Ongoing transcription of the Ig gene coupled with temporary pausing within the targeted region facilitates somatic hypermutation.
Abstract
Somatic hypermutation (SHM) of Ig genes is initiated by the activation-induced cytidine deaminase (AID), and requires target gene transcription. We previously proposed that AID may associate with the RNA polymerase II (Pol). Here, to determine aspects of the transcription process required for SHM, we knocked-in a transcription terminator into an Ig gene variable region in DT40 chicken B cell line. We found that the human β-globin terminator was an efficient inhibitor of downstream transcription in these cells. The terminator reduced mutations downstream of the poly(A) signal, suggesting that the process of transcription is essential for efficient SHM and that AID has better access to its target when Pol is in the elongating rather than terminating mode. Mutations upstream of the poly(A) site were almost doubled in the active terminator clones compared with an inactivated terminator, and this region showed more single-stranded DNA, indicating that Pol pausing assists SHM. Moreover, the nontranscribed DNA strand was the preferred SHM target upstream of the active terminator. Pol pausing during poly(A) site recognition may facilitate persistence of negative supercoils, exposing the coding single strand and possibly allowing the nascent RNA intermittent reannealing with the template strand, for prolonged access of AID.
The processes of somatic hypermutation (SHM) and class-switch recombination (CSR) of Ig genes are initiated by the activation-induced cytidine deaminase (AID). In SHM, AID creates cytidine (C) to uridine (U) mutations, starting ∼100–200 bp from the promoter within the variable (V) region and in flanking DNA sequences and extending for ∼2 kb (Longerich et al., 2006; Storck et al., 2011). Error-prone lesion-bypass polymerases introduce mutations at the U and sequences within a dozen or so nucleotides in the vicinity of the U, and both mismatch repair (MMR) and base excision repair (BER) proteins are involved in the error processes (Rada et al., 2002, 2004; Shen et al., 2006).
AID-initiated SHM and CSR are essential for generating the antibody repertoire that is necessary to acquire resistance to infections (Wei et al., 2011). We have also found that the protooncogene BCL6 is mutated in human memory B cells (Shen et al., 1998). Additionally, other protooncogenes have been reported to become active and mutated in transformed germinal center B cells (Pasqualucci et al., 2001). In fact, it appears that AID can mutate many genes expressed in germinal center B cells undergoing SHM (Liu et al., 2008; Storb et al., 2009; Tanaka et al., 2010). Absence of AID results in immunodeficiencies (Conley et al., 2009); however, AID is also a potentially dangerous mutator. Finally, removal of methylC from DNA via AID may be essential for normal early development (Rai et al., 2008; Hochedlinger and Plath, 2009; Popp et al., 2010). Thus, the study of the molecular mechanisms of AID action is highly significant in the effort to understand its importance in various physiological processes such as, immunity, oncogenesis, development, and the production of multipotent stem cells for organ and tissue replacement.
The precise molecular mechanism by which AID targets Ig genes is not clear. However, we and others have previously shown that the process of SHM requires transcription without necessitating an SHM-specific promoter, but is linked to transcription initiation (Peters and Storb, 1996), and requires an Ig enhancer (Klotz and Storb, 1996) or certain motifs that are present in Ig enhancers (Betz et al., 1994; Tanaka et al., 2010). We have proposed an SHM model that postulates that a mutator factor, now known to be AID, assembles with the transcription complex at the promoter and travels with the elongating RNA polymerase (Pol; Peters and Storb, 1996). Moreover, transcription is also required for Ig class switch recombination (CSR), and the Ig enhancer motifs are essential to both SHM and CSR (Di Noia et al., 2007; Storb et al., 2007; Peled et al., 2008; Stavnezer et al., 2008). In fact, AID was shown to be associated with Pol (Nambu et al., 2003). However, the role of transcription in SHM and CSR remains unclear.
We have now investigated the influence of transcription termination and pausing on mutability in vivo. To do this we introduced a strong transcription terminator into a variable Ig gene region to assess its impact on SHM.
RESULTS
Controlling transcription within the IgL locus
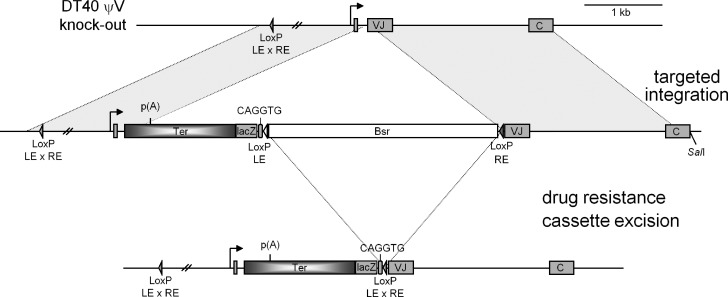
To test the requirement for ongoing transcription and how a transcriptional terminator affects SHM within the Ig gene, we placed a strong human transcription terminator into the active lambda gene of mutating chicken B cells, DT40, by homologous recombination (Fig. 1). The cell line used in this study is a variant of DT40 cells that is an AID knock-out and expresses AID as a transgene (AID-IRES-GFP) and all 25 ψV IgL genes are deleted (Arakawa et al., 2004) to make sure that these cells do not undergo IgL gene conversion.
Figure 1.
Map of the rearranged Ig light chain locus in the chicken B cell line DT40 ψV knockout. (Not to scale.) The locus contains a leader, VJ, and C region of the IgL gene. The strategy of knocking in the human β-globin transcription terminator (∼1.4 kb) by the targeted integration is shown. The terminator is knocked-in at +166 bp from the transcription start site, and 60 bp of DNA fragment containing part of leader – VJ intron and 8 bp of V region was deleted (from +166 to +225 from transcription start site).
The knock-in plasmids were constructed by cloning genomic sequences that flank the integration site in the IgL gene (Fig. 1). We chose to introduce an ∼1.4-kb human β-globin transcription terminator (Dye and Proudfoot, 2001), which contains three AATAAA sequences, into the intron between the V region leader and V, 165 NTs from the start of transcription (Figs. S2 and S3). To make the β-globin terminator a better target for AID activity we added a 34 bp DNA segment with two CAGGTG motifs, which we have recently shown to be essential for attracting SHM to Ig genes in DT40 cells (Tanaka et al., 2010). As a control, we created an inactivated terminator by mutating the three poly(A) addition sites located at 320 bp, 637 bp and 834 bp, respectively, from the start of the terminator sequence (Fig. S1). The first AATAAA hexamer located at 320 bp in the human β-globin transcription terminator (Fig. S1) is referred as the poly(A) site in this study. We mutated the three AATAAA sites by PCR-based mutagenesis to GGATCC, TCTAGA, and GGTACC, respectively, to create an “inactivated terminator” control plasmid.
We chose a surface IgM-positive clone of DT40 ψV knock-out cells for transfection so that targeted integration into the IgL locus was easily detected by the loss of surface IgM expression. The targeted integration of the terminator and control were confirmed by Southern blotting with the terminator region as probe (unpublished data). To remove potentially repressive bacterial sequences, the blasticidin drug marker gene present between two LoxP sites (Fig. 1) was excised by treating the cells with tamoxifen. Incidentally, the AID-IRES-GFP transgene is also flanked by two LoxP sites and is likely to be excised after treatment with tamoxifen. To retain a fraction of cells that contain the AID-IRES-GFP transgene, we used a very low concentration of tamoxifen (25 nM) for 24 h. Cell clones that were GFP-positive and had excised the blasticidin drug marker gene present in LoxP sites were selected, and the blasticidin drug marker gene excision was confirmed by PCR amplification (unpublished data).
