Abstract
Bacterial whole-genome sequencing (WGS) of human pathogens has provided unprecedented insights into the evolution of antibiotic resistance. Most studies have focused on identification of resistance mutations, leaving one to speculate on the fate of these mutants once the antibiotic selective pressure is removed. We performed WGS on longitudinal isolates of Acinetobacter baumannii from patients undergoing colistin treatment, and upon subsequent drug withdrawal. In each of the four patients, colistin resistance evolved via mutations at the pmr locus. Upon colistin withdrawal, an ancestral susceptible strain outcompeted resistant isolates in three of the four cases. In the final case, resistance was also lost, but by a compensatory inactivating mutation in the transcriptional regulator of the pmr locus. Notably, this inactivating mutation reduced the probability of reacquiring colistin resistance when subsequently challenged in vitro. On face value, these results supported an in vivo fitness cost preventing the evolution of stable colistin resistance. However, more careful analysis of WGS data identified genomic evidence for stable colistin resistance undetected by clinical microbiological assays. Transcriptional studies validated this genomic hypothesis, showing increased pmr expression of the initial isolate. Moreover, altering the environmental growth conditions of the clinical assay recapitulated the classification as colistin resistant. Additional targeted sequencing revealed that this isolate evolved undetected in a patient undergoing colistin treatment, and was then transmitted to other hospitalized patients, further demonstrating its stability in the absence of colistin. This study provides a unique window into mutational pathways taken in response to antibiotic pressure in vivo, and demonstrates the potential for genome sequence data to predict resistance phenotypes.
Acinetobacter baumannii is a significant cause of hospital-acquired infections among immune compromised patients, accounting for 5% of Gram-negative infections (Lockhart et al. 2007). The threat posed by A. baumannii to hospital patients has increased in recent years because of the widespread dissemination of multidrug resistant (MDR) strains (Dijkshoorn et al. 2007; Perez et al. 2007; Munoz-Price and Weinstein 2008): 30% of A. baumannii isolates from United States hospitals were identified as MDR in data from 2004 (Lockhart et al. 2007). The acquisition of drug resistance in A. baumannii has culminated with recent reports of strains resistant to all commonly used antibiotics (Valencia et al. 2009). The positively charged peptide antibiotic colistin is the last drug for which widespread resistance has not been observed in A. baumannii (Gordon and Wareham 2010). Colistin is also considered the drug of last resort to treat other Gram-negative MDR bacteria, such as carbapenem-resistant Klebsiella pneumoniae.
While still rare, colistin resistance has begun to emerge in A. baumannii and other Gram-negative pathogens (Gales et al. 2011; Cai et al. 2012). Previous studies of colistin resistance in A. baumannii have found resistance to be mediated either by the complete loss of lipopolysaccharide (LPS) (Moffatt et al. 2010), or by mutations at the pmr locus (Adams et al. 2009; Beceiro et al. 2011; Park et al. 2011; Rolain et al. 2012). The pmr locus is an auto-regulated two-component signal transduction system, which in addition to a sensor-kinase and response-regulator, also includes an ethanolamine transferase. The ethanolamine transferase contributes to colistin resistance by adding ethanolamine moieties to the lipid A component of LPS, which reduces the negative charge of the bacterial membrane, and thereby decreases binding of positively charged colistin (Beceiro et al. 2011).
Mutations conferring resistance to antibiotics often come with a fitness cost in the absence of the drug (Andersson and Levin 1999; Andersson and Hughes 2010). Multiple studies have reported a significant fitness cost associated with colistin resistance (Fernández-Reyes et al. 2009; López-Rojas et al. 2011; Rolain et al. 2011), leading to the hypothesis that this fitness cost may explain the lack of widespread resistance (López-Rojas et al. 2011). With other antibiotics, it has been found that the cost of resistance can be overcome either by acquiring low-cost mutations or by accumulating compensatory mutations that diminish the fitness cost (Andersson and Hughes 2010). However, low-cost resistance to peptide antibiotics such as colistin may be difficult to achieve, as evidenced by the continued effectiveness of positively charged human anti-microbial peptides, despite their role in innate immunity over millions of years (Brogden 2005).
Advances in genome sequencing technology have revolutionized the study of the evolution of antibiotic resistance. The ability to sequence entire bacterial genomes has afforded the opportunity to sequence in vitro and in vivo evolved drug-resistant isolates, along with their susceptible ancestors, to identify mutations associated with resistance (Mwangi et al. 2007; Arias et al. 2011; Howden et al. 2011; Saunders et al. 2011). Strikingly, several studies have noted that the most prevalent resistance genotypes are those associated with the lowest fitness cost (Mwangi et al. 2007), pointing to the long-term instability of high-cost mutations in the absence of compensatory mutations (Gagneux et al. 2006; Comas et al. 2012; Nielsen et al. 2012).
