Abstract
The regions bound by sequence-specific transcription factors can be highly variable across different cell types despite the static nature of the underlying genome sequence. This has been partly attributed to changes in chromatin accessibility, but a systematic picture has been hindered by the lack of large-scale data sets. Here, we use 456 binding experiments for 119 regulators and 84 chromatin maps generated by the ENCODE in six human cell types, and relate those to a global map of regulatory motif instances for these factors. We find specific and robust chromatin state preferences for each regulator beyond the previously reported open-chromatin association, suggesting a much richer chromatin landscape beyond simple accessibility. The preferentially bound chromatin states of regulators were enriched for sequence motifs of regulators relative to all states, suggesting that these preferences are at least partly encoded by the genomic sequence. Relative to all regions bound by a regulator, however, regulatory motifs were surprisingly depleted in the regulator's preferentially bound states, suggesting additional non-sequence-specific binding beyond the level predicted by the regulatory motifs. Such permissive binding was largely restricted to open-chromatin regions showing histone modification marks characteristic of active enhancer and promoter regions, whereas open-chromatin regions lacking such marks did not show permissive binding. Lastly, the vast majority of cobinding of regulator pairs is predicted by the chromatin state preferences of individual regulators. Overall, our results suggest a joint role of sequence motifs and specific chromatin states beyond mere accessibility in mediating regulator binding dynamics across different cell types.
Although the genome sequence of each human cell is invariant across nearly all cell types of the human body, the morphology and function of each cell is dramatically different owing to their differential regulation and gene expression patterns. At the molecular level, the binding landscape of a given regulator can be extremely dynamic, although its sequence specificity remains unchanged (Harbison et al. 2004; Zhong et al. 2010; Mullen et al. 2011; Trompouki et al. 2011). This is attributed at least in part to the dynamic chromatin landscape of each cell via active and repressed regions that can then be epigenetically maintained (Lam et al. 2008; Essien et al. 2009; Segal and Widom 2009; John et al. 2011; Li et al. 2011; Lickwar et al. 2012). The chromatin landscape is itself thought to be driven at least in part by the regulators active in each cell type (Lefterova et al. 2008; Lupien et al. 2008; Steger et al. 2010; Siersbaek et al. 2011). For example, transient overexpression of a small number of transcription factors has been shown sufficient for stable epigenetic reprogramming, which is now commonplace in the generation of induced Pluripotent Stem (iPS) cells (Takahashi and Yamanaka 2006; Meissner 2010). However, a systematic study of the interplay between regulator binding, including both general and sequence-specific regulators, chromatin accessibility, and chromatin states defined with histone modification marks, has been unfeasible due to the lack of systematic genome-wide regulator binding experiments in multiple cell types with matched chromatin data sets.
This situation changed with the scale-up of the ENCODE project (The ENCODE Project Consortium 2012). First, the genome-wide binding locations of more than 100 regulators have been mapped in one or multiple cell types (Supplemental Tables 1, 2), identifying thousands of constitutive and variable target locations for each experiment. Second, the chromatin accessibility landscape of matched cell types has been mapped using DNase hypersensitivity and formaldehyde-based FAIRE (Hesselberth et al. 2009; Song et al. 2011). Third, at least eight histone modification marks have been mapped in the same cell types that can be used to pinpoint distinct chromatin functions such as enhancer and promoter regions. These data have individually highlighted the remarkable fact that in a given cell type, only a very small percentage of the 3 billion bases of the genome have robustly detectable regulator binding, accessible chromatin, or histone marks denoting active regulatory elements. Strong relationships between each pair of data types have been previously reported, and regulatory motifs have been shown to be over-represented (enriched) within both active chromatin marks and regions of regulator binding (Heintzman et al. 2007, 2009; Xi et al. 2007; Boyle et al. 2008; Hon et al. 2008; Lupien et al. 2008; Robertson et al. 2008; Ernst and Kellis 2010; Ernst et al. 2011; The modENCODE Consortium 2010; John et al. 2011; Wu et al. 2011). However, the dynamic changes in regulator binding and active regulatory elements across cell types are far from understood at the systems level.
In this paper, we integrate this vast collection of histone modification, chromatin accessibility, and regulator binding, including both transcription factor and general binding as well as regulatory motif information, to systematically study dynamic regulatory binding across multiple cell types. We leverage 84 genome-wide data sets of chromatin-related mark patterns and 456 regulator binding experiments for 119 different regulators generated by the ENCODE project in six human cell types and 2.76 million total motif instances from 51 different positional weight matrices (The ENCODE Project Consortium 2012; P Kheradpour and M Kellis, in prep.). We use a chromatin state model that summarizes biologically meaningful combinations of chromatin marks (Hoffman et al. 2013), shown to more directly correlate with diverse functional elements (Thurman et al. 2007; Jaschek and Tanay 2009; Ernst and Kellis 2010). The model used here integrates both histone modification and chromatin accessibility into a joint 25-state model, which enables us to distinguish diverse classes of open chromatin. We use this rich chromatin annotation to study the interplay between chromatin state, regulator binding, and regulatory motifs across multiple cell types, revealing numerous new insights:
We find several distinct classes of chromatin state preferences for different regulators, which are stable across experimental conditions and across different cell types.
We find different chromatin state preferences for locations of cell type unique, shared, and excluded binding, respectively enriched in enhancer, promoter, and repressed states.
We find that the chromatin states preferentially bound by regulators show strong enrichment for their cognate regulatory sequence motifs.
Surprisingly, however, relative to all regions bound by a given regulator, binding events within preferred chromatin states are more often depleted for regulatory motif instances, suggesting additional nonspecific binding that does not rely on specific regulatory motif instances.
Moreover, we find nonspecific binding primarily in those accessible chromatin states that also contain active histone modification marks, suggesting that permissive binding is associated with specific modifications rather than open chromatin alone.
Lastly, we find that previously reported pairwise enrichment in the binding locations of regulator pairs can potentially be explained to a large extent by similar chromatin state preferences.
Overall, our results suggest a previously unappreciated diversity in the chromatin state preferences of different transcription factors that likely underlies nonpermissive binding, potentially mediates interactions between multiple regulators, and facilitates the cell type-specific and cell type-restricted regulator binding.
Results
Chromatin landscape reveals diverse classes of accessible chromatin regions
To study the dynamic nature of regulator binding and chromatin states, we focused on six human cell types, consisting of lymphoblastoid (Gm12878), cervix adenocarcinoma (HeLa-S3), liver carcinoma (HepG2), umbilical vein endothelial cells (Huvec), myelogenous leukemia (K562), and embryonic stem cells (H1-hESC). These were prioritized as Tier 1 and Tier 2 cells in the ENCODE project and thus benefit from extensive experimentation across all the ENCODE groups, enabling integration across histone modifications, chromatin accessibility, transcription factor binding, and gene expression data sets, although these were generated by different the ENCODE production groups.