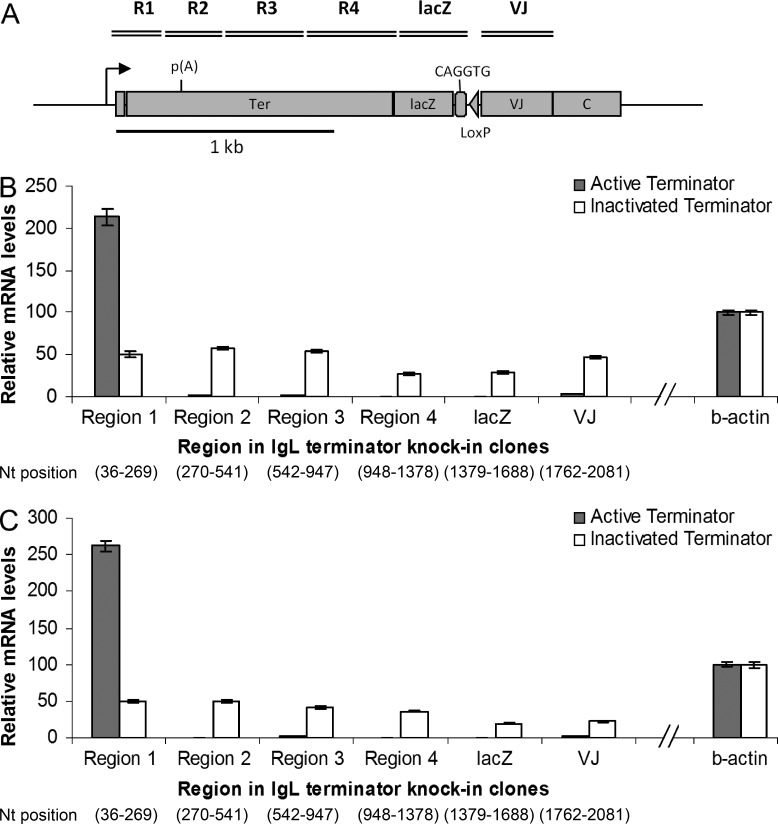
To test whether the presence of an active or inactivated terminator influenced transcription through the IgL gene, we performed RTQ-PCR in the terminator and VJ region of the IgL gene (Fig. 2 A) in active and inactivated terminator knock-in clones, respectively, and observed that knocking in a strong human β-globin terminator significantly decreased stable transcripts through the IgL gene compared with those in the inactivated terminator knock-in clones (Fig. 2, B and C). We found that transcripts were greatly reduced downstream of the poly(A) site in the active terminator but not the inactive terminator (Fig. 2, B and C). This suggests that the human β-globin terminator designed by the Proudfoot laboratory (Dye and Proudfoot, 2001) works very efficiently in chicken DT40 cells.
Figure 2.
Transcription levels at the β-globin transcription terminator. (A) Region 1– Region 4, lacZ, and IgL VJ region in the active and inactivated terminator knock-in clones (not in scale). B and C are bar graphs of two independent cell clones each of the active and inactivated terminator, respectively, showing relative mRNA levels reverse transcribed with random hexamer primers and PCR amplified with region-specific primers; the values are normalized with chicken β-actin levels and primer efficiencies. The numbers at the bottom indicate start and end nucleotide positions of the respective regions. The data represent means and SDs of three independent experiments.
Transcription termination significantly influences the mutation frequencies
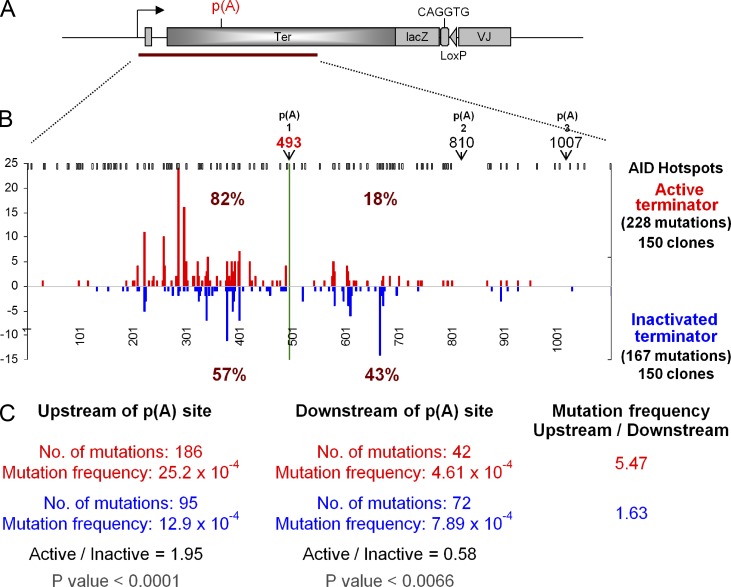
The active and inactivated terminator knock-in clones were cultured for 5 wk to acquire mutations in the IgL and terminator locus. A 1.2-kb PCR product of genomic DNA was amplified and sequenced, encompassing the region 48 bp upstream of the promoter of the IgL gene, as well as the terminator region (Fig. 3 A). Fig. 3 B shows the SHM pattern in the active and inactivated terminator knock-in clones. Out of 228 mutation events in the active terminator knock-in clones in which the stable transcript levels downstream of the first poly(A) site were significantly reduced, 82% of total mutations were upstream of the poly(A) site and only 18% downstream. In contrast, in the inactivated terminator knock-in clones, 43% of the total mutations were downstream of the poly(A) site. In this region the mutation frequency in the active terminator knock-in clones was 4.6 × 10−4/bp, which is a >70% reduction (P < 0.0066) as compared with the inactivated terminator knock-in clones (7.9 × 10−4/bp; Fig. 3 C). This suggests that the process of ongoing transcription is essential for SHM and that AID may get better access to its target DNA when Pol is in the elongation phase rather than in the termination phase. However, SHM eventually ceased altogether, with the active terminator and the inactivated terminator; we had introduced a lacZ gene into the Ig locus in the hope of easy quantitation of the mutability of the two types of constructs, however there were essentially no mutations that far from the promoter (1.57-kb). There were also no mutations in the VJ region. Overall, the distribution of mutations is determined by the position from the promoter (as always in SHM of Ig genes) and not by the primary sequence: there are very few, if any, mutations in the first 100 NTs after the transcription start, and then the mutations rise steeply to a short plateau and, after a prolonged decline, the mutations end before 1.5 kb from the promoter.
Figure 3.
Ig light chain sequence analysis of the transcription terminator knock-in clones. (A) Map of lg gene with transcription terminator (not to scale); the triangle represents the two recombined loxP sites. (B) Mutations in 1.1 kb from the start of transcription (= 1). Numbers on y-axis represent point mutations at the indicated positions in active terminator (red) and inactivated terminator (blue). The data are combined mutation numbers for two cell clones for the active and inactivated terminator, respectively. AID hotspots (WRC and GYW) are shown on the top in black. Three poly(A) sites (pA) in the human β-globin transcription terminator are marked by arrows and numbers (493, 810, and 1,007) from the transcription start. (C) Summary of mutations.
