Abstract
Despite increasing concerns over inappropriate use of antibiotics in medicine and food production, population-level resistance transfer into the human gut microbiota has not been demonstrated beyond individual case studies. To determine the “antibiotic resistance potential” for entire microbial communities, we employ metagenomic data and quantify the totality of known resistance genes in each community (its resistome) for 68 classes and subclasses of antibiotics. In 252 fecal metagenomes from three countries, we show that the most abundant resistance determinants are those for antibiotics also used in animals and for antibiotics that have been available longer. Resistance genes are also more abundant in samples from Spain, Italy, and France than from Denmark, the United States, or Japan. Where comparable country-level data on antibiotic use in both humans and animals are available, differences in these statistics match the observed resistance potential differences. The results are robust over time as the antibiotic resistance determinants of individuals persist in the human gut flora for at least a year.
When exposed to antibiotic compounds, bacteria evolve resistance mechanisms. These include polymorphisms in antibiotic targets that reduce vulnerability, as well as genes encoding efflux systems, drug modifiers, or proteins that fortify target sites (Wright 2007; Davies and Davies 2010). Resistance determinants can be transferred via mobile genetic elements, such as plasmids, prophages, or transposons, allowing horizontal transfer within and between bacterial species (Davies and Davies 2010), particularly in environments such as the gut microbiome (Salyers et al. 2004; Schjørring and Krogfeldt 2011; Smillie et al. 2011), and have collectively been dubbed the antibiotic resistome (Wright 2007; Marshall and Levy 2011). The transfer of resistance genes into the gut can come from diverse environments, for example, from soil bacteria (Forsberg et al. 2012). Previous studies have explored the pig gut resistome (Looft et al. 2012), as well as that of two human donors (Sommer et al. 2009), but population-scale studies are still lacking. Since antibiotics are widely used in medicine (Goossens et al. 2005) and food production (Barton 2000; Davies and Davies 2010; Marshall and Levy 2011; Aarestrup 2012), understanding the variation of the resistome within the population is crucial.
Results
To assess the resistome of an individual or a group of individuals, we collected 380 known antibiotic resistance determinants (Liu and Pop 2009) for 68 broad or narrow classes and subclasses (which share chemical structure and resistance mechanisms) (see Supplemental Methods) of antibiotics, accounted for known cross-resistances (Supplemental Table S1; Supplemental Fig. S1), and identified homologs in public metagenomes derived from 252 fecal samples of 207 individuals from three countries (6.37 Gbp, per individual on average; see Methods). To compare their relative abundances across samples, we down-sampled each individual to 726 Mbp, the size of the smallest sample, and found resistance genes for 50 of the 68 antibiotic classes and subclasses in our cohort (Fig. 1A; Supplemental Fig. S2), at an average of 21 per sample.
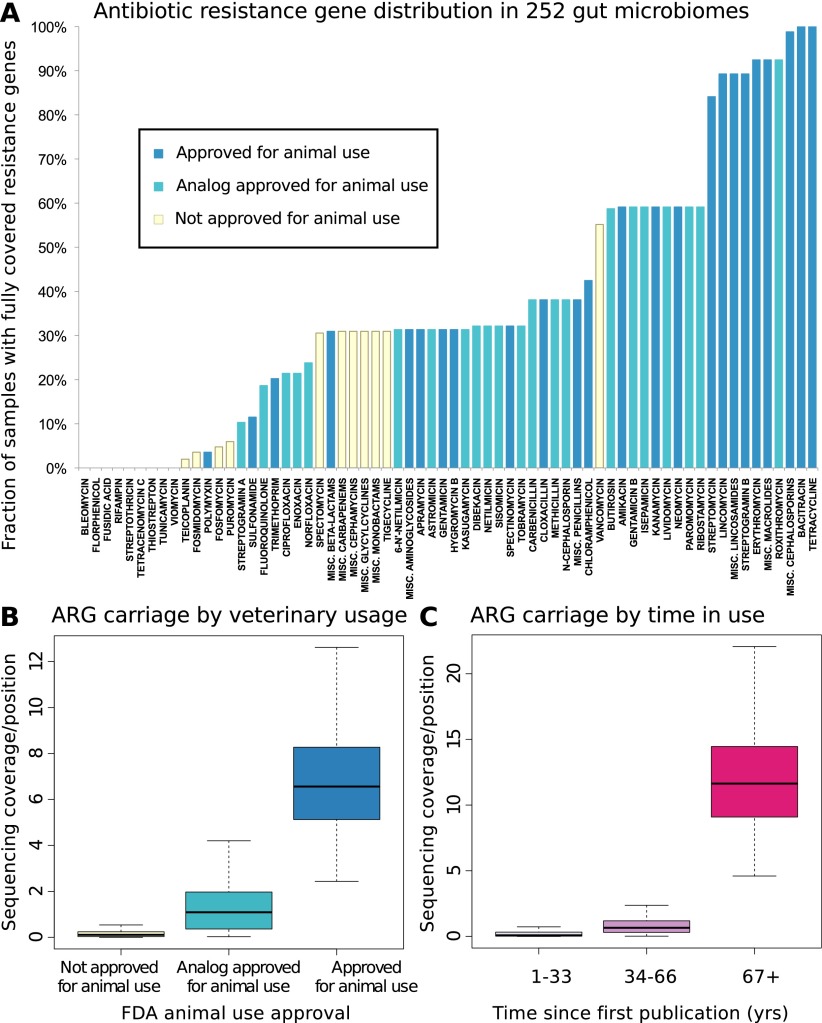
Figure 1.
