Abstract
Chromosomal translocations are signatures of numerous cancers and lead to expression of fusion genes that act as oncogenes. The wealth of genomic aberrations found in cancer, however, makes it challenging to assign a specific phenotypic change to a specific aberration. In this study, we set out to use genome editing with zinc finger (ZFN) and transcription activator-like effector (TALEN) nucleases to engineer, de novo, translocation-associated oncogenes at cognate endogenous loci in human cells. Using ZFNs and TALENs designed to cut precisely at relevant translocation breakpoints, we induced cancer-relevant t(11;22)(q24;q12) and t(2;5)(p23;q35) translocations found in Ewing sarcoma and anaplastic large cell lymphoma (ALCL), respectively. We recovered both translocations with high efficiency, resulting in the expression of the EWSR1–FLI1 and NPM1–ALK fusions. Breakpoint junctions recovered after ZFN cleavage in human embryonic stem (ES) cell–derived mesenchymal precursor cells fully recapitulated the genomic characteristics found in tumor cells from Ewing sarcoma patients. This approach with tailored nucleases demonstrates that expression of fusion genes found in cancer cells can be induced from the native promoter, allowing interrogation of both the underlying mechanisms and oncogenic consequences of tumor-related translocations in human cells. With an analogous strategy, the ALCL translocation was reverted in a patient cell line to restore the integrity of the two participating chromosomes, further expanding the repertoire of genomic rearrangements that can be engineered by tailored nucleases.
Despite the wide range of recurrent chromosomal translocations identified in various cancers (more than 300 genes implicated) (Mitelman et al. 2007), the direct path from translocation formation to tumorigenesis is not always clear. In mouse and human cells, studies are mostly based on either ectopically expressing or silencing the fusion gene induced by the translocation. In the case of fusion protein expression from a cDNA (often randomly integrated into the genome), the choice of the fusion transgenic promoter is crucial because the level of fusion protein expression must often be tightly controlled to recapitulate endogenous levels or risk spurious results from overexpression. And in silencing strategies, even low levels of expression remaining for the fusion protein could mask to some extent the full cellular consequences of the translocation.
DNA double-strand breaks (DSBs) are considered to be causative lesions for many genomic rearrangements, including chromosomal translocations (Richardson and Jasin 2000; Mani and Chinnaiyan 2010). With the development of tailored endonucleases like zinc finger nucleases (ZFNs) (Urnov et al. 2010; Carroll 2011) and more recently transcription activator-like effector nucleases (TALENs) (Doyon et al. 2011; Mussolino and Cathomen 2012), it is now possible to create a DSB in the genome of a human cell at any locus of interest for the purposes of gene correction and gene disruption. In addition, contemporaneous expression of two ZFNs targeting loci has led to the induction of translocations at model loci in human multipotent and stem cells (Brunet et al. 2009) and intrachromosomal rearrangements (e.g., deletions) in transformed cell lines (Lee et al. 2010, 2012). This approach to study translocation formation obviates the need for prior genetic manipulation or cloning of cells, significantly expanding the repertoire of human cells that can be interrogated for translocation formation.
In this study, we now investigate the formation of two specific translocations, one frequently observed in Ewing sarcoma and one found in anaplastic large cell lymphoma (ALCL) using both types of nucleases (ZFNs and TALENs). Ewing sarcoma is a prototype of a solid tumor carrying a specific chromosomal translocation; it enables the transcription of the EWSR1–FLI1 chimeric protein corresponding to the in-frame fusion of the EWSR1 amino terminus with the FLI1 carboxyl terminus. It is well accepted that the EWSR1–FLI1 fusion protein acts as a transcriptional factor, but target genes induced or repressed by the fusion protein are not fully identified yet (Chansky et al. 2004; Prieur et al. 2004; Smith et al. 2006; Riggi et al. 2010). ALCL is an aggressive T-cell non-Hodgkin lymphoma, accounting for as much as 10%–15% of children with the disease. About half of tumors exhibits the specific translocation t(2;5)(p23;q35) resulting in NPM1–ALK expression and constitutive ALK tyrosine kinase activity (Morris et al. 1994; Elmberger et al. 1995; Kuefer et al. 1997).
Results
Inducing Ewing sarcoma specific translocations with ZFNs
To target reported Ewing sarcoma breakpoints, two ZFN pairs were designed within the EWSR1 and FLI1 genes on chromosomes 22 and 11, respectively, to induce t(11;22)(q24;q12) translocations (Fig. 1A). ZFNEWS targets EWSR1 intron 7 and ZFNFLI targets FLI1 intron 5 (Fig. 1B), which contain breakpoints for the most common type of EWSR1–FLI1 translocation (Plougastel et al. 1993). In particular, the ZFNs target sequences at breakpoint junctions reported in two tumors, T60 for EWSR1 and T64 for FLI1 (Supplemental Fig. S1; Zucman-Rossi et al. 1998).
Figure 1.
Induction of t(11;22)(q24;q12) translocations in hES-MP cells with ZFNs. (A) Ewing sarcoma translocations involve breakpoints within the EWSR1 and FLI1 genes on chromosomes 22 and 11, respectively, creating an EWSR1–FLI1 fusion gene on der(22). To induce t(11;22)(q24;q12), ZFNs are expressed in hES-MP cells to create DSBs (scissors) in both genes. (B) ZFNEWS and ZFNFLI cleavage sites are within EWSR1 and FLI1 introns, respectively, relevant to the EWSR1–FLI1 translocation. Zinc fingers in the ZFNs are designed to bind to the shaded sequences. (Arrows) The presumed DSB sites after FokI nuclease domain cleavage. ZFN cleavage activity in hES-MP cells is monitored by a T7-endonuclease assay (Guschin et al. 2010). The region around the ZFN site is amplified; the amplified product is then denatured, reannealed, and then subjected to T7 endonuclease cleavage. Insertions and deletions (indels) characteristic of imprecise DSB repair by NHEJ give rise to T7 endonuclease-cleavable DNA. (C) Nested PCR to detect derivative chromosomes der(11) and der(22) in hES-MP cells. Translocation breakpoint junctions are only detected after expression of both ZFNEWS and ZFNFLI. (D) RT-PCR detection of the EWSR1–FLI1 fusion transcript after ZFNEWS and ZFNFLI expression in hES-MP cells. The forward primer overlaps the exon 2/3 junction of EWSR1, and the reverse primer is within exon 9 of FLI1, amplifying most of the EWSR1–FLI1 coding sequence.
