from six patients, for a total of 31 samples. By using somatic mutations as lineage markers we built trees that relate the tissue samples within each patient. On the basis of these lineage trees we inferred the order, timing, and rates of genomic events. In four out of six cases, an early neoplasia and the carcinoma share a mutated common ancestor with recurring aneuploidies, and in all six cases evolution accelerated in the carcinoma lineage. Transition spectra of somatic mutations are stable and consistent across cases, suggesting that accumulation of somatic mutations is a result of increased ancestral cell division rather than specific mutational mechanisms. In contrast to highly advanced tumors that are the focus of much of the current cancer genome sequencing, neither the early neoplasia genomes nor the carcinomas are enriched with potentially functional somatic point mutations. Aneuploidies that occur in common ancestors of neoplastic and tumor cells are the earliest events that affect a large number of genes and may predispose breast tissue to eventual development of invasive carcinoma.
The cells of a multicellular organism are related to one another by a bifurcating lineage tree whose root is the zygote. DNA replication, chromosome segregation, and cell division during development from the zygote to the adult introduces point mutations and other DNA changes into the genome, which persist in the descendants of the cells in which they occurred. Germ-line point mutations occur at a rate of approximately one per diploid genome per cell division (Kong et al. 2012), but the rate of somatic changes is less well-understood, and is likely to vary by tissue type. Large-scale genomic changes such as aneuploidies are generally thought to be extremely rare in normal tissue.
Cancers, in contrast to normal tissue, accumulate much larger numbers of genomic changes, as illustrated by genome sequencing of late-stage tumors (Ley et al. 2008; Stratton et al. 2009; Bignell et al. 2010; Pleasance et al. 2010a; Chapman et al. 2011; Stratton 2011; Banerji et al. 2012; Nik-Zainal et al. 2012a,b). Solid tumors are highly mutated by several mechanisms, such as point mutations, copy-number variations, and chromothripsis (Greenman et al. 2007; Leary et al. 2008; Beroukhim et al. 2010; Liu et al. 2011; Meyerson and Pellman 2011; Stephens et al. 2011; Crasta et al. 2012; Maher and Wilson 2012); relapses or metastases exhibit further mutational evolution (Ding et al. 2010, 2012; Yachida et al. 2010; Navin et al. 2011; Mardis 2012; Turajlic et al. 2012; Walter et al. 2012; Wu et al. 2012). The state of an individual advanced cancer genome sheds little light on the order of genomic changes, however, except in analyses of subclone evolution (Nik-Zainal at al. 2012a; Shah et al. 2012). In an advanced tumor, the earliest driver changes that had predisposed ancestral cells to eventual carcinoma development are confounded with later changes. As a consequence, our understanding of early tumor evolution is still in its infancy.
The historically proven approach to understanding evolution is comparative analysis of extant species, whose power was greatly increased by whole-genome sequencing in recent years. Analogous to species comparisons, which are based on evolutionary (bifurcating) lineage trees, comparisons of somatic genomes from a single individual could, in principle, shed light on somatic evolution, but in normal tissue the number of mutations is low. However, given the large number of genomic changes during tumor evolution, it may be possible to dissect the evolutionary history of a cancer by comparing its genome to clinically recognized precursor lesions. In this context, breast cancers provide a proof-of-principle opportunity, due to their frequent association with early neoplastic lesions that are readily identified by morphology (Simpson et al. 2005; Abdel-Fatah et al. 2007; Lopez-Garcia et al. 2010; Bombonati and Sgroi 2011), and whose genomes may provide windows into the earliest stages of tumor evolution.
Using whole-genome sequencing of histologically characterized archival (formalin-fixed, paraffin-embedded) samples, we determine lineage relationships of early neoplasias with carcinomas, quantify mutational load and mutation spectra during progression from normal tissue to neoplasia to carcinoma, and find the earliest detectable mutations and aneuploidies in cell lineages ancestral to the lesions. A subset of these early events may have provided the initial oncogenic potential and helped trigger the first clonal expansion. Our analyses reveal variation among the six cases in the specific evolution of neoplasia and tumor, as would be expected for an evolutionary process dominated by stochasticity. The mechanistic commonalities among the cases, however, bear significant implications for our conceptualization of tumor origins and progression.
Results
Whole-genome sequencing of early neoplasias and related carcinomas from archival material
Our workflow (Supplemental Fig. S1) began with the screening of histopathological sections of archival estrogen receptor-positive invasive ductal carcinoma (IDC) resection specimens for the presence of concurrent early neoplasias, which are microscopic in size (typically 1–3 mm). We selected cases in which early neoplasia with or without atypia (“EN” or “ENA”; a spectrum of usual ductal hyperplasia, columnar cell lesions, and flat epithelial atypia), and in some cases ductal carcinoma in situ (DCIS) were present in addition to the IDC. Areas of high neoplasia or carcinoma content were cored and histologically re-evaluated for lesion purity. Six cases were chosen, in which each sample met criteria for purity and had enough DNA for whole-genome sequencing. Each case had at least one early neoplasia sample from the same side in which the carcinoma was found, and five also had a contralateral early neoplasia (Supplemental Fig. S2). Each had at least one control sample (lymph, normal breast tissue, or both), and three cases also had a DCIS in addition to the IDC, yielding a total of 31 samples that belong to seven classes of normal and neoplastic tissue (Fig. 1A).
Figure 1.
Lineage tree and alternate allele frequencies. (A) The samples in this study by type (rows) and patient (columns). (B) Model of neoplastic progression on the basis of organismal tissue and cell lineage. For simplicity, only one possible scenario of the progression from normal to neoplasia to carcinoma is shown. Mutations that arise in ancestors are propagated through subsequent divisions to all descendants. Depending on the ancestors in which they arise, they will be found in one or more samples of the patient, with varying prevalence. For example, mutations that arise in the B branches will be found in all cells of the neoplasia and of the carcinoma; in contrast, mutations that arise on the C branch will be present only in a subset of the neoplasia cells and mark the neoplastic subpopulation from which the carcinoma arose. Mutations that arise on the F branch mark a clonal expansion within the neoplasia, after the last common ancestor with the carcinoma. Note that if there are no mutations found that define branches B and C, it is not possible to infer a specific relationship of the carcinoma with the neoplasia. (NS) Not sampled. In the expanded box are alternate allele frequency comparisons relevant to neoplasias and carcinomas. The two starred comparisons require independent estimates of the proportion of normal cells in each sample, as they compare AAFs across different samples. All other comparisons are either within samples, or the AAF is zero, thus requiring no independent estimate of the proportion of normal cells in the sample. (C–F) Alternate allele frequencies as a function of the class and sample for each patient with phylogenetically informative SNV-sharing classes. The number of SNVs in each class and the branch in the lineage tree of A are listed below each plot. For Patient 1, the only phylogenetically informative class was where the IDC shared SNVs with ENA. For the other patients, the AAFs of informative classes are grouped together and the mutation pattern for each class is represented by a series of zeros and ones directly above the sample labels (a “1” indicates that the SNVs were present in the corresponding sample and a “0” indicates that they were not). (EN) Early neoplasia; (EN_cl) early neoplasia contralateral; (ENA) early neoplasia with atypia. Subscript in lineage-tree branch of patient 6 denotes whether the neoplasia in the lineage tree is this patient's EN or ENA, and whether the carcinoma is DCIS or IDC.