(A) Resistance gene penetration is higher for antibiotics approved for use in animals or with analogs that have such approval. Shown is the fraction of 252 gut samples where at least one resistance-associated gene is fully covered by sequencing, for members of 66 classes or subclasses (with the most narrow subclasses being single compounds, see Methods) of antibiotics represented in the Antibiotic Resistance Genes Database (ARDB) (Supplemental Table S1), and for which the time since introduction and animal usage approval information was available. The colors of the bars represent whether or not animal use has been approved by the U.S. FDA according to the “Green Book” database (Shields 2009), and whether or not such use is approved for any close analogs of each antibiotic. (B) Antibiotics approved for animal use have significantly (Kruskal-Wallis test for categories having same median, P < 2 × 10−16) higher resistance potential in our data set. The figure shows base coverage per site for resistance genes assigned to categories based on animal use approval. To control for different numbers of known resistance genes targeting each antibiotic (Supplemental Table S1), the average over all resistance gene families are taken. The box plots represent the 252 Illumina samples. (C) Antibiotics that have been longer in use have significantly (Kruskal-Wallis test for categories having same median, P < 2 × 10−16) higher resistance potential in our data set. The figure shows base coverage per site for resistance genes assigned to categories based on how long the antibiotics they protect against have been in use, estimated from the time since first publication for each compound. If an antibiotic has analogs, the age of the oldest analog is used to account for cross-resistances (Supplemental Table S1). The box plots represent the 252 Illumina samples.
We first analyzed some general trends such as the connection between use in animal husbandry and the spread of resistance, previously suggested from studies of one or a few antibiotics at a time (Levy et al. 1976; Holmberg et al. 1984; Hummel et al. 1986; Bager et al. 1997; Fey et al. 2000). We observe a clear and significant increase in resistance gene abundance both for antibiotics approved for animal use by the U.S. FDA (Fig. 1B) and for older antibiotics that have been longer in the market (Kruskal-Wallis P < 2.2 × 10−16 in both cases) (Fig. 1C). These effects are independent (Supplemental Fig. S3) and hold even when controlling for differences in number of genes active against each antibiotic class or subclass (Supplemental Figs. S4, S5), as well as separately for different countries (Supplemental Fig. S6) and at varying degrees of down-sampling (Supplemental Fig. S7). Thus, we conclude that the use of antibiotics in animals contributes to resistance development in human commensal bacteria.