In each of these cell lines, we characterize the chromatin landscape by integrating 14 genome-wide chromatin tracks. These include eight histone modification marks: mono-, di-, and trimethylation of histone 3 lysine 4 (H3K4me1, H3K4me2, and H3K4me3), typically associated with enhancer and/or promoter regions; acetylation of histone 3 lysine 9 and 27 (H3K9ac and H3K27ac) that mark active regulatory elements; the repressive mark H3K27me3; H3K36me3; and H4K20me1 associated with gene bodies (Barski et al. 2007; Wang et al. 2008; Ernst et al. 2011). These also include three tracks of chromatin accessibility, typically associated with increased regulator binding: single-cut DNase (Song et al. 2011), double-cut DNase (Hesselberth et al. 2009), and sonication (Song et al. 2011) assays. Lastly, they include binding of two general regulators: CTCF, associated with insulator and other functions; and RNA polymerase POL2; and an input control.
We used chromatin states (Ernst and Kellis 2010) from ChromHMM (Ernst and Kellis 2012) to summarize biologically meaningful combinations of those marks into 25 chromatin states (Hoffman et al. 2013), which were consistently defined across cell types. Briefly, the states consist of the following: Transcription start site (1_Tss), Tss flanking (2_TssF), promoter flanking (3_PromF), poised (Bernstein et al. 2006) promoter (4_PromP); strong and weak enhancers (Heintzman et al. 2007) (5_Enh, 8_EnhW) and enhancer flanking (6_EnhF, 7_EnhWF); three types of open chromatin states that lack active histone modification marks (9_DNaseU, 10_DNaseD, 11_FaireW preference for double-, single-cut DNase, and FAIRE, respectively) and also lie distal to active histone modification states; CTCF in open and closed chromatin (12_CtcfO, 13_Ctcf); gene body-associated (14–19) including 5′, 3′, and elongating; specific repression (20-22); low signal (23_Low) and quiescent (24_Quies) states; and possible artifacts (25_Art).
These states are defined by the frequency of each mark in each chromatin state (emission probabilities) and the frequency with which states are found adjacent to each other (transition probabilities) (Supplemental Fig. 1). The state definitions are constant across cell types because the model was learned jointly by a virtual concatenation of the six cell types, but the state assignments are cell type-specific because they depend on the specific combination of chromatin marks observed in a given cell type. Different states cover very different fractions of the genome: individual promoter, enhancer, open chromatin, and CTCF states usually cover <1% of the genome, the set of transcribed states (14–19) on average cover ∼10% of the genome, and the low and quiescent states 23 and 24 together cover ∼70% of the genome (Supplemental Fig. 2).
Although transcription factor binding is known to be generally associated with regions of open chromatin (Song et al. 2011), our chromatin state annotations suggest a much more complex picture with several types of open chromatin. In fact, at least nine chromatin states showed DNase hypersensitivity emission parameter frequencies ∼50% or greater (Supplemental Fig. 1), including promoter states (1_Tss, 4_PromP), enhancer states (5_Enh, 8_EnhW), DNase-only regions lacking other histone marks (9_DNaseU, 10_DNaseD), CTCF binding regions (12_CtcfO), the specific repression state frequently enriched in promoter regions (20_ReprD), and the artifact state (25_Art). This diversity of open chromatin states suggests that a more complex relationship may exist between transcription factor binding and chromatin beyond simply a general preference for accessible chromatin regions, which we explore next by studying the preferences of each transcription factor for each chromatin state.
Regulators show multiple distinct chromatin state enrichment patterns
As expected, we find that regulators are typically most enriched for states with open chromatin. In fact, after collapsing multiple experiments for the same regulator and excluding CTCF and POLR2A, we find that 104 of 115 regulators (90%) with cell type matched chromatin data available showed a maximum enrichment in either the active promoter state 1_Tss (59 regulators) or the strong enhancer state 5_Enh (45 regulators). However, the extent of each regulator's preference for the two states varied substantially for different regulators, as did their enrichment for additional open chromatin states.
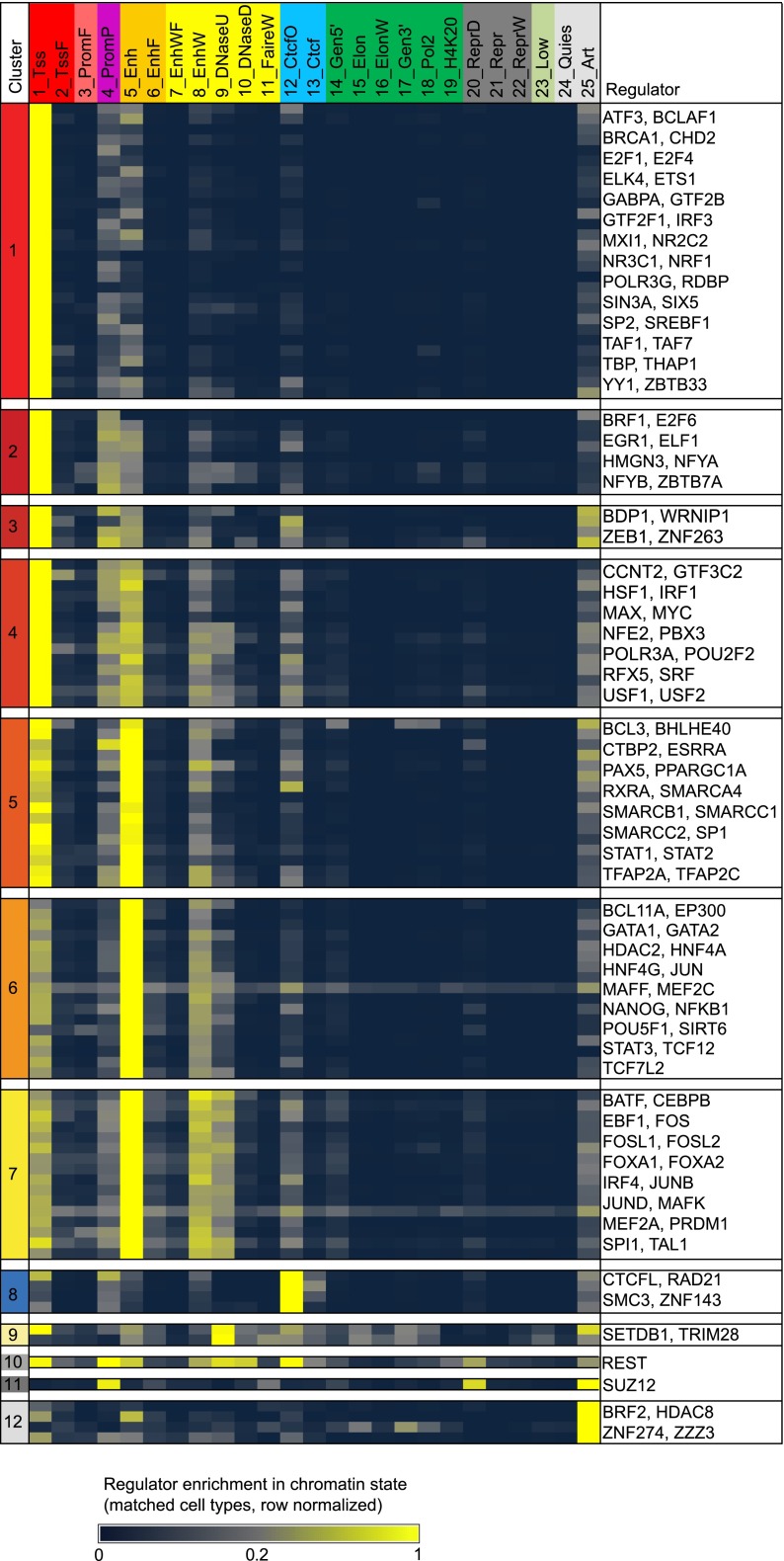
We recognized common patterns of chromatin state preference (Fig. 1; Supplemental Fig. 3), using a k-means clustering algorithm and selecting 12 clusters (Supplemental Table 3; Supplemental Fig. 4; see Methods). Regulators profiled in multiple cell types and conditions showed generally consistent enrichment patterns with two-thirds of the individual regulator experiments showing highest similarity to the cluster center where the corresponding regulator was assigned (Supplemental Fig. 5).