Here we applied whole-genome sequencing to isolates of A. baumannii from patients undergoing colistin treatment to determine the fate of colistin-resistance mutations. Longitudinal isolates of A. baumannii were sequenced from four patients in whom colistin resistance evolved during treatment and was subsequently lost after termination of colistin. Genomic comparisons revealed the pathways leading to both the gain and subsequent loss of resistance in each patient. We find that genetic alterations to modulate initial antibiotic sensitivity can constrain the future evolution of resistance. While mutations conferring antibiotic resistance are frequently unstable due to the associated high fitness cost, genomic analysis identified a low-cost mutation, which went undetected by clinical susceptibility assays. The potential stability of this resistant genotype was indicated by its transmission to other patients in the hospital.
Results
Fitness cost of colistin resistance leads to the return of drug susceptibility after colistin is withdrawn from patients
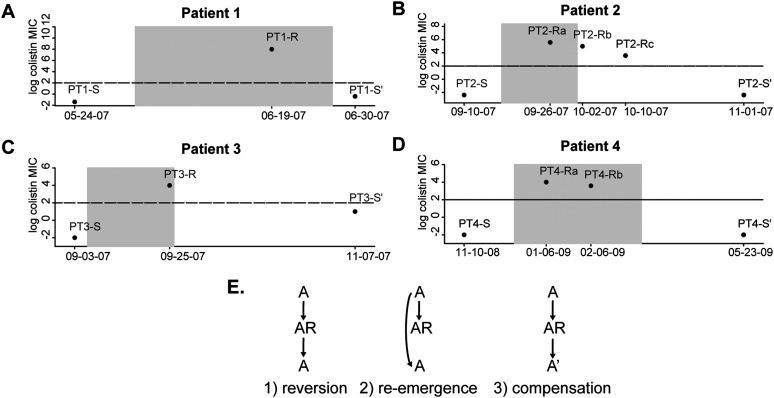
The National Institutes of Health Clinical Center (NIHCC) has had two outbreaks of multidrug-resistant Acinetobacter baumannii (MDRAB) in recent years (Palmore et al. 2011; Snitkin et al. 2011). The four strain types observed during these outbreaks were all highly drug resistant, but each was initially susceptible to colistin (see Methods and Supplemental Table S2). Because of this uniform susceptibility to colistin, it was heavily used during these outbreaks to treat patients with MDRAB infections. Among the 37 patients treated with colistin for MDRAB during these outbreaks, colistin-resistant strains were isolated from five patients. Resistance appears to have evolved independently in each patient, as resistant isolates were only detected after initiation of colistin treatment (Fig. 1; Supplemental Fig. S4A). The failure to observe a patient whose initial isolate was colistin resistant supports the fitness cost associated with colistin resistance limiting its spread. Further evidence for a fitness cost of colistin resistance comes from the in vitro observation of decreased growth rates (Supplemental Fig. S2). Even more striking was the in vivo observation that in each of the four patients for whom colistin treatment was stopped, susceptible isolates returned soon after the termination of colistin treatment (Fig. 1). The duration of colistin treatment ranged from <1 mo (PT-3) to 3 mo (PT-4), indicating that longer drug exposure did not result in mutations compensating for the fitness cost of resistance. Note that colistin was not withdrawn from the fifth patient and his/her case is not included for further studies.
Figure 1.
Gain and loss of colistin resistance in A. baumannii. (A–D) The colistin minimal inhibitory concentrations (MICs) of A. baumannii isolates from the four study patients are shown. The x-axis represents time, with the shaded box indicating the time period during which the patient received colistin. The y-axis shows the colistin MICs, with the horizontal line drawn at an MIC value of 4, which is the clinical cutoff for colistin resistance. Patients' isolates include their initial susceptible isolate (S), at least one resistant isolate (R), and a post-colistin susceptible isolate (S′). (E) Models illustrating the potential evolutionary paths underlying the loss of colistin resistance in patients 1 through 4.