To screen the cohort for differences in resistance adaptations between gut samples from different countries, we developed a measure for the “antibiotic resistance potential” of a microbial community based on the abundance of its resistance genes relative to its species composition (see Methods). We then determined the antibiotic resistance potential in 142 American (The Human Microbiome Project Consortium 2012; Schloissnig et al. 2012), 39 Spanish (Qin et al. 2010), and 71 Danish (Qin et al. 2010) samples that were previously Illumina-sequenced and tested for influence of possible confounding factors (disease status, gender, enterotype [Arumugam et al. 2011], BMI, or geographic origin of samples) (see Supplemental Figs. S8–S12), but we only found an effect of enterotypes, for which we controlled (see Methods). We tested each antibiotic for significant country differences in resistance potential at a false-discovery rate (FDR) of 5%, using an FDR correction robust to the interdependencies of the resistance potentials (Benjamini and Yekutieli 2001). In almost every such case, the Spanish samples exhibit both higher antibiotic resistance gene penetration in the population and higher relative resistance potential (Fig. 2A) than either the Danish or U.S. samples. The U.S. and Danish samples are similar, though the U.S. individuals have somewhat higher resistance potentials for macrolides, lincosamides, and streptogramins, common agricultural growth promoters (Barton 2000), as well as for cephalosporins, which are used prophylactically in food animals. The Danish antibiotic resistome, on the other hand, has a relative bias toward bacitracin and vancomycin and to a lesser extent toward streptomycin, spectomycin, and chloramphenicol. Notably, a vancomycin analog (avoparcin) has been previously administered to animals in Europe but not in the United States (Barton 2000), and was subsequently banned as its use was linked to a rapid European increase in vancomycin-resistant enterococci (VRE) (Kjerulf et al. 1996; Bager et al. 1997; Barton 2000; Aarestrup 2012). The presence of VRE in Danish fecal samples has previously been linked to imported turkey meat (Agersø et al. 2008). The resistance determinant (vanA) reported in that study was not found in any of the samples analyzed in the present work, but instead other vancomycin resistance gene families were detected, with the genes of the vanG vancomycin resistance operon most often being enriched. We also verified that these trends hold independently of resistance mechanism and found that resistance genes that can increase cross-protection within the community through antibiotic modification shows the most extreme country bias (Fig. 2B; Supplemental Figs. S13–S15).
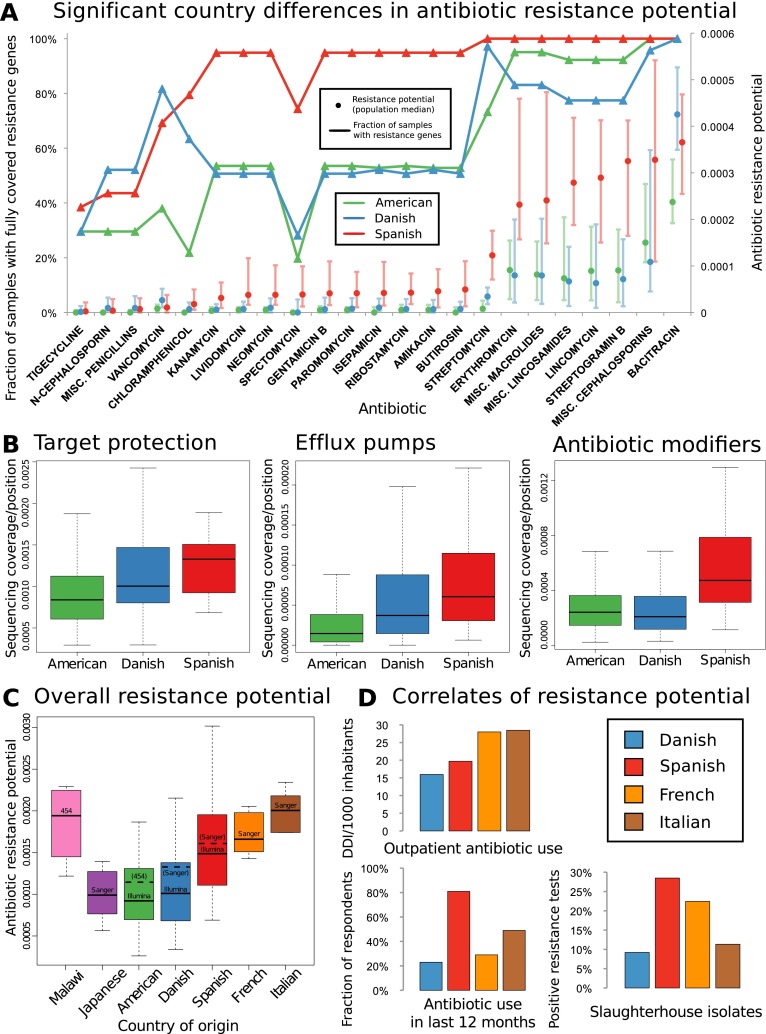
Figure 2.