Although the number of mutations is dramatically reduced directly downstream of the poly(A) site in the active terminator knock-in clone, we did observe some mutations in this region. If AID were associated only with the processive, elongating RNA polymerase, one would expect a complete absence of mutations after the poly(A) site. The presence of mutations in this region is presumably due to Pol remaining engaged with the DNA for some distance beyond the poly(A) site.
The status of Pol and the distribution of poly(A) RNAs in the terminator knock-in loci
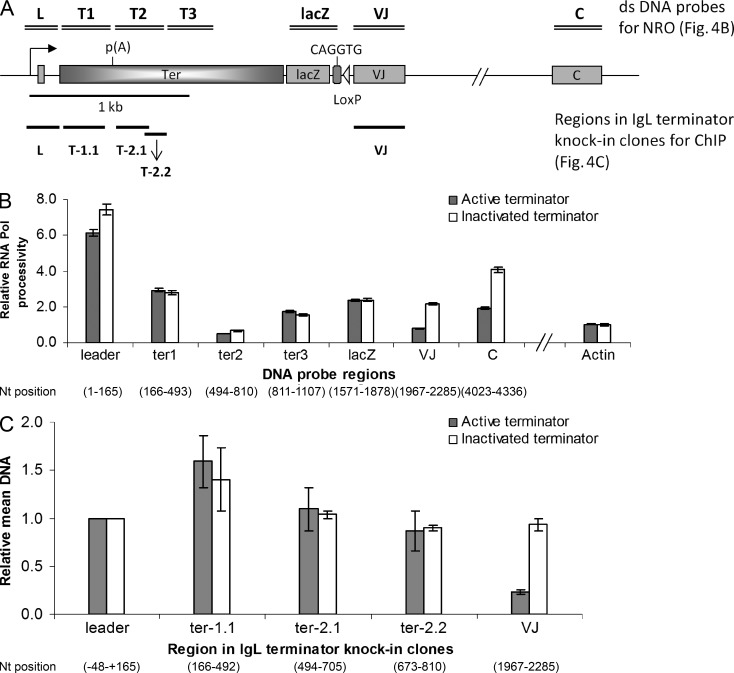
We wanted to check whether Pol is present on the region downstream of the poly(A) site by performing nuclear run-on (NRO) assays for both the active and inactivated terminator knock-in clones (Fig. 4 A). Fig. 4 B shows relative Pol occupancy in the IgL locus in the active and inactivated terminator knock-in clones. The RNA Pol occupancy is very similar in the terminator region until ∼1.5 kb downstream from the poly(A) (lacZ Fig. 4 B), and is reduced only in the VJ and C regions. The terminator region we have analyzed for SHM shows almost identical RNA Pol occupancy with the active and inactivated terminators. To confirm the findings of the NRO assays, we performed chromatin immunoprecipitation (ChIP) assays for the active and inactivated terminator knock-in clones using antibody against Pol. Fig. 4 C shows relative mean levels of immunoprecipitated DNA in active and inactivated terminator knock-in clones normalized for the respective input levels. ChIP results show no significant differences in the RNA Pol levels in the terminator region, however, Pol levels are sharply reduced in the Ig VJ region, corroborating the NRO results. Thus, the residual mutations observed downstream of the poly(A) site might be caused by the presence of transcriptionally active Pol in the terminator immediately downstream of the poly(A) site. To determine whether the poly(A) site in the active terminator cell lines did indeed terminate the elongation phase of Pol, we analyzed the poly(A) RNAs in the active and inactivated terminator cells (Fig.5, A and B). Strikingly, close to 100% of the poly(A) RNAs of the active terminator are processed at the first poly(A). There are none from the other two poly(A) sites of the terminator, and only a few contain a polyadenylated constant region. The latter may represent read-through by Pol of the first poly(A) site or possibly another weak promoter upstream of the constant region. All poly(A) RNAs from the inactivated terminator cells are apparently full-length transcripts, terminated and polyadenylated 3′ of the constant region.
Figure 4.
NRO and ChIP assays. NRO and ChIP analysis of the active and inactivated terminator knock-in clones. (A) Map of the rearranged Ig light chain locus in the chicken B cell line DT40. The dsDNA probes used in the NRO assays are shown by double bars on the top, and the regions analyzed with the ChIPed samples are shown by single bars at the bottom of the map (not drawn to scale). (B) NRO: Bar graphs show relative Pol transcriptional activity at the various regions in the active and inactivated terminator knock-in clones. The values are normalized for chicken β-actin levels, background hybridization, and incorporation of α-[32P]-UTP. (C) ChIP with antibody against Pol. Bar graphs indicate relative mean levels of immunoprecipitated DNA over input DNA by quantitative PCR, normalized for β-actin. The data represent means and SDs of three independent experiments with one of the active and inactivated terminator cell lines.
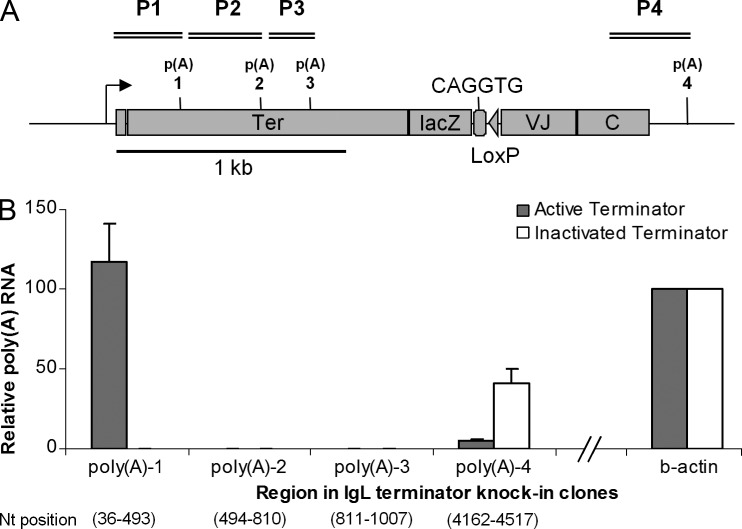
Figure 5.
Poly(A) RNAs. (A) Region P1– Region P4 in the active and inactivated terminator knock-in clones (not to scale). (B) Polyadenylated RNA Assay. Bar graphs show relative poly(A) RNA levels reverse transcribed with oligo-dT primers and PCR amplified with region-specific primers; the values are normalized with chicken β-actin levels and primer efficiencies. The numbers at the bottom indicate start and end nucleotide positions of the respective regions. The data represent means and SDs of three independent experiments with two of the active and inactivated terminator cell lines.
These NRO, ChIP, and poly(A) RNA findings suggest that the ongoing process of transcription is required for SHM and furthermore, that AID is preferentially associated with the elongating RNA polymerase (compare Fig. 3 with Figs. 4 and 5).
The terminator alters the mutation pattern within the IgL locus
The mutation frequency upstream of the poly(A) site in the active terminator knock-in clones is 25.2 × 10−4/bp, which is around twice (P < 0.0001) that in the inactivated terminator knock-in clones (12.9 × 10−4/bp; Fig. 3 C). This suggests that the transcriptional pausing because of the poly(A) site has facilitated SHM in the Ig locus (see Discussion).