Whole-genome sequencing reveals a common mechanism of resistance in all four patients
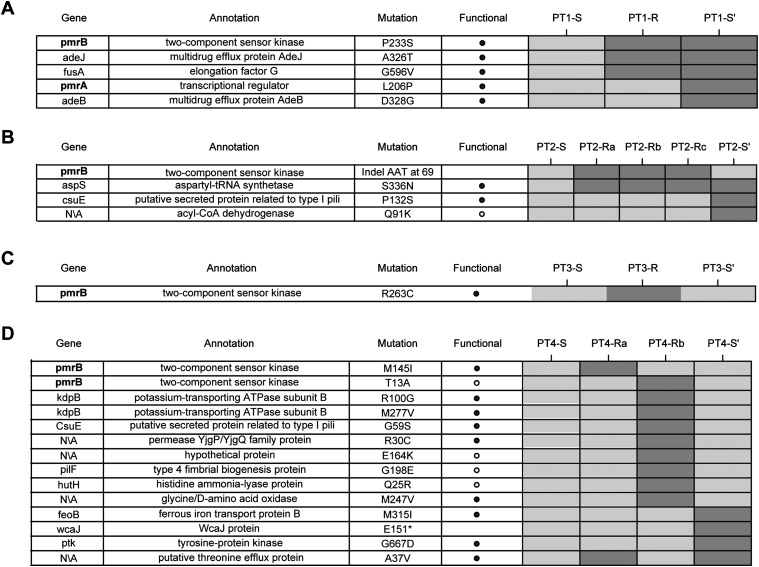
To provide insight into the gain, and subsequent loss, of colistin resistance in MDRAB isolated from these four patients, whole-genome sequencing was performed on longitudinal isolates taken from each patient (Fig. 1; Supplemental Table S1). Whenever possible, isolates from each patient were taken from the same anatomical site, in hopes of sampling from the same bacterial population over time (Supplemental Table S1). Longitudinal isolates started with each patient's initial colistin susceptible MDRAB isolate (e.g., PT1-S), included at least one resistant isolate (e.g., PT1-R), and ended with an isolate after resistance to colistin had been lost (e.g., PT1-S′). These genomic time courses provided clear insight into the mechanism of resistance (Fig. 2; Supplemental Fig. S4B). In all patients, resistant isolates showed mutations at the pmr locus, supporting previous reports of the role of the pmr locus in colistin resistance (Adams et al. 2009; Park et al. 2011). More specifically, each resistant isolate had either a nonsynonymous substitution or a small indel in the pmrB gene, which encodes the sensor-kinase in the pmr two-component system. The observation of distinct mutations in each isolate supports the existence of many mutations at this locus conferring resistance. We hypothesized that these pmrB mutations constitutively activated the sensor-kinase, leading to overexpression of the pmrC ethanolamine transferase (Beceiro et al. 2011). Supporting this hypothesis, the pmrB mutations in each resistant isolate were associated with increased expression of the pmr transcript (Fig. 3; Supplemental Fig. S4C). Furthermore, extrapolating from mutagenesis studies done on the Escherichia coli EnvZ gene, which is a homolog of pmrB, revealed that several of the pmrB mutations observed in resistant isolates mapped to EnvZ mutation sites, causing constitutive kinase activity (Supplemental Table S5; Hsing et al. 1998). In addition to recurrent mutations at the pmr locus, mutations in the translational machinery were observed in multiple patients (Fig. 2; Supplemental Material). We hypothesized that these mutations might play a role in compensating for the fitness cost associated with colistin resistance (see Supplemental Material).
Figure 2.
Genome-wide single-nucleotide variants in longitudinal patient isolates. (A–D) Rows in each table represent all nonsynonymous mutations identified across the genomes of isolates derived from patients 1 through 4. Isolates from each patient are shown in temporal order, in the columns of the corresponding patient's heatmap. Dark/light gray boxes in the heatmap represent the presence/absence of mutations in patient isolates. The pmr genes appear in boldface, to emphasize their mutation in the resistant isolates from each patient. The putative functional impact of mutations was predicted using SIFT. Note that for PT3-S′ unique mutations are not shown, as this is a different strain type from PT3-S and PT3-R and, therefore, has several hundred unique variants.
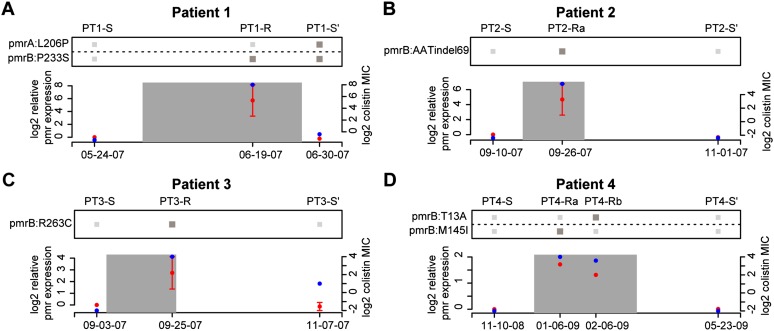
Figure 3.
Mutations at pmr locus account for altered pmr expression and colistin MICs. (A–D) The relationship between pmr genotype, pmr transcript level, and colistin MICs is shown for patients 1 through 4. The x-axis of each plot represents time, with the date of each isolate labeled. The left y-axis of each plot represents the pmr expression relative to the initial patient isolate and corresponds to red points. The expression values and error bars are based on the mean and standard error of three replicate experiments. The right y-axis of each plot represents the isolates MICs and corresponds to blue points. The heatmap above represents the presence/absence (dark/light gray) of mutations to the pmr observed in each patient's isolates.
Whole-genome sequencing demonstrates different evolutionary pathways leading to loss of resistance in different patients
In addition to providing insight into the genetic basis for the gain of colistin resistance, the patterns of mutations in each patient's isolates also allowed us to distinguish among the different scenarios leading to the loss of resistance: reversion, compensation, or re-emergence (Fig. 1E). Patient 1 shows genetic evidence for a compensatory mutation accounting for the loss of resistance. In addition to having the pmrB mutation from his/her resistant isolate (PT1-R), the post-colistin susceptible isolate (PT1-S′) also has a mutation in the DNA-binding domain of the pmrA transcriptional regulator. We hypothesized that the pmrA mutation ablated resistance by preventing the hyper-activation of the pmrB kinase from having a downstream impact on pmr transcription. This hypothesis is supported by the return of the pmr transcript to wild-type levels in the PT1-S′ isolate (Fig. 3). Furthermore, mutagenesis studies in the E. coli ompR gene, a homolog of pmrA, found that mutation of the equivalent residue resulted in loss of DNA binding (Supplemental Table S5; Martínez-Hackert and Stock 1997).