We optimized DNA isolation from archival samples to obtain sufficient quantities of preparative material, and honed the generation of robust libraries. For each sample, a single library was built and sequenced with paired-end reads (2 × 101 bp) on the Illumina HiSeq platform. Library complexity was sufficient to support deep whole-genome sequencing, with the vast majority of sequence data coming from independent DNA fragments as opposed to PCR duplicates. The samples from the first patient were sequenced to higher coverage (average of 84.6×) to calibrate the tradeoff between cost and sensitivity in variation calling. Coverage of each sample by confidently mapped reads ranged from 46.7× to 105.7×, with a median of 53.4× (Supplemental Fig. S3).
Somatic SNVs fall into a limited and highly structured set of classes
Detection of somatic single nucleotide variants (SNVs), such as those occurring during cancer evolution, requires a methodology with high specificity, because inherited (germline) variants are orders of magnitude more numerous, and even a low rate of miscalling inherited variants as somatic results in low accuracy. Our high sequence coverage and purity of samples allowed us to pursue highly sensitive and specific somatic SNV identification. Because we sequenced several samples from each patient, we identified the total set of SNVs in each patient with a multi-sample strategy using GATK (McKenna et al. 2010; DePristo et al. 2011). For each patient we called variants using reads from all samples simultaneously, and then assigned genotypes to each sample. The vast majority of SNVs were present in all samples, as expected from germline variants. Standard quality control metrics confirmed the high quality of our variant calls. The total number of high-confidence germline variants ranged from 2,650,714 (Patient 5) to 2,973,005 (Patient 1). Between 97.91% and 98.06% of these were present in dbSNP. On average, 59,697 SNVs per patient were present in all samples, but not in dbSNP, and therefore represent novel SNPs of low population-allele frequency (Table 1).
Table 1.
Variant call statistics
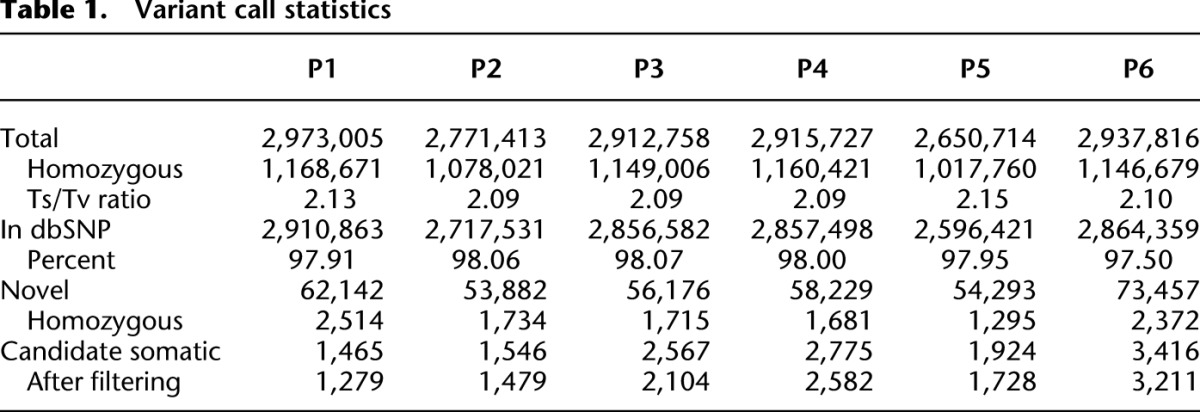
Between 1465 (Patient 1) and 3416 (Patient 6) SNVs were candidate somatic variants, as they were not detected in at least one sample of that patient (Table 1). If the samples are related by a tree, then only some sharing classes are possible and the total number of observed classes is much lower than the number of possible classes. For example, in Patient 1, from whom we sequenced six samples, there are 26 − 1 = 63 possible classes to which an SNV can belong. In this patient, 1766 SNVs were absent from at least one sample, and excluding those present in lymph we retain 1465 candidate somatic SNVs (Supplemental Table 1; Supplemental Material). Only six of the classes, containing 1279 out of the initial 1465 candidate SNVs (87%), survived filtering. Those SNVs removed during filtering were either germline SNVs where one allele was poorly covered, or somatic SNVs whose class membership we could not confidently establish. PCR-based targeted validation of 388 SNVs in Patients 2 and 6 revealed a call accuracy of 100% and 92%, respectively (Supplemental Fig. S4; Supplemental Material).
Across the six cases, we retained 82%–96% (median = 91%) of SNVs and 19%–43% (median = 27%) of classes, revealing substantial structure in the data. The final number of confident somatic SNVs ranges from 1279 in Patient 1 to 3211 in Patient 6, for a total of 12,392 in all six patients. A total of 8950 (72%) of these are private to only one sample in only one patient, and the number of such private SNVs increases as a function of the severity of the cancer phenotype: the IDCs harbor the most private mutations (average of 601 per sample, n = 7, range 46–1809), the DCISs have an average of 470 SNVs per sample (n = 3 range 70–978), early lesions 229 per sample (n = 14, range 123–387), and normal have the fewest (n = 2, range 39–89). On average, the IDCs accumulated 2.6-fold more private mutations than the early neoplasias, and almost 10-fold more than normal breast tissue. This may be due to a larger number of cell divisions or an increased mutation rate in the ancestral cell lineage of the IDC.
Allele frequencies of somatic SNVs support common ancestral relationships
Somatic SNVs that are not private to individual samples define phylogenetically informative classes. A total of 3442 SNVs define such classes, ranging from 0 SNVs in Patient 4 to 1054 SNVs in Patient 3, with a per-case average of 574 and a per-class (n = 7) average of 492. To illustrate the logic of phylogenetic inference using informative classes, we consider a hypothetical lineage tree that relates non-breast somatic, normal breast, neoplastic, and carcinoma cell lineages (Fig. 1B). Mutations that occurred in ancestral cells are present in specific subsets of samples, with the lineage tree constraining the set of possible classes.