We observed a predominance of single nucleotide substitutions with few insertions and deletions in both active and inactivated terminator knock-in clones (Figs. S2 and S3). Both types of knock-in clones show the usual pattern of SHM in DT40 (Arakawa et al., 2004): very few mutations at A/T bases and a preference for transversion mutations (Figs. S2 and S3 and Fig. 6). Two independent cell clones for the active or inactivated terminator, respectively, showed these properties: the data are combined in all relevant Figures. In the total 1.1 kb sequenced for both cell types, generally the numbers of mutations at G bases were lower than at C (C/G ratios 0.31, 0.58, and 0.67; Fig. 6). However, in the active terminator knock-in clones upstream of the poly(A) site the number of mutations at C was 1.36 times higher than at G (Fig. 6). This relative increase in mutations at C suggests that the coding strand is targeted preferentially in the active terminator knock-in clones (see Discussion).
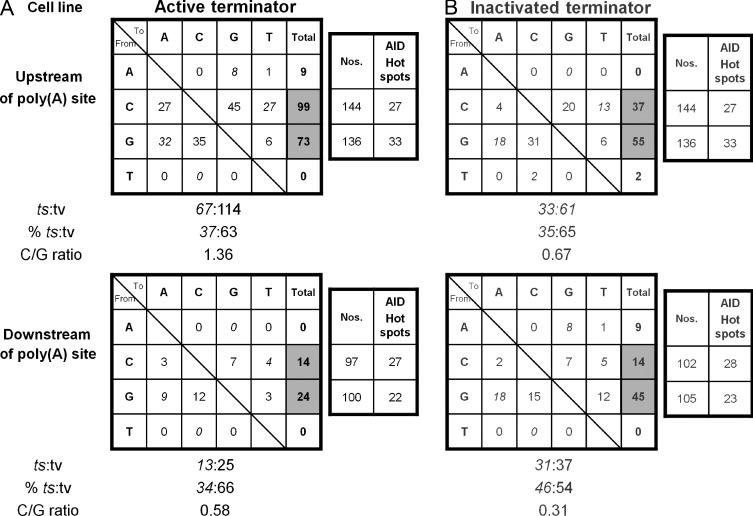
Figure 6.
Mutation events in the IgL and human β-globin transcription terminator sequences. Mutation events in the 1.1-kb DNA sequence in the active (A) and inactivated (B) terminator knock-in clones. The data are combined mutation numbers for two cell clones for the active and inactivated terminator, respectively. Mutation events in the region upstream of poly(A) site (top) and downstream of poly(A) site (bottom) are analyzed separately. The number of C and G nucleotides, and the number of AID hotspots (WRC and GYW) in the respective regions are shown next to each region. The numbers of transition (ts) and transversion (tv) mutations and their percentages are shown below each region. C/G ratio = ratio of mutations at C and G.
More sequences 5′ of the poly(A) site are single-stranded in cells with the active terminator
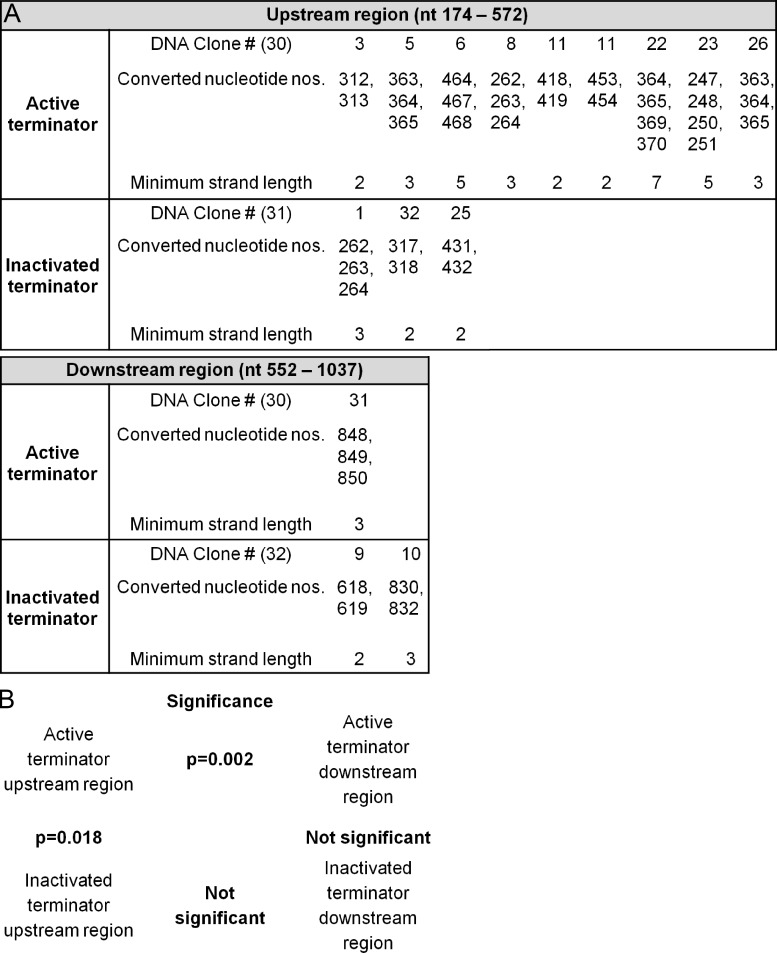
Because AID is known to require single-stranded DNA sequences (Dickerson et al., 2003), and because these likely arise by negative supercoils in the wake of Pol, we determined single-stranded DNA upstream and, as a control, downstream of the poly(A) site. This was investigated by using a bisulfite analysis of conversions of C and G nucleotides as previously described (Parsa et al., 2012). Bisulfite cannot access cytosines unless they are single-stranded. Comparing patches of at least two consecutive C or G conversions in the active and inactivated terminator containing cells, we found more patches of single-stranded DNA in the 5′ region with the active terminator (Fig. 7, S2). The analyses show that there is a significant difference in the upstream sequences between active and inactivated (P = 0.018), and in the active sequences between upstream and downstream (P = 0.002).
Figure 7.
Sodium bisulfite analysis to determine the presence of single-stranded DNA patches. (A) The DT40 cells with either the active or inactivated terminator were fixed and treated with sodium bisulfite (Parsa et al., 2012). The regions upstream and downstream of the poly(A) site of the terminator were amplified, sequenced, and analyzed for single-strand patches with at least 2 consecutive Cs converted to Ts or two consecutive Gs converted to As (minimum strand length). The DNA clone # is shown; the total number of DNA clones analyzed for each region are shown in parentheses. The control DT40 cells that were fixed but not treated with sodium bisulfite did not show any single-stranded DNA patches in any of the four regions. (B) The significant differences (or lack of) are indicated between the respective regions of the active and inactivated terminators.
DISCUSSION
The process of SHM of Ig genes (and non-Ig genes) requires that these genes are active. The question we addressed by placing a transcription terminator into the VJ region was whether the process of transcription itself was required for SHM or whether an active gene region was a target independent of transcript elongation by the RNA polymerase II (Pol). Eukaryotic Pol termination includes transcription of a poly(A) site followed by pausing of Pol and then endoribonucleotide cleavage of the nascent RNA (Colgan and Manley, 1997; Kuehner et al., 2011). The poly(A) addition signal, AAUAAA, is identified by the cleavage polyadenylation specificity factor that signals Pol to pause (Nag et al., 2007). Then the cleavage stimulation factor takes over and cleaves the nascent RNA between the AAUAAA and a G/U rich downstream element. However, after the cleavage Pol is still associated with the DNA and continues to polymerize RNA. The terminating uncapped nascent RNA molecule is removed by the action of the protein XRN2 (5′–3′ exoribonuclease 2; Kuehner et al., 2011), finally causing the release of Pol.