(A) Geographic differences in antibiotic resistance potential. For several antibiotics, strong and significant country differences in the respective resistance gene penetration and taxonomy-adjusted resistance potential are observed, whereby mostly those of Spanish individuals are higher than those of U.S. or Danish individuals. Antibiotics with significantly different resistance distribution between Danish (N = 71), Spanish (N = 39), and American (U.S.) (N = 142) samples are displayed, with general resistance to broad classes represented by including “miscellaneous” or “generic” members of those classes subject only to resistance from the genes with the more general annotation (see Methods). To account for cross-resistances, a multiple testing correction procedure was chosen which does not assume independence between the resistance potentials of different antibiotics (Benjamini and Yekutieli 2001). Most antibiotics that show significant country differences are approved for animal use or have analogs that are, although this is not a significant enrichment over the full set of antibiotics. All samples were stochastically down-sampled to the size of the smallest sample (∼726 Mbp) prior to the analysis. The lines/triangle markers represent the fraction of samples from each country where at least one resistance gene is fully covered by sequencing. The dot/bar markers represent median and 25%/75% quartiles for resistance potential, measured as the total resistance gene abundance for each antibiotic relative to the amount of genetic material in each sample that comes from species where any resistance genes are found. (B) Significant country differences are seen separately for antibiotic resistance genes acting by different mechanisms (Kruskal-Wallis test for countries having same median, P [drug modification] < 1.98 × 10−8, P [efflux] < 6.68 × 10−9, P [target protection] < 4.17 × 10−5). The figures show base coverage per site for resistance genes assigned to categories based on whether they operate by modifying antibiotic molecules, protecting cellular target sites, or exporting antibiotic molecules from bacterial cells. The average is taken over the resistance genes in each category, with the boxes representing the 142 American, 71 Danish, and 39 Spanish samples, respectively. (C) The higher resistance potential in Spanish than in Danish samples is also seen in other samples from southern Europe (France, Italy). The distributions are significantly different between these four countries (Kruskal-Wallis P < 1.07 × 10−5). Results are broadly in agreement between different sequencing platforms (see Supplemental Text). The samples were stochastically down-sampled to 50 Mbp prior to the analysis. (D) Gut resistance potential coincides with antibiotic exposure when comparing Denmark with southern Europe. The bar charts show comparative statistics from the literature: outpatient antibiotic consumption in 2008 from the same four countries (European Surveillance of Antimicrobial Consumption [ESAC] survey) (Goossens et al. 2005) measured in defined daily doses (DDDs) per 1000 inhabitants, frequency of antibiotic resistance in bacterial isolates from slaughterhouses in a 2011 comparative study (de Jong et al. 2012), and fraction of approximately 1000 respondents in each country that had taken antibiotics during the last 12 mo (Borg 2012). Resistance potential correlates significantly with outpatient antibiotic use (Pearson r = 0.97; t-test [N = 4, df = 2]; Bonferroni-corrected P < 0.08).
To better understand the observed country differences, we included additional smaller metagenomes from other European countries. Since these were generated with different sequencing technologies, and also to allow testing of the robustness of the results, we down-sampled the Illumina data further and included additional Sanger or 454 Life Sciences (Roche) sequencing data sets from all three countries analyzed above (Fig. 2C; Supplemental Fig. S16; Arumugam et al. 2011; Yatsunenko et al. 2012). Simulations show that neither down-sampling to 50 Mbp per sample nor reducing the number of samples to four eliminates the country differences we see (Supplemental Figs. S17–S19). Thus, at the price of lower resolution, we could include smaller metagenomes from individuals of several other countries (Fig. 2C; Supplemental Fig. S16) and found that individuals from two additional southern European countries, Italy and France, have resistance potentials comparable to those of Spanish individuals (Fig. 2C).
A linkage of fecal carriage of resistant bacteria to human antibiotic consumption has been demonstrated (Levy et al. 1988; Barbosa and Levy 2000), and previous comparative studies have also revealed higher human antibiotic consumption in southern Europe than in Denmark (Fig. 2D, top and bottom left; Goossens et al. 2005; Borg 2012), as well as higher proportions of resistant bacteria isolated from food-producing animals (Fig. 2D, bottom right; de Jong et al. 2012), suggesting greater use of antibiotics in food production in southern Europe (fair comparisons are presently only possible within Europe as there are no antibiotic use or sales statistics available that were compiled in the same way there and in the United States). Each of these country-level measures of antibiotic exposure is positively correlated with the median resistance potential of samples from these countries, and despite data being available only for four countries, the correlation between resistance potential and total outpatient antibiotic use is significant (Pearson r = 0.97, Bonferroni-corrected P < 0.08).
To further investigate the effect of agricultural use of antibiotics on the human gut resistance potential, we collected data on 2009 veterinary antibiotic sales in Denmark and Spain and normalized by livestock biomass (Supplemental Table S2). We then modeled resistance potential using a mixed-effects framework. Biomass-normalized antibiotic exposure significantly (P < 0.023) increases median resistance potential across the 11 antibiotic classes tested in this manner (Supplemental Fig. S20).
The higher transmission of resistance genes for antibiotics used in food-producing animals in Mediterranean countries might also be linked to diet as it has been reported that raw or cured food products, common in southern Europe (aged cheeses, cured sausages, olive fermentations), can carry high bacterial loads (Franz et al. 2011). Indeed, we find that resistance was detected in fermented food products (see Supplemental Information) for some of the antibiotics where the resistance potential also differs most between countries.