As demonstrated in recent studies of subclone evolution in IDC (Nik-Zainal et al. 2012a,b; Shah et al. 2012), alternate allele frequency (AAF) is a powerful metric for understanding tumor evolution. The “alternate allele” is the allele that does not match the reference base, and which in the vast majority of cases is the somatic mutation. Its frequency is estimated from its sequence coverage divided by the coverage of the alternate base plus that of the reference base. Depending on the ancestral lineage in which a collection of mutations arose, their AAF distributions in each sample vary. For example, if a variant arose in a common ancestor of a subset of lesional cells in the sample, its AAF is lower than that of an earlier mutation that is present in all lesional cells of the sample (Fig. 1B).
For each SNV class of each patient, we obtained estimates of AAF distributions with highly consistent class patterns (Fig. 1C–F). For example, in Patient 1 the AAFs of the SNVs that are present in ENA and IDC and absent everywhere else are higher than the AAFs of the ENA-only or the IDC-only classes. The same patterns hold for Patients 2 and 6. The patterns in Patient 5 are complicated by the presence of two IDCs and by low numbers of SNVs in relevant classes. Note that the mean AAFs are always <50% due to unavoidable contamination of the lesional tissue with normal cells that derive from lineages that branched off before the lesional ancestors accumulated their somatic mutations.
Mutated neoplasias are evolutionarily related to carcinomas
Each case represents an independent evolution; therefore, common patterns across the cases may be of general significance. We first asked to what extent the early neoplasias and the carcinomas share mutations that are not present in other samples, pointing to shared ancestral cell lineages. In four cases (Patients 1, 2, 5, and 6) (Fig. 1C–F; Supplemental Table 1), the phylogenetically informative SNV classes indicate that a neoplasia shares a common ancestor with the carcinoma. In each of these cases, a neoplasia and the carcinoma share a significant number of SNVs. For example, in Patient 1, 775 SNVs are shared between ENA and IDC, and in Patient 2, 681 SNVs are shared among the EN, DCIS, and IDC, with additional SNVs shared between the EN and IDC. There are no well-supported classes (in terms of number of SNVs and their AAFs) that are in conflict with each other, and none in which normal tissue or contralateral EN share SNVs with the carcinomas (Supplemental Table 1). The aforementioned PCR-based targeted validation showed 94% and 98% accuracy in assigning SNVs to the correct phylogenetic class (Supplemental Fig. S4; Supplemental Material).
In three of these four cases (Patients 1, 2, and 6) the number of SNVs in common between a neoplasia and carcinoma suggests the existence of a common ancestor that had already accumulated many somatic SNVs. Strikingly, in two cases (Patients 1 and 2) the number of mutations in the ancestor is greater than the number of mutations that subsequently occurred in the ancestral lineage private to the carcinoma.
In three cases (Patients 2, 3, and 6) DCIS was concurrent with IDC, and in one case (Patient 5) two independent IDC lesions were present. These four cases provided us the opportunity to ask whether the carcinoma phenotype arose once or multiple times independently. In Patient 3, the DCIS and IDC share a mutated common ancestor, suggesting that the carcinoma phenotype arose in the ancestral lineage, and that the IDC subsequently acquired the invasive phenotype. In Patients 2 and 6, there is no well-supported class of SNVs that unites the two carcinomas to the exclusion of a neoplasia. Instead, in both patients, the DCIS and the IDC each share separate classes of SNVs with a neoplasia, suggesting independent origins of the carcinoma phenotype from neoplastic ancestors.
These results suggest that some early neoplasias harbor a predisposition to spawning a carcinoma that later acquires an invasive phenotype (Patients 1, 2, 6). The chance of acquiring a carcinoma phenotype, given the predisposition provided by the neoplasia, is sufficiently high to allow for concurrent and independent development of carcinomas (DCIS and IDC in Patients 2 and 6).
Point-mutational mechanisms are evolutionarily stable and reproducible among cases
SNVs result from mutations that occurred in ancestral cells, and if a specific molecular mechanism were primarily responsible for the mutations, the distribution of the SNVs among the various types of change (the “mutation spectrum”) would carry that mechanism's signature (Pleasance et al. 2010b). To investigate the cause of the ancestral accumulation of mutations, we analyzed the mutational spectrum as a function of the samples in which SNVs were found. The mutational spectrum in our cases is remarkably consistent from patient to patient (Fig. 2A) and is also stable across SNVs in different types of samples and in different patterns (Fig. 2B). Transitions outnumber transversions about 1.5-fold in a pattern that is typical for replication errors and not indicative of any specific type of DNA damage or failed repair mechanism. C-to-T changes (or G-to-A, which are the same due to base pairing) are most numerous. Converted to substitution rates, this bias is even more pronounced because there are only roughly two C's for every three T's in the human genome. The consistency across patients implies a common mechanism, and the consistency among the three SNV groups (SNVs in early lesions only, in carcinoma only, and shared between early lesions and carcinoma) implies that the common mechanism acts throughout neoplastic and tumor evolution.
Figure 2.
Mutation spectra and rates of somatic SNVs. (A) Mononucleotide substitution frequencies by patient. (B) Mononucleotide substitution frequencies by SNV class. (C) Dinucleotide substitution rates of SNVs private to early neoplasias. (D) Dinucleotide substitution rates of SNVs private to carcinomas. (E) Dinucleotide substitution rates of SNVs shared among neoplasias and carcinomas. For C–E, SNVs are pooled across patients. The mutated dinucleotide is indicated in the inner circle, and the substitution occurring within it is color coded. Rate is defined as mutations per dinucleotide of that class.
To further shed light on the mutational mechanism we turned to analysis of dinucleotide substitution patterns. Because dinucleotide frequencies vary by an order of magnitude in the human genome, with AA/TT being most common and CG least common, we converted mutation counts to rates. Truly random substitutions would have the same rates for each of the 60 possible mutations (10 dinucleotides with six possible changes each, not counting changes in both bases because they are exceedingly rare). A dinucleotide-unaware process would recapitulate the mononucleotide rates, with the average transition having an about fourfold higher rate than the average transversion. In contrast, we detect an approximately eightfold higher rate of C-to-T transitions in the CpG context. This higher mutation rate is due to methylation of the C in a CpG dinucleotide, which upon deamination becomes a TpG. If the repair machinery catches this event it is reversed, but if the replication fork passes first it leads to a C-to-T transition in one of the daughter strands. The relative rate of C-to-T transitions in CpGs versus C-to-T transitions in the other dinucleotide contexts and versus all other changes provides an internal calibration as to whether DNA damage processes or defective repair mechanisms have disproportionally affected the genome.