We had previously postulated that SHM is restricted to the 5′ VJ region and generally does not occur in the downstream constant region because the mutator factor AID associates with the transcription complex at or near the promoter, travels with Pol, and dissociates permanently from the transcription complex when AID deaminates C (Peters and Storb, 1996). We had reasoned that a terminator placed within the leader-VJ region might terminate both transcription and SHM. Our results show that polyadenylated transcripts 3′ of the poly(A) site are drastically reduced in active terminator knock-in clones as compared with inactivated terminator knock-in clones (Fig. 5 B). Thus, the human β-globin terminator works efficiently in the DT40 cells. In the presence of the active terminator, SHM is clearly reduced 3′ of the poly(A) site, almost twofold compared with the inactivated terminator (Fig. 3). However, there are still mutations in this downstream region, which is identical in the active and inactivated terminator constructs, except for two additional poly(A) sites that were changed in the inactivated terminator. Since this region is occupied by Pol in the active terminator construct to the same extent as in the inactivated terminator construct (Fig. 4), the terminating Pol is allowing less access of AID than the elongating Pol.
In cell-free assays, AID can act only on single-stranded DNA substrates and not on double-stranded DNA when in a relaxed linear fragment (Bransteitter et al., 2003, 2004; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003; Chaudhuri et al., 2004) and not on a relaxed circular plasmid (Shen and Storb, 2004). However, supercoiled DNA in a circular plasmid is an excellent target for AID (Shen and Storb, 2004). The negative supercoils generated upstream of a transcription bubble do have single-stranded DNA properties (Liu and Wang, 1987), and we had proposed that AID acts in these regions (Peters and Storb, 1996; Shen and Storb, 2004). Surprisingly, upstream of the active terminator, mutations were increased almost twofold compared with the inactivated terminator (Fig. 3). To assess how the 5′ region could be a more favored target for AID, we determined whether there was more single-strand DNA using bisulfite conversion of single-stranded C-to-T patches. This showed that upstream of the active terminator significantly more single-stranded DNA (SSD) patches exist than in the same sequence with the inactivated terminator (Fig. 7). We postulate that the SSD patches are located in negative supercoils that arise behind Pol that is temporarily pausing at the poly(A) site. Such a relationship between Pol pausing and SSD patches has been observed in the V region of B cells undergoing SHM (Parsa et al., 2012). Enhancement of the mutation frequency because of Pol briefly pausing due to the poly(A) site supports the model that Pol pausing in the V region might be a cause of AID’s preferred access to the V region (Peters and Storb, 1996). Convincing evidence for increased SHM by AID when transcription is paused has also been shown by Canugovi et al. (2009).
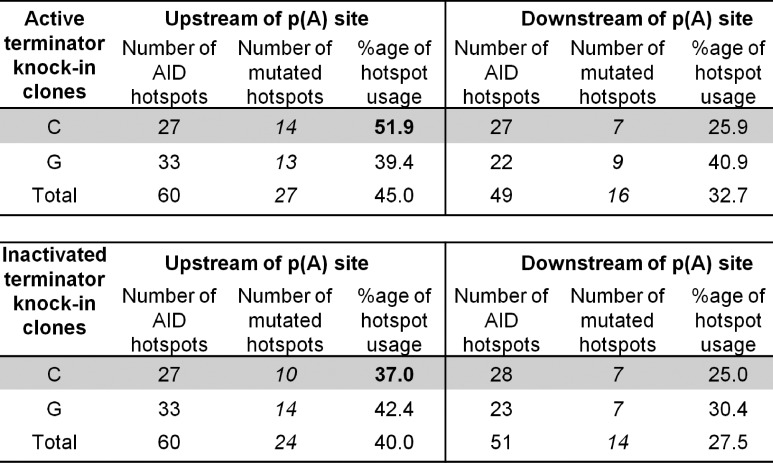
Thus, it appears that the molecular details of transcription itself are important for SHM. Furthermore, SHM appears to be more active with an elongating Pol than with a terminating Pol. With the active terminator the mutation frequency 5′ of the poly(A) site is 5.47 fold higher than 3′ of the AATAAA (Fig. 3 C); however, with the inactivated terminator mutations in the 5′ region are only 1.63 fold higher than 3′ (P < 0.0066; Fig. 3 C). Thus, the elongating Pol allows a 3.36 (5.47:1.63) times higher AID access than the terminating Pol. At the same time, SHM in the region upstream of the terminator shows a strand bias with a significant increase of mutations at C (C/G = 1.36; Fig. 6) whereas the same region in the cells with the inactive terminator and the downstream regions of both cell types show a bias for mutations at G (C/G = 0.67; 0.58; 0.31, respectively; Fig. 6). The higher frequency of mutations at C upstream of the poly(A) site in the active terminator knock-in clones coincides with increased targeting of C at AID hotspots (51.9%) in this region compared with the identical region in the inactivated terminator knock-in clones (37.0%; Fig. 8). Because all mutations at C and G are the effect of C deamination by AID, the C bias suggests that the nontranscribed strand was the preferred target in the upstream region.
Figure 8.
AID hotspot targeting in the active and inactivated terminator knock-in clones. Summary of AID hotspot (WRC and GYW) targeting in the active and inactivated terminator knock-in clones. The data are combined mutation numbers for two cell clones for the active and inactivated terminator, respectively.
We can only speculate why there were more single-stranded regions upstream of the active terminator: 3′ of the poly(A) site (position 493 from the start of transcription) nascent polyadenylated RNA is released in the active terminator cells. These ∼500 nt polyadenylated RNAs represent >90% of the total transcripts from the unique IgL promoter (Fig. 5). With the help of the single-strand DNA-binding protein, RPA, occupying the nontranscribed strand (Chaudhuri et al., 2004) and Spt5, a factor associated with paused Pol and single-stranded DNA, interacting with AID and facilitating AID recruitment to Ig genes (Pavri et al., 2010), this RNA may remain associated with the transcribed strand intermittently for a slightly extended time. When the transcribed strand is less single-stranded because of hybridized nascent RNA, AID would have less access to that strand and the nontranscribed strand would be favored.
These considerations would explain how AID is recruited during transcription of the active terminator containing gene. But how would AID be prevented from deaminating cytosines beyond ∼2 kb from the promoter here and during SHM in general? In SHM of Ig genes in general, there is no poly(A) site 5′ of the constant region. We have previously investigated SHM in Ung/Msh6 double-negative mice in which SHM shows an AID footprint, presenting only C > T and G > A transitions because secondary error-prone events of BER and MMR cannot occur (Shen et al., 2006). In these mice, the decrease of SHM beyond ∼2 kb is the same as in wild-type mice, indicating that AID acts in the 5′ region of Ig genes but not beyond. The same conclusion was evident in ung−/− mice (Longerich et al., 2005).