We chose to test this system in human mesenchymal precursor cells, in particular, those derived from human embryonic stem cells (hES-MP) (Barberi et al. 2005), because of the presumed mesenchymal origin of this sarcoma (Tirode et al. 2007; Riggi et al. 2008). Both ZFNs efficiently generated DSBs at the respective target sites in these cells, as evidenced by the frequent formation of indels (Fig. 1B). To detect translocations, we used a nested PCR strategy on genomic DNA from cells transfected with the ZFN expression vectors (Fig. 1C). Translocation breakpoint junctions for both derivative chromosomes, der(11) and der(22), were detected upon expression of both ZFNEWS and ZFNFLI (Fig. 1C). Expression of a single ZFN, however, was not sufficient to give rise to translocations. We next asked whether the EWSR1–FLI1 fusion transcript was expressed from der(22). Strikingly, we could detect the EWSR1–FLI1 fusion transcript by RT-PCR of RNA extracted from the bulk hES-MP cell population treated with ZFNEWS and ZFNFLI, but not from treatment with either ZFNEWS or ZFNFLI alone (Fig. 1D).
The detection of EWSR1–FLI1 fusion transcripts from the transfected cell population suggested efficient t(11;22)(q24;q12) translocation. To quantify translocations, we applied a high-throughput 96-well PCR screen for translocation junctions using small pools of cells (∼800 cells per well) (Brunet et al. 2009). Translocation breakpoint junctions were detected in this small pool PCR at a frequency of 3.1 ± 0.7 × 10−4 for der(22) and 4.2 ± 0.45 × 10−4 for der(11). Reciprocal translocations were frequent: ∼30% of wells with one derivative chromosome also contained the other derivative chromosome. The lack of a reciprocal product in the remaining wells may indicate nonreciprocal translocation formation, as observed in a portion of tumors (Zucman-Rossi et al. 1998) or, in some cases, lack of amplification (Brunet et al. 2009).
Breakpoint junctions recovered after ZFN cleavage fully recapitulate the genomic characteristics found in Ewing tumor cells
The occurrence of translocation breakpoints within intronic sequences means that joining of DNA ends can occur with a variety of DNA end modifications without compromising the ability to express the EWSR1–FLI1 fusion protein. To gain insight into the joining mechanism, we analyzed 174 t(11;22)(q24;q12) breakpoint junctions detected by small pool PCR after ZFN expression and compared the joining characteristics (deletions, microhomology, insertions) with those from patient-derived translocations (Zucman-Rossi et al. 1998). Small deletions were observed in most ZFN-induced translocations: 74.5% of the derivative chromosomes had deletions of <30 bp total from the two DNA ends forming the junction (Fig. 2A; Supplemental Fig. S2). Exact breakpoints are not known for patient-derived events; however, for the 36 cases in which junctions from both derivative chromosomes were sequenced, the total deletion from each chromosome could be determined and was found to be similarly small, with 71% of deletions also <30 bp (Supplemental Fig. S3). Except in one tumor junction (T23 with a deletion of 8.3 kb), all deletions from patient-derived translocations were <200 bp. Thus, our PCR-based screening approach, which detects deletions of <250 bp, appears to be appropriate for detecting most translocations. Overall, no significant difference in deletion length was found between ZFN-induced and patient-derived translocations (medians of 6.5 and 19 bp, respectively, P = 0.188, Mann–Whitney test, with the 8.3-kb deletion removed from the analysis as an outlier).
Figure 2.
Analysis of t(11;22)(q24;q12) breakpoint junctions. (A) Deletion lengths for der(11) and der(22) junctions from ZFN-induced translocations from hES-MP cells. Each value represents the combined deletion from both ends of an individual junction. The median deletion length is indicated by a bar on the graph, and the value is given below the graph. (B) Microhomology length distributions for junctions from ZFN-induced and tumor translocation (Zucman-Rossi et al. 1998). [Der(11) and der(22) are grouped.] Only junctions with simple deletions (i.e., without an insertion) are included. The probability that a junction will have X nucleotides of microhomology by chance assumes an unbiased base composition and is calculated as previously described (Roth et al. 1985). (C) Examples of complex breakpoint junctions with insertions derived from sequences near the DSB sites. Schematics of the derivative and parental chromosomes are shown. The DSB sites on the unrearranged chromosomes are represented by scissors. Segments that are duplicated, deleted, or added are represented as independent blocks and in the orientation relative to that found on the parental chromosomes (see Supplemental Fig. S4 for the sequences).