Figure 1.
Regulator enrichments for each chromatin state in matched cell types. Different regulators show distinct chromatin state preferences. For each regulator with matching chromatin data, the average enrichment is shown for each chromatin state (columns). Enrichments have been row-normalized, scaling by the largest enrichment value for each experiment. K-means clustering with 12 clusters produced the clusters labeled C1–C12.
Four of these clusters (C1–C4) had their strongest relative preference for promoter states:
Regulators in cluster C1 showed almost exclusive binding preference in the 1_Tss promoter state. This cluster included all six factors annotated as ‘general Pol II associated factor, not site specific’ (P < 0.001) (Wang et al. 2012), consistent with nonspecific binding that may be partly mediated by the chromatin landscape at transcription start sites.
Regulators in clusters C2 and C3 also had a preference for the 1_Tss state, but relative to C1 had a stronger preference for the more repressive 4_PromP state (both C2 and C3) and for 25_Art (C3 only). Cluster C2 contained both NFYA and NFYB, which we had previously predicted to have repressive activity in enhancer states (Ernst et al. 2011), suggesting they may also show repressive roles in poised promoters.
Regulators in C4 showed the strongest preference for 1_Tss but also a weaker preference for 5_Enh. This cluster contained both helix-loop-helix heterodimers MAX-MYC and USF1-USF2. The presence of TFIID-interacting USF1 and USF2 in this cluster suggests a possibly underappreciated role in distal as well as proximal regulatory regions (Rada-Iglesias et al. 2008).
Three clusters (C5–C7) had strong relative enrichment for enhancer states and varying levels of promoter enrichment:
Regulators in C5 had a balanced preference for both state 1_Tss and 5_Enh and likely play dual roles in enhancers and promoters. C5 includes all four regulators that are part of the SWI/SNF chromatin remodeling complex (P < 0.05), consistent with previous reports suggesting both enhancer and promoter roles (Euskirchen et al. 2011).
Cluster C6 had regulators with a much stronger preference for enhancers than promoters or other states. Of the 17 regulators in this cluster, 13 are known to be involved in the regulation of developmental processes (P < 0.01). The cluster also enriched for genes involved in intracellular transport (P < 0.01). Among the C6 genes was EP300 a common coregulator found in large numbers of enhancers (Visel et al. 2009).
C7 regulators showed the strongest preference for enhancer state 5_Enh but also a weaker preference for 8_EnhW, 9_DNaseU, and 1_Tss. This cluster contained eight of 12 leucine zipper domain regulators (P < 0.001) and was also significantly enriched for positive regulation of cell differentiation genes (P < 0.01).
The remaining clusters (C8–C12) contained a number of regulators that did not fit the usual enhancer or promoter pattern:
Cluster C8 regulators had the strongest enrichment for 12_CtcfO and contained RAD21 and SMC3, both members of the cohesin complex, which is known to interact with CTCF (Wendt and Peters 2009). It also contained the CTCF paralog CTCFL and ZNF143, which have been previously implicated with CTCF (Gerstein et al. 2012).
Cluster C9 regulators were most enriched in the 9_DNaseU open-chromatin state that lacks any activating histone marks and contained two regulators, TRIM28 and SETDB1, suggesting a potential repressive role upon binding. Indeed, both are known to be associated with chromatin gene-silencing (Schultz et al. 2002).
Cluster C10 regulators were associated with multiple states with DNase but lacking active histone marks and consisted of a single regulator, REST (NRSF), which has a known role in gene silencing.
Cluster C11 also consisted of a single regulator SUZ12, a polycomb protein, which had a strong enrichment for the 4_PromP and 20_ReprD, which is expected given that these states are associated with high levels of H3K27me3.
Lastly, C12 regulators had relatively high enrichment for the ‘artifact’ state 25_Art; and indeed, three of the four regulators (BRF2, HDAC8, and ZZZ3) were independently flagged by the ENCODE Consortium as being of medium quality (Landt et al. 2012; A Kundaje, LY Jung, PV Kharchenko, BJ Wold, A Sidow, S Batzoglou, and PJ Park, in prep.). The remaining regulator, ZNF274, also had a notable enrichment for the 3′ gene body state 17_Gen3′, consistent with its involvement in recruiting a methlytransferase to the 3′ end of ZNF genes (Frietze et al. 2010).
Dynamic enrichment patterns across cell types are both regulator and cell-type driven
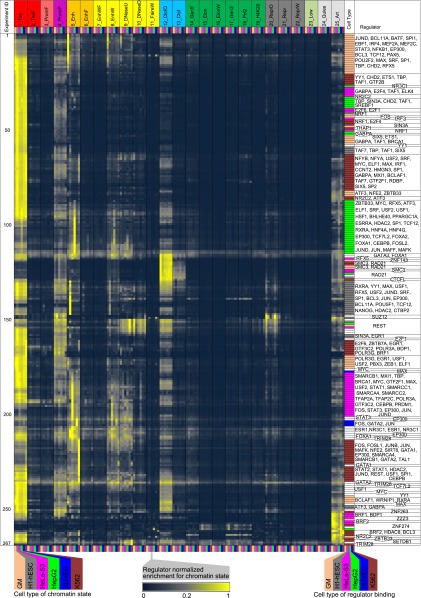
We next expanded our chromatin state enrichment analysis to incorporate multiple cell types. For each regulator, we directly compared the chromatin state enrichment across different cell types (Fig. 2), extending our analysis focusing on matched regulator-chromatin state experiments to study vectors of enrichments across six cell types (see Methods). We ordered the resulting matrix to minimize the total correlation-based distance between neighboring rows by using an instance of the traveling salesman problem (see Methods), which showed greater coherence in both cell type and regulator streaks relative to optimal leaf ordering of a hierarchical clustering solution (Supplemental Fig. 6; Bar-Joseph et al. 2001). We also generated an unbiased ordering at the individual experiment level to study whether common cell types or regulators were preferentially consecutively ordered and to highlight unexpected similarities in relative or absolute enrichment patterns (Supplemental Figs. 7–10).