Although based on smaller data sets, generated using different sequencing technologies, we also measured the antibiotic resistance potential in Japanese and Malawian individuals (Fig. 2C; Supplemental Fig. S16). The Japanese cohort (Kurokawa et al. 2007) has an overall antibiotic resistance potential similar to that of U.S. and Danish donors, whereas the Malawian cohort (Yatsunenko et al. 2012), representing a rural population in a nonindustrialized nation, has an even higher overall resistance potential than those of southern European countries. The Malawian samples are highly skewed toward cephalosporin and tetracycline resistance genes (data not shown) when compared to the rest of the data set. This might indicate extensive use of old, broad-spectrum antibiotics, a known problem in many developing countries (Hart and Kariuki 1998; Nordberg et al. 2005), although this hypothesis would need to be further validated with more samples and antibiotic consumption information.
To assess persistence of individual resistomes over time, we utilized a subset of 43 U.S. samples (The Human Microbiome Project Consortium 2012) taken at two or three separate time points from each donor. Samples from the same individual are, on average, more similar in their antibiotic resistance potential than samples from different individuals, and this similarity does not decrease noticeably with time (Fig. 3). This is consistent with earlier research on individual antibiotics (Shoemaker et al. 2001; Salyers et al. 2004; Johnsen et al. 2011) showing that resistance determinants, once introduced into the microbial flora, can persist for a long time at low abundances, which might also explain the high vancomycin resistance potential in the Danish population despite its animal-use analog being banned since 1995 (Aarestrup 2012).
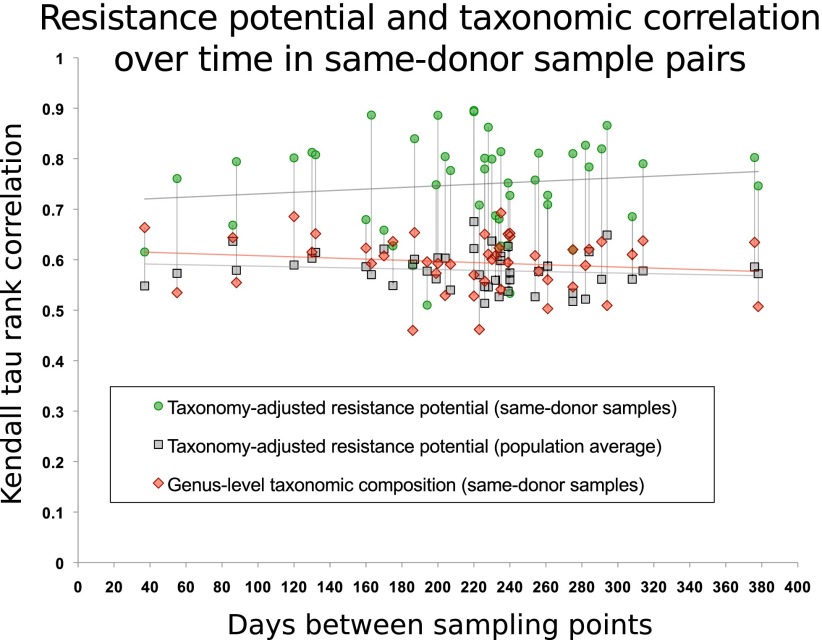
Figure 3.
Antibiotic resistance potential persists over time in the human gut flora for at least a year. For 43 U.S. individuals, two or three time points were sampled (The Human Microbiome Project Consortium 2012). Each interval (measured in days) between two sampled time points is represented by a green dot. For each such pair of samples, they are compared with respect to carriage of 99 resistance gene families from ARDB that are detectable in the gut samples, using the Kendall τ correlation coefficient, following compensation for sequencing depth and differences in species composition. The gray dots represent the average correlation between either of the two samples and the remaining 137 HMP samples in the data set. Red markers show the Kendall τ correlation coefficient for genus-level taxonomic composition profiles. Vertical lines connect values for each sample. Linear regression of similarity of same-donor sample pairs with respect to the time interval yields no notable decrease in resistance potential similarity within the time spans considered here (R2 < 0.015). Previous studies (Costello et al. 2009; The Human Microbiome Project Consortium 2012) have shown that the composition of a person's gut microbiome as a whole is self-similar during a year compared with that of other people. In almost every case, resistance potentials are better conserved than overall taxonomic composition.