Breakpoint junctions can occur at microhomologies (short sequence identities) between DNA ends more frequently than that expected by chance. The involvement of microhomology for joining is considered to be more typical of the alternative pathway of NHEJ than of the canonical pathway (Weinstock et al. 2007; Yan et al. 2007; Simsek and Jasin 2010; Simsek et al. 2011). We determined the microhomology present at t(11;22)(q24;q12) junctions from both ZFN-induced and patient-derived events and compared the microhomology distributions with that expected from chance joining of two random sequences (Fig. 2B; Supplemental Figs. S2, S3). Microhomologies of ≥3 bp were observed in only a fraction of junctions from both ZFN-induced events and tumors, although somewhat more frequently than that expected by chance. Only one particularly long microhomology of 8 bp was found in one tumor (T44) (Supplemental Fig. S3). These results imply that substantial preexisting microhomology is not required for translocation formation in human cells. Microhomology of 1 bp was often present in junctions involving the ZFN overhangs (e.g., asterisks, Supplemental Fig. S2), whereas for tumor translocation junctions, which derive from a variety of DNA breakpoint sequences and types of ends, the major class of events showed no microhomology at the junctions, as would be expected if microhomology was not driving these events. Despite these differences, overall the difference in microhomology length distributions between the ZFN-induced and patient-derived events was not significant (P = 0.076, Mann–Whitney test), supporting the ability of ZFN induction of translocations to model events seen in patients. They also point out that subtle design changes, i.e., avoiding sites where ZFN overhangs have the possibility to anneal at even 1 base, could even more faithfully recapitulate events in tumors.
As seen in some tumors from Ewing sarcoma patients (Zucman-Rossi et al. 1998), a portion of the ZFN-induced translocation breakpoints contained inserted sequences. Although many were only a few base pairs, several longer insertions were also detected that were derived from a variety of sources: nearby the DSB sites on chromosomes 11 and 22, other chromosomes, and exogenous DNA (i.e., the ZFN expression plasmids). For example, a patient der(11) junction was reported to have a 140-bp insertion from chromosome 1, with otherwise little modification to the chromosome 11 and 22 ends (T89) (Supplemental Fig. S3). Likewise, a ZFN-induced der(11) junction was identified with a 180-bp insertion from chromosome 5, also with little other modification to the DNA ends.
Furthermore, insertions were also identified in ZFN-induced translocations where different pieces of DNA were assembled and/or duplicated in a more complex way (Fig. 2C; Supplemental Fig. S4). For duplications, der(22)-D duplicated 139 bp from chromosome 11 that is derived from 363 bp downstream from the DSB, and der(11)-A and der(11)-B duplicated chromosome 22 sequences derived from upstream and downstream of the DSB, respectively. A more complicated insertion is found in der(22)-B, which has a duplication of 153 bp from chromosome 11 derived from directly upstream of the DSB, but in inverted orientation; this type of insertion has previously been observed in patient translocations and has been termed a locally derived inverted sequence (LDIS) (Zucman-Rossi et al. 1998). Der(22)-B additionally contains an insertion from the ZFN plasmid. The longest insertion, in der(22)-C, is 994 bp and is derived from four DNA segments from three different chromosomes (Supplemental Fig. S4), suggestive of iterative processing of DNA ends prior to the ligation step (Simsek and Jasin 2010).
Inducing ALCL specific translocations with TALENs
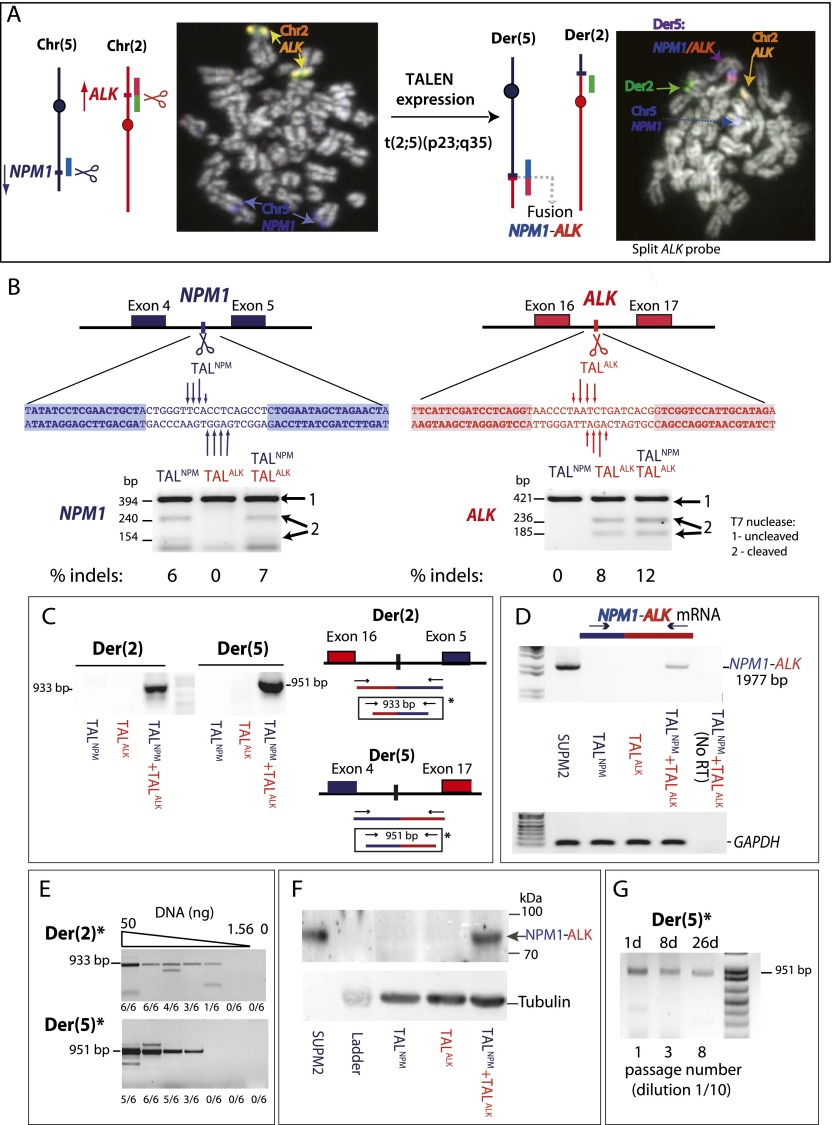
With ZFNs, we managed to recapitulate Ewing sarcoma translocations in human stem cells, but a limitation of extending this method to other translocations is that ZFN design is not trivial. The recent development of TALENs provides a potentially more advantageous strategy to generate DSBs at endogenous loci in human cells, given the simpler code of base recognition (Miller et al. 2011). We designed TALENs to target DSBs to the NPM1 and ALK genes on chromosomes 5 and 2, respectively, to induce t(2;5)(p23;q35) translocations found in ALCL patients (Fig. 3A; Morris et al. 1994). TALNPM targets NPM1 intron 4 and TALALK targets ALK intron 16, both of which contain breakpoints found in patient-derived NPM1–ALK translocations. In this manner, TALEN-induced chromosomal translocations would result in expression of the oncogenic NPM1–ALK fusion gene, preserving the N-terminal dimerization domain of NPM1 and the C-terminal cytoplasmic domain of ALK.