We next wondered if the compensatory pmrA mutation resulting in the loss of resistance in patient 1 might have had implications on the future evolution of colistin resistance in this patient. We hypothesized that, for the PT1-S′ isolate to re-evolve colistin resistance, the function of the DNA-binding domain of pmrA must be restored, which may only be achievable by a small number of mutations. We tested this hypothesis by performing fluctuation tests (Rosche and Foster 2000) on the PT1-S and PT1-S′ isolates to determine the rates of colistin resistance mutations in each isolate (Supplemental Fig. S5). Fluctuation tests confirmed that PT1-S′ had a significantly lower rate of colistin resistance mutations than PT1-S (uPT1-S = 1.47 × 10−8, uPT1-S′ = 3.12 × 10−9, Spearman rank P = 0.007). These assays confirmed that patient 1's A. baumannii isolate evolved colistin resistance by mutating the pmrB locus and with the removal of colistin, a secondary mutation in the pmrA locus restores colistin sensitivity, but at the cost of significantly decreasing the future potential to re-evolve resistance.
Patients 2 and 3 show evidence for the loss of resistance being due to the re-emergence of ancestral susceptible strains. For patient 2, the pmr genotype of the post-colistin susceptible isolate (PT2-S′) matched their pre-colistin susceptible ancestor (PT2-S) (Fig. 3). While in principle a reversion mutation is also consistent with the data, we deemed this less likely due to the likely low probability of the 3-bp insertion that would have been required to return to the ancestral genotype. Loss of resistance in patient 3 is clearly due to a resistant strain being outcompeted by a susceptible competitor, as the post-colistin susceptible strain is of a different strain type (PT3-S′) (pusoltype A in Supplemental Fig. S1) from the resistant strain (PT3-R) (pusoltype B in Supplemental Fig. S1). It is possible that patient 3 had an undetected mixed infection with both strain types A and B, or that s/he acquired a strain of type A after discontinuation of colistin treatment.
Functional, genetic, and epidemiological evidence suggests existence of low-cost resistance mutation that went undetected by clinical assay
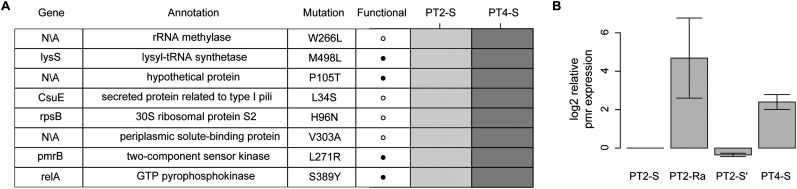
Patient 4 also shows evidence for re-emergence, as the pmr genotype of the pre- and post-colistin isolates are identical (PT4-S vs. PT4-S′) (Fig. 3). As in the other patients, we took the re-emergence of the pretreatment genotype as evidence of the fitness cost of colistin resistance leading to population instability upon removal of colistin. However, unlike in the other patients, for patient 4 we also found several lines of evidence indicating population instability in the presence of colistin. First, unlike in the other patients, in patient 4 there was not a rapid stabilization of an initial resistance mutation. Rather, the two resistant isolates sequenced have distinct pmrB mutations (PT4-Ra and PT4-Rb) (Fig. 2). Second, when the two resistant isolates were cultured from patient 4, both resistant and susceptible isolates were identified in the same sample. This heterogeneity was initially uncovered based on the observation of different colony morphologies in the patient sample, with subsequent resistance testing demonstrating that smaller colonies were colistin resistant and larger ones colistin susceptible (see Methods). Distinct colony morphologies were not observed for any other patient sample. Sequencing of the pmr locus in these co-isolated susceptible isolates identified the same genotype as the initial PT4-S isolate. These observations indicated that the initial isolate from patient 4 was able to remain in the population in the presence of colistin, despite its apparent colistin susceptibility, as evaluated by E-test. To determine if there was a genetic basis for the persistence of the PT4-S's genotype during colistin treatment we compared the genomes of the initial isolates from patients 2 and 4 (PT2-S and PT4-S), which were of the same strain type, and whose common ancestor initiated the outbreak. Most variants between these two genomes were surrounding a recombinant region containing a putative iron acquisition gene locus (see Supplemental Material). Filtering out variants in this recombinant region left eight nonsynonymous SNPs (Fig. 4A), one of which was pmrBL271R, and was predicted to affect protein function based on SIFT analysis and mapping to envZ mutagenesis studies (Supplemental Table S6). Comparison of pmr transcript levels revealed that PT4-S indeed expressed the pmr transcript at higher levels than PT2-S (Fig. 4B).
Figure 4.