We did not discriminate here between pathogens and commensals but rather considered the gut microbiome in its entirety and as a common antibiotic resistance reservoir (Barton 2000; Goossens et al. 2005), which potentially enables pathogens to become resistant through lateral gene transfer or enables rare resistant commensals or opportunistic pathogens to gain dominance in the case of antibiotic treatment. This can disturb normal gut function, such as in antibiotic-associated diarrhea and Clostridium difficile–associated symptoms (Gerding 2004). As our approach characterizes the whole of this reservoir, it might thus also allow making individualized recommendations on which antibiotics to avoid. For many antibiotics, some individuals have extremely high resistance potentials. For example, for vancomycin, tigecycline, chloramphenicol, and several aminoglycosides, more than 20 individuals in our data set have a resistance potential 10-fold higher than the population median, which might indicate the presence of resistant strains which in turn could transfer this resistance further upon antibiotic exposure of the community (Supplemental Table S3). One U.S. individual has on average more than 30 times the population median resistance potential for nine aminoglycosides due to antibiotic modification systems. Another extreme case is a Spanish individual with more than 30 times the population median resistance potential for cephalosporins and more than 100 times the population median resistance potential for macrolides, lincosamides, and streptogramin B. While our sequence-based analysis method only captures the potential for antibiotic resistance, studies show high correlation between the presence of resistance determinants and results of in vitro susceptibility tests (Martineau et al. 2000; Zhu et al. 2007; Eitel et al. 2012), suggesting clinical utility of these findings.
Discussion
We have measured the content and variation of the human gut antibiotic resistome across large-scale data sets from different populations. While our analysis can detect only the potential for resistance stemming from previously characterized resistance determinants, and only to the extent that the target spectrum of each such gene is correctly annotated, it is based on a comprehensive collection of resistance gene families and therefore at least unlikely to miss any major trends.
Comparing different antibiotics, we have shown that resistance gene abundance and penetration on average are higher for drugs used in animals, even when compensating for differences in how many resistance genes are known. This is consistent with expectations from previous research into a “farm-to-fork” connection (Marshall and Levy 2011).
By analyzing a large cohort of individuals, we can compare their resistomes and point to resistance potentials elevated beyond what is expected from the microbial community composition. The determination of individual profiles of resistance potentials, which can be increased for particular antibiotics, may eventually find use in clinical settings.
Beyond individual variation, we see robust differences in gut resistance potential between countries. Presently available data sets lack individual-level metadata on diet or antibiotic use history, and so the causes for these differences cannot yet be conclusively determined. However, we note that the observed differences largely coincide in scope and direction with country-level statistics of veterinary and human antibiotic use, for countries where comparable statistics are available. While future studies need to quantify the individual contributing factors, the causes suggested by this and previous studies are all, in principle, open to intervention: policies, use practices, or food consumption habits, whereby it remains to be seen whether the observed persistence over time of the already acquired individual resistomes can be overcome too.
This outcome of our global, metagenomic-based approach, mapping variation within and between populations and covering a vast range of antibiotics, should provide a profound molecular basis for the ongoing debate on the appropriate use of antibiotics in agriculture and medicine. While interpretation of our results is limited by availability of comparable antibiotic exposure data on both an individual and a national level, we are confident that further efforts in large-scale antibiotic resistance surveillance and molecular characterization of resistance genes will eventually allow the resistance potentials introduced in this study to be measured at an increased resolution.
Methods
Stool samples were previously taken from volunteers as part of the MetaHIT (71 Danish and 39 Spanish samples) (Qin et al. 2010) and HMP (139 U.S. samples) (The Human Microbiome Project Consortium 2012) projects, with an additional three U.S. samples from another project (Schloissnig et al. 2012), and sequenced as part of other projects using Illumina technology (Supplemental Table S4). A collection of 3496 fully sequenced reference genomes was compiled (Supplemental Table S5), and a catalog of gut metagenomic reference genes was generated from the stool samples. The MOCAT pipeline (Kultima et al. 2012) was used to map reads to the set of reference genes, producing gene abundance estimates (Supplemental Table S6). Simulations were carried out to verify that no artifacts were introduced by this pipeline (Supplemental Table S7). Species composition of the samples was estimated using a curated set of 40 marker genes that are usually found in single copy in microbial genomes (Supplemental Table S8). Samples were stochastically down-sampled by randomly discarding sequenced bases in short read-sized blocks. Previously described Sanger and 454 Life Sciences (Roche) samples (Kurokawa et al. 2007; Arumugam et al. 2011; Yatsunenko et al. 2012) were converted into the same short-read format as the larger data set, and all samples were then stochastically down-sampled to a common size of 50 Mbp, before repeating the analysis.
