Notch1 signaling sustains the proinflammatory behavior of Th1 cells, implicated in the development of aplastic anemia in humans and mice.
Abstract
Severe aplastic anemia (AA) is a bone marrow (BM) failure (BMF) disease frequently caused by aberrant immune destruction of blood progenitors. Although a Th1-mediated pathology is well described for AA, molecular mechanisms driving disease progression remain ill defined. The NOTCH signaling pathway mediates Th1 cell differentiation in the presence of polarizing cytokines, an action requiring enzymatic processing of NOTCH receptors by γ-secretase. Using a mouse model of AA, we demonstrate that expression of both intracellular NOTCH1IC and T-BET, a key transcription factor regulating Th1 cell differentiation, was increased in spleen and BM-infiltrating T cells during active disease. Conditionally deleting Notch1 or administering γ-secretase inhibitors (GSIs) in vivo attenuated disease and rescued mice from lethal BMF. In peripheral T cells from patients with untreated AA, NOTCH1IC was significantly elevated and bound to the TBX21 promoter, showing NOTCH1 directly regulates the gene encoding T-BET. Treating patient cells with GSIs in vitro lowered NOTCH1IC levels, decreased NOTCH1 detectable at the TBX21 promoter, and decreased T-BET expression, indicating that NOTCH1 signaling is responsive to GSIs during active disease. Collectively, these results identify NOTCH signaling as a primary driver of Th1-mediated pathogenesis in AA and may represent a novel target for therapeutic intervention.
Severe aplastic anemia (AA) is an acquired BM failure (BMF) syndrome (Young et al., 2008). Evidence in the majority of cases suggests that a breakdown in self-tolerance leads to infiltration of destructive T helper type-1 (Th1) cells into the BM where they target hematopoietic stem cells and compromise stromal cells through bystander effects (Chen et al., 2005; Young et al., 2008). As a result, the population of self-renewing progenitors in the BM is destroyed. Without the ability to replenish platelets and red and white blood cells, patients with AA are at increased risk of bleeding episodes, hypoxia, and infection. If left untreated, AA is uniformly fatal (Dezern and Brodsky, 2011).
Although the pathology of AA is well defined, there is a significant gap in our understanding of the molecular mechanisms that drive disease progression. Unlike other autoimmune diseases, such as multiple sclerosis (MS) or type I diabetes, an inciting self-antigen has not been identified for AA; thus, there are no true autoimmune animal models of the disease. However, mouse models of immune-mediated BMF have been successfully generated by transferring parental splenocytes or lymph node cells into minor histocompatibility– or major histocompatibility–mismatched recipients, and the ensuing immune response robustly targets BM cells (Bloom et al., 2004; Chen, 2005; Chen et al., 2007). The utility of these AA mouse models is well established; mice exhibit many of the clinical features of AA, and they provide an excellent system with which to study underlying mechanisms of disease and to test the efficacy of potential therapeutics (Bloom et al., 2004; Chen et al., 2007). Furthermore, the modulation of disease severity in these mice achieved using immunosuppressive therapies (ISTs) validates the immune-mediated pathology of the model.
The NOTCH family is an evolutionarily conserved group of transmembrane receptors and ligands. In mammals, it is comprised of four receptors, NOTCH1–4, and five ligands, designated Jagged1 and -2 and Delta-like-1, -3, and -4. Signaling is initiated when NOTCH receptors engage cognate ligands, leading to sequential cleavage events that culminate in release from the cell membrane of the intracellular, signaling-competent form of NOTCH (NOTCHIC; Osborne and Minter, 2007). Once liberated from the inner membrane, NOTCHIC translocates to the nucleus and regulates the transcription of numerous genes either through its canonical nuclear binding partner, CBF1/Suppressor of Hairless/Lag1 (CSL), or through interaction with noncanonical partners, such as members of the NF-κB family of transcriptional regulators (Minter and Osborne, 2012). The final cleavage that untethers NOTCH receptors from the cell membrane is mediated by the enzymatic action of γ-secretase and can be blocked with pharmacological inhibitors (De Strooper et al., 1999; Shih and Wang, 2007). γ-Secretase inhibitors (GSIs) successfully prevent the final enzymatic step required for NOTCH cleavage and activation and can block NOTCH signaling in vitro and in vivo (Minter et al., 2005; Wei et al., 2010). As a result, the use of GSIs as a therapeutic modality is the focus of substantial and growing interest. Numerous clinical trials are under way with several chemically distinct GSIs, primarily in oncology. Despite initial concerns that dose-limiting toxicity may reduce the clinical usefulness of these drugs, intermittent administration of GSIs is safe and well tolerated. Two phase 1 clinical trials in early-stage and metastatic ER+ breast cancer with endocrine therapy/GSI combinations have shown safety, tolerability, and preliminary suggestions of efficacy (Albain et al., 2011. Thirty-Fourth Annual CTRC-AACR San Antonio Breast Cancer Symposium. Abstr. S1-5.; Means-Powell et al. 2012. Thirty-Fifth Annual CTRC-AACR San Antonio Breast Cancer Symposium. Abstr. P2-14-04).
NOTCH1IC has been shown to directly regulate genes involved in T cell activation, cell cycle progression, differentiation, cytokine production, and effector cell function (Adler et al., 2003; Palaga et al., 2003; Osborne and Minter, 2007; Joshi et al., 2009). We and others have demonstrated that NOTCH signaling can facilitate differentiation of naive T cells to a Th1 phenotype in mice (Maekawa et al., 2003; Minter et al., 2005; Skokos and Nussenzweig, 2007; Zhang et al., 2011). One way NOTCH1 drives Th1 polarization is by influencing expression of the T-box transcription factor, Tbx21, the gene encoding T-BET, which is a master transcriptional regulator of Th1 cell differentiation (Szabo et al., 2000). T-BET is often up-regulated in patients with AA, and mouse models using cells deficient in Tbx21 show less severe disease induction (Tang et al., 2010). Furthermore, T-BET expression may serve as a biomarker for response to IST because high levels of T-BET have been observed in patients who are refractory to IST, whereas, for those who respond to IST and remain in remission, T-BET expression in circulating PBMCs is below detectable limits (Solomou et al., 2006).
Given evidence for a Th1-mediated pathology of AA, a defined role for T-BET in promoting Th1-mediated immune responses, and previous observations that NOTCH1 influences T-BET levels, we asked whether NOTCH1 signaling contributes to disease pathogenesis in AA. Using a mouse model of AA, we show that the cleaved, active form of NOTCH1 (NOTCH1IC) is increased in the T cells of mice with AA and abrogating NOTCH1 signaling through genetic or pharmacological inhibition attenuates disease. Importantly, at the efficacious dose, extended GSI treatment showed no adverse effects on engraftment or long-term hematopoiesis, as assessed by serial BM transplantation. We further demonstrate that in PBMCs from patients with untreated AA, NOTCH1IC is increased, can be detected bound to the TBX21 promoter, and is lost from the promoter after GSI treatment. Collectively, our findings demonstrate that NOTCH1 is a critical mediator of Th1 pathology in AA through its direct regulation of TBX21 and is responsive to the inhibitory actions of GSIs, both in vitro and in vivo.
RESULTS
NOTCH1 is activated in a mouse model of immune-mediated BMF
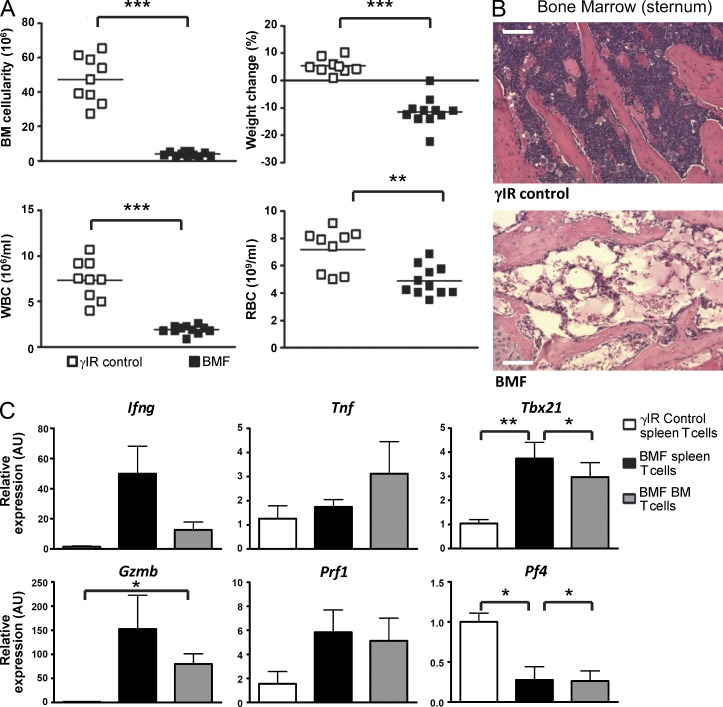
We previously demonstrated a CD4+ T cell–intrinsic role for NOTCH1 in promoting Th1 cell differentiation (Minter et al., 2005). However, a role for NOTCH signaling in the Th1-mediated human BMF disease, severe AA, has not previously been examined. To explore the mechanisms by which NOTCH signaling may contribute to disease progression, we optimized a murine model of immune-meditated BMF (AA mice) that is highly representative of human AA. This MHC-mismatched lymphocyte transfer model combines the bulk splenocyte transfer method of one model (Yada et al., 2005) with the parental and F1 hybrid genetic backgrounds used in another model (Chen et al., 2005). Donor-derived, destructive T cells infiltrate the BM of AA mice causing hypoplastic BM, pancytopenia, increased levels of IFN-γ and TNF, and a gene expression profile consistent with that found in patients with AA (Franzke et al., 2006; Fig. 1). As with the human disease, mice left untreated uniformly succumb to BMF by 25 d after disease induction.
Figure 1.
Induction of BMF in AA mice recapitulates human disease. (A) BM cellularity (top left), weight loss (top right), and peripheral pancytopenia (bottom) at day 17 after disease induction for control mice (open boxes; n = 9) or AA mice (closed boxes; n = 11). (B) Representative hematoxylin and eosin staining of sternum from one AA mouse (bottom) compared with one irradiation control (top). Bars, 200 µm. (C) The relative expression of Ifng, Tnf, Tbx21, Gzmb, Prf1, and Pf4 in T cells isolated from spleens and BM of control and AA mice was determined by real-time PCR and normalized to naive T cells isolated from irradiation controls (n = 3–5 samples of pooled T cells, with each sample generated from four to eight mice). Data represent the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
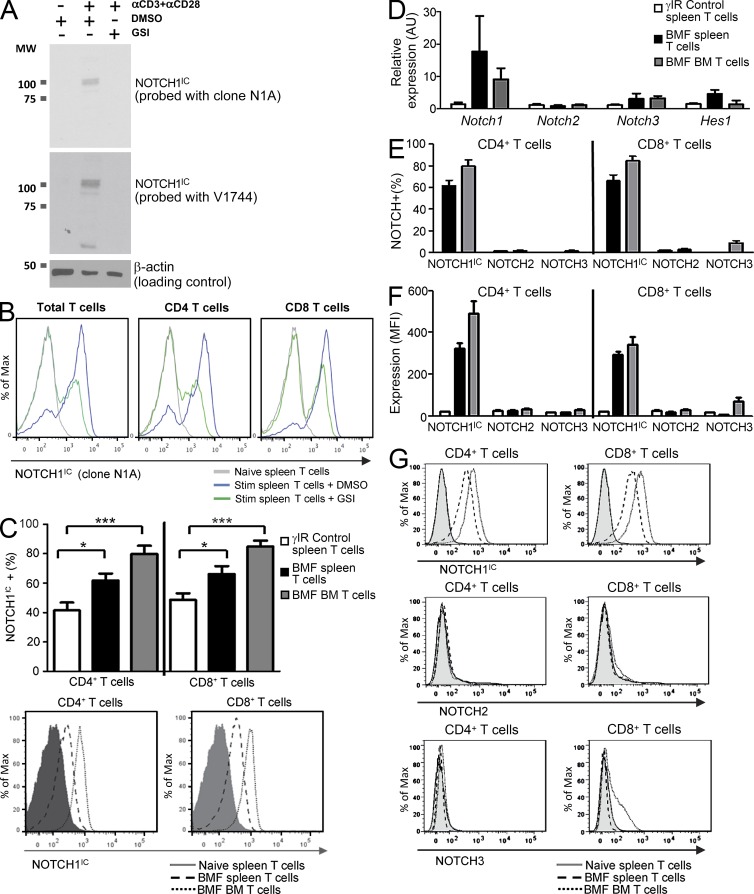
Assessing protein expression and intracellular localization using a limited number of cells presents a technical challenge, as is the case with cells recovered from murine BM or from patient PBMCs. Therefore, we validated a flow cytometric approach for determining levels of intracellular NOTCH1 against conventional immunoblotting techniques, using two distinct antibodies shown to recognize NOTCH1IC (Fig. 2, A and B).
Figure 2.
NOTCH1 is increased during immune-mediated BMF. Flow cytometric detection of NOTCH1IC was validated using conventional immunoblotting methods. (A) CD4+ and CD8+ T cells from C57BL/6 mice were left unstimulated (first lane) or stimulated for 48 h with anti-CD3ε plus anti-CD28 in the absence (second lane) or presence (third lane) of GSIs. Whole cell lysates were generated from one half of the sample, separated by SDS-PAGE, and probed with clone N1A (top), stripped, and reprobed with Val1744 (middle), both of which recognize NOTCH1IC. Blots were stripped again then reprobed with anti–β-actin to verify equal loading. Data are representative of three independent replicates. (B) The other half of the sample was used to assess NOTCH1IC by flow cytometry using anti-NOTCH1IC, clone N1A, in total T cells (left), CD4+ T cells (middle), and CD8+ T cells (right). Data are representative of three independent replicates. (C) Percentages of NOTCH1IC-expressing cells in spleens and BM of control and AA mice were determined by flow cytometry; a representative histogram shows NOTCH1IC expression in CD4+ and CD8+ T cells isolated from naive spleens and from spleens and BM of mice with BMF (n = 9–11 mice/group). (D) Expression of Notch1, Notch2, Notch3, and Hes1 in T cells isolated from control spleens and from spleens and BM of AA mice was determined by real-time PCR (n = 3–5 samples, each pooled from four to eight mice). (E and F) Percent positive (E) and MFI (F) of NOTCH1IC (n = 9–11)-, NOTCH2 (n = 4)-, and NOTCH3 (n = 4)-expressing CD4+ and CD8+ T cells from spleens and BM of control and AA mice. (G) Representative histograms show NOTCH1IC, NOTCH2, and NOTCH3 expression in CD4+ and CD8+ T cells isolated from naive spleens and from spleens and BM of mice with BMF (n = 4–11 mice/group, as noted in F). Data represent the mean ± SEM. *, P < 0.05; ***, P < 0.001.
After validation, we used flow cytometry to detect NOTCH1IC in a significantly greater percentage of CD4+ and CD8+ T cells isolated from spleens of AA mice compared with spleens isolated from mice receiving only irradiation (Fig. 2 C). Naive T cells from spleens of unmanipulated mice showed negligible NOTCH1IC expression (Fig. 2 C, histogram; and not depicted). In addition, percentages of NOTCH1IC-expressing CD4+ and CD8+ T cells that migrated to and infiltrated the BM were higher in AA mice (P < 0.001) and showed increased NOTCH1IC (Fig. 2 C) compared with control mice. When we analyzed gene expression of NOTCH receptors, we found a 10-fold greater expression of Notch1 compared with Notch2 (unchanged) and Notch3 (2-fold increase; Fig. 2 D). This finding supports previous observations that NOTCH1 drives its own expression (Deftos et al., 1998) and correlates with a concomitant increase in NOTCH1 but not NOTCH2 or NOTCH3 proteins (Fig. 2, E–G). These data document for the first time that NOTCH1IC is increased in the T cells of mice with immune-mediated BMF and, importantly, that these AA mice represent an excellent model with which to assess its contribution to disease pathology.
Abrogating NOTCH1 signaling reduces expression of AA proinflammatory proteins
A hallmark of AA is the expression of a signature profile of proinflammatory molecules (Young et al., 2008). These include IFN-γ, a potent Th1-associated cytokine, GRANZYME B, an effector molecule expressed in cytotoxic CD8+ T cells, and T-BET, a transcription factor deemed to be the master regulator of CD4+ Th1 cell differentiation (Kook et al., 2001; Young et al., 2008; Tang et al., 2010). Patients often present with high levels of T-BET at the time of diagnosis, and expression of T-BET seems to correlate with patient response to IST (Solomou et al., 2006). Because studies previously linked NOTCH signaling to regulation of Th1-associated molecules, including T-BET (Maekawa et al., 2003; Minter et al., 2005), we asked whether expression of the proinflammatory proteins that drive AA pathology requires intact NOTCH1 signaling.
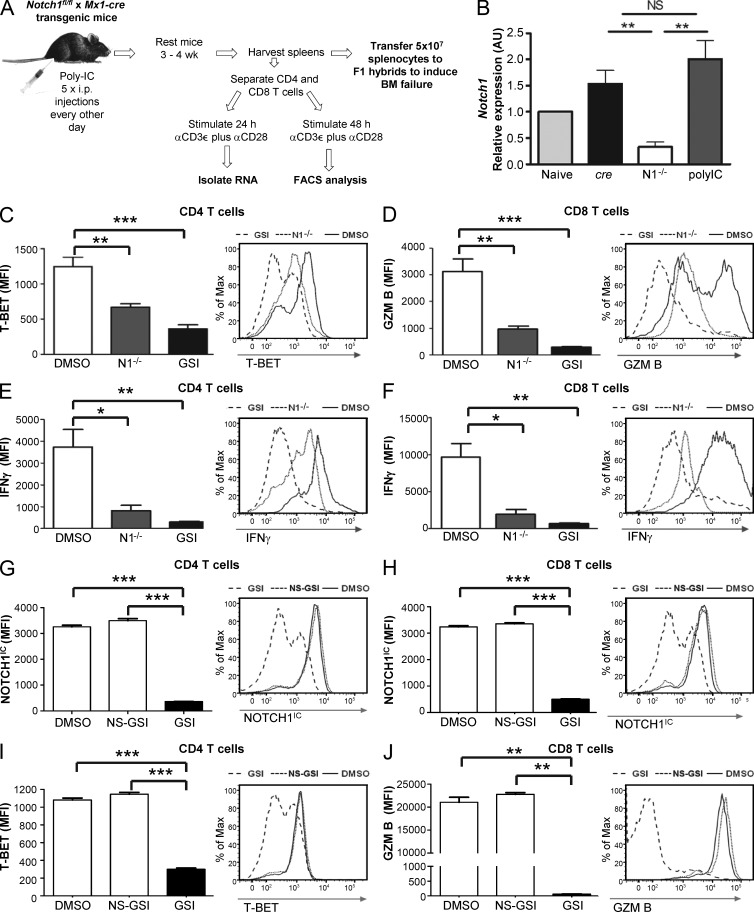
To do this, we generated mice in which Notch1 was functional during T cell development but which could be conditionally deleted from mature peripheral T cells. NOTCH signaling is required for T cell development. When NOTCH1 is deleted early in thymocyte development, i.e., when cre expression is driven by the CD4 promoter, NOTCH2 can substitute in a redundant fashion and generate mature T cells with intact IFN-γ production (Auderset et al., 2012). Thus, we chose to induce deletion of NOTCH1 from IFN-responsive, mature T cells to examine the specific effects of NOTCH1 signaling in peripheral T cells that had developed under normal thymic conditions. We crossed Notch1fl/fl mice (Notch1tm2Rko/GridJ) to IFN-responsive Mx1-cre+/− mice (B6.Cg-Tg(Mx1-cre)1Cgn/J) to generate Notch1fl/fl Mx1-cre+/− animals. We then induced deletion of floxed Notch1 alleles by treating these mice with serial injections of the IFN-inducing compound, polyinosinic:polycytidylic acid (polyIC; Fig. 3 A). PolyIC-treated Notch1fl/fl Mx1-cre+/− donor mice (abbreviated as N1−/− mice) expressed significantly reduced Notch1 compared with control mice (Fig. 3 B).
Figure 3.
Abrogating NOTCH1 signaling reduces expression of signature proinflammatory proteins. (A) Schematic of experimental approach for generating and evaluating N1−/− mice. (B) Relative expression of Notch1 in T cells isolated from the spleens of N1−/− mice was determined by real-time PCR and normalized to naive T cells isolated from control littermates (data are the mean ± SEM of at least three replicates). (C–F) CD4+ and CD8+ T cells from C57BL/6 mice were treated with vehicle only (DMSO) or with the GSI IL-CHO (GSI) before being stimulated for 72 h with anti-CD3ε plus anti-CD28. CD4+ and CD8+ T cells from N1−/− mice were stimulated for 72 h with anti-CD3ε plus anti-CD28. After 72 h, the level of T-BET (C) and GRANZYME B (D) was determined. (E and F) After a further 5-h restimulation with anti-CD3ε, in the presence of GolgiPlug, IFN-γ was determined by intracellular staining and flow cytometric methods (for C–F, data represent the mean ± SEM of at least three independent replicates). (G–J) Human CD4+ and CD8+ T cells were treated with vehicle only (DMSO), with the NS-GSI JLK-6 (NS-GSI), or with the NOTCH-inhibiting GSI IL-CHO (GSI) before being stimulated for 72 h with anti-CD3ε plus anti-CD28, under Th1-polarizing conditions. After 72 h, expression of NOTCH1IC (G and H), T-BET (I), and GRANZYME B (J) were determined by flow cytometry (for G–J, data are the mean ± SEM of at least three replicates). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
We isolated T cells from the spleens of N1−/− mice and cultured them in vitro for 72 h under conditions favoring Th1 cell differentiation. When we compared CD4+ and CD8+ T cells from N1−/− mice with T cells from wild-type controls, we found that the median fluorescence intensity (MFI), an indicator of protein expressed on a per cell basis, of T-BET expressed in CD4+ T cells (Fig. 3 C), GRANZYME B expressed in CD8+ cells (Fig. 3 D), and IFN-γ in both subsets (Fig. 3, E and F) was significantly reduced. Treating wild-type T cells with GSIs also robustly inhibited expression of these proteins (Fig. 3, C–F) and phenocopied results obtained using N1−/− cells.
Numerous substrates of γ-secretase, in addition to NOTCH receptors, have been identified in immune cells (Haapasalo and Kovacs, 2011). To discern whether other substrates might be responsible for the observed suppression of Th1-associated proteins during GSI treatment, we incubated cells with JLK-6, a NOTCH-sparing GSI (NS-GSI) which inhibits enzymatic processing of a broad range of γ-secretase substrates but which does permit cleavage of NOTCH receptors (Petit et al., 2001, 2003; Hellström et al., 2007). In contrast to GSI treatment, when NS-GSI was added to cells, Th1-associated proteins were expressed at levels equivalent to vehicle-only (DMSO)–treated cells (Fig. 3 G-J), suggesting that these signaling and effector molecules are regulated by NOTCH1 and not by another γ-secretase target. Collectively, these data support a role for NOTCH1 signaling in driving expression of Th1-associated proinflammatory proteins that are a hallmark of AA.
Conditionally deleting Notch1 ameliorates disease in AA mice
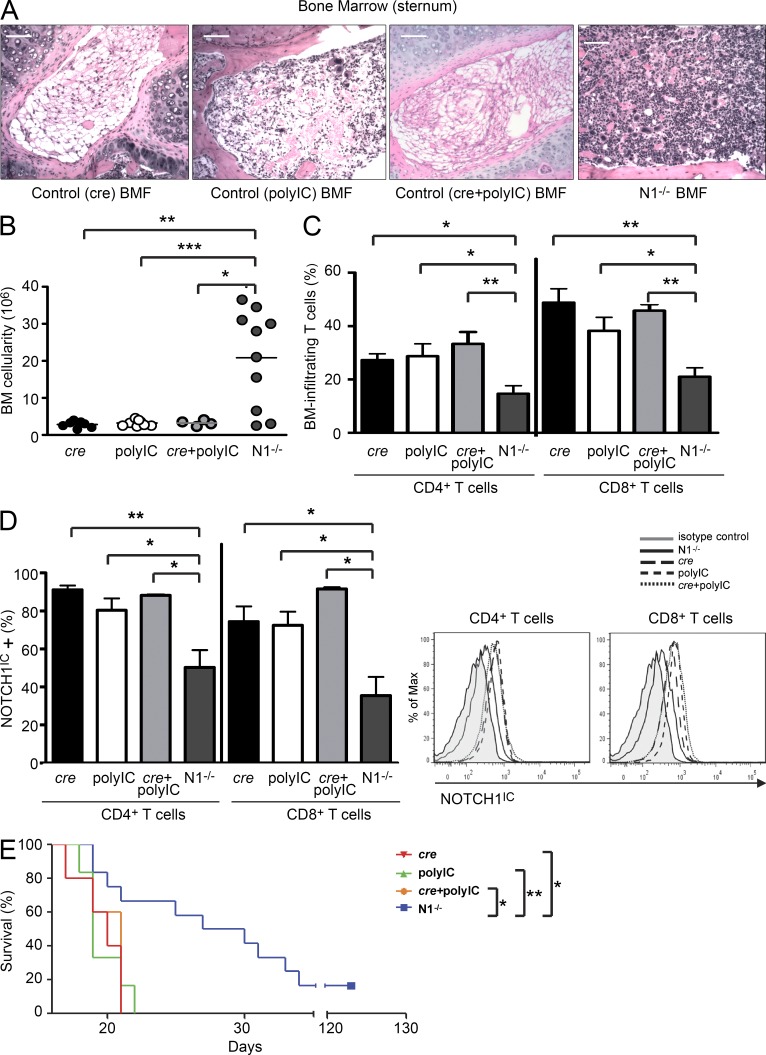
To confirm that the elevated NOTCH1IC observed in splenocytes and BM-infiltrating T cells in AA mice was contributing to disease pathology (Figs. 1 and 2), we used donor splenocytes from N1−/− mice to induce BMF in AA mice. Compared with transferring control splenocytes, we found that transferring N1−/− splenocytes induced significantly less severe disease, as measured by BM cellularity (Fig. 4, A and B), peripheral blood counts, weight loss, and plasma IFN-γ and TNF levels (not depicted). Mice receiving N1−/− splenocytes had fewer BM-infiltrating CD4+ and CD8+ T cells compared with wild-type–induced animals (Fig. 4 C), and of those T cells that did migrate to the BM, the percentage that expressed NOTCH1IC was significantly reduced (Fig. 4 D). In survival experiments, this translated into median survival times of 28.5 d (range = 19–123 d) for mice whose BMF was induced with N1−/− splenocytes (Fig. 4 E), compared with 21 d for mice induced with splenocytes from control mice that expressed cre but did not receive polyIC treatments (cre controls; range = 19–25 d), 19 d for control mice that did not express cre but received injections of polyIC (polyIC control; range = 18–22 d), and 20 d for mice that lacked Notch1fl/fl alleles but expressed cre and received polyIC (cre + polyIC control; range = 12–25 d). Altogether, these observations support a previously unknown role for NOTCH1 signaling in immune-mediated BMF.
Figure 4.
Conditionally deleting Notch1 ameliorates disease in AA mice. (A) Representative hematoxylin and eosin staining of BM from one AA mouse each whose BMF was induced with cre control, polyIC control, cre + polyIC control, or Notch1 conditional KO (N1−/−) splenocytes. Bars, 200 µm. (B and C) BM cellularity (B) and percentages of BM-infiltrating T cells (C) were determined in AA mice induced with N1−/− splenocytes (n = 10) and compared with AA mice induced with cre control (n = 4), polyIC control (n = 6), or cre + polyIC control (n = 4) splenocytes. (D) Flow cytometric analysis of NOTCH1IC in T cells isolated from the BM of cre control (n = 4), polyIC control (n = 6), cre + polyIC control (n = 4), or N1−/− mice after treatment with polyIC. (E) Kaplan–Meier survival estimates of AA mice induced with N1−/− splenocytes (n = 12) compared with animals induced with cre control (n = 6), polyIC control (n = 6), or cre + polyIC control (n = 7) splenocytes. Data represent the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test; log-rank test for survival estimates.
GSI treatment attenuates lethal BMF in AA mice
Evidence suggests that GSIs can provide therapeutic relief from symptoms of experimental autoimmune encephalomyelitis in a mouse model used to study the human autoimmune condition MS (Minter et al., 2005). Although tissues targeted for destruction differ between patients with MS and patients with AA, both autoimmune conditions exhibit a strong Th1 component. Therefore, we asked whether GSIs could be used therapeutically to ameliorate BMF in a mouse model of AA.
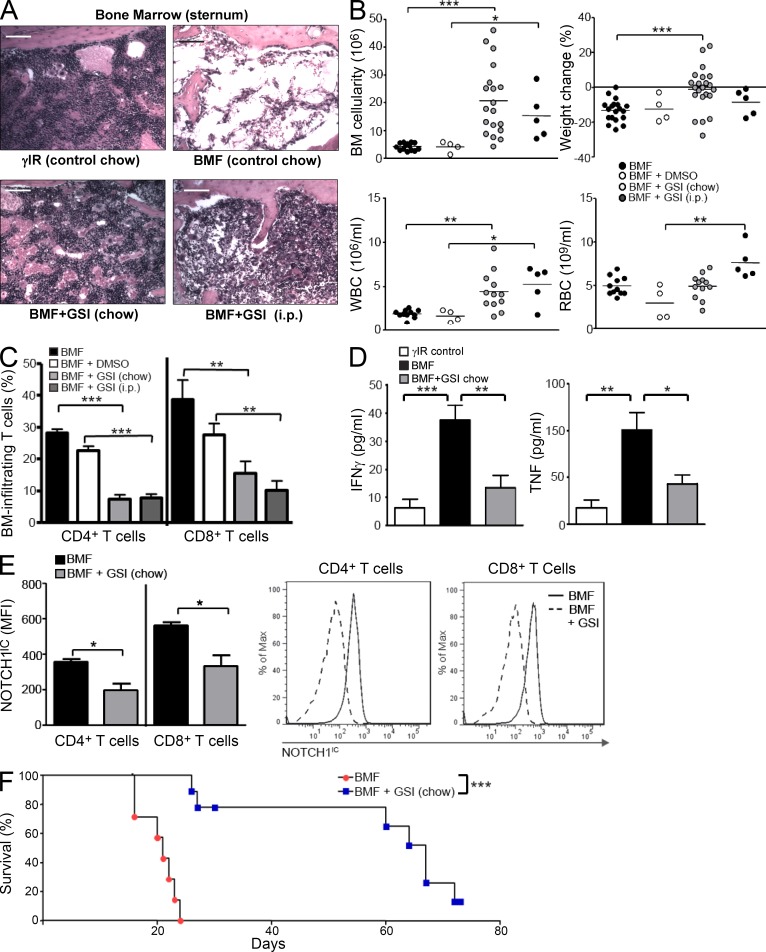
We began GSI treatment either at day −14, relative to disease induction, by feeding mice GSIs formulated in rodent chow, or at day −3, by administering GSIs to mice via daily i.p. injections. Disease course proceeded for 17 d, at which time animals were humanely euthanized to assess disease severity. We found that regardless of route of administration, GSI treatment lessened disease severity by protecting BM and peripheral blood cells and decreasing weight loss (Fig. 5, A and B). The BM of GSI-treated AA mice showed significantly diminished infiltration of destructive T cells (Fig. 5 C) and expression of NOTCH1IC compared with control-treated animals (Fig. 5 E). Levels of circulating IFN-γ and TNF were also significantly reduced (Fig. 5 D). Most notably, compared with control-treated mice for which the median survival time was 21 d (range = 20–24 d), mice treated with GSIs achieved a median survival time of 67 d (range = 26–73 d; P < 0.001; Fig. 5 F) with one of nine mice (11%) fully rescued from lethal BMF. Thus, in proof-of-concept experiments, inhibiting NOTCH signaling attenuated immune-mediated BMF in AA mice.
Figure 5.
GSI treatment attenuates lethal BMF in AA mice. (A) Representative hematoxylin and eosin staining of BM from one control chow–fed AA mouse (top right) compared with BM from one irradiation control mouse (top left) or from one each, GSI-treated AA mouse (bottom). Bars, 200 µm. (B) BM cellularity, weight change, and peripheral white and red blood cell counts were assessed in AA mice left untreated (n = 11–14) or treated with vehicle alone (DMSO; n = 4) or with GSIs administered in rodent chow, beginning 14 d before BMF induction (n = 12–22), or by i.p. injection, beginning 3 d before BMF induction (n = 5). (C) Percentages of BM-infiltrating CD4+ and CD8+ T cells were quantified in untreated (n = 9), vehicle-treated (DMSO; n = 4), and GSI-treated animals (n = 5–22). (D) IFN-γ (left) and TNF (right) were measured in the plasma of control (n = 9), AA mice (n = 11–14), and AA mice receiving GSI in rodent chow (n = 12–22). (E) NOTCH1IC in BM-infiltrating T cells (n = 11–22 mice/condition) was determined by flow cytometry; representative histograms of NOTCH1IC staining within BM-infiltrating CD4+ and CD8+ T cells from one control and one GSI-treated AA mouse. (F) Kaplan–Meier survival estimates for AA mice fed control chow or GSI chow (P < 0.001, as determined by the log-rank test). Data represent the mean ± SEM (n = 4–22 animals). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
Therapeutic administration of GSIs prolongs survival of AA mice
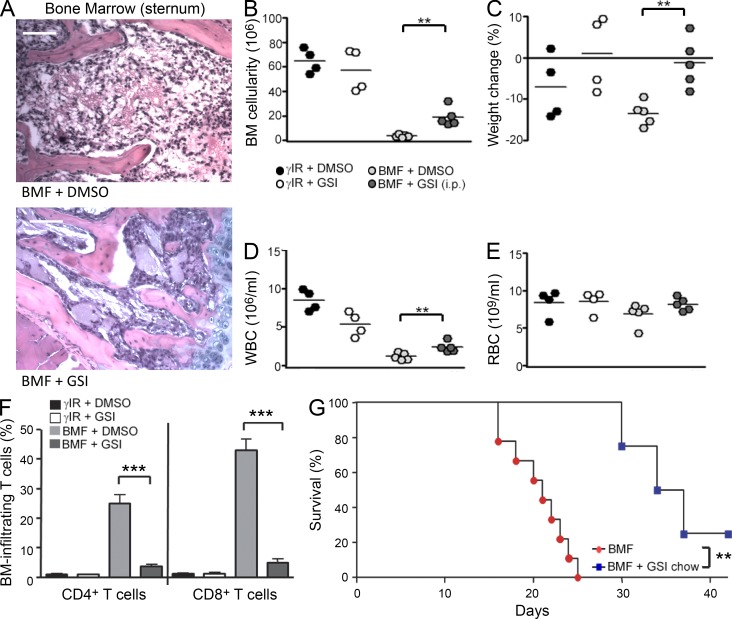
Previous studies have evaluated the therapeutic potential of treatment modalities or neutralizing antibodies by administering them to AA mice 1 h after disease induction (Bloom et al., 2004; Chen et al., 2005). However, we sought to assess the effectiveness of GSI administration (Fauq et al., 2007) under more clinically relevant conditions. To do this, we induced disease in AA mice and began GSI treatment via daily i.p. injections or by placing AA mice on GSI chow 5 d after BMF induction. Beginning GSI administration at this time ameliorated disease symptoms to an extent similar to that seen in mice treated with GSIs before BMF induction. Delayed GSI treatment protected BM cellularity and peripheral blood counts and reduced T cell infiltration to the BM (P < 0.001; Fig. 6, A–F). Remarkably, even when GSI treatments were started on day 5, they conferred a significantly increased survival benefit to AA mice compared with untreated controls (P = 0.002; Fig. 6 G). These studies are the first to provide compelling evidence that GSI is efficacious in attenuating the lethal outcome of immune-mediated BMF under clinically relevant conditions. They further indicate that in BM-infiltrating T cells, its actions are likely to be modulating NOTCH1 signaling, a principal target of GSIs in the immune system.
Figure 6.
Therapeutic administration of GSIs prolongs survival of AA mice. (A) Representative hematoxylin and eosin staining of BM from one AA mouse each whose treatment with vehicle only (DMSO; top) or GSIs (bottom) was begun 5 d after BMF induction. Bars, 200 µm. (B–E) BM cellularity (B), weight change (C), and circulating white (D) and red cells (E) were determined in vehicle-only–treated AA mice (n = 5) and GSI-treated (i.p.) AA mice (n = 5) beginning 5 d after disease induction. (F) Percentages of BM-infiltrating T cells were determined for control (γIR + DMSO [n = 4] and γIR + GSI [n = 4]), vehicle-treated (BMF + DMSO; n = 5), and GSI-treated (BMF + GSI; n = 5) AA mice. (G) Kaplan–Meier survival estimates for AA mice fed control chow or GSI chow beginning 5 d after disease induction (P = 0.002, log-rank test). Data represent the mean ± SEM (n = 4 to 8 mice/group). **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
NOTCH1IC is increased in PBMCs of patients with untreated AA and binds to the TBX21 promoter
The contribution of aberrant NOTCH1 signaling to the pathology of human T cell acute lymphocytic leukemia (T-ALL) has been well documented (Weng et al., 2004; Lee et al., 2005; Grabher et al., 2006). However, a role for NOTCH signaling in autoimmunity is just beginning to be rigorously explored (Palaga and Minter, 2012).
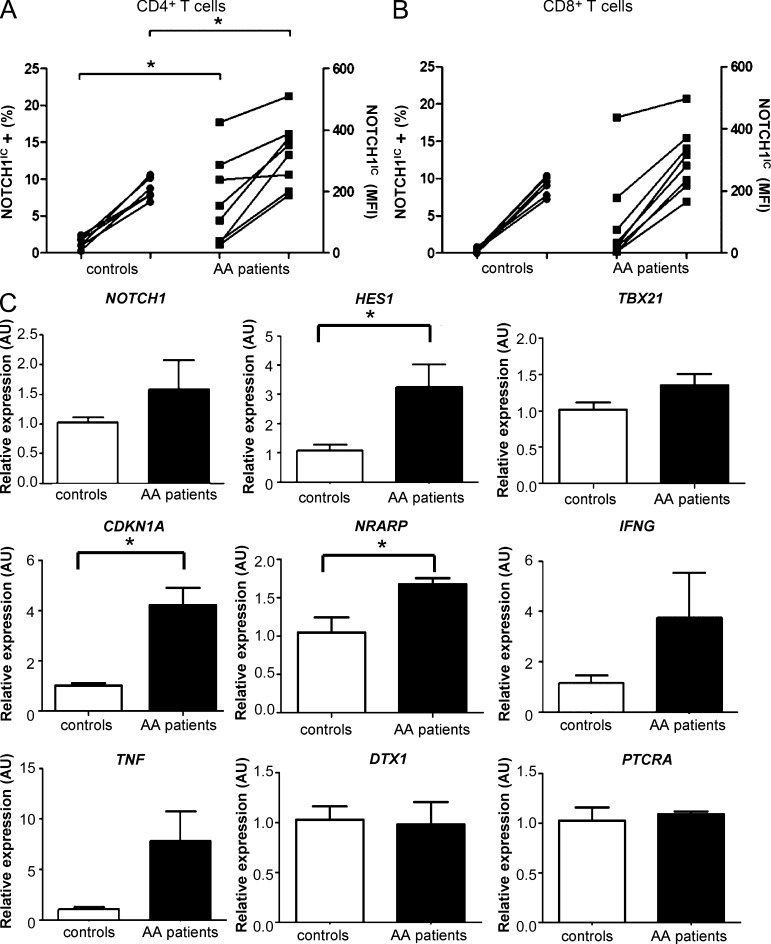
To determine whether NOTCH1 signaling is involved in mediating human autoimmune BMF, we evaluated its expression in the peripheral blood of a cohort of patients with AA who had not received prior IST (n = 9). When compared with samples from a group of healthy donors (n = 6), NOTCH1IC in patient samples was detected in a significantly greater percentage of CD4+ T cells (Fig. 7 A). NOTCH1IC MFI was also significantly elevated in patient CD4+ T cells (P = 0.042) compared with healthy controls. The CD8+ T cell population from patients showed a greater percentage of cells expressing NOTCH1IC, as well as a higher MFI, compared with healthy donors, although these differences did not reach statistical significance (Fig. 7 B). We next examined the expression of a panel of genes known to be regulated by NOTCH1. We found that HES1, CDKN1A, and NRARP, all transcriptional targets of NOTCH1, were significantly higher in patient samples, indicating that NOTCH1 signaling in patients’ T cells was functionally active (Fig. 7 C). Transcripts of the proinflammatory cytokines IFNG and TNF were also up-regulated in patients’ samples (Fig. 7 C), reflecting the Th1 phenotype of the disease.
Figure 7.
NOTCH1IC is increased in PBMCs of patients with untreated AA and is functionally active. (A and B) Flow cytometric analyses of NOTCH1IC in CD4+ (A) and CD8+ (B) T cells from healthy controls (n = 6) and patients with AA (n = 9) who had not received prior IST. Percentages of NOTCH1IC-positive cells are indicated on the left-hand y axes, whereas NOTCH1IC protein expression is indicated by MFI on the right-hand y axes. (C) Expression of NOTCH1 and NOTCH-regulated genes HES1, TBX21, CDKN1A, NRARP, IFNG, TNF, DTX1, and PTCRA in PBMCs from healthy controls (n = 4–5) and AA patients (n = 6) was determined by real-time PCR. Data represent the mean ± SEM. *, P < 0.05; unpaired Student’s t test with Welch’s correction applied when variances were significantly different.
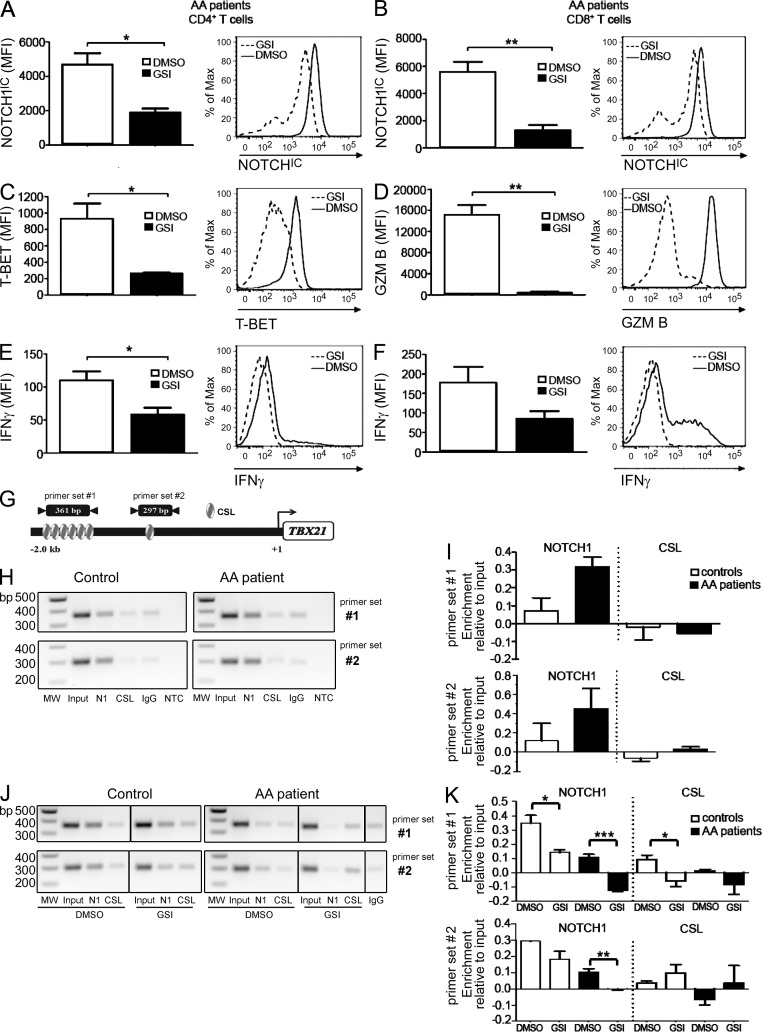
Next, we stimulated T cells from patients with AA with antibodies specific for CD3ε and CD28 and evaluated the effects of GSIs on NOTCH1IC and other proinflammatory proteins. Incubating patient T cells with GSIs before stimulation significantly blunted NOTCH1IC accumulation (Fig. 8, A and B), as well as the expression of T-BET in CD4+ T cells (Fig. 8 C) and GRANZYME B in CD8+ T cells (Fig. 8 D). IFN-γ expression was also reduced in both subsets after GSI treatment (Fig. 8, E and F). These data have important clinical relevance as they indicate that, although NOTCH1IC is increased in AA patients’ samples during active disease, it is responsive to the inhibitory actions of GSIs.
Figure 8.
NOTCH1 regulates Th1-associated molecules through its direct regulation of the TBX21 promoter. Peripheral T cells from AA patients (n = 3–6) were treated with vehicle only (DMSO) or with 40 µM GSIs and then stimulated in vitro for 72 h. (A–F) CD4+ T cells were analyzed by intracellular staining and flow cytometry for expression of NOTCH1IC (A), T-BET (C), and IFN-γ (E); CD8+ T cells were analyzed for expression of NOTCH1IC (B), GRANZYME B (D), and IFN-γ (F); representative histograms from one patient each are shown to the right of collated data for each protein evaluated. (G) Schematic representation of the TBX21 promoter showing relative location of CSL-binding sites and regions amplified by primer sets #1 and #2 (not shown to scale). (H) Representative negative image of agarose gel showing two amplified regions of the TBX21 promoter immunoprecipitated using antibodies specific for NOTCH1 and CSL from PBMCs of one healthy control (left) and one AA patient who had not received prior IST (right). (I) Quantification of band intensities of three healthy control samples and two AA patient samples subjected to ChIP. (J) Representative negative image of agarose gel showing two amplified regions of the TBX21 promoter immunoprecipitated using antibodies specific for NOTCH1 and CSL from PBMCs of one Th1-polarized healthy control (left) and one stimulated AA patient (right) after 48 h of treatment with vehicle only (DMSO) or with GSIs. (K) Quantification of band intensities of three Th1-polarized healthy control samples and three stimulated AA patient samples treated with vehicle only (DMSO) or with GSIs for 48 h before ChIP. Data represent the mean ± SEM of three independent replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
To further determine whether NOTCH1 was playing a direct role in mediating expression of these Th1-promoting molecules, and given our earlier observation that NOTCH1 influences T-BET levels in murine T cells, we asked whether NOTCH1 was directly regulating T-BET expression in patient samples. To do this we used chromatin immunoprecipitation (ChIP) to examine a 2-kb region upstream of the start site in the promoter of TBX21, the gene which encodes human T-BET (Fig. 8 G). In PBMCs from healthy controls, cultured under Th1-polarizing conditions, NOTCH1IC could be found bound to putative CSL-binding sites at two separate regions of the TBX21 promoter. Remarkably, in two out of three samples of PBMCs from patients who had not received prior IST, we could also detect NOTCH1IC resident at these same CSL-binding sites (Fig. 8, H and I). When we cultured Th1-polarized PBMCs from healthy controls or stimulated PBMCs from AA patients with GSIs for 48 h, we observed a dramatic decrease in NOTCH1IC binding to the TBX21 promoter (Fig. 8, J and K). This finding correlated with a concomitant decrease in T-BET and T-BET–regulated proinflammatory molecules after GSI treatment (Fig. 8, A–F), suggesting that reducing NOTCH1 binding at the TBX21 promoter could effectively reduce the proinflammatory phenotype of PBMCs from patients with AA.
Extended GSI treatment does not compromise hematopoietic stem cell engraftment or long-term hematopoiesis
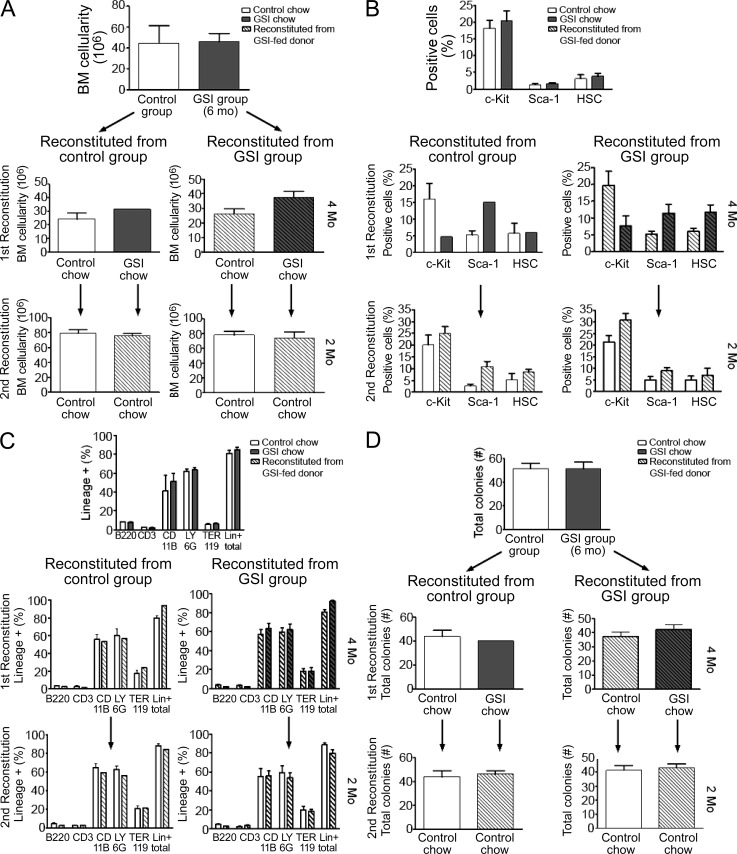
For patients with AA, a BM transplant from an HLA-identical sibling, or HLA-matched unrelated donor, is the preferred treatment option (Dezern and Brodsky, 2011). However, one impediment to successful transplantation is failure of the hematopoietic stem and progenitor cells to engraft the BM, preventing full restoration of the hematopoietic compartment in the host. Furthermore, patients with AA may receive IST for an extended period of time before transplantation; thus, it is imperative that immunosuppressive regimens do not negatively impact BM engraftment. To evaluate whether prolonged GSI treatment had adverse effects either on engraftment or long-term reconstitution of hematopoietic stem cells, we performed serial BM transplants using donor mice that constitutively expressed GFP (GFP+). This approach allowed us to track engraftment and hematopoietic potential of BM stem cells during the sequential transplant procedures.
BM donor mice were fed control chow or GSI chow (5 mg/kg/day), for 6 mo, at which time the animals were sacrificed to assess any differences in total BM cellularity (Fig. 9 A). We also compared percentages of CD34lo/−, Lin−, Sca-1+, and c-kit+ (LSK) cells in both groups of mice to determine how extended GSI administration affects these progenitor cell populations (Wilson and Trumpp, 2006; Fig. 9 B). We transplanted 2 × 106 total BM cells from GSI chow– or control chow–fed GFP+ mice into lethally irradiated recipients (first reconstitution). Some mice from each group were placed on GSI chow; all animals were followed for 4 mo. At the end of the reconstitution period, all mice were sacrificed and BM was analyzed for cellularity and percentages of LSK cells. Next, we transplanted 2 × 106 total BM cells from each group of GSI chow– or control chow–fed mice into new, lethally irradiated recipients (second reconstitution) and followed these mice for an additional 2 mo. At the end of the second reconstitution period, all mice were sacrificed and BM cells were analyzed. For mice serially transplanted and maintained on GSIs throughout the duration of the experiments, the transplanted BM cells were exposed to systemic effects of GSIs for a total of 10 mo. Of note, we did not observe any significant differences in BM cellularity, expression of lineage markers or the distribution of CD34lo/−, Lin−, Sca-1+, or c-kit+ cells between control- and GSI-treated animals at any of the time points analyzed (Fig. 9, A–C). We also did not observe any gut-associated toxicity in mice at any time during their extended treatment with GSIs.
Figure 9.
Extended GSI treatment does not compromise hematopoietic stem cell engraftment or long-term hematopoiesis. (A–D) Donor mice were fed control (open bars) or GSI chow (gray bars) for 6 mo before harvesting and transplanting total BM cells into lethally irradiated recipients (first reconstitution; n = 1–8 mice/group). Some mice were fed GSI chow for an additional 4 mo (diagonal stripe bars) before all mice (n = 4) were harvested and total BM cells from each group were transplanted into a second group of lethally irradiated recipients (second reconstitution; n = 1–8 mice/group). Mice were followed for an additional 2 mo. At each harvest, BM cellularity (A), distribution of stem and progenitor cells (B), lineage markers (C), and clonogenic potential (D) were assessed. Data represent the mean ± SEM.
To confirm our flow cytometric analyses of the lineage distribution of BM cells from GSI- or control-treated mice, we used an in vitro clonogenic assay to determine the potential of BM cells to give rise to erythroid, myeloid, and lymphoid lineages. We observed that regardless of the treatment, an equivalent frequency of colonies formed (Fig. 9 D), and all lineages were equally represented (not depicted). We also did not observe myeloproliferative disorder. This can be caused by loss of function of NOTCH1 in the epidermis, which induces a systemic cytokine response, including TSLP-1 and G-CSF (Dumortier et al., 2010). We conclude from these data that administering GSIs, even for extended periods of time at doses that effectively reduce NOTCH1IC expression in T cells and rescue lethal BMF, does not adversely affect the engraftment of or long-term repopulating capacity of hematopoietic progenitor cells. This indicates that, in this mouse model, there exists a therapeutic window in which efficacy can be observed without unacceptable toxicity.
DISCUSSION
NOTCH1IC expression is increased in T cells of mice and patients with AA. Using a mouse model of immune-mediated BMF, we show that NOTCH1 signaling contributes to disease pathology, as inducing BMF with NOTCH1-deficient splenocytes results in attenuated symptoms and significantly increased survival in diseased mice. Our findings indicate a GSI can similarly protect BM cellularity, prevent pancytopenia, and greatly increase survival in AA mice through its inhibitory actions on NOTCH1 cleavage and activation. GSI treatment also effectively reduces, to control levels, expression of multiple Th1-associated molecules that are otherwise up-regulated in T cells of mice and patients with AA. This correlated with the concomitant down-regulation of NOTCH1IC and, in patient samples, with loss of NOTCH1 from the promoter of TBX21 and indicates that although NOTCH1 is increased in patient T cells, its activity can be successfully modulated with GSIs.
In mice, genetically deleting Itch, a HECT E3 ubiquitin ligase which negatively regulates NOTCH1, produces a progressive autoimmune phenotype that is exacerbated when Itch−/− mice are crossed with mice carrying an activated Notch1 transgene (Matesic et al., 2006). Additionally, in a mouse model of autoimmune and lymphoproliferative disease (ALPS), treating mice with the GSI DAPT significantly decreased NOTCH1IC expression and reduced hyperproliferation (Teachey et al., 2008). In the present study, we extend the link between NOTCH1 expression and aberrant immune responses to include a mouse model highly representative of the human autoimmune BMF syndrome AA.
NOTCH1 signaling is required for T cell development (Deftos et al., 2000). When Notch1 is conditionally deleted early in thymocyte development, signaling through Notch2 can substitute in a redundant fashion, specifically in regulating production of IFN-γ (Auderset et al., 2012). We conditionally deleted NOTCH1 from mature T cells to avoid redundant contributions of NOTCH2 signaling to disease pathology. In doing so, we provide genetic evidence that NOTCH1 contributes to disease because AA mice induced with N1−/− splenocytes showed less severe symptoms and increased survival compared with mice induced with NOTCH1-sufficient, wild-type cells. These data are in keeping with results from another study that used a different genetic approach to reduce NOTCH signaling during BMF. In that study, splenocytes from transgenic mice expressing a dominant-negative form of the NOTCH nuclear coactivator mastermind-like (DN-MAML) were used to induce disease (Zhang et al., 2011). Although less severe disease symptoms were observed using DN-MAML splenocytes, NOTCH1 levels in these cells were not measured.
Pharmacologically inhibiting NOTCH1 by treating mice with GSIs before disease induction, or therapeutically, 5 d after transferring splenocytes, resulted in robust attenuation of disease and significantly increased survival. Collectively, our results parallel previous studies that have linked dysregulated NOTCH1 signaling and certain autoimmune phenotypes in mice and treatment with GSIs to alleviate disease symptoms. However, ours is the first to use combined genetic and inhibitor studies to clearly define a role for NOTCH1 in the pathology of immune-mediated BMF.
Deregulated NOTCH signaling has been linked to multiple human diseases (Talora et al., 2008). Accumulating evidence supports an association between aberrant NOTCH signaling and autoimmunity and, furthermore, between autoimmunity and hematological malignancies (Stern et al., 2007). Increased expression of NOTCH1IC and HES1 has been noted in patients with idiopathic thrombocytopenic purpura, an autoimmune condition which targets circulating platelets and possibly megakaryocytes for destruction through T cell– and B cell–dependent mechanisms (Ma et al., 2010). This finding is consistent with our analyses of CD4+ T cells from patients with autoimmune AA, which revealed elevated levels of transcriptionally active NOTCH1IC.
Our in vitro experiments provide compelling evidence for a T cell–intrinsic contribution of NOTCH1 to the expression of Th1-associated, proinflammatory molecules. Reducing NOTCH1 signaling in CD4+ and CD8+ T cells through conditional deletion in N1−/− mice suppressed expression of intracellular NOTCH