Figure 2.
Dynamics of regulator enrichment in different chromatin states across cell types. Each row corresponds to one regulator in a given cell type. Each column corresponds to a state-cell type combination. The columns are organized first by state and then by cell type in the following order: Gm12878, H1-hESC, HeLa-S3, HepG2, HUVEC, and then K562. The rows have been automatically ordered computationally using a traveling salesman problem instance solver, and reveal both regulator and cell type groups. The fold enrichments have been row-normalized, scaled to the maximum enrichment in the row. In the six columns of each group, yellow indicates higher enrichment values and blue lower enrichment values. The next-to-last column indicates the cell type of the experiment color-coded, with all GM cell types colored the same and all other non-Tier 1 and 2 cell types colored white. The last column indicates the regulators of the experiments listed consecutively within the same cell type block.
Most cell types exhibited substantial coherence in grouping together different regulators (Fig. 2; Table 1). For example, 85% of the regulators in HepG2 fell into one of two groups preferentially enriched in states 1_Tss or 5_Enh. However, we also observed cases in which multiple experiments of the same regulator profiled in different cell types were grouped together (Table 2). Notable among these was eight of the nine REST experiments and all four ZNF274 experiments in different cell types, both of which play repressive roles.
Table 1.
Different regulators in the same cell type ordered consecutively

Table 2.
Different cell types with the same regulator ordered consecutively
We also observed constitutive regulator enrichments across cell types, particularly in states 1_Tss and 12_CtcfO, known to be less dynamic than enhancer states (Heintzman et al. 2009). We confirmed at the individual experiment level that this was associated with invariant chromatin states rather than simply independent enrichments at similar levels in each cell type (Supplemental Fig. 7). Ordering experiments by cell type shows that each regulator is most enriched in enhancers active in the cell type in which it was profiled (Supplemental Fig. 11), and ordering by regulator highlights dynamic changes in enrichment across cell types (Supplemental Fig. 12) primarily for enhancer states and to a lower degree for promoter states.
Motif depletion suggests abundant nonspecific binding in permissive chromatin states
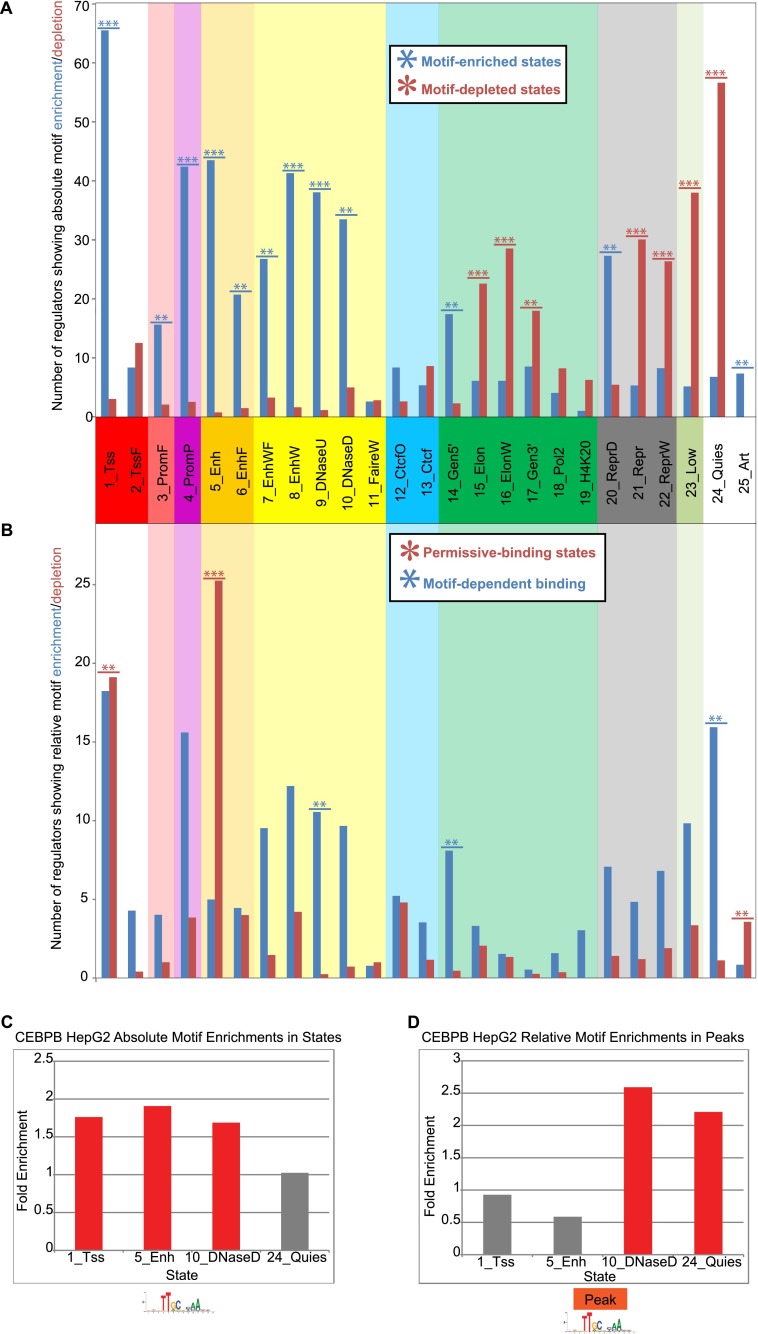
To understand the potential role of DNA sequence in guiding observed transcription factor-chromatin associations, we studied the absolute motif enrichment in each chromatin state for regulators with a known regulatory motif compared to the rest of the genome using control motifs (Supplemental Fig. 13; see Methods). The states for which the greatest number of regulators show significant absolute motif enrichment were 1_Tss, 4_PromP, 5_Enh, 8_EnhW, 9_DNaseU, and 10_DNaseD (Fig. 3A), corresponding well to the states showing the greatest transcription factor binding enrichment (with the exception of 10_DNaseD) (Supplemental Fig. 14). This property also held when considering motifs instances in aggregate across all regulators (Supplemental Fig. 15). Moreover, the chromatin state enrichments for regulatory motifs mirrored their chromatin state binding preferences for the corresponding transcription factor clusters (Supplemental Fig. 16). Together, these results suggest that the chromatin state preferences of sequence-specific regulators are at least in part encoded by genome sequence.
Figure 3.