To identify antibiotic resistance determinants, the ARDB reference protein collection (Liu and Pop 2009) was augmented with homologs from the 3496 reference genomes, using family-specific inclusion thresholds and, where available, gene symbol annotation. These sequences were used to search the gut metagenomic reference gene catalog to yield a set of resistance genes present in the gut samples (Supplemental Tables S9, S10). These were further curated by searching also the NCBI nonredundant database to verify the top annotated matches were matched to the correct resistance gene families. For each resistance gene family, its distribution among the reference genomes was determined (Supplemental Tables S11, S12).
The resistance potential was defined as the average genome fraction encoding resistance genes for a particular antibiotic or category of antibiotics, across all bacteria in a sample that could potentially carry such resistance genes, based on the known taxonomic ranges of these resistance gene families. This potential was determined from the total number of bases sequenced from these genes and species, respectively. To compensate for stochastic effects in the case of very small abundances of applicable species, the resistance gene base count was down-sampled, separately for each antibiotic, by randomly removing average read-sized blocks of bases such that all samples analyzed have the same resulting proportion of bases mapping to the carrier species. The ratio of the down-sampled resistance gene base count to this “target number” of bases from potentially resistance-carrying species was taken as the final resistance potential. Results were similar if raw ratios were used instead of down-sampled (Supplemental Fig. S12). By measuring resistance gene abundances relative to the pooled abundances of potential carrier species, this taxonomy-adjusted resistance potential aims to broadly disentangle, to the extent that current metagenomic data allows, the effects of selection toward higher relative prevalence of resistance determinants from the effects of broad changes in gut microbial species composition. Simulations show that a method not considering taxonomic composition in this manner will falsely conclude country differences, whereas our resistance potential analysis will not (Supplemental Table S13). The term “resistance potential,” rather than simply “resistance,” was chosen to reflect how differences in resistance gene expression and regulation also affect phenotypic resistance in vivo, in addition to the role the resistome plays.
Significant country differences in antibiotic resistance were concluded if a likelihood-ratio test comparing a linear model of the resistance potential with country and enterotype as independent variables with a null model omitting country achieved Benjamini-Hochberg-Yekutieli FDR ≤ 5%.
Acknowledgments
We thank Julien Tap for assistance with sample metadata, Matt Rogon for help with graphics, and members of the Bork group for valuable suggestions and comments. K.F. was supported by the CancerBiome (ERC project reference 268985) project and by the Alexander von Humboldt Foundation. The project was also supported by the European Commission 7th Framework Programme (International Human Microbiome Standards grant, HEALTH-F4-2010-261376).
Author contributions: P.B. proposed and K.F., M.A., and P.B. conceived the study. S.S. and D.M. compiled the reference genome collection, and D.M. carried out simulations to test the specificity of the mapping pipeline. S.S. and J.R.K. compiled the reference gene catalog and calculated the gene and taxa abundances. K.F. developed the down-sampling and antibiotic resistance potential analysis procedures, compiled all antibiotic resistance and use data, and performed the main analysis, the control experiments, and all statistical tests. K.F. and P.B. interpreted the results with help from A.T. and wrote the manuscript with assistance from A.T., S.S., J.R.K., and M.A. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.155465.113.