This restricted mutability is still a major puzzle concerning SHM. We propose the following possibility: during SHM many transcription complexes on Ig genes or, depending on the enhancer, non-Ig genes (Tanaka et al., 2010), do not carry AID and are responsible for the production of full-length mRNAs; stochastically, only some complexes carry AID. This suggestion is supported by cell-free experiments with SHM substrates and AID where only a small proportion of the substrate target molecules suffer deamination of C, but most of the molecules that are mutated have multiple mutations (Pham et al., 2003; Shen et al., 2005; Storb et al., 2009). We further consider two, not mutually exclusive, possibilities for the restriction of SHM to the region upstream of ∼2 kb: (a) Those transcription complexes that are associated with AID are terminating before and up to 2 kb from the transcription start because AID binds factors that can interact with Pol and the nascent RNA to cause the degradation of the RNA somewhere 5′ of 2 kb from the start of transcription. Indeed, recent findings have implicated the RNA exosome in SHM (Basu et al., 2011). It is possible that the RNA exosome, a cellular RNA-processing–degradation complex that was shown to interact with AID during SHM (Basu et al., 2011) acts efficiently in the upstream region of Ig genes. The “chewing” away of the nascent RNA would cause Pol to be released (Kuehner et al., 2011) so that termination of transcription and thus SHM would occur before transcription has reached ∼2 kb. (b) Pol has a natural tendency for pausing that can lead to stalling (Davenport et al., 2000; Galburt et al., 2007). Presumably, natural pausing would have the same result as the terminator effect observed in the current study. Thus, the chance for transcribing beyond ∼2 kb would be rare in transcription complexes associated with AID, and the gradual decrease in SHM of Ig genes in B cells and also with the inactivated terminator (Fig. 3), would be explained.
In summary, we conclude that ongoing transcription of the Ig gene coupled with temporary pausing within the targeted region is necessary for SHM, and that this facilitates SHM upstream of the pause. These observations are consistent with our previous model (Peters and Storb, 1996) in which active AID becomes associated with the Pol complex at the promoter or after the transition from the initiation to the elongation state.
MATERIALS AND METHODS
Cell culture and transgenic clones.
DT40 ψV knock-out cells derived from avian leukosis virus–induced chicken bursal B cells were a gift from H. Arakawa and J.M. Buerstedde (Institute of Molecular Radiology, Neuherberg, Germany; Arakawa et al., 2004). The cells were cultured in RPMI 1640 with 1% penicillin/streptomycin, 1% l-glutamine (Invitrogen), 1% chicken serum, and β-mercaptoethanol (Sigma-Aldrich) at 39.5°C with 5% CO2. The active and inactivated terminator knock-in constructs were linearized with SalI (Fig. 1) and transfected as previously described for DT40 cells (Arakawa et al., 2004). After 12 h, transfected cells were treated with 20 µg/ml blasticidin for selection of blasticidin resistance and single clones were isolated by subsequent limiting dilutions. Single clones from limiting dilutions were expanded to perform FACS analysis for surface-IgM negative clones and subsequently collect their genomic DNA for Southern blotting. Genomic DNA was digested with MluI or EcoNI–ApaLI, respectively, and a radioactive probe for the human β-globin transcription terminator region was used for Southern blot analysis. Two independent clones of the active and inactivated terminator knock-in clones, respectively, were selected for the study. Subsequently, the blasticidin drug marker gene present between two LoxP sites (Fig. 1) was excised by treating the cells with tamoxifen as discussed in the results section. The excision of blasticidin drug marker gene from the active and inactivated terminator knock-in clones was confirmed by PCR analysis, using primers pk07 and pk65 (Table S1). From six sub-clones, two sub-clones of each clone of the active and inactivated terminator knock-in clones, respectively, were selected for the somatic mutation analysis.
Flow-cytometric analysis and cell sorting.
DT40 knock-in clones for active and inactivated terminator were stained with PE-conjugated anti–chicken IgM antibody (Santa Cruz Biotechnology, Inc.) and were analyzed for loss of surface IgM and presence of AID-IRES-GFP expression of 50,000 live cells on an LSR II (BD) using DT40 CL18 cells and GFP+ ψV KO (a gift from H. Arakawa and J.M. Buerstedde; Arakawa et al., 2004) as gating controls. DT40 knock-in clones treated with tamoxifen were sorted for AID-IRES-GFP+ single cells on a cell sorter (FACSAria; BD) at the University of Chicago Flow Cytometry Facility.
RT-PCR analysis of transcripts in active and inactivated terminator knock-in clones.
Total RNA was made from DT40 cells with RNA STAT-60 (Tel-Test Inc.), recovered in 50 µl, and stored at −80°C. For Fig. 2, equal amounts of RNA were used for making complementary DNA (cDNA) by the SuperScript III First-Strand Synthesis System (Invitrogen) for RT-PCR using random hexamers. Real-time PCRs were run and analyzed on a MYiQ system with SYBR Green SuperMix (both from Bio-Rad Laboratories). Primers used were pk87 and pk85 for the region 1, pk154 and pk22 for the region 2, pk155, and pk10 for the region 3, pk93 and pk94 for the region 4, pk48 and pk50 for the lacZ region, pk12 and pk24 for the IgL VJ region, and gg-actin1 and gg-actin2 for the chicken β-actin gene (see Table S1 for primer sequences). PCR conditions were 95°C for 30 s, 64°C for 45 s, and 72°C for 60 s for 40 cycles. The data from chicken actin were used as a reference for the relative quantification of transcript levels using the Pfaffl method (Pfaffl, 2001).
Polyadenylated RNA assay.
Total RNA was made from DT40 cells with RNA STAT-60 (Tel-Test Inc.), recovered in 50 µl, and stored at −80°C. Equal amounts of RNA were used for making cDNA by the SuperScript III First-Strand Synthesis System (Invitrogen) for RT-PCR using oligo-dT primers. Real-time PCRs were run and analyzed on a MYiQ system with SYBR Green SuperMix (both from Bio-Rad Laboratories). Primers used were pk87 and pk182 for the region P1, pk103 and pk183 for the region P2, pk107 and pk184 for the region P3, pk174 and pk185 for the region P4, and gg-actin1 and gg-actin2 for the chicken actin region (see Table S1 for primer sequences). PCR conditions were 95°C for 30 s, 64°C for 45 s, and 72°C for 60 s for 40 cycles. The data from chicken actin were used as a reference for the relative quantification of poly(A) RNA levels using the Pfaffl method (Pfaffl, 2001).
Sodium bisulfite analysis.
The presence of single-stranded DNA patches in the active and inactivated terminators were determined by following the protocol of Parsa et al. (2012). In brief, 30 × 107 DT40 cells transfected with either the active terminator or the inactivated terminator were treated with 1% formaldehyde for 5 min at room temperature to cross-link the DNA and proteins. Of these formaldehyde-treated cells, 1 × 107 cells were treated with sodium bisulfite and 1 × 107 cells were treated as control in the absence of sodium bisulfite. The cells (both bisulfite-treated and control) were then reverse cross-linked and DNA was extracted. The upstream region from 174 to 572 bp was amplified using primers pk186 and pk187, and the downstream region from 552 to 1,037 bp was amplified with primers pk188 and pk189 (Table S1), using Taq polymerase at 95°C for 30 s, 57°C for 30 s, and 72°C for 60 s for 30 cycles in 50-µl reactions. The PCR products were cloned in TOPO TA vectors (Invitrogen) and sequenced with the M13 forward primer by the University of Chicago, Cancer Research Center DNA sequencing facility. The DNA sequences were analyzed for the presence of single-stranded patches where at least two consecutive Cs or Gs were converted to Ts or As respectively. The patches observed were only C-to-T or G-to-A conversions. In Fig. 7 the DNA clone number shows that the data were from different DNA clones that were distinguished by the presence of different mutations outside of the C and G patches. We compared the number of single-stranded DNA patches between the (a) upstream region of the active and inactivated terminator, (b) downstream region of the active and inactivated terminator, (c) upstream and downstream region of the active terminator, and (d) upstream and downstream region of the inactivated terminator.