In our patients, the rate increase of C-to-T transitions in the CpG context and in the dinucleotide mutation spectrum in general is similar to germline evolution (Sved and Bird 1990; Hwang and Green 2004), and is consistent across patients (Supplemental Fig. S5) as well as among classes of SNVs (private to neoplasias, private to IDCs, and shared among neoplasias and carcinomas) (Fig. 2C–E). This implies that the sources of the somatic SNVs are mutations that accumulated during many rounds of DNA replication (many ancestral cell divisions), and that cancer- or neoplasia-specific point mutational mechanisms, if present at all, did not substantially affect the mutation spectrum. Taken together, these lines of evidence support a model of mutation accumulation that is gradual and largely a function of the number of cell divisions, as opposed to recurring DNA damage events or mutational storms.
The somatic SNVs are randomly distributed in each patient with no enrichment of exonic or nonsynonymous changes, regardless of the phylogenetic class to which they belong. We also detect very little clustering of mutations that might be indicative of localized mutagenic events (Nik-Zainal et al. 2012b; Supplemental Figs. S6–S11). Across all cases, 159 out of the 12,392 high-confidence somatic SNVs fall into coding regions, with 2/3 (106) being nonsynonymous, which is what is expected by chance. This holds true for any biological subdivision of the data (e.g., neoplasias vs. IDC). The affected genes exhibit no enrichment for pathways by GO analysis (Ashburner et al. 2000; Huang et al. 2009). One point mutation, H1047R in PIK3CA, which has been previously implicated in cancer (Samuels et al. 2004; Ellis et al. 2012) and early neoplasias (Troxell et al. 2012), was recurrent in our cases (Patients 1, 3, 4, and 5, in various samples) at varying allele frequencies. Common cancer loci such as TP53 and BRCA1 were not mutated.
Aneuploidies are the dominant evolutionary feature of progression
The paucity of candidate driver mutations and overall random distribution of point mutations in our cases suggest that other genomic events may be contributing to the initial neoplastic phenotype and its progression to carcinoma. We therefore devised a multistep strategy to identify chromosome arm-scale losses and gains in each patient, utilizing those germline variants for which the patients were heterozygous. Each patient was heterozygous for between 1.56 and 1.74 million SNPs, ensuring substantial statistical power to detect subchromosomal-sized aneuploidies and copy-number variations.
We quantified, in each somatic sample separately, the fraction of reads that support the allele with the fewer number of reads (the lesser allele fraction, or LAF). We then ordered the SNVs according to their position in the genome and identified transition points where the LAF abruptly changes. In one case (Patient 5), the 20 large-scale copy-number variations which are confined to this patient's two IDC samples are suggestive of chromothripsis (Liu et al. 2011; Meyerson and Pellman 2011; Stephens et al. 2011; Crasta et al. 2012; Maher and Wilson 2012). In the other five patients, we identified a total of 46 large-scale copy-number variations, 43 of which involve whole chromosomes or whole chromosome arms.
None of the normal breast and contralateral neoplastic samples, some of the ipsilateral neoplasias, and all of the carcinomas exhibit aneuploidy. Four of the seven IDCs exhibit evidence for the presence of a subclone population in which additional chromosomes have undergone aneuploidy events (Supplemental Table 2).
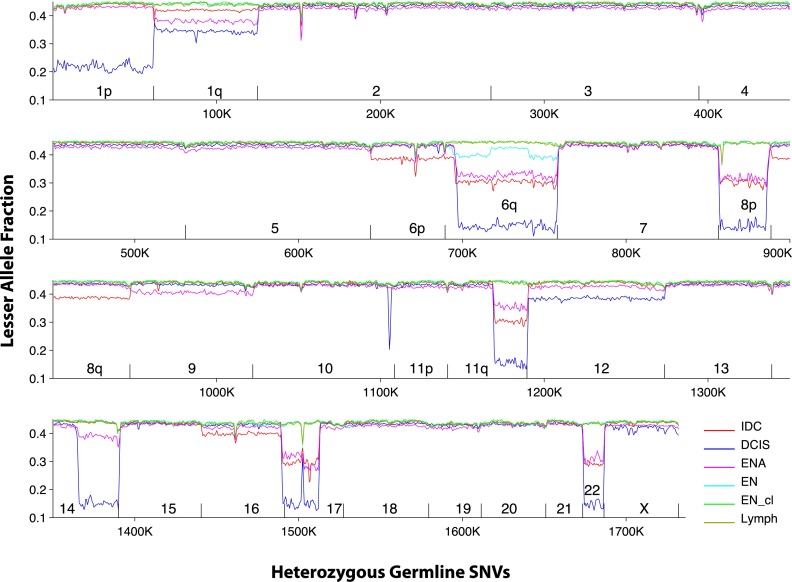
In Patients 1, 2, and 6, aneuploidy events are shared among early neoplasias and carcinomas. All aneuploidies that are present in the neoplasias are also present in the carcinomas. Plotting the LAFs of all samples from a patient powerfully illustrates both the chromosome scale of these events as well as the sharing of the same aneuploidies among certain samples. In Patient 6, for example, the aneuploidies involving chromosomes 1q, 6q, 8p, 17 and 22 are shared among both carcinomas and the EN (Fig. 3). The plot also reveals the aneuploidies of many other chromosomes that are present in a subclone population that makes up about 30% of the IDC sample. Examination of the corresponding plots of all patients reveals the extraordinary prevalence of aneuploidies in these cases (Supplemental Figs. S12–S17).
Figure 3.
Lesser allele fraction plot of Patient 6. SNVs are arranged by their order in the genome, and LAF is plotted for each sample in windows of 1000 SNVs with 500 SNV overlap. Aneuploidies are visible as precipitous drops in the LAF, which are often shared between samples. Chromosome boundaries are indicated by short vertical lines. All samples are plotted and give highly consistent LAFs for chromosomes that are euploid.