Figure 3.
Induction of t(2;5)(p23;q35) translocations with TALENs. (A) ALCL translocations have breakpoints within the NPM1 and ALK genes on chromosomes 2 and 5, respectively, creating an NPM1–ALK fusion gene on der(5). To induce t(2;5)(p23;q35), TALENs are expressed to create DSBs (scissors) in both genes. FISH demonstrates the t(2;5)(p23;q35) translocation after TALEN expression in RPE-1 cells. Red and green signals are from an ALK probe that “breaks apart” upon translocation. The blue signal is from an NPM1 probe. Of 70 metaphases screened, two exhibited translocations and three showed breaks with the ALK break-apart probe, likely due to remaining TALEN expression at this time. (B) TALNPM and TALALK cleavage within NPM1 and ALK introns, respectively, relevant to the NPM1–ALK translocation. DNA binding domains of TALENs are designed to bind the shaded sequences. (Arrows) DSB sites with different tail lengths representing the efficiency of cleavage in vitro, as assayed by in vitro expression of the TALENs (see Supplemental Fig. S6). TALEN cleavage activity in Jurkat cells is monitored by the T7-endonuclease assay, as shown below the sequences. (C) Nested PCR to detect derivative chromosomes der(2) and der(5) in Jurkat cells. Translocation breakpoint junctions are only detected after expression of both TALNPM and TALALK. (D) RT-PCR detection of the NPM1–ALK fusion transcript after TALNPM and TALALK expression in Jurkat cells and in ALCL cell line SUP-M2. The forward primer is within exon 2 of NPM1, and the reverse primer is within exon 29 of ALK, amplifying most of the NPM1-ALK coding sequence. (E) Single-round PCR to detect derivative chromosomes der(2) and der(5) in Jurkat cells. Translocation breakpoint junctions are detected after expression of both TALNPM and TALALK by PCR of the fragment marked (*) in C on serial dilutions of genomic DNA (50, 25, 12.5, 6.25, 3.125, and 1.56 ng). The number of times the PCR was positive for each dilution from six total experiments is indicated. The markers for 933 and 951 bp correspond to der(2) and der(5) junctions, respectively, without end modification. The larger and smaller fragments seen in some of the lanes likely correspond to junctions with large insertions or deletions. (F) Detection of the NPM1–ALK fusion protein in Jurkat cells after TALNPM and TALALK coexpression and in the ALCL cell line SUPM2. The signal from 1 μg of cell extract from SUPM2 cells was compared with 40 μg from Jurkat cells. (G) Expanding pools of RPE-1 cells treated with TALENs. The PCR product corresponding to der(5) was detected when cells were split 1 to 10 every 3 d. The number of passages at each time point is indicated below the gel.
Two different human cell lines were tested in this system. We first expressed the TALENs in a human acute T-cell leukemia line, Jurkat, because ALCL is a lymphoma of T-cell origin (Gillis and Watson 1980). Jurkat cells are pseudodiploid with two normal NPM1 and ALK loci (Mathas et al. 2009). Both TALENs efficiently generated DSBs at the NPM1 and ALK target sites in these cells (Fig. 3B). Using a similar nested PCR strategy as for ZFN induction of EWSR1–FLI1 translocations, we detected translocation breakpoint junctions for both derivative chromosomes when the TALENs were coexpressed, while no PCR product was detected when a single TALEN was expressed (Fig. 3C). Applying the same high-throughput 96-well format used to detect EWSR1–FLI1 translocations, we detected NPM1–ALK1 translocations at a frequency of ∼1 × 10−3 per cell (see Methods). Given this relatively high frequency of translocations, we attempted direct PCR detection of translocation junctions using only one round of PCR and found that der(2) and der(5) breakpoint junctions were readily apparent (Fig. 3E). In serial dilutions of genomic DNA, both breakpoint junctions were obtained with as little as 6.25 ng of DNA (representing ∼1000 cells), corroborating that translocations arose at a frequency of ∼10−3 per cell.
Confirming the generation of translocations, the NPM1–ALK fusion transcript was detected by PCR from the bulk Jurkat cell population following coexpression of TALNPM and TALALK (Fig. 3D), and the NPM1–ALK fusion protein was detected by Western blot analysis using an antibody directed against ALK (Fig. 3F). The NPM1–ALK fusion protein comigrated with that found in the ALCL tumor SUPM2 cells and was ∼1/40th the level of protein seen in the SUPM2 cells (Fig. 3F). Expanding pools of cells, junctions were observed 25 d following TALEN expression (data not shown), indicating maintenance of the translocation.