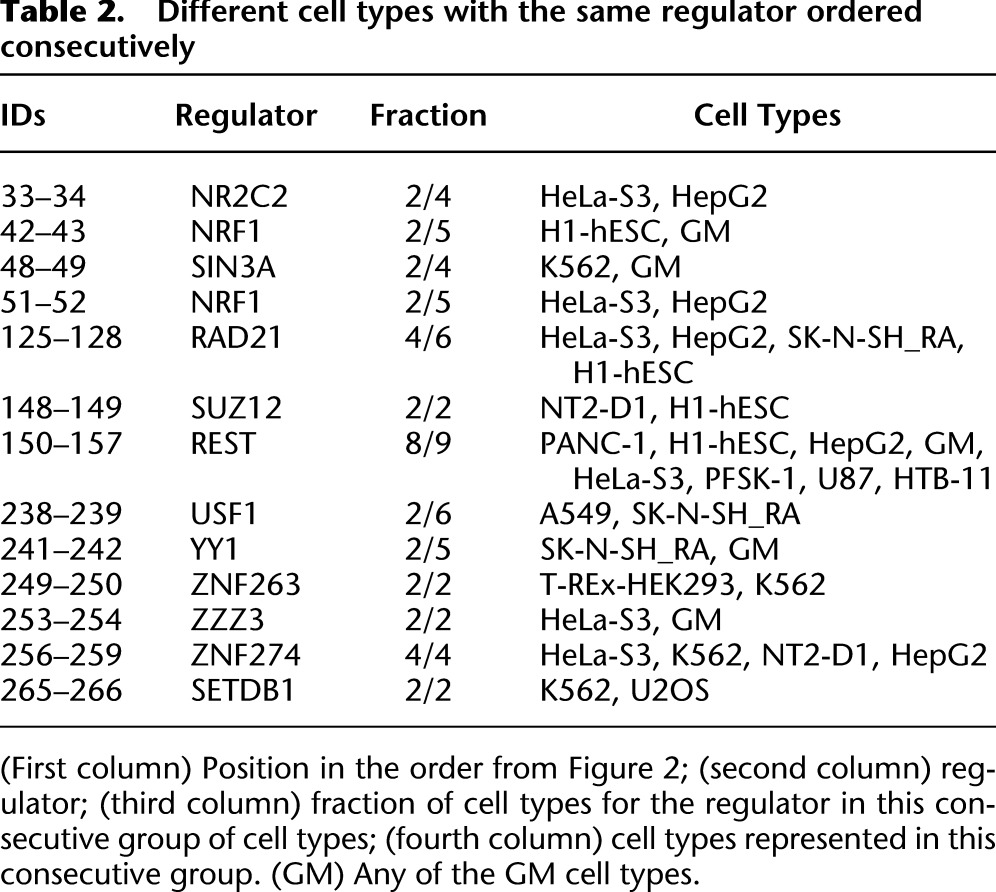
Motif enrichment and depletion variation across chromatin states. (A) Number of transcription factors with significantly enriched or depleted motif instances in each state at a P-value of 0.001 (see Methods). The maximum value for the y-axis was 79, corresponding to the number of transcription factors considered with regulatory motif instances available. If a transcription factor was profiled multiple times, each experiment was counted inversely proportional to the number of times it was profiled. Stars indicate if the number of transcription factors with significantly enriched (depleted) motifs in a state is significant based on a binomial distribution with the number of samples equal to the total number of significant enrichments or depletions in the state and the probability of success equal to the proportion of significant enrichments (depletions) of all significant enrichments or depletions across all states. Fractional values were first rounded to the nearest integer for the calculation. The P-value cutoff for triple stars was 10−6 and for double stars was 0.01. (B) Number of transcription factors with significantly enriched or depleted motif instances in each state conditioning on regions falling within a peak. Stars were computed the same way as in A except a 0.05 P-value cutoff was used for double stars. (C) Fold enrichment for CEBPB motifs in four different HepG2 chromatin states. (D) Fold enrichments for CEBPB motifs within peaks in the same four states relative to the baseline motif enrichment in peaks.
Surprisingly however, the two most often maximally preferentially bound states, 1_Tss and 5_Enh, showed a depletion of regulatory motif instances relative to all bound regions (Fig. 3B; Supplemental Fig. 15). This relative depletion implies that regulator binding in these regions exceeds the level predicted by regulatory motifs, suggesting that in addition to motif-driven binding, these states are conducive to nonspecific binding. A similar depletion of strong CTCF motifs was noted in the presence of active modifications (Essien et al. 2009) and of many regulator motifs in hotspots of regulator binding in fly (The modENCODE Consortium 2010) and human (Yip et al. 2012). Indeed, states 1_Tss, 5_Enh, and 25_Art contained on average about half hotspot regions (Supplemental Fig. 17). However, even among hotspot regions, those overlapping states 1_Tss, 5_Enh, and 25_Art were less likely to contain motifs (Supplemental Fig. 18), suggesting that chromatin states contain additional properties facilitating transcription factor binding than explained by high-occupancy binding alone. Enhancer state 5_Enh, which showed the most dynamic binding across cell types, also showed one of the strongest motif depletion signals (Fig. 3B; Supplemental Fig. 15), consistent with permissive binding facilitating condition specificity.
In contrast, relative motif depletion was not found in states lacking active marks. Instead, repressive states (20_ReprD, 21_Repr, 22_ReprW) and low-activity states (23_Low, 24_Quies) showed motif enrichments relative to all bound regions. This suggests that for binding to occur in these regions, a regulatory motif is more often required; and thus, regulator binding within these regions is more likely motif-dependent. In other words, binding appears to be more frequently sequence-mediated in regions that are not permissive or chromatin mediated.
Importantly, the two DNase-associated open-chromatin states that lack active chromatin marks showed a signature of nonpermissive, motif-dependent binding characteristic of repressive states. Recall that States 9_DNaseU and 10_DNaseD showed signatures of open chromatin but had limited H3K4 methylation marks characteristic of enhancer and promoter regions and also lacked the H3K9 and H3K27 acetylation marks associated with activation of both enhancer and promoter regions. These states showed strong relative motif enrichments compared to all bound regions across all factors and showed more factors with relative enrichment than with relative depletion, suggesting that they are strongly motif-dependent.
In summary, promoters, enhancers, and open chromatin states lacking active histone modifications all show strong absolute enrichments for regulatory motifs (e.g., CEBPB motif for CEBPB-bound sites in HepG2) (Fig. 3C), suggesting that motifs at least partially determine regulator binding preferences in different chromatin states. However, motifs are depleted in promoter and enhancer peaks, relative to all peaks, suggesting permissive binding in active states restricted to open chromatin regions that also show active histone modification marks (Fig. 3D, for CEBPB).