Initial isolate from patient 4 has elevated pmr expression. (A) Rows in the table represent all nonsynonymous mutations in nonrecombinant regions identified between the initial isolates of patients 2 (PT2-S) and 4 (PT4-S). Dark/light gray boxes in the heatmap represent the presence/absence of mutations in patient isolates. The pmr gene mutation is bolded to emphasize its hypothesized role in colistin resistance of PT4-S. (B) The expression of the pmr transcript relative to PT2-S is shown for PT2-R, PT2-S′, and PT4-S. The expression values are the mean and standard error for three replicates
The observation of elevated pmr expression in patient 4's initial isolate led us to wonder if this isolate was in fact colistin resistant, despite demonstrating susceptibility by E-test. E-tests have been previously shown to be reliable indicators of colistin resistance in A. baumannii, demonstrating good correlation with other standard susceptibility tests (Arroyo et al. 2005). Here, however, we found that despite susceptibility by E-test (MIC [minimal inhibitory concentration] = 0.125), broth-microdilution (BMD) experiments showed growth of PT4-S well into the range of colistin concentrations associated with resistance (MIC = 64 μg/mL) (Supplemental Table S4). PT4-S was the only isolate for which colistin resistance differed between E-test and BMD, with the accepted cutoff at 4 μg/mL.
We next sought to identify PT4-S's origin, and specifically if the pmrBL271R mutation had evolved in a previous patient receiving colistin, who might then have transmitted this allele to PT4. To this end we sequenced the pmr locus from isolates in patients who received colistin prior to patient 4, and found that the pmrBL271R had indeed appeared in a patient during the course of his/her colistin treatment (Supplemental Fig. S3). Specifically, this patient's original isolate had the wild-type pmrB sequence and evolved the pmrBL271R mutation during colistin treatment. We identified an additional patient whose initial isolate had the pmrBL271R, further demonstrating the stability of this genotype in the absence of colistin (Supplemental Fig. S3). Thus, pmrBL271R appears to provide a sufficient level of resistance to survive in patients receiving colistin, while being fit enough in the absence of colistin to be transmitted and not be outcompeted by susceptible strains.
Discussion
Here we tracked the evolution of colistin resistance in A. baumannii by sequencing longitudinal isolates from four patients, and found a common in vivo mechanism of resistance in all patients. Following the fate of A. baumannii isolates in these patients after the selective pressure of colistin was removed, allowed us to observe the long-term evolutionary consequences of different mutational trajectories. In some patients, the evolution of resistance did not impact future pathogen evolution, as the original susceptible isolate was able to re-emerge after removal of the drug. It is unclear if the re-emergence of susceptible ancestors is a result of the presence of a dormant persister population (Lewis 2007) or bacterial populations in locations inaccessible to cytotoxic drug concentrations (Hermsen et al. 2012). Both of these issues are major concerns in effective patient treatment. While the current study was retrospective, and therefore limited by samples that were collected, it will be interesting in the future to attempt to gain insight into the impact of spatial population heterogeneity on the evolution of resistance. For instance, by sequencing entire bacterial populations from multiple patient sites during the course of treatment, the presence of distinct evolutionary trajectories within a single patient could be monitored (Sun et al. 2012).
In one patient, resistance was lost via the acquisition of a secondary mutation, which compensated for the fitness cost of drug resistance. Following up on computational predictions, we performed in vitro experiments that demonstrated that this mutation moved the population to a new part of the fitness landscape, in which re-acclimation to colistin was more difficult. This observation suggests that for this individual patient, reintroduction of colistin might have been effective. It is intriguing to wonder whether, for drugs with high frequencies of high-cost resistance mutations, cycling of the drug may prove effective in ultimately driving the population to a state where resistance mutations are inaccessible.
Despite the fitness cost associated with most observed colistin resistance mutations, we ultimately found evidence of a stably resistant strain that could navigate the fluctuations in drug exposure during patient-to-patient transmission. It is important to note that while we believe that this isolate has some level of resistance, it does not appear to have complete resistance to physiological concentrations of colistin, as demonstrated by the accumulation of additional pmr mutations in PT4, when treated with colistin. We therefore hypothesize that this mutation provides enough resistance to the cytotoxic effects of colistin to remain in the population during treatment, and subsequently re-emerges after colistin is withdrawn. Thus, more than a resistance mutation, it may be more aptly classified as a mutation that predicts resistance upon exposure to colistin. It will be of great interest in the future to determine the long-term viability of this strategy for surviving fluctuations in drug concentrations, while circumventing the fitness cost associated with resistance.
More broadly, we believe that our results provide further support for the future power of bacterial genomics to make inferences into both current and future resistance. There has been a great deal of work in characterizing the machinery associated with drug resistance (Alekshun and Levy 2007; Kohanski et al. 2010). However, the presence of this machinery in a bacterial genome is not sufficient to confer resistance, as it must be both functional (e.g., lack inactivating mutations) and regulated in a way that it is deployed in the correct place and time. Here, applying both background knowledge and genomic insights, we were able to correctly identify (1) colistin resistance missed by laboratory assay in PT4S, and (2) the acquisition of a compensatory mutation, which impedes future evolution of resistance. Much work is needed to understand the mutational landscape associated with resistance to colistin and other drugs, such that predictions can be made with high confidence. However, advances in massively parallel directed bacterial evolution (Wang et al. 2009) and sequencing (Loman et al. 2012) provide the necessary tools to determine the local fitness landscape of known resistance loci. The capacity to sequence bacterial populations directly from the patient, and rapidly determine current resistance spectrums and the probabilities of future resistance, can be a powerful tool in limiting the evolution of resistance and maintaining the effectiveness of available drugs.