References
- Aarestrup F 2012. Sustainable farming: Get pigs off antibiotics. Nature 486: 465–466 [DOI] [PubMed] [Google Scholar]
- Agersø Y, Lester CH, Porsbo LJ, Orsted I, Emborg H-D, Olsen KEP, Jensen LB, Heuer OE, Frimodt-Møller N, Aarestrup FM, et al. 2008. Vancomycin-resistant Enterococcus faecalis isolates from a Danish patient and two healthy human volunteers are possibly related to isolates from imported turkey meat. J Antimicrob Chemother 62: 844–845 [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager F, Madsen M, Christensen J, Aarestrup FM 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med 31: 95–112 [DOI] [PubMed] [Google Scholar]
- Barbosa TM, Levy SB 2000. The impact of antibiotic use on resistance development and persistence. Drug Resist Updat 3: 303–311 [DOI] [PubMed] [Google Scholar]
- Barton MD 2000. Antibiotic use in animal feed and its impact on human health. Nutr Res Rev 13: 279–299 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Borg MA 2012. National cultural dimensions as drivers of inappropriate ambulatory care consumption of antibiotics in Europe and their relevance to awareness campaigns. J Antimicrob Chemother 67: 763–767 [DOI] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Davies D 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74: 417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A, Thomas V, Simjee S, Godinho K, Schiessl B, Klein U, Butty P, Vallé M, Marion H, Shryock TR 2012. Pan-European monitoring of susceptibility to human-use antimicrobial agents in enteric bacteria isolated from healthy food-producing animals. J Antimicrob Chemother 67: 638–651 [DOI] [PubMed] [Google Scholar]
- Eitel Z, Soki J, Urban E, Nagy E 2012. Investigation of the main antibiotic resistances and their correlation with the presence of antibiotic resistance genes in clinical Bacteroides strains. In The 11th Biennial Congress of the Anaerobe Society of the Americas, p. 163. Anaerobe Society of the Americas, San Francisco, CA [Google Scholar]
- Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradford PA, Angulo FJ, Hinrichs SH 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med 342: 1242–1249 [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CMAP, Huch M, Abriouel H, Holzapfel W, Gálvez A 2011. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151: 125–140 [DOI] [PubMed] [Google Scholar]
- Gerding DN 2004. Clindamycin, cephalosporins, fluoroquinolones, and Clostridium difficile-associated diarrhea: This is an antimicrobial resistance problem. Clin Infect Dis 38: 646–648 [DOI] [PubMed] [Google Scholar]
- Goossens H, Ferech M, Stichele RV, Elseviers M, Group ESACP 2005. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 365: 579–587 [DOI] [PubMed] [Google Scholar]
- Hart CA, Kariuki S 1998. Antimicrobial resistance in developing countries. BMJ 317: 647–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg SD, Wells JG, Cohen ML 1984. Animal-to-man transmission of antimicrobial-resistant Salmonella: Investigations of U.S. outbreaks, 1971–1983. Science 225: 833–835 [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel R, Tschäpe H, Witte W 1986. Spread of plasmid-mediated nourseothricin resistance due to antibiotic use in animal husbandry. J Basic Microbiol 26: 461–466 [DOI] [PubMed] [Google Scholar]
- Johnsen PJ, Townsend JP, Bøhn T, Simonsen GS, Sundsfjord A, Nielsen KM 2011. Retrospective evidence for a biological cost of vancomycin resistance determinants in the absence of glycopeptide selective pressures. J Antimicrob Chemother 66: 608–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjerulf A, Pallesen L, Westh H 1996. Vancomycin-resistant enterococci at a large university hospital in Denmark. APMIS 104: 475–479 [DOI] [PubMed] [Google Scholar]
- Kultima JR, Sunagawa S, Li J, Chen W, Chen H, Mende DR, Arumugam M, Pan Q, Liu B, Qin J, et al. 2012. MOCAT: A metagenomics assembly and gene prediction toolkit. PLoS ONE 7: e47656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14: 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, FitzGerald G, Macone A 1976. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N Engl J Med 295: 583–588 [DOI] [PubMed] [Google Scholar]
- Levy SB, Marshall B, Schluederberg S, Rowse D, Davis J 1988. High frequency of antimicrobial resistance in human fecal flora. Antimicrob Agents Chemother 32: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Pop M 2009. ARDB: Antibiotic Resistance Genes Database. Nucleic Acids Res 37: D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, et al. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci 109: 1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BM, Levy SB 2011. Food animals and antimicrobials: Impacts on human health. Clin Microbiol Rev 24: 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau F, Picard FJ, Lansac N, Menard C, Roy PH, Ouellette M, Bergeron MG 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 44: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg P, Stålsby-Ludborg C, Tomson G 2005. Consumers and providers—could they make better use of antibiotics? Int J Risk Saf Med 17: 117–125 [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Gupta A, Wang Y 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12: 412–416 [DOI] [PubMed] [Google Scholar]
- Schjørring S, Krogfeldt KA 2011. Assessment of bacterial antibiotic resistance transfer in the gut. Int J Microbiol 2011: 312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. 2012. Genomic variation landscape of the human gut microbiome. Nature 493: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields T 2009. Animal drugs @ FDA. J Med Libr Assoc 97: 158 [Google Scholar]
- Shoemaker NB, Vlamakis H, Hayes K, Salyers AA 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol 67: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480: 241–244 [DOI] [PubMed] [Google Scholar]
- Sommer MOA, Dantas G, Church GM 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325: 1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GD 2007. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat Rev Microbiol 5: 175–186 [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486: 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LX, Zhang ZW, Wang C, Yang HW, Jiang D, Zhang Q, Mitchelson K, Cheng J 2007. Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among staphylococcal clinical isolates. J Clin Microbiol 45: 3514–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]