For regulators profiled in multiple cell types, we also analyzed the chromatin state enrichments in a given cell type for commonly bound and differentially bound sites. We defined: (1) “shared” sites, bound in the cell type and at least one other of the six primary cell types; (2) “unique” sites, bound only in the considered cell type and none of the other primary cell types considered; and (3) “excluded” sites, that were bound in at least one other cell type but not the one considered (Fig. 4; Supplemental Figs. 19, 20). For example, the transcription factor CEBPB shows very different chromatin state enrichments for these different classes of bound sites (Fig. 4A): for 1_Tss, enrichment is strongest for shared sites, consistent with constitutive binding in promoter regions; for 5_Enh, enrichment is strongest for unique sites, consistent with dynamic binding in enhancer regions; for 10_DNaseD, enrichment is strongest for excluded sites, consistent with specific repression; for 24_Quies, enrichment is weak throughout, consistent with lack of binding. Considering all regulators and all states, uniquely bound locations are preferentially found in States 5_Enh and 8_EnhW, “shared” locations that are bound in multiple cell types are preferentially found in state 1_Tss, and “excluded” locations that are bound only in other cell types are more enriched than the other two classes in states 9_DNaseU, 10_DNaseD, and 20_ReprD, suggesting they are specifically repressed (Fig. 4B). We also found that regions of shared binding were more likely to contain regulatory motifs (Fig. 4C), consistent with sequence-driven binding, whereas dynamically bound regions showed lower enrichments, as expected for dynamic binding since motifs are by definition invariant across cell types.
Figure 4.
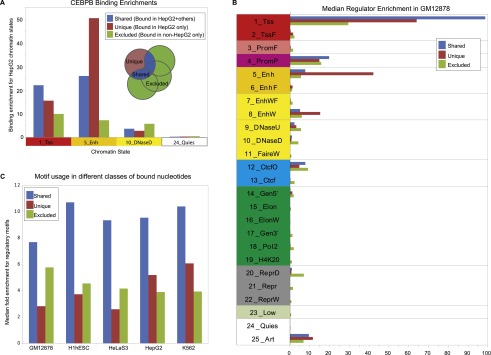
Dynamic binding enrichments. (A) The enrichment in four HepG2 chromatin states for locations of the genome that are bound by CEBPB in HepG2 and another cell type, only in HepG2, and in another cell type but not HepG2. (B) The median enrichment over all regulators in Gm12878 for the three different classes of dynamic binding (for other cell types, see Supplemental Figs. 19, 20). (C) The median enrichment of regulatory motifs in bound regions for the different classes of dynamic binding.
Common chromatin state preferences predictive of regulator cobinding
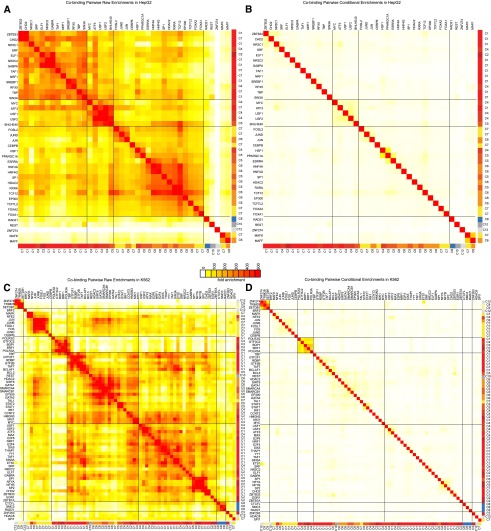
We next studied the role of chromatin in the pairwise binding of regulators. As reported in previous studies (Moorman et al. 2006; Li et al. 2008; Gerstein et al. 2012), we found extensive cobinding enrichments ranging from 50-fold to 500-fold on average for most pairs of regulators (Fig. 5A), which held for both pairs of regulators and pairs of individual experiments (Supplemental Figs. 21, 22). The computationally ordered pairwise enrichment matrix showed strong internal structure, revealing several subgroups of regulators with even higher pairwise overlap to each other, as reported in previous studies (Negre et al. 2011), but whose origin has remained unclear.
Figure 5.
Pairwise regulator cobinding enrichments are captured by chromatin state preferences. (A) Pairwise regulator cobinding enrichment for all pairs of regulators in HepG2 show strong groups of cobinding. The regulators in each group are typically assigned to the same set of chromatin state cluster preferences from Figure 1. Enrichment levels up to 500-fold are found for the full pairwise enrichment table. Rows of the table have been ordered to maximize correlation of neighboring rows. Black lines correspond to groups of highly enriched pairs of regulators that emerge from this ordering. (B) After conditioning on the chromatin state preferences for each regulator (see Methods), the pairwise regulator enrichments are dramatically reduced. (C,D) The same as A and B except for K562. Other cell types can be found in Supplemental Figure 21.
The chromatin state preferences of regulators in each co-occupancy group revealed a potential chromatin state basis of co-occupancy groups because regulators in the same subgroup showed preferences for the same set of chromatin states. For example, among the regulators mapped in HepG2 (Fig. 5A), a large cobinding group was dominated by regulators almost exclusively in cluster C1 of Figure 1 that bind primarily promoter states, whereas another large group consisted of regulators in clusters C5, C6, and C7 that primarily bind enhancer states.
Remarkably, after controlling for chromatin state preferences, genome-wide regulator coassociations were dramatically reduced (Fig. 5B). Cobinding enrichments were reduced approximately 30-fold on average, and the strong structure of cobinding associations was lost (Fig. 5; Supplemental Figs. 21, 22; see Methods). Previous work has noted extensive cobinding within DNase I hypersensitive regions (Kaplan et al. 2011; Li et al. 2011). However, we find that conditioning on DNase accessibility still leaves substantial block structures in the coassociation that are eliminated when including chromatin state information (Supplemental Fig. 23). Only small groups of coassociations remained that could not easily be explained by common chromatin state preferences for the corresponding regulators. For example, these factors included MAFF and MAFK proteins in HepG2, which can be involved in repression (Blank 2008) and were found together in heterochromatic regions; SETDB1, KAP1, and ZNF274 in K562, three factors known to interact (Frietze et al. 2010); and POL3-associated factors in K562 and HeLa-S3 (Fig. 5; Supplemental Figs. 21, 22). Even when cobinding groups mirrored chromatin state preferences, the direction of causality remains unclear. It is possible that cobinding regulators help define chromatin states or that chromatin states facilitate regulator interactions.
Discussion
We undertook a global analysis of the relationship between regulator binding across multiple cell types, the dynamic chromatin landscape, and regulatory motifs. We integrated histone modifications, DNase hypersensitivity, FAIRE, CTCF, and POL2 to define chromatin states, revealing several classes of open chromatin, some of which were surprisingly devoid of histone modification marks. All chromatin states with open chromatin are enriched in regulator binding, but different regulators show distinct preferences for specific subsets of chromatin states: some bind primarily promoter regions such as NRF1 and TAF1; others bind in enhancers regions, such as JUN and FOXA1; others bind equally in both, as members of the SWI/SNF chromatin remodeling complex; and a smaller number of regulators bind insulators such as RAD21, SMC3, and ZNF143 or other regions such as SETDB1, TRIM28, SUZ12, and REST.