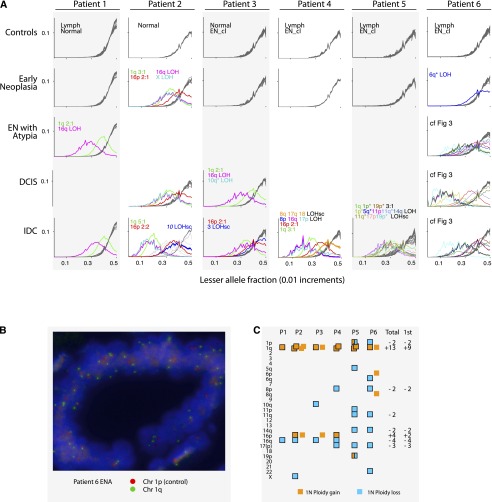
Graphing the distribution of LAFs for each LAF-derived section of the genome separately (usually a whole chromosome or arm) further supports the robustness of LAF as a metric to identify aneuploidies (Fig. 4A). However, a reduction of LAF can be a result of ploidy gains as well as losses. Therefore, we calculated the actual ploidy changes in a two-step process: first, we estimated the contribution of normal cells to the sample using chromosome losses, and then we calculated the additional number of chromosome copies for those chromosomes that exhibited increased ploidy. We validated a subset of these calls using FISH (Fig. 4B) and found all LAF-based calls that we tested to be correct.
Figure 4.
Aneuploidy summary. (A) LAF distributions for each chromosome across all patients and samples. In each sample-by-patient panel, the LAF distributions of all chromosomes are superimposed. In the absence of aneuploidy, the plot lines of all chromosomes are well-aligned, as is evident in the control plots and some EN plots. Control panels often contain plots from two samples (indicated) and so there are sometimes 46 lines superimposed, revealing the robustness of the LAF metric across samples and chromosomes. A chromosome's plot line is gray when it does not deviate from the typical distribution. The line is colored when the chromosome's LAF is skewed. Distinct colors are assigned to represent aneuploid regions that recur in different samples and patients. Colors are labeled in the panel in which they first appear. For Patient 6 please see Figure 3. (B) FISH of chromosome 1 in ENA of Patient 6. (C) Distribution of aneuploidies by patient, excluding those in IDC subclones. Each square denotes a unit gain (orange) or loss (blue). In Patients 2, 3, and 6, two phases of aneuploidies occurred, with those of the second phase not surrounded by a border. (Total) The total number of chromosomes lost (−) or gained (+) across all patients; (1st) the number during the first detected phase. Only recurrent events are listed. In Patient 5 (which exhibits hallmarks of chromothripsis), different pieces of chromosomes 1p and 19 underwent simultaneous losses or gains.
The distribution of aneuploidies across chromosomes among the six patients is highly nonrandom (Fig. 4C). Gain of chromosome 1q is by far the most common event, with a total of 13 extra copies accumulated in these patients, not considering the IDC subclones. All cases exhibit 1q gain, and it is the only event that is shared by all three early neoplasias in which we could detect aneuploidy. In three cases (Patients 2, 3, and 6), the IDC underwent gains of 1q in addition to previous ones, increasing 1q ploidy to 6, 4, and 4, respectively. This suggests that the selective advantage conferred by 1q gain increases with further gains of 1q during tumor evolution.
Like the shared SNVs, the shared aneuploidies support specific lineage relationships among the samples of each patient. We therefore built lineage trees using the somatic SNVs as phylogenetic markers, and then asked whether the shared aneuploidies are consistent with these trees (Fig. 5). All aneuploidies are unambiguously and parsimoniously assigned to specific branches in the SNV-based lineage trees.
Figure 5.

Genome evolutions of all patients (P1–P6). Vertical black lines are ancestral lineages whose lengths are proportional to the number of SNVs that occurred in each (except Patient 4, which is 50% shorter for fit). Cones represent tissue samples; cone width represents approximate amount of tissue; cone height is constrained at the top by the position of the last common ancestral cell of the sample, which is determined by the ancestral branch lengths, and on the bottom by the time of surgery, which is the same for all samples. The ratio of cone width to height is an approximation of the rate of cell division in each sample since the last common ancestral cell. Chromosome ploidy changes are indicated with the chromosome number; stand-alone numbers in italics indicate the number of chromosomes affected by subclone evolution (or putative chromothripsis in Patient 5). Thick branches are the earliest branches for which we are able to infer genomic events. Circles at the end of thick branches are ancestors with the colors denoting their inferred neoplasia-like, DCIS-like, or IDC-like phenotypes.
The order of aneuploidies during the evolution of each case is also unambiguous and highly suggestive of a small number of aneuploidies being first drivers of the neoplastic phenotype. In all cases, gain of 1q was among the events that occurred first, including in the three cases in which genomic crises occurred in a common ancestor of neoplasias and carcinomas (Patients 1, 2, and 6). Loss of 16q occurred four times, and loss of 17 three times, as part of the first set of aneuploidies. Gain of 16p occurred three times. The remaining aneuploidies occurred once or twice in all trees, and none were recurrent in the earliest stages of evolution.
In order to time the occurrence of aneuploidies relative to SNVs, we identified the branch in the lineage tree of each patient where the first ploidy gains of chromosome 1q occurred and considered SNVs that occurred on this branch. AAF spectra of SNVs that occurred before the ploidy gains and located on the chromatid that was duplicated should be enriched for higher AAF in the progeny samples. In each of the six patients, statistical tests rejected the null hypothesis that there are no such SNVs (Fisher's exact test, P-values ranging from 0.5 × 10−2 to 0.8 × 10−36; Supplemental Table 3). This pattern is reproducible between different samples of the same case, and the SNVs that exhibit high AAF largely overlap. The same pattern holds for the ploidy gain in chromosome 16p, but due to fewer SNVs the statistical signal is less strong. Overall, the AAF distributions of 1q SNVs are consistent, with some mutations occurring before the ploidy gain, and some mutations occurring after the ploidy gain (Supplemental Fig. S18). This suggests gradual accumulation of point mutations as a function of the number of cell divisions, as opposed to mutational bursts.
Because the aneuploidies and SNVs independently support the lineage tree topologies, the genotypes and phenotypes of the common ancestors can be confidently inferred in each case. The aforementioned mutated common ancestors of neoplasias and carcinomas in Patients 1, 2, and 6 bore extensive aneuploidy, as did the mutated common ancestor of the DCIS and IDC in Patient 3. In all four cases, therefore, genomic crises occurred in an ancestral cell or in consecutive daughter cells of the ancestral cell lineage. The phenotypes of these ancestors likely included nuclear atypia and increased rate of cell division, but no invasive capabilities. Their genomes were predisposed to further genomic change, and as a result the subsequent lineages leading to IDC accumulated numerous additional SNVs and aneuploidies.