We also performed similar experiments in retinal pigment epithelial cells, RPE-1, expressing hTERT to determine if the efficiency of t(2;5)(p23;q35) induced by TALENs is seen in other cells or may be related to the lymphocytic origin of the Jurkat cells. As in Jurkat cells, both TALENs efficiently generated DSBs at the target sites (Supplemental Fig. S5A). We recovered translocation junctions for der(2) and der(5) from RPE-1 cells coexpressing both TALNPM and TALALK (Supplemental Fig. S5B); using one round of PCR on dilutions of genomic DNA from the transfected cells, translocation frequency was estimated at 0.5 × 10−2 to 1 × 10−2 (Supplemental Fig. S5C). The NPM1–ALK fusion transcript and fusion protein were detected from the bulk-transfected cell population (Supplemental Fig. S5D,E). Fluorescence in situ hybridization confirmed the presence and the high frequency of the translocation (two of 70 metaphases) (Fig. 3A). Junctions were observed at 8 and 26 d following TALEN expression (Fig. 3G), indicating maintenance of the translocation. These results demonstrate that TALENs can efficiently induce t(2;5)(p23;q35) in different human cell types and that the translocation can be propagated.
TALENs and ZFNs both cleave DNA using the FokI nuclease domain, but the left and right DNA binding domains have a different spacing from each other in the two types of nucleases, with the potential to influence the site(s) of DNA cleavage. For ZFNEWS and ZFNFLI, the spacers are 6 bp; cleavage occurs at two sites, adjacent to the DNA binding site and 1 bp into the spacer, leaving 5′ overhangs of 6 and 4 bp, respectively (Fig. 1B; Smith et al. 2000), consistent with our junction analysis (Supplemental Fig. S2). In contrast, the spacers in TALNPM and TALALK are 18 bp (Fig. 3B). To map the TALEN cleavage sites, TALNPM and TALALK were expressed in vitro, and cleavage assays were performed on double-stranded DNA fragments labeled at either 5′ end (Supplemental Fig. S6). For both TALNPM and TALALK, we identified a region of cleavage spanning 6–7 bp that overlapped the center of the spacer (Fig. 3B). TALEN cleavage results primarily in 5′ overhangs, potentially of different lengths, although a fraction of blunt ends are also possible.
To examine joining mechanisms from TALEN-induced translocations, we analyzed 161 breakpoint junctions arising from t(2;5)(p23;q35) in Jurkat cells (Supplemental Fig. S7A). Because the TALEN cleavage sites may vary in cells as in vitro, we considered the longest 5′-overhang encompassing the full set of in vitro cleavage sites. Consistent with the in vitro cleavage assays, bases predicted to be in an overhang were incorporated into some of the junctions, and deletions emanating from the predicted DSB sites were frequent. As seen in the ZFN-induced translocations, small deletions were most common in the TALEN-induced translocations: 81% of junctions had deletions <30 bp, while <5% of junctions had deletions of >100 bp.
One feature of TALEN-induced breakpoint junctions was the frequent presence of short insertions (Supplemental Fig. S7A). Almost half of breakpoint sequences showed insertions of <10 bp (47.5%), compared with 7% in the ZFN-induced translocations. A few insertions could be identified as duplications of sequences close to the DSB sites, but most of them were not readily evident as being templated. As shown for ZFN-induced translocations, some longer insertions were observed that were derived from a variety of sources: Duplications of regions located near the DSB sites on chromosomes 2 and 5 as well as sites more distant from the DSBs (Mb), and the TALEN expression plasmids. Complex insertions were also identified. For example, one der(5) junction had tandem duplications from both chromosomes 2 and 5 in addition to a deletion from each DSB and another der(5) junction had duplications from chromosome 5 from both upstream and downstream of the DSB (junctions 19 and 54, respectively) (Supplemental Fig. S7A).
The involvement of microhomology in the joining mechanism of TALEN-induced translocations is more problematic to analyze because of the large number of short insertions. Nevertheless, when considering only sequences without insertions, microhomologies of 3–6 bp were observed in only a small fraction of sequences (6.2%), suggesting a minimal dependence on microhomology in formation of these translocations as found in ZFN-induced and tumor translocations.
All of the joining characteristics observed in Jurkat cells were recapitulated in translocations from hES-MP cells, including a large fraction of junctions with short insertions (24/38 total) (Supplemental Fig. S7B). Frequent small insertions were previously reported for intrachromosomal repair after expression of a single TALEN in hES cells (Hockemeyer et al. 2011), although not in another study using U2OS cells (Reyon et al. 2012).
The homodimerizing FokI domains in the TALENs leave open the possibility that newly formed translocation junctions could be recleaved by mixed TALENs (i.e., TALNPM/ALK and TALALK/NPM). To circumvent this possibility, TALENs were also constructed with FokI heterodimerization domains (Doyon et al. 2011). Translocations were observed in Jurkat cells with the heterodimeric TALENs (Supplemental Fig. S8A). Junctions showed similar characteristics as with the homodimeric TALENs (Supplemental Fig. S8B), including frequent small insertions, indicating that cleavage of newly formed translocation junctions are not substantially affecting characteristics.
Reversion of the NPM1–ALK translocation
The ability to specifically induce translocations suggested that it should also be possible to reverse translocations with a similar approach (Fig. 4A). Genetic reversion permits the analysis of phenotypic consequences of fusion protein loss, such as whether tumor cells are “addicted” to the oncogene. To revert the t(2;5)(p23;q35) translocation, TALNPM and TALALK were expressed in SUDHL-1 patient-derived cells, with efficient cleavage of both the NPM1 and ALK loci (Fig. 4B). Due to the position of the TALEN cleavage sites relative to the translocation breakpoint junctions, restoration of the intact chromosomes leaves a small segment of DNA from the partner chromosome that serves as a tag to differentiate the revertant chromosomes from the wild-type chromosomes 2 and 5 (Fig. 4C). By PCR, both revertant chromosomes were detected (Fig. 4C), and sequencing confirmed that the translocation had been reversed (Fig. 4D; Supplemental Fig. S9). We also tested the reversion strategy in SUPM2 cells, another ALCL patient-derived cell line, and also obtained revertant breakpoint junctions (Supplemental Fig. S9). As with translocations, many of breakpoint junctions for both revertant chromosomes from both ALCL cell lines had small insertions. Overall then, frequent small insertions were observed at four different junctions (two translocation and two reversion junctions), suggesting that they do not derive from a specific sequence context from the TALEN pair we used.