Chromatin state preferences are highly stable for a given regulator across different cell types and conditions and are reflected in the underlying regulatory motif enrichments for the corresponding regulators. However, specific enhancer and promoter states that showed enrichment for transcription factor binding were also least likely to contain regulatory motifs relative to all bound regions. This suggests that regulatory sequence motifs may help define promoter and enhancer states; but once these chromatin states are established, they in turn provide a permissive environment for additional binding that does not require regulatory motifs. Cooperative and non-sequence-specific binding within such a generally permissive environment can also explain occurrences of regulator binding peaks that lack a sequence motif. Consistent with this latter possibility, we find that bound locations lacking a motif interact with locations bound by the same transcription factor and containing a motif, based on 5C interaction data (Supplemental Table 4; see Methods). Importantly, open chromatin states that lack histone modifications did not show evidence of nonspecific regulator binding, suggesting that active histone marks, rather than open chromatin, may enable permissive regulator binding.
Although cobinding patterns between pairs of regulators have garnered much attention in previous studies, we find that most of these patterns can potentially be explained by similar chromatin state preferences for individual regulators. Once chromatin state preferences of each regulator are accounted for, cobinding enrichments are reduced by more than an order of magnitude, revealing only few instances of cobinding regulators that are difficult to explain by chromatin preferences alone. These results do not imply that their regulator co-occurrence patterns are not meaningful. In fact, chromatin may simply provide a mechanism for guiding functionally related regulators to the same locations of the genome and thus enabling their joint activity even in the absence of any direct protein–protein interactions between them. Conversely, interactions between regulators may underlie common chromatin state preferences by mutual recruitment. In either case, it is important to recognize that regulator cobinding should be studied in the context of chromatin that may explain, or facilitate, regulator interactions.
Going forward, a systematic understanding of the joint role of DNA sequence information and epigenetic modifications will be paramount in understanding the molecular basis of human disease. On one hand, single-nucleotide polymorphisms (SNPs) have been shown to result to highly pronounced changes in regulator binding, chromatin accessibility, and gene expression across individuals (Montgomery et al. 2010; Degner et al. 2012). On the other hand, epigenetic changes in the lifetime of an individual, and sometimes spanning decades and generations, can lead to reproducible effects on metabolism and health even without underlying genomic alterations. Understanding how genome sequence and chromatin act jointly to specify the dynamic landscape of active and repressive regulatory elements across individuals and cell types will be needed to decipher the regulatory, molecular, and organismal phenotypes that underlie human disease in the context of genetic and epigenomic variation.
Methods
Inferring chromatin states
The chromatin state model is the 25-state ChromHMM (Ernst and Kellis 2012) model described in Hoffman et al. (2013). Details on the data processing and model learning can be found in that paper.
Computing regulator binding enrichments
For binding peak calls, we used the standardized peak calls produced by the ENCODE Consortium (Gerstein et al. 2012; A Kundaje, Q Li, J Rozowsky, JB Brown, A Harmanci, SP Wilder, M Gerstein, S Batzoglou, A Sidow, E Birney, et al., in prep.) using the SPP peak caller (Kharchenko et al. 2008). To compute the enrichment for a peak call-data set in a specific chromatin state and cell type, s, we computed the enrichment for transcription factor binding as (as/b)/(cs/d), where as is the total number of bases in a peak call in s; b is the total number of bases in a peak call; cs is the total number of bases in s; and d is the total number of bases for which the segmentation was defined.
Clustering of cell-type matched enrichments
We clustered a single vector of enrichment values for each regulator with matched cell type chromatin data, excluding CTCF and POLR2A. If multiple experiments were available for the regulator, we averaged them using the median. If there was an even number of enrichment values, we used the geometric mean to average the two middle values. To cluster the experiments based on cell type matched enrichments of chromatin state and binding, we used the k-means function in MATLAB. We used the correlation distance function, 10 random restarts, and singleton as the empty action. We tried between two and 20 clusters and focused on 12 clusters since the patterns were largely homogenous and peaked for enrichment in protein–protein interactions (Supplemental Fig. 4).
Ordering rows of the dynamic enrichment heatmap
We ordered the rows of the dynamic enrichment heatmap to minimize the distance between rows. The distance between two rows was defined using the distance metric sqrt[1-corr(x,y)], where x and y are the vectors of the 150-fold enrichment values for two experiments. Finding such an ordering can be made trivially equivalent to the computational traveling salesman optimization problem (Biedl et al. 2001; Applegate et al. 2006) of finding a minimum cycle that visits every city once by adding a dummy city with zero distance to every other city (Supplemental Fig. 24). We applied a specially designed optimal traveling salesman problem solver, Concorde (http://www.tsp.gatech.edu/concorde.html) (Applegate et al. 2006), which despite the general problem being NP-hard was able to find an optimal solution to our instance of the problem in less than a minute using a single CPU. When forming the rows at the matrix at the regulator level, we averaged enrichments for experiments on the same regulator in the cell type using the median and treating the various GM cell types as the same.
Motif analysis
The motif instance were obtained from http://compbio.mit.edu/encode-motifs (The ENCODE Project Consortium 2012; P Kheradpour and M Kellis, in prep.). The motif instance enrichments were computed relative to a set of selected permuted control motif instances based on the approach described in Kheradpour et al. (2007) without using motif conservation. If a regulator had multiple known motifs associated with it, the motif that had the greatest chromatin state fold enrichment in any cell type was selected and used consistently in the analysis. The P-value significance of motif instance overlap with a chromatin state was computed using a binomial distribution. The null probability of a motif falling into a specific chromatin state was the ratio of the number of control motif instances overlapping the state to the total number of control motif instances, denoted by fs. For determining the significance after first conditioning on a peak call, there generally were too few control motif instances overlapping peak calls to obtain robust null estimates based on the frequency of control motifs within peaks. Instead, to determine the frequency, the assumption was made that conditioned on a chromatin state the random chance expectation of a motif within a peak or outside a peak was uniform after controlling for the number of bases considered in the motif scanning. The null probability of motifs in peaks for a state, s, was computed by first computing the ratio of the number of bases considered in motif scanning overlapping a peak call within state, s, to the total number of such bases overlapping a peak call, denoted by ps. This was then adjusted by fs and the total fraction of bases included in the motif scanning falling in the state, denoted by es, using the formula:
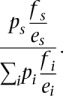 |
The summation is over all states. For computing both the motif enrichments in chromatin states and conditioned on peaks, a P-value significance of 0.001 was used for testing separately enrichment and depletion. The cell type of the chromatin state corresponds to the cell type in which the regulator was profiled. If multiple experiments on the same regulator were conducted, then each experiment was counted inversely proportional to the number of experiments conducted on the transcription factor.
The aggregate motif fold enrichment for a chromatin state was computed as (as/b)/(cs/d), where as is the total number of motif bases in state s; b is the total number of motif bases; cs is the total number of control motif bases in state s; and d is the total number of control bases. The enrichment conditioned on a peak call was computed the same way except restricting as, bs, c, and d to only bases that fell within a peak call. For computing the cluster motif enrichments, each motif was counted once for each cell type in which there was a corresponding regulator experiment assigned to the cluster. The geometric mean of the enrichments after adding a pseudocount of one was used. For the motif usage in different classes of bound regions, the median fold enrichment relative to control motifs across regulators was reported excluding CTCF from the analysis.