Discussion
Evolutionary studies of cancer have so far focused on the inference of clonal evolution within the cancer (e.g., Nik-Zainal et al. 2012a) or analyses of the relationship of metastases with the primary tumor (e.g., Navin et al. 2011). Here we addressed a different perspective, namely that of the early origins of the cancer phenotype. These three approaches can be thought of as mimicking progression, at least as far as solid tumors are concerned: Studies of metastatic evolution are about the terminal stages of the cancer; studies of within-cancer subclone diversity are about the Darwinian process of faster versus slower growing cell populations and the evolution of the primary tumor mass; and studies of early neoplasias and their relationships to the diagnostic tumors are about early origins of cancer.
Our understanding of these early origins will be greatly enhanced by molecular evolutionary analyses similar to those that have advanced our understanding of organismal evolution. Cells within concurrent lesions are analogous to extant organisms: they are related to one another by bifurcating lineage trees and have accumulated genomic changes over the course of evolution. In our study of multiple lesions in six cases of ductal breast carcinoma, we found that the genomes of ancestors of some early neoplasias and carcinomas were already aneuploid and harbored a modest number of point mutations. By comparing mutational spectra of somatic SNVs across patients and samples we inferred that somatic SNVs accumulated gradually as a result of a large number of ancestral cell divisions and not during saltatory mutational crises. In two cases, the carcinoma phenotype originated twice independently from an ancestral neoplastic phenotype, suggesting a substantial predisposition of the ancestor to generate cancerous progeny.
All of the neoplasias with aneuploidies shared common cellular ancestors with the carcinomas; in all of these cases the neoplasia and carcinoma shared these aneuploidies as well as somatic SNVs. In contrast, none of the neoplasias that were devoid of aneuploidies (all contralateral ENs and five ipsilateral ENs) were closely related to a carcinoma. Among the aneuploidies, gain of chromosome 1q was most dramatically recurrent, which is consistent with its prevalence among late-stage breast cancers (Curtis et al. 2012, cf. Fig. 4). 1q harbors more than a thousand genes, and while the increased dosage alone is not sufficient for a carcinoma phenotype (some of our neoplastic samples carry the increased 1q ploidy), it is likely to be predisposing to further genomic change. Initially, such change may be catalyzed primarily by an increased rate of cell division, as the mutation spectrum of the early neoplasias is indistinguishable from that of the IDCs in every patient examined. Additional aneuploidies accumulate, however, and at some point a combination of dosage imbalances and mutational load, and perhaps epigenetic or stromal changes as well, results in an invasive carcinoma phenotype.
We anticipate that the evolution of a diverse set of breast and other cancers will soon be studied similarly and with complementary approaches (Shah et al. 2009; Navin et al. 2011; Gerlinger et al. 2012; Nik-Zainal et al. 2012a; Shah et al. 2012). Current practice in clinical diagnosis of cancer facilitates studies on archival material because of the low cost and superior quality of histopathological examination of formalin-fixed, paraffin-embedded samples. We show that high-quality, large-scale genome sequence can be obtained from archival material, and show by validation that the data from such material can be highly robust. Evolutionary inference based on many samples of such material opens a new dimension for analysis of cancer origins and progression. In the future, phylogenetic analysis of carcinomas and concurrent lesions will suggest drugs that attack both carcinoma and early lesions by targeting genomic changes common to all lesions, removing not only the carcinoma, but also the reservoir of related cells from which a carcinoma might recur.
Methods
Identification and processing of neoplasias
All patients except one had opted for mastectomies, and all of the available breast tissue had been formalin-fixed, which allowed for the discovery of multiple sites of neoplastic lesions in each case by examination of large sets of tissue sections. Neoplastic lesions were classified according to a standard set of criteria that included nuclear morphology, cell shape, and tissue organization. Once a lesion was identified and characterized, we estimated the extent of the neoplastic tissue by taking cores and performing further sectioning and histology. We then dissected the material to minimize the proportion of normal breast tissue in the final sample. Our goal was to achieve 50% or more neoplastic or tumor content, but we could not rigorously quantify this number until after sequencing had been performed.
Library construction and sequencing
DNA extraction from each dissected sample was performed using procedures optimized for archival material. FFPE cores were cut into 20-μm slices. Paraffin was dissolved in Xylene and removed (four repeats of 5 min incubation with rotation in 1 mL of Xylene and microcentrifugation for 3 min) and followed by washing with ethanol (four repeats of 5 min incubation with rotation in 1 mL of ethanol and microcentrifugation for 3 min). Tissue was then lysed with Proteinase K and crosslinks reversed by overnight incubation at 56°C. After brief digestion with RNase A (Qiagen), DNA was purified with a column-based method (Qiagen QIAamp DNA Mini Kit). For each sample, one Illumina library was built with an average insert size of between 300 and 400 bases, depending on the quality of the DNA. Half to 1 μg of genomic DNA (depending on the availability of the material) was sheared to 400 bp with Covaris S2, end-repaired, ligated to Illumina adapter, size selected, and amplified with eight cycles of PCR to generate the final library. Standard Illumina 2 × 101 paired-end sequencing on the HiSeq2000 platform was performed such that the final sequence coverage of confidently aligned reads was nearly 100× for each sample in the first patient, and 50× for the samples of Patients 2–6. Analysis of the mapped reads confirmed high library quality (very low duplicate read-pair fraction, almost normally distributed fragment size, and highly uniform genome coverage) that was indistinguishable from that of comparable libraries constructed from fresh DNA.
Read mapping and BAM file processing
Raw Illumina reads were uploaded to DNAnexus (https://dnanexus.com/) and aligned to the human reference genome (UCSC build hg19) using the DNAnexus read mapper, a hash-based probabilistic aligner that incorporates paired read information. We used standard quality-control metrics, such as percent confidently mapped reads and insert size distribution, to discard problematic Illumina lanes prior to subsequent analysis. Successfully aligned reads from high-quality lanes were labeled using read group tags and then merged into sample-level BAM files. Lane-level read group tags improve the performance of downstream BAM processing and variant calling with the Genome Analysis Toolkit (GATK) (McKenna et al. 2010; DePristo et al. 2011).
We followed GATK's best practices guidelines (v3) to perform sample-level BAM processing using the Picard java utilities (http://picard.sourceforge.net/) and GATK tools (McKenna et al. 2010). This protocol has three steps that are executed in the following order: duplicate read marking, local realignment, and base quality score recalibration. We used the Picard MarkDuplicates utility to mark duplicate reads based upon the read position and orientation of read pairs. Marked duplicates were ignored in subsequent processing and variant calling steps. GATK local realignment was performed with standard parameters and the recommended known indel sets (Mills et al. 2006 and 1000 Genomes indels from the GATK v1.2 bundle). GATK base quality score recalibration was performed with the standard set of covariates. The realigned, recalibrated BAM files produced by these processing steps were used for multisample SNV calling and for all alignment-related statistics such as allele counts.