Figure 4.
Reversion of the t(2;5)(p23;q35) translocation in SUDHL-1 cells. (A) Patient-derived SUDHL-1 cells carry the t(2;5)(p23;q35) translocation and express the NPM1–ALK fusion gene from der(5). To reverse the translocation, TALNPM and TALALK are expressed to create DSBs in both fusion genes; repair between the derivative chromosomes restores intact chromosomes 5 and 2. (B) TALNPM and TALALK efficiently cleave target loci in SUDHL-1 cells. Cleavage is monitored by the T7-endonuclease assay directed to the NPM1 (left) and ALK (right) loci. (C) PCR detection of revertant chromosomes 2 and 5. Reversion is only detected after expression of both TALNPM and TALALK. Because the TALENs cleave to the side of the translocation breakpoint junctions in the SUDHL-1 cells, segments of the other chromosome remain to “tag” the revertant chromosomes to distinguish them from chromosomes that did not participate in the translocation. (D) Sequences of representative revertant chromosomes 5 and 2, which restore the NPM1 and ALK genes, respectively.
Discussion
In this report, we could reproduce cancer relevant translocations and revert them by targeting concurrent DSBs to intronic sequences in human cells using two types of designed nucleases, ZFNs and TALENs. With this approach, fusion genes created during translocation formation, which are potential oncogenes, are expressed from their endogenous promoters. In addition, translocation disrupts a copy of each participating gene, reproducing the heterozygosity (and potential haploinsufficiency) found in tumor cells. Therefore, this strategy provides a more accurate model for translocation-related tumor formation than ectopic expression of a fusion protein to study effects on proliferation and other cellular phenotypes of newly translocated cells and, further, provides an approach to revert the translocation.
Despite the fact that a ZFN induces cohesive 5′-overhangs that should be easily repaired by direct ligation or by polymerase fill-in followed by ligation, breakpoint junctions of ZFN-treated cells recapitulated the features found in Ewing sarcoma cells: deletions, short insertions, complex insertions from other chromosomes and/or nearby the break regions, and an absence of long microhomology. Overall ZFN-treated cells showed a comparable proportion of each type of junction modification as patient cells, with the exception of complex duplications, which are overrepresented in tumor cells (Zucman-Rossi et al. 1998). It is possible that these latter events in patients arise from multiple breaks in the same intronic regions (fragile sites). Translocation junctions obtained with TALENs frequently showed short deletions and other modifications seen in patient translocation junctions. Short insertions were overrepresented, although it is not clear if this is specific to the TALEN pair.
Relevant to Ewing sarcoma, the strategy presented here has the potential to resolve inconsistencies in previous studies that have attempted to identify target genes using ectopic expression or knockdown of the EWSR1–FLI1 fusion protein and also those in studies using mouse cells as a tumor model (for review, see Lessnick and Ladanyi 2012). Der(5) NPM1–ALK-driven ALCL tumorigenesis appears to be more defined, since the fusion protein is known to inhibit apoptosis and promote cell proliferation through the constitutively active ALK tyrosine kinase moiety (Kuefer et al. 1997; Greenland et al. 2001; Kasprzycka et al. 2006). Nonetheless, it must be noted that NPM1 is a multifunctional protein involved in many cellular processes (Colombo et al. 2006), including centrosome duplication through a C-terminal interaction with the CRM1 complex, such that der(2) expressing the NPM1 C terminus has the potential to participate in the lymphomagenesis of ALCL in addition to der(5) (Wang et al. 2005). And while normal levels of NPM1 induce senescence and/or apoptosis under UV damage conditions by binding TP53 (Kurki et al. 2004), NPM1 overexpression can lead to cellular transformation (Li et al. 2008), although paradoxically, loss of one allele of NPM1 leads to DNA instability allowing possible cellular transformation as well (Grisendi and Pandolfi 2005). Thus, formation of both derivative chromosomes with loss of at least one allele of NPM1 may all be critical to recapitulate the disease initiation state faithfully.
The present study provides a novel approach to understand translocation-related tumorigenesis and, more broadly, demonstrates that precise genome rearrangements can be achieved in relevant cell types with tailored nucleases. This study also expands the repertoire of genome engineering accomplished with these nucleases beyond gene mutation/correction and intrachromosomal rearrangements (Lee et al. 2010, 2012) and translocations in model systems (Brunet et al. 2009; Simsek et al. 2011). Unlike site-specific recombinases, endonucleases are designed to cleave endogenous sites, abrogating the need for prior genome manipulation. And because they introduce DSBs, endonucleases provide more accurate models for studying the etiology of genomic rearrangements and their potential for reversion, as well as providing more accurate cellular models for further functional investigation.
Methods
Cell lines, transfections, FISH
hES-MP cells, derived from hES strain H1, were provided by the SKI Stem Cell Research Facility (MSKCC), and cultivated in α-MEM supplemented with 10% FBS and 50 ng/mL recombinant human FGF (R&D Systems). Jurkat and SUDHL-1 and SUPM2 (DSMZ, Germany) cells were cultivated in RPMI-1640 medium supplemented with 10% heat inactivated FBS. The hTERT-immortalized retinal pigment epithelial cell line, RPE-1, was cultured in DMEM:F12 medium supplemented with 10% FBS.