Methods
Isolation of strains from patient samples
Clinical isolates of A. baumanii were obtained from respiratory, blood, or surveillance specimens as part of standard of care patient treatment. The strains used in this study originated from the NIH Department of Laboratory Medicine, Microbiology Department. The NIH microbiology laboratory stores all bacterial isolates tested for antibiotic susceptibility. Samples were not collected specifically for this study or research purposes.
Isolates were identified at the species level by MALDI-TOF mass spectrometry. Antibiotic susceptibility testing was carried out with the Microscan except for colistin, which was performed by E-test (Biomerieux). If different colony morphologies of A. baumannii were visualized, each was isolated, identified, tested for antibiotic susceptibilities, and frozen separately. For a list of strains see Supplemental Table S1.
Colistin susceptibility testing
Minimal inhibitory concentrations (MICs) were determined by the E-test method according to the manufacturer's guidelines (AB Biodisk). Strains were plated and grown overnight, and then used to inoculate 5 mL tryptic soy broth to a turbidity of 0.5 McFarland. Mueller-Hinton agar plates were swabbed with the innocula in three directions to ensure uniform growth. Once the agar surface was completely dry, an E-test colistin strip was applied to each plate with sterile forceps, and the plates were incubated at 37°C for 16–20 h in an incubator without CO2.
For comparison, we determined MIC values by broth microdilution. Colistin sulfate powder (hereafter referred to as colistin) was obtained from Sigma Chemical Co., and a stock solution of 10 mg/μL was created. Broth microdilution was performed in accordance with NCCLS guidelines. Briefly, strains were plated and grown overnight on Mueller-Hinton plates. Plates were used to inoculate 5 mL tryptic soy broth to a turbidity of 0.5 McFarland, and then diluted 1:20 to achieve an inoculum of 5 × 106 CFU/mL. A solution of 128 μg/mL colistin in Mueller-Hinton broth was created, followed by 1:2 serial dilutions to obtain solutions with colistin concentrations ranging from 0.025 to 128 μg/mL. Ninety microliters of colistin solutions were placed in microtiter plates, followed by the addition of 10 μL of the appropriate innocula to each well. The MIC was defined as the lowest concentration with which no growth was visible after incubation of plates at 35°C for 16–20 h.
Calculation of in vitro growth rates
Growth curves were performed for all isolates from a given patient in a single experiment. Overnight cultures for each strain were grown in 2 mL of LB media by inoculating with a single colony from a fresh plate. Thirty milliliters of fresh LB media was inoculated the next morning to an OD of 0.015. Cultures were placed on orbital shaker at 37°C and shaken at 160 rpm. OD measurements were taken every 30 min between 1 and 4 h, and then every hour up until 8 h. Growth rates were determined as the average slope of the log of the OD vs. time between 1.5 and 3 h. All growth curves were performed in triplicate on separate days, from fresh overnight cultures.
Measurement of pmr transcript levels
Half a milliliter of culture was taken during mid-log phase (t = 2.5 h) of growth and combined with 1.0 mL of RNA protect (Qiagen). The sample was then spun down and the pellet stored at −80°C until subsequent experiments. Total RNA was extracted from cell lysates using the RNeasy kit (Qiagen). RT-PCR was performed using the QuantiTect SYBR Green RT-PCR (Qiagen). The 16S rRNA gene was used for control and sample normalization. Primers for the pmrA and the 16S rRNA were taken from reference (Adams et al. 2009). In each run a blank sample (distilled water) and a no reverse-transcriptase control were included to exclude DNA contamination. Technical replicates of each sample were run, and the mean of these two readings was used.
RT-PCR reactions were run on an ABI 7300 System (Applied Biosystems). Relative gene expression differences were calculated by first normalizing Ct values by subtracting out the 16S rRNA control, and then comparing with the appropriate reference sample (initial patient sample for Fig. 3 and PT2-S for Fig. 4).
Pulsed-Field Gel Electrophoresis (PFGE)
Isolates were grown overnight in Brain Heart Infusion broth at 35°C in 5% CO2, concentrated by centrifugation, and resuspended in 1 M NaCl, 10 mM Tris-Cl (pH 7.6) to achieve approximately the turbidity of a 4.0 McFarland turbidity standard. Extraction of genomic DNA in 0.8% InCert agarose plugs (Cambrex Corp.) was performed by standard methods and included 2 mg of lysozyme/mL followed by proteinase K digestion (100 μg/mL) for bacterial lysis. DNA was digested with 125 U of the restriction endonuclease ApaI at 250°C. Pulsed-Field Gel Electrophoresis (PFGE) was performed in a CHEF DR III (Bio-Rad Laboratories) apparatus with ramped pulse times from 7 to 21 sec for 24 h at 6 V/cm. Size standards and an A. baumannii control strain were included in each run for gel-to-gel comparisons.