Multisample SNV calling
Multisample SNV calling was performed on processed, sample-level BAM files with the GATK Unified Genotyper (DePristo et al. 2011). Multisample runs were grouped by patient such that BAM files from different patients were run separately. Notable parameters for the Unified Genotyper include standard call confidence of 50.0 (-stand_call_conf 50.0) and minimum base quality score of 20 (-mbq 20). To reduce SNV false discovery rate, raw variant calls were filtered using GATK variant quality score recalibration tools (VQSR) with the recommended training sets. The following annotations were used for training: FS (strand bias), MQ (mapping quality), DP (depth), HaplotypeScore, MQRankSum, and ReadPosRankSum. Replacing the recommended QD annotation (call quality divided by depth) with DP greatly improves sensitivity for low-frequency somatic variants.
We used pass-filter SNVs to create a set of high-confidence germline calls and a set of high-confidence somatic calls for each patient. For a given patient, we defined germline SNVs as calls meeting the following multisample criteria: (1) depth 20 or greater in every sample, where depth is defined as the sum of alternate and reference base counts, and (2) non-reference GATK genotype (GT) in every sample. These high-confidence germline calls were used for aneuploidy analyses (below). Somatic SNVs were defined using a similar set of criteria: (1) depth 20 or greater in every sample, (2) fewer than two reads supporting the alternate allele in at least one sample, and (3) absence in dbSNP 132. We excluded SNVs in dbSNP 132 in order to reduce the number of false-negative germline calls in our somatic SNV call set.
Three out of four Patient 2 genomic libraries were contaminated with mouse DNA, with ∼15% of DCIS reads aligning to the mouse genome. Approximately 1% of reads from Normal and 0.65% of reads from EN aligned to mouse; these fractions were significantly above background levels for unaffected libraries. To remove contamination-related mapping artifacts from our SNV data, we added additional filtering steps to the SNV calling protocol for Patient 2. Prior to variant calling with the Unified Genotyper, we eliminated all reads lacking confidently mapped mates. After variant calling and VQSR, we removed all novel pass-filter SNVs positioned in areas of the genome with significant homology with the mouse genome. Homology was assessed by mapping tiled 75-mer reference sequences, surrounding each position of interest, to the mouse genome (mm9). This second step dramatically reduced spurious calls in DCIS while eliminating only 1% germline dbSNP positions used as controls.
Determination of somatic SNV class patterns and of robust sharing classes
Multisample somatic SNV calls were further analyzed to determine patterns of SNV-sharing across samples within the same patient. Although GATK provides sample genotype calls based on genotype likelihood calculations, these calls lack sensitivity when applied to cancer samples with substantial normal contamination or subclonal tumor populations. To further enhance sensitivity of SNV detection beyond GATK multisample calls, we applied a simple but sensitive metric to determine each sample's mutation status. At each somatic SNV position predicted by GATK in at least one sample, we considered any sample with two or more reads supporting the alternate allele to harbor the mutation (i.e., mutation present). Samples with fewer than two reads supporting the alternate allele were labeled as reference (i.e., mutation absent). Our rationale was that given that a specific SNV is detected in some samples, reads supporting this SNV in other samples have a significant prior to be true rather than sequencing errors. We call this criterion “evidence of presence” of an SNV in a given sample. These patterns of mutation presence and absence define mutation classes for lineage construction and other somatic SNV analyses. We note that a small but important number of SNVs were reallocated by this method from candidate somatic SNVs with inconsistent patterns of sharing among samples to germline events, and that very few single-sample (“private”) SNVs were reallocated to sharing classes, underscoring the high-sequence and alignment quality of our datasets.
A case with n samples has 2n possible class patterns. For example, for a case with five samples, the patterns are 00000 to 11111. No case has the 00000 class, because an SNV has to be present in at least one sample, and the 11111 class is that of germline variants. Classes that are private to one sample are 10000, 01000, 00100, 00010, and 00001. Candidate classes that are possibly phylogenetically informative are defined by SNVs that are present in two or more, but not all, samples. To identify the subset of robust phylogenetically informative classes, we applied the following steps:
Eliminate classes with the SNV present in the lymph sample (applicable to Patients 1, 4, 5, and 6). These classes consisted of lymph-only SNVs (presumably somatic mutations in the lymph sample) and germline SNVs, where one or very few samples were missing the alternate allele presumably due to sampling variance.
Retain the classes that, when ranked in decreasing order of the number of SNVs present within them, together contain 95% of all candidate somatic SNVs. This eliminated all spurious classes that were not supported by an overall substantial number of SNVs, most of which were missing from just one sample, presumably due to sampling variance.
Eliminate classes with a large fraction of SNVs whose mutation-absent samples exhibit one alternate-allele supporting read, suggestive of systematic false-negative calls. This also constituted a small number of classes with SNVs whose alternate alleles were missing from just one sample presumably due to sampling variance.
PCR-based validation of SNVs and accuracy assessment of whole-genome calls
Please see the Supplemental Material for methodology used and results.
Aneuploidy and tumor purity
To identify aneuploidies we selected a subset of the germline SNVs identified by GATK. These “sgSNVs” were defined, separately for each patient, as a patient's multisample germline SNVs that had dbSNP132 entries, were heterozygous, and had minor allele frequencies in the control sample of at least 0.25. We define the “lesser allele” as the one supported by fewer reads than the other allele (which is the “prevalent allele”). Three metrics were calculated for each SNV: the lesser allele coverage, the prevalent allele coverage, and the lesser allele fraction (LAF). The LAF was used to identify aneuploidies, whose “sign” (loss or gain) was then set by the two coverage metrics.
In all patients except 5, the vast majority of chromosomal copy-number transitions coincided with the centromere, or the whole chromosome was involved (Supplemental Figs. S12–S17). Fine mapping of the transition points was therefore not usually necessary. In the handful of cases where a transition point did not coincide with a centromere, we found the window of the plot (Supplemental Figs. S12–S17) at which the event either started or ended (window i). As discussed in Figure 3, each window spans 1000 SNVs, with an overlap of 500 SNVs between adjacent windows. We then plotted the frequency of the heterozygous variants in the three relevant windows (i-1,i,i+1, totaling 2000 variants) in that sample. The variant at which the frequency shifted was easily detected by eye, and it was not necessary to deploy segmentation methods. The resolution of this analysis is low (determined by what can be seen by eye on the plots) and we did not attempt to identify events that involved regions smaller than about a third of a chromosome arm. We also note that we did not attempt to identify structural rearrangements that do not result in copy-number changes, such as inversions.