Transfections were done as previously described (Brunet et al. 2009). Typically, 7.5 × 105 (for hES-MP and RPE-1) or 2 × 106 (for Jurkat, SUDHL-1, and SUPM2) cells were transfected by Amaxa technology (Lonza) using the cell line nucleofector kit V (program B-16 for hES-MP cells and X-001 for Jurkat, SUDHL-1, SUPM2, and RPE-1 cells) with 7.5 μg of each ZFNEWS and 5 μg of each ZFNFLI1; or 5 μg of each TALNPM and 2 μg (for Jurkat, SUDHL-1, and SUPM2 cells) to 2.5 μg (for RPE-1 cells) of each TALALK.
FISH was performed on metaphases of RPE-1 cells 48 h after transfection with TALNPM and TALALK using ALK break apart probes (LPS019 Amplitech) and an NPM1 BAC probe (RP11-117L6). Images were taken with the Zeiss Axio Observer.Z1 system.
PCR-based translocation detection, frequency determination, and sequencing of breakpoint junctions
Primer sequences are presented in Table 1. For detection of translocations in pooled cells, 100 ng of genomic DNA from cells transfected with ZFNs or TALENs was amplified by nested PCR 72 h post-transfection. Where noted, the transfected cell population was kept in culture for the indicated length of time to monitor maintenance of the translocation. For translocation frequency determination, a high-throughput 96-well screen using small pools of cells and nested PCR to amplify translocation junctions was used 48 h post-transfection, as described (Brunet et al. 2009). The number of PCR-positive and negative wells for translocation junctions was determined to calculate the translocation frequency. Amplified products from positive wells were directly sequenced to verify translocations and determine the junction sequences. For hES-MP cells, 96-well plates contained 8 × 104 transfected cells per plate, resulting in 17–33 positive wells per plate in four experiments for each derivative chromosome. For Jurkat cells, 96-well plates contained 2 × 105 transfected cells per plate. Centrifugation of plates with the nonadherent Jurkat cells, followed by media removal, resulted in cell loss, and thus cells were requantified prior to lysis, demonstrating ∼1 × 105 cells remaining. After transfection of the TALEN homodimer expression plasmids, two 96-well plates were analyzed for each derivative chromosome; 61 ± 2 positive wells were obtained per plate, which adjusted to a β distribution is estimated to represent about 95 translocations per plate, for a frequency of ∼1 × 10−3. Sequencing of PCR products from positive wells showed mixed sequences in a single well from almost half of the wells corresponding to multiple junctions, consistent with the high frequency of translocations, and were removed from the junction analysis. For TALEN heterodimer experiments, the translocation frequency was half that of the homodimer experiments.
Table 1.
Primer sequences

Translocation frequency was also determined for Jurkat cells and for RPE-1 cells by serial dilution of genomic DNA from cells transfected with the TALEN expression vectors followed by one round of PCR (i.e., non-nested) indicated in Figure 3C. For Jurkat cells, genomic DNA dilutions were 1:2, for 50, 25, 12.5, 6.25, 3.125, and 1.56 ng of DNA. Considering that one cell contains ∼6 pg of DNA (Jurkat cells being a pseudodiploid human cell line), 6.25 ng of represents ∼103 cells. PCR amplification with 6.25 ng of DNA was seen in half of the experiments with the homodimeric TALENs, corresponding to a frequency of ∼10−3 per cell. For the heterodimeric TALENs, junction detection was less efficient (one of three experiments with 6.25 ng of DNA and two of three experiments with 12.5 ng of DNA), implying about half the translocation efficiency as with homodimeric TALENs. Thus, translocation frequency determination by the 96-well format and by serial dilution of genomic DNA gave similar results. For RPE-1 cells, genomic DNA dilutions were 1:2, for 5, 2.5, 1.25, 0.625, and 0.31 ng of DNA. Amplification was detected at 1.25 ng of genomic DNA (two of three experiments) and 0.625 (one of three experiments), indicating a frequency of 0.5 × 10−2 to 1 × 10−2 per cell. The detection of two translocations by FISH from 70 total metaphases is in agreement with the high translocation frequency estimated for RPE-1 cells.
PCR for revertant chromosome detection and sequencing of breakpoint junctions
Both Chr(2) and Chr(5) revertant chromosomes were detected by PCR amplification 72 h post-transfection of the TALEN expression vectors. Chr(2)- and Chr(5)-amplified products were cloned with the TOPO-TA cloning system (Invitrogen) and sequenced.
T7 endonuclease I assay
Forty-eight hours after transfection, genomic DNA was extracted with QIAamp DNA Mini Kit (QIAGEN). The genomic region encompassing the ZFN and TALEN target sites was amplified with the primers listed below, typically using 50 ng of genomic DNA, melted, and annealed (5 min at 95°C, 95°C to 25°C at −0.5°C/30 sec, and 15 min at 4°C) to form heteroduplex DNA. The annealed DNA was treated with 1.5 units of T7 endonuclease I (New England BioLabs) for 10 min at 37°C and run on a 2.4% agarose gel.
RT-PCR and cDNA amplification
RNA was extracted from transfected cells with absolutely RNA Microprep Kit (Agilent Technologies) and RT-PCR was done following manufacturer directions (SuperScript II First-Strand Synthesis System; Invitrogen). Primers used for cDNA amplification of EWSR1–FLI1 and NPM1–ALK are listed below.
Western blotting
Whole-cell extracts were prepared with protein lysis buffer (50 mM Tris-HCl at pH 7.4, 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT prepared from a 1 M DTT stock), with addition of cocktail protease inhibitor tablets (Complete, Roche). Typically 40 μg of protein extract from TALEN-treated cells were run on an 8% (w/v) Tris-HCl SDS page gel, blotted, and then probed with ALK (31F12) Mouse Antibody (Cell Signal). Alpha-tubulin mouse antibody (Sigma-Aldrich) was used to detect those proteins as loading controls.