Genome sequencing and assembly
Genomic libraries were constructed using the Roche 454 Titanium Kit (Roche 454 LifeScience). Sequencing was performed on the Roche/454 XLR instrument with an average N50 contig size of 44,836 bp and an average of 246 large contigs. Sequencing assembly statistics for each isolate are provided in Supplemental Table S3. Contig assembly was executed using the gsAssembler v.2.5. Genome annotation was performed using the publicly available Prokaryotic Genomes Automatic Annotation Pipeline.
Genome alignment and mutation detection
Large contigs from each genome assembly were ordered and oriented relative to a finished reference A. baumannii genome (ACICU/Accession Number NC_010611 for pulsotypes A, B, and C and AB0057/Accession Number NC_011586 for pulsotype D), and then stitched together to form a pseudo-chromosome with the Mauve contig mover (Rissman et al. 2009). For each patient lineage, each genome was then aligned to the initial isolate using MUMmer (Delcher et al. 2002), with single-base variants and small indels being extracted from these alignments using the show-snps function. Single-nucleotide variants (SNVs) were filtered to remove those SNVs that were likely to be due to alignment or sequencing errors. SNVs were filtered out if (1) they resided in genes annotated as phage, transposase, or integrase, (2) they resided in genomic regions annotated as phage by the Phage Finder program (Fouts 2006), (3) they resided within 20 bp of the start or end of a contig, (4) they resided in tandem repeats of total length >20 bp, as determined by the exact-tandem program associated with MUMmer (Delcher et al. 2002), (5) they resided in large inexact repeats as determined by nucmer, (6) they were within two positions of a second putative SNV, (7) the SNV position was ambiguous or low quality in any of the aligned genomes, (8) the 10-bp window surrounding the putative SNV contained more than two ambiguous or low-quality base calls, or (9) the 10-bp window surrounding the putative SNV contained a A/T homopolymer run of length five or longer. The putative functional impact of nonsynonymous mutations was determined using SIFT (Ng and Henikoff 2003).
Fluctuation tests
Fluctuation tests were used to determine the rate of colistin resistance mutations in PT1-S and PT1-S′. For each strain, a single colony was inoculated into 2 mL of Mueller-Hinton broth and grown overnight at 37°C. The following day, overnight cultures were diluted 1:100 in 10 mL of fresh Mueller-Hinton broth and grown overnight at 37°C and 160 rpm on an orbital shaker. The next day, each overnight culture was diluted 1:106, to a concentration of roughly 5000 cells/mL. These diluted cultures were used to create 15 × 200 μL cultures for each strain. After 24 h of growth at 37°C/160 rpm, 12 of the 200 μL cultures were plated onto Mueller-Hinton agar plates containing 8 μg/mL colistin, and three were diluted by a factor of 1:106 and plated on Mueller-Hinton agar plates lacking colistin. Plates were incubated at 37°C in the presence of CO2 for 18 h, after which the number of colonies on each plate was counted. The FALCOR online tool (Hall et al. 2009) was used to calculate the rate of colistin resistance mutations using the MSS maximum likelihood method (Rosche and Foster 2000).
Sequencing of pmr locus
The pmr locus from several strains of A. baumannii was sequenced using amplicons as templates. Four overlapping amplimers (Supplemental Table S6) spanning 4 kb of the targeted region were designed using Consed (Gordon et al. 1998) and the genome sequence of PT1-R. Each amplimer ranged from 1 to 1.5 kb in length and overlapped by ∼50–100 bp. Amplicons were generated from purified genomic DNA using Sprint Advantage (Clontech) as directed by the manufacturer. Each amplicon was sequenced from each end using the respective amplimer primer and Big Dye Terminator chemistry (Applied Biosystems), and then analyzed on an AB3130xl sequencing instrument. To improve data quality additional sequence was generated using nested primers (Supplemental Table S7). Sequence reads were aligned and analyzed in Consed.
Data access
The sequence data from this study have been submitted to NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject) under BioProject accessions: PRJNA73903/APAW00000000, PRJNA73905/APAX00000000, PRJNA73907/APAY00000000, PRJNA73913/APAZ00000000, PRJNA73915/APBA00000000, PRJNA73919/APBB00000000, PRJNA73921/APBC00000000, PRJNA73923/APBD00000000, PRJNA73925/APBE00000000, PRJNA73927/APBF00000000, PRJNA73929/APBG00000000, PRJNA73931/APBH00000000, PRJNA73933/APBI00000000, PRJNA73937/APBJ00000000, PRJNA73939/APBK00000000, PRJNA73941/APBL00000000, PRJNA172976/APBM00000000, PRJNA172977/AOGD00000000.
Acknowledgments
We thank David Lipman and Heidi Kong for their thoughtful discussions and advice. Research support came from NHGRI and NIH CC Intramural Research Programs and from an NIH Director's Challenge Award for genome sequencing. E.S.S. is supported by a Pharmacology Research Associate Training Fellowship, NIGMS.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.154328.112.