Statistical analysis of single-strand patches by bisulfite conversion.
To assess the significance of differences in conversion patterns between inactive and active clones, we used the following procedure: We calculated the difference in proportions of converted mutations between active and inactivated sequences. We then constructed an empirical null distribution for this statistic by permuting clones between the two conditions. The statistical analysis on the differences observed was done by D. Nicolae, Departments of Human Genetics and Statistics, University of Chicago.
Identification of somatic mutations.
Mutations in knock-in clones were detected by PCR cloning using Pfu polymerase (Agilent Technologies), and primers pk64 and pk10 (Table S1) were used for PCR cloning with Pfu polymerase at 95°C for 30 s, 67°C for 30 s, and 72°C for 160 s for 25 cycles, and cloned with a PCR cloning kit (Zero Blunt TOPO; Invitrogen). DNA sequencing was performed by the University of Chicago Cancer Research Center DNA Sequencing Facility. Sequences with identical mutations at the same positions are not included.
NRO assay probes.
The double-stranded (ds) DNA probes for the NRO assay were made by PCR amplification using the active terminator knock-in construct as the template. Primers used were pk95 and pk96 for the IgL leader region, pk99 and pk100 for the ter1 region, pk103 and pk104 for the ter2 region, pk107 and pk108 for the ter3 region, pk111 and pk112 for the lacZ region, pk113 and pk114 for the IgL VJ region, pk117 and pk118 for the IgL C region and gg-actin1 and gg-actin2 for the chicken actin region (see Table S1 for primer sequences). PCR conditions were 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 30 cycles.
NRO assay.
Cells were harvested by centrifugation, washed with PBS, and resuspended in 1 ml of ice-cold NRO wash buffer (10 mM Tris-HCl, pH 8.0, 0.3 M sucrose, 3 mM CaCl2, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.15 mM spermine, 0.5 mM spermidine and protease inhibitor) plus 0.5% Nonidet P-40. After incubation on ice (5 min), nuclei were washed with 1 ml ice-cold NRO wash buffer. Nuclei were resuspended in 290 µl of transcription buffer (10 mM Tris-HCl pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.1 mM MnCl2, 1 mM SAM, 10% glycerol, 40 U RNasin, 10 mM DTT, and 0.5 mM AGC mix [rATP, rGTP, rCTP]) plus 60 µCi[α-32P]-UTP (800 Ci/mmol; GE Healthcare). Transcription reactions were performed at 37°C for 30 min and terminated by centrifugation for 30 s. Nuclei were resuspended in HSB (10 mM Tris-HCl, pH 7.5, 0.5 M NaCl, and 10 mM MgCl2) containing 10 U RNase-free DNase and incubated at 37°C for 5 min. Total RNA was made from DT40 cells with RNA STAT-60 (Tel-Test Inc.), recovered in 50 µl, and incubated in DNaseI buffer (10 mM Tris-HCl, pH 7.5, and 10 mM MgCl2) containing 10 units of DNaseI for 30 min at 37°C. RNA was again extracted, ethanol precipitated, and resuspended in water. After partial RNA hydrolysis by 0.2 M NaOH on ice for 5 min, the RNA was neutralized with 0.1 M Tris/0.2 M HCl. Hybridization of the labeled RNA to a preblocked filter was performed overnight at 42°C in 50% formamide, 6× SSPE, 5× Denhardt’s solution, 0.1% SDS, and 100 µg/ml of tRNA. Filters were made with a dot blot (Schleicher & Schuell Bioscience, Inc.) with 500 ng of dsDNA. DNA was denatured at 95°C for 10 min, snap chilled on ice, and then applied to each slot. Filters were washed in 1× SSPE/0.1% SDS at room temperature for 20 min and in 0.1× SSPE/0.1% SDS at 42°C for 10 min, and then exposed to x-ray films for 24 h. Signals were quantitated with the ImageJ program (National Institutes of Health).
ChIP.
Cells (107) were cross-linked with formaldehyde 1% for 10 min at 37°C. The reaction was stopped by adding glycine to final concentration 0.125 M for 5 min at room temperature. Fixed cells were rinsed twice with PBS plus protease inhibitor and resuspended in Lysis Buffer 1 (50 mM Tris-HCl pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100), rocked for 10 min at 4°C to isolate nuclei. Samples were centrifuged at 1,350× g and pellet washed with Lysis Buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, pH 8.0, and 0.5 mM EGTA) by gently rocking for 5 min at room temperature. Nuclei were resuspended in Lysis Buffer 3 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, pH 8.0, 0.5 mM EGTA, and 0.5% N-lauroylsarcosine) and incubated on ice for 10 min. The lysate was sonicated for 18 min (30 s on/30 s off) in a Bioruptor (Diagenode) water bath sonicator and centrifuged at 13,200 rpm at 4°C for 10 min. The cleared supernatant was used immediately in ChIP experiments. 50–150 μg of sonicated chromatin was diluted 10 times in ChIP Dilution Buffer (16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, pH 8.0, and 1.1% Triton X-100) and precleared for 1 h, rotating at 4°C, with 50 μl blocked beads (rProtein G Agarose; Invitrogen) before the overnight incubation with 20 μl of the anti RNA Pol antibody (C-18; Santa Cruz Biotechnology, Inc.). The bound material was recovered with 50 μl blocked rProtein G Agarose beads, rotating at 4°C for 1 h. The beads were washed twice for 5 min in ChIP Wash Buffer 1 (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, pH 8.0, and 1% Triton X-100), twice in ChIP Wash Buffer 2 (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, pH 8.0, and 1% Triton X-100), and twice in TE. ChIPed material was eluted by two 15-min incubations at room temperature with 250 µl Elution Buffer (0.1 M NaHCO3, 1% SDS). Chromatin was reverse-cross-linked by adding 25 μl of 4 M NaCl and incubated at 65°C for 4 h, and DNA was submitted to RNase and proteinase K digestion and extracted by phenol-chloroform. Primers used were pk64 and pk03 for the IgL leader region, pk07 and pk85 for the region ter-1.1, pk31 and pk22 for ter-2.1, pk155 and pk33 for ter-2.2, and pk12 and pk24 for the IgL VJ region (see Table S1 for primer sequences). PCR conditions were 95°C for 30 s, 64°C for 45 s, and 72°C for 60 s for 40 cycles. The data presented are mean levels of immunoprecipitated DNA to the input DNA, normalized for the IgL leader region using the Pfaffl method (Pfaffl, 2001).
Online supplemental material.
The human β-globin transcription terminator sequence is shown in Fig. S1. Figs. S2 and S3 shows mutations in the active and inactivated terminator knock-in clones. Primers used in this study are listed in Table S1. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121523/DC1.