The identified loss of heterozygosity (LOH) chromosomes were then used to estimate the fraction of the sample that is due to normal cells (lymphocytes, myocytes, etc.), as follows: All cancer cells contribute zero copies of an allele that was lost due to LOH, and the normal cells contribute one copy of the LOH allele times the contamination fraction n. Note that in all of our patients, the control samples were free of LOH chromosomes (Fig. 4A). The LOH allele is almost always the one with fewer reads. Therefore, the LAF l should, on average, be equal to the lost-chromosome fraction that is contributed by the normal contamination. Some arithmetic shows that n = l / (1 - l). Once n was estimated from l, the exact ploidy p for those chromosomes that had gains was calculated according to the formula P = (1-2nl)/(l(1-n)).
Sequence-based n's roughly matched estimates of n by histology. The histology-based estimates are necessarily an approximation because they are based on limited sampling, by sectioning of the tissue core mass from which DNA is obtained.
SNV mutation spectra
Mutation spectra for patient samples were aggregated in two ways: (1) combined across patients to form three “superclasses'” of SNVs based on lesion class (private in early neoplasias, private in carcinomas, and shared between neoplasias and carcinomas); (2) combined within each patient, ignoring lesion class, to form six groups. Complementary mutations were pooled, reducing the number of possible mononucleotide mutations from 12 to 6, and the number of single-base substitution classes in dinucleotides from 16 × 6 = 96 to 10 × 6 = 60.
Mononucleotide mutation spectra were simply estimated from the frequency of the mutation type (Fig. 2, cf. A and B, where the bars of each color add up to 1). For dinucleotides, we calculated rates by dividing the number of events of each of the 60 changes by the genome-wide count of the dinucleotide that was mutated.
Tree inference
Tree topology was defined by the phylogenetically informative SNV classes (Supplemental Table 1). The data are unambiguous and we therefore used parsimony to establish which samples shared common ancestors in which configuration. Once the SNV-based trees were built, aneuploidy events could be mapped onto them, and again the data were unambiguous. Even successive gains of ploidy of the same chromosome, most prominently among them 1q (e.g., Fig. 5F), could be ordered without conflicts.
Ordering SNVs vs. chromosome 1q ploidy gain in ancestral branches
We devised a statistical test to ask whether some SNVs occurred before copy gain in aneuploidy regions. For each patient, we identified the branch in the lineage tree responsible for the first copy-number changes in chromosome 1q, which consistently represents the earliest aneuploidy event in our patients. We then analyzed the AAF spectra of SNVs occurring in that branch. The test below is based on the idea that SNVs that occur on a 1q chromatid prior to gain of a copy of that chromatid should have higher AAF than SNVs occurring on a 1q chromosome after copy gain.
We used SNVs on all diploid chromosomes on the same branch as our control set. Sequence coverage is scaled with respect to the aneuploidy and controls for contamination of the sample by normal cells (lymphocytes, etc.):
where p is the estimated ploidy and n is the estimated normal contamination. In order to find outliers indicative of events prior to copy gain, we calculated a Z-score. SNVs with AAFs with Z-score > 3 were labeled as “high” and SNVs falling below threshold were labeled as “low.” For each patient, we used Fisher's exact test to compare the distribution of SNV labels in the control chromosomes vs. 1q. In each of the patients, we reject the null hypothesis that the 1q distribution is equal to or less extreme than the control distribution (Supplemental Table 3).
Data access
The sequence data from this study have been submitted to NCBI (http://www.ncbi.nlm.nih.gov/bioproject) under BioProject identifier PRJNA193652.
Acknowledgments
This work was supported by the Sequencing Initiative of the Stanford Department of Pathology, grants from the California Breast Cancer Research Program and NIH/NCI to R.B.W. and a grant from KAUST to S.B. D.K.H. was supported by a STMicroelectronics Stanford Graduate Fellowship, and D.E.N. by a training grant from NIH/NLM and a Bio-X Stanford Interdisciplinary Graduate Fellowship. This study is the result of an equal collaboration among the Batzoglou, Sidow, and West groups. Listed order of corresponding authors was determined by a series of coin flips.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.151670.112.
References
- Abdel-Fatah TM, Powe DG, Hodi Z, Lee AH, Reis-Filho JS, Ellis IO 2007. High frequency of coexistence of columnar cell lesions, lobular neoplasia, and low grade ductal carcinoma in situ with invasive tubular carcinoma and invasive lobular carcinoma. Am J Surg Pathol 31: 417–426 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. 2012. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486: 405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. 2010. Signatures of mutation and selection in the cancer genome. Nature 463: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombonati A, Sgroi DC 2011. The molecular pathology of breast cancer progression. J Pathol 223: 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. 2011. Initial genome sequencing and analysis of multiple myeloma. Nature 471: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. 2010. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by Multiregion sequencing. N Engl J Med 366: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci 101: 13994–14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al. 2012. Rate of de novo mutations and the importance of father's age to disease risk. Nature 488: 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, Jones S, Sjöblom T, Park BH, Parsons R, et al. 2008. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci 105: 16224–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchió C, Reis-Filho JS 2010. Breast cancer precursors revisited: Molecular features and progression pathways. Histopathology 57: 171–192 [DOI] [PubMed] [Google Scholar]
- Maher CA, Wilson RK 2012. Chromothripsis and human disease: Piecing together the shattering process. Cell 148: 29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2012. Genome sequencing and cancer. Curr Opin Genet Dev 22: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Pellman D 2011. Cancer genomes evolve by pulverizing single chromosomes. Cell 144: 9–10 [DOI] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE 2006. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 16: 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012a. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. 2012b. The life history of 21 breast cancers. Cell 149: 994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Sloane JP, Hanby A, Pinder SE, Lee AH, Humphreys S, et al. 2005. Columnar cell lesions of the breast: The missing link in breast cancer progression? A morphological and molecular analysis. Am J Surg Pathol 29: 734–746 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR 2011. Exploring the genomes of cancer cells: Progress and promise. Science 331: 1553–1558 [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci 87: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell ML, Brunner AL, Neff T, Warrick A, Beadling C, Montgomery K, Zhu S, Corless CL, West RB 2012. Phosphatidylinositol-3-kinase pathway mutations are common in breast columnar cell lesions. Mod Pathol 25: 930–937 [DOI] [PubMed] [Google Scholar]
- Turajlic S, Furney SJ, Lambros MB, Mitsopoulos C, Kozarewa I, Geyer FC, MacKay A, Hakas J, Zvelebil M, Lord CJ, et al. 2012. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res 22: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. 2012. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 366: 1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. 2012. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 482: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]