In vitro mapping of TALEN cleavage sites
The TNT SP6 Quick Coupled Transcription/Translation System (Promega) was used to express TALNPM and TALALK (Mussolino et al. 2011). Sixty-eight-bp fragments containing target sites of TALNPM and TALALK were 5′-radiolabeled on one strand. Typically, 0.5 μL of each TNT lysate containing one TALEN subunit was incubated overnight at 37°C with 10 nM target duplex in NEB4 buffer (New England Biolabs) supplemented with 100 mM NaCl and 0.4 μg of salmon sperm (DNA competitor) in 10 μL. The reaction was analyzed on a denaturing 12% PAGE gel.
Competing interest statement
G.E.K., L.Z., M.C.H., and Y.D. are past (G.E.K.) or present (L.Z., M.C.H., Y.D.) employees of Sangamo BioSciences, Inc.
Acknowledgments
We thank Francesca Cole and other members of the Jasin laboratory, Patrizia Alberti and Loic Ponger from the laboratory of “Regulation and Dynamics of Genomes” at MNHN, and Alexei Morozov in Malcolm Moore’s laboratory at MSKCC for helpful discussions. We also thank Fyodor Urnov (Sangamo BioSciences, Inc.) for his help in initiating the project. M.P. is supported by a “Le Canceropole IDF” PhD grant and H.G. is supported by a “La Ligue Nationale contre le Cancer” PhD grant. This work was supported by CNRS, Inserm, MNHN, IBiSA (Infrastructures for Biology), and ANR-12-JSV6-0005-01 (E.B.) and by grant R01 NIHGM54668 (M.J.).
Author contributions: E.B. and M.J. designed the research. M.P., H.G., A.D.C., L.G., M.T., L.P., O.R., and E.B. performed the experiments. G.E.K., L.Z., M.H., and Y.D. designed new ZFN reagents. A.D.C., J.-P.C., and C.G. designed new TALEN reagents and did analytic analysis. E.B. and M.J. did statistical analysis of data. E.B. and M.J. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.147314.112.
Freely available online through the Genome Research Open Access option.
References
- Barberi T, Willis LM, Socci ND, Studer L 2005. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med 2: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D 2011. Genome engineering with zinc-finger nucleases. Genetics 188: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chansky HA, Barahmand-Pour F, Mei Q, Kahn-Farooqi W, Zielinska-Kwiatkowska A, Blackburn M, Chansky K, Conrad EU III, Bruckner JD, Greenlee TK, et al. 2004. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J Orthop Res 22: 910–917 [DOI] [PubMed] [Google Scholar]
- Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, Volorio S, Bernard L, Pruneri G, Alcalay M, et al. 2006. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res 66: 3044–3050 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79 [DOI] [PubMed] [Google Scholar]
- Elmberger PG, Lozano MD, Weisenburger DD, Sanger W, Chan WC 1995. Transcripts of the npm–alk fusion gene in anaplastic large cell lymphoma, Hodgkin’s disease, and reactive lymphoid lesions. Blood 86: 3517–3521 [PubMed] [Google Scholar]
- Gillis S, Watson J 1980. Biochemical and biological characterization of lymphocyte regulatory molecules. V. Identification of an interleukin 2-producing human leukemia T cell line. J Exp Med 152: 1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland C, Touriol C, Chevillard G, Morris SW, Bai R, Duyster J, Delsol G, Allouche M 2001. Expression of the oncogenic NPM–ALK chimeric protein in human lymphoid T-cells inhibits drug-induced, but not Fas-induced apoptosis. Oncogene 20: 7386–7397 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Pandolfi PP 2005. NPM mutations in acute myelogenous leukemia. N Engl J Med 352: 291–292 [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649: 247–256 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzycka M, Marzec M, Liu X, Zhang Q, Wasik MA 2006. Nucleophosmin/anaplastic lymphoma kinase (NPM/ALK) oncoprotein induces the T regulatory cell phenotype by activating STAT3. Proc Natl Acad Sci 103: 9964–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuefer MU, Look AT, Pulford K, Behm FG, Pattengale PK, Mason DY, Morris SW 1997. Retrovirus-mediated gene transfer of NPM–ALK causes lymphoid malignancy in mice. Blood 90: 2901–2910 [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5: 465–475 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS 2012. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res 22: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessnick SL, Ladanyi M 2012. Molecular pathogenesis of Ewing sarcoma: New therapeutic and transcriptional targets. Annu Rev Pathol 7: 145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Boone D, Hann SR 2008. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci 105: 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Chinnaiyan AM 2010. Triggers for genomic rearrangements: Insights into genomic, cellular and environmental influences. Nat Rev Genet 11: 819–829 [DOI] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. 2009. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci 106: 5831–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT 1994. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Mussolino C, Cathomen T 2012. TALE nucleases: Tailored genome engineering made easy. Curr Opin Biotechnol 23: 644–650 [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plougastel B, Zucman J, Peter M, Thomas G, Delattre O 1993. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics 18: 609–615 [DOI] [PubMed] [Google Scholar]
- Prieur A, Tirode F, Cohen P, Delattre O 2004. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 24: 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK 2012. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Jasin M 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405: 697–700 [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, et al. 2008. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 68: 2176–2185 [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suva D, et al. 2010. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 24: 916–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Porter TN, Wilson JH 1985. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol 5: 2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M 2010. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4–ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 17: 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. 2011. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res 28: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, Lessnick SL 2006. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 9: 405–416 [DOI] [PubMed] [Google Scholar]
- Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O 2007. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 11: 421–429 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Wang W, Budhu A, Forgues M, Wang XW 2005. Temporal and spatial control of nucleophosmin by the Ran–Crm1 complex in centrosome duplication. Nat Cell Biol 7: 823–830 [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M 2007. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 9: 978–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449: 478–482 [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Legoix P, Victor JM, Lopez B, Thomas G 1998. Chromosome translocation based on illegitimate recombination in human tumors. Proc Natl Acad Sci 95: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]