References
- Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53: 3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekshun M, Levy S 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050 [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D 2010. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat Rev Microbiol 8: 260–271 [DOI] [PubMed] [Google Scholar]
- Andersson DI, Levin BR 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2: 489–493 [DOI] [PubMed] [Google Scholar]
- Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, et al. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365: 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo LA, Garcia-Curiel A, Pachon-Ibanez ME, Llanos AC, Ruiz M, Pachon J, Aznar J 2005. Reliability of the E-test method for detection of colistin resistance in clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43: 903–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, et al. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55: 3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA 2005. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3: 238–250 [DOI] [PubMed] [Google Scholar]
- Cai Y, Chai D, Wang R, Liang B, Bai N 2012. Colistin resistance of Acinetobacter baumannii: Clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67: 1607–1615 [DOI] [PubMed] [Google Scholar]
- Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S 2012. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44: 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30: 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkshoorn L, Nemec A, Seifert H 2007. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5: 939–951 [DOI] [PubMed] [Google Scholar]
- Fernández-Reyes M, Rodríguez-Falcón M, Chiva C, Pachón J, Andreu D, Rivas L 2009. The cost of resistance to colistin in Acinetobacter baumannii: A proteomic perspective. Proteomics 9: 1632–1645 [DOI] [PubMed] [Google Scholar]
- Fouts DE 2006. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34: 5839–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312: 1944–1946 [DOI] [PubMed] [Google Scholar]
- Gales AC, Jones RN, Sader HS 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: Results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 66: 2070–2074 [DOI] [PubMed] [Google Scholar]
- Gordon NC, Wareham DW 2010. Multidrug-resistant Acinetobacter baumannii: Mechanisms of virulence and resistance. Int J Antimicrob Agents 35: 219–226 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P 1998. Consed: A graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma C-X, Liang P, Singh KK 2009. Fluctuation AnaLysis CalculatOR: A web tool for the determination of mutation rate using Luria–Delbrück fluctuation analysis. Bioinformatics 25: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R, Deris JB, Hwa T 2012. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc Natl Acad Sci 109: 10775–10780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, McEvoy CRE, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, et al. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7: e1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KK, Silhavy TJ 1998. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol 180: 4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ 2010. How antibiotics kill bacteria: From targets to networks. Nat Rev Microbiol 8: 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5: 48–56 [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, Quinn JP, Doern GV 2007. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol 45: 3352–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Constantinidou C, Chan JZM, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ 2012. High-throughput bacterial genome sequencing: An embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10: 599–606 [DOI] [PubMed] [Google Scholar]
- López-Rojas R, Domínguez-Herrera J, McConnell MJ, Docobo-Peréz F, Smani Y, Fernández-Reyes M, Rivas L, Pachón J 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203: 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hackert E, Stock AM 1997. The DNA-binding domain of OmpR: Crystal structures of a winged helix transcription factor. Structure 5: 109–124 [DOI] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St. Michael F, Cox AD, et al. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54: 4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price LS, Weinstein RA 2008. Acinetobacter infection. N Engl J Med 358: 1271–1281 [DOI] [PubMed] [Google Scholar]
- Mwangi MM, Wu SW, Zhou Y, Sieradzki K, De Lencastre H, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, et al. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci 104: 9451–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Pedersen TM, Udekwu KI, Petersen A, Skov RL, Hansen LH, Hughes D, Frimodt-Møller N 2012. Fitness cost: A bacteriological explanation for the demise of the first international methicillin-resistant Staphylococcus aureus epidemic. J Antimicrob Chemother 67: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Palmore TN, Michelin AV, Bordner M, Odom RT, Stock F, Sinaii N, Fedorko DP, Murray PR, Henderson DK 2011. Use of adherence monitors as part of a team approach to control clonal spread of multidrug-resistant Acinetobacter baumannii in a research hospital. Infect Control Hosp Epidemiol 32: 1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Choi JY, Shin D, Ko KS 2011. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents 37: 525–530 [DOI] [PubMed] [Google Scholar]
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51: 3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT 2009. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics 25: 2071–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain J-M, Roch A, Castanier M, Papazian L, Raoult D 2011. Acinetobacter baumannii resistant to colistin with impaired virulence: A case report from France. J Infect Dis 204: 1146–1147 [DOI] [PubMed] [Google Scholar]
- Rolain J-M, Diene SM, Kempf M, Gimenez G, Robert C, Raoult D 2012. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan-drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob Agents Chemother 57: 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche WA, Foster PL 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NJ, Trivedi UH, Thomson ML, Doig C, Laurenson IF, Blaxter ML 2011. Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background. J Infect 62: 212–217 [DOI] [PubMed] [Google Scholar]
- Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, NISC Comparative Sequence Program, Murray PR, Segre JA 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci 108: 13758–13763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Luo T, Yang C, Dong X, Li J, Zhu Y, Zheng H, Tian W, Wang S, Barry CE, et al. 2012. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis 206: 1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia R, Arroyo LA, Conde M, Aldana JM, Torres M, Fernández-Cuenca F, Garnacho-Montero J, Cisneros JM, Ortíz C, Pachón J, et al. 2009. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol 30: 257–263 [DOI] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460: 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]