Supplementary Material
Acknowledgments
We thank N.J. Proudfoot and M.J. Dye (University of Oxford, Oxford, England, UK) for the human β-globin terminator vector and sequence. We thank H. Arakawa and J.M. Buerstedde (Institute of Molecular Radiology, Neuherberg, Germany) for the DT40 ψV knock-out cells, W. Buikema and C. Hall for DNA sequencing, and R. Duggan for flow cytometric cell sorting. We are grateful to D. Nicolae for statistical advice and B. Kee for technical advice and use of her instrument for real-time PCR.
P. Kodgire thanks the Lady Tata Memorial Trust, UK, and the Cancer Research Institute, USA, for postdoctoral fellowships, and Department of Science and Technology, Government of India for the Ramanujan Fellowship. This work was supported by the National Institutes of Health (AI081167 and AI047380 to U. Storb).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- cDNA
- complementary DNA
- ChIP
- chromatin immunoprecipitation
- CSR
- class switch recombination
- NRO
- nuclear run-on
- Pol
- RNA polymerase SHM, somatic hypermutation
References
- Arakawa H., Saribasak H., Buerstedde J.M. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179 10.1371/journal.pbio.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U., Meng F.L., Keim C., Grinstein V., Pefanis E., Eccleston J., Zhang T., Myers D., Wasserman C.R., Wesemann D.R., et al. 2011. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 144:353–363 10.1016/j.cell.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A.G., Milstein C., González-Fernández A., Pannell R., Larson T., Neuberger M.S. 1994. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 77:239–248 10.1016/0092-8674(94)90316-6 [DOI] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Scharff M.D., Goodman M.F. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107 10.1073/pnas.0730835100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Calabrese P., Goodman M.F. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612–51621 10.1074/jbc.M408135200 [DOI] [PubMed] [Google Scholar]
- Canugovi C., Samaranayake M., Bhagwat A.S. 2009. Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J. 23:34–44 10.1096/fj.08-115352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730 10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Khuong C., Alt F.W. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 430:992–998 10.1038/nature02821 [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Manley J.L. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766 10.1101/gad.11.21.2755 [DOI] [PubMed] [Google Scholar]
- Conley M.E., Dobbs A.K., Farmer D.M., Kilic S., Paris K., Grigoriadou S., Coustan-Smith E., Howard V., Campana D. 2009. Primary B cell immunodeficiencies: comparisons and contrasts. Annu. Rev. Immunol. 27:199–227 10.1146/annurev.immunol.021908.132649 [DOI] [PubMed] [Google Scholar]
- Davenport R.J., Wuite G.J., Landick R., Bustamante C. 2000. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 287:2497–2500 10.1126/science.287.5462.2497 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Williams G.T., Chan D.T., Buerstedde J.M., Baldwin G.S., Neuberger M.S. 2007. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 204:3209–3219 10.1084/jem.20071768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296 10.1084/jem.20030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye M.J., Proudfoot N.J. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 105:669–681 10.1016/S0092-8674(01)00372-5 [DOI] [PubMed] [Google Scholar]
- Galburt E.A., Grill S.W., Wiedmann A., Lubkowska L., Choy J., Nogales E., Kashlev M., Bustamante C. 2007. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 446:820–823 10.1038/nature05701 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Plath K. 2009. Epigenetic reprogramming and induced pluripotency. Development. 136:509–523 10.1242/dev.020867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz E.L., Storb U. 1996. Somatic hypermutation of a lambda 2 transgene under the control of the lambda enhancer or the heavy chain intron enhancer. J. Immunol. 157:4458–4463 [PubMed] [Google Scholar]
- Kuehner J.N., Pearson E.L., Moore C. 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12:283–294 10.1038/nrm3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Wang J.C. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 84:7024–7027 10.1073/pnas.84.20.7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Longerich S., Tanaka A., Bozek G., Nicolae D., Storb U. 2005. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. J. Exp. Med. 202:1443–1454 10.1084/jem.20051604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S., Basu U., Alt F., Storb U. 2006. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 18:164–174 10.1016/j.coi.2006.01.008 [DOI] [PubMed] [Google Scholar]
- Nag A., Narsinh K., Martinson H.G. 2007. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat. Struct. Mol. Biol. 14:662–669 10.1038/nsmb1253 [DOI] [PubMed] [Google Scholar]
- Nambu Y., Sugai M., Gonda H., Lee C.G., Katakai T., Agata Y., Yokota Y., Shimizu A. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 302:2137–2140 10.1126/science.1092481 [DOI] [PubMed] [Google Scholar]
- Parsa J.Y., Ramachandran S., Zaheen A., Nepal R.M., Kapelnikov A., Belcheva A., Berru M., Ronai D., Martin A. 2012. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID-mediated mutagenesis. PLoS Genet. 8:e1002518 10.1371/journal.pgen.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Küppers R., Dalla-Favera R. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346 10.1038/35085588 [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 143:122–133 10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S., Kalis S.L., Goodman M.F., Scharff M.D. 2008. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26:481–511 10.1146/annurev.immunol.26.021607.090236 [DOI] [PubMed] [Google Scholar]
- Peters A., Storb U. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 4:57–65 10.1016/S1074-7613(00)80298-8 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Popp C., Dean W., Feng S., Cokus S.J., Andrews S., Pellegrini M., Jacobsen S.E., Reik W. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 463:1101–1105 10.1038/nature08829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755 10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 135:1201–1212 10.1016/j.cell.2008.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos P., Jankovic M., Nussenzweig M.C. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456 10.1038/ni920 [DOI] [PubMed] [Google Scholar]
- Shen H.M., Storb U. 2004. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. USA. 101:12997–13002 10.1073/pnas.0404974101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.M., Peters A., Baron B., Zhu X., Storb U. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 280:1750–1752 10.1126/science.280.5370.1750 [DOI] [PubMed] [Google Scholar]
- Shen H.M., Ratnam S., Storb U. 2005. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 25:10815–10821 10.1128/MCB.25.24.10815-10821.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.M., Tanaka A., Bozek G., Nicolae D., Storb U. 2006. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J. Immunol. 177:5386–5392 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U., Shen H.M., Longerich S., Ratnam S., Tanaka A., Bozek G., Pylawka S. 2007. Targeting of AID to immunoglobulin genes. Adv. Exp. Med. Biol. 596:83–91 10.1007/0-387-46530-8_8 [DOI] [PubMed] [Google Scholar]
- Storb U., Shen H.M., Nicolae D. 2009. Somatic hypermutation: processivity of the cytosine deaminase AID and error-free repair of the resulting uracils. Cell Cycle. 8:3097–3101 10.4161/cc.8.19.9658 [DOI] [PubMed] [Google Scholar]
- Storck S., Aoufouchi S., Weill J.C., Reynaud C.A. 2011. AID and partners: for better and (not) for worse. Curr. Opin. Immunol. 23:337–344 10.1016/j.coi.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Shen H.M., Ratnam S., Kodgire P., Storb U. 2010. Attracting AID to targets of somatic hypermutation. J. Exp. Med. 207:405–415 10.1084/jem.20090821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Shinkura R., Doi Y., Maruya M., Fagarasan S., Honjo T. 2011. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 12:264–270 10.1038/ni.1991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.