We compared the motif usage within peaks within states 1_Tss, 5_Enh, and 25_Art to the other 22 states restricted to High Occupancy of Transcription related factors (HOT) regions defined by (Yip et al. 2012) in Gm12878, H1-hESC, HeLa-S3, HepG2, and K562. For each experiment in one of these five cell types corresponding to a regulator with a motif defined, we computed the proportion of bases included in motif scanning that fell within peaks within HOT regions in states 1_Tss, 5_Enh, and 25_Art to all bases included in the motif scanning that fell within peaks within HOT regions. We then determined, based on a binomial test using this proportion at a P < 0.01, if there was a significant number of motifs in states 1_Tss, 5_Enh, and 25_Art or the other 22 states out of the total number of motifs that fell within peaks within HOT regions. If multiple experiments were conducted for the same regulator, each was counted inversely to the number of experiments conducted on it.
Pairwise enrichment calculations
We computed the raw pairwise enrichments based on peak overlap at the base level. Let a and b be the number of bases in peaks for transcription factors A and B, respectively. Let c be the number of bases in their intersection. Let d be the size of the genome and ɛ be a pseudocount of 100 bases for smoothing. In this case, the pairwise enrichment is (c + ɛ)/[(a × b)/d + ɛ]. To compute the conditional pairwise enrichments, let as and bs denote the number of bases in peaks for transcription factor A and B, respectively, in state s, and ds is the total number of bases in state s, then the conditional enrichment is (c + ɛ)/{[∑s(as × bs)/ds] + ɛ}. When computing pairwise enrichments between pairs of regulators, we averaged using the median of all pairs of experiments involving both regulators. To compute the conditional enrichments based on DNase data, we used peak calls from one replicate of the University of Washington DNase data (Thurman et al. 2012). The same formulas above applied where conditioning on DNase was effectively equivalent to a two-state model with the states corresponding to being in a DNase peak or not. When also including state information with DNase, the same formulas also applied, but the number of states effectively doubled by partitioning each state into the portion overlapping a DNase peak or not. The ordering of the heatmaps was determined using the traveling salesman formulation as was also used for ordering the dynamic enrichment matrix.
Protein–protein interaction data
The protein–protein interactions used were the Biogrid version 3.1.87 physical interaction data sets (Stark et al. 2011). Fold enrichments were computed two ways. Let n be the total number of known protein–protein interactions that fall into a cluster; N is the total number of known protein–protein interactions involving regulators considered; K is the number of clusters; r is the number of regulator pairs that are in the same cluster; and R is the total number of pairs of regulators. Under the uniform expectation enrichment was K(n/N), and when conditioned on the cluster size, it is (n/N)/(r/R).
Dynamic binding analysis
For a given cell type, nucleotides of “unique,” “shared,” and “excluded” binding for a regulator were defined if they were profiled in two or more of the Gm12878, K562, HepG2, HeLa-S3, H1-hESC, and Huvec cell types. If multiple experiments were conducted on the same regulator in a cell type, they were first combined by taking the union.
5C-interaction analysis
We used 5C data available on the ENCODE pilot 1% regions (Sanyal et al. 2012) to investigate if there was a preferential enrichment for sites with a transcription factor peak without a motif to interact with locations with a peak containing a motif. We restricted our analysis to transcription factors that had peaks in one of the four cell types with 5C data available: Gm12878, K562, HeLa-S3, and H1-hESC. We separately considered peaks without motifs on the forward primers (not specific to TSS) interacting with peaks containing motifs on the reverse primers (covering TSS) and vice versa. Only regulators with a minimum of ten possible detected interactions were evaluated. When evaluating the significance of interaction of forward sites without motifs with reverse sites with motifs, we compared the count of observed interactions to the number of interactions when randomizing the interacting reverse primer for 1000 randomizations. When randomizing the reverse primers, they were required to be selected from the same regions as the original interactions, thus, to only generate randomized interacting pairs that could have been observed in the real data. A similar analysis was conducted for bound sites without a motif on the reverse primers to interact with forward primers.
Acknowledgments
We thank Pouya Kheradpour for assistance with the motifs and Bradley Bernstein for useful discussions related to this work. We thank Anshul Kundaje for data processing; Michael Hoffman, Bill Noble, and Ross Hardison for discussions on chromatin states; and the ENCODE Project Consortium for sharing data sets. We acknowledge funding for this work provided by NIH grants R01HG004037 and RC1HG005334, NSF Fellowship 0905968 to J.E., and the David Geffen School of Medicine at UCLA.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.144840.112.
Freely available online through the Genome Research Open Access option.
References
- Applegate DL, Bixby RE, Chvatal V, Cook WJ 2006. The traveling salesman problem: A computational study. Princeton University Press, Princeton, NJ [Google Scholar]
- Bar-Joseph Z, Gifford DK, Jaakkola TS 2001. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics 17: S22. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Biedl T, Brejová B, Demaine ED, Hamel AM, Vinar T 2001. Optimal arrangement of leaves in the tree representing hierarchical clustering of gene expression data. Tech. Rep. 2001, Department of Computer Science, University of Waterloo, Ontario, Canada, p. 14 [Google Scholar]
- Blank V 2008. Small Maf proteins in mammalian gene control: Mere dimerization partners or dynamic transcriptional regulators? J Mol Biol 376: 913–925 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, De Leon S, Michelini K, Lewellen N, Crawford GE, et al. 2012. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M 2012. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods 9: 215–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 10: R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ 2010. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS ONE 5: e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, et al. 2013. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 41: 827–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Ren B, Wang W 2008. ChromaSig: A probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol 4: e1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaschek R, Tanay A 2009. Spatial clustering of multivariate genomic and epigenomic information. In Proceedings of the 13th Annual International Conference on Research in Computational Molecular Biology, RECOMB 2009, pp. 170–183. Springer-Verlag, Berlin [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T, Li XY, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet 7: e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M 2007. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res 17: 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, et al. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22: 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol 12: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD 2012. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A 2010. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 28: 1079–1088 [DOI] [PubMed] [Google Scholar]
- The modENCODE Consortium 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci 103: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C 2008. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res 18: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. 2008. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res 18: 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. From DNA sequence to transcriptional behaviour: A quantitative approach. Nat Rev Genet 10: 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J 30: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. 2011. The BioGRID interaction database: 2011 update. Nucleic Acids Res 39: D698–D704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ Jr, Lazar MA 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA 2007. Identification of higher-order functional domains in the human ENCODE regions. Genome Res 17: 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Peters JM 2009. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res 17: 201–214 [DOI] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. 2011. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res 21: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. 2007. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. 2012. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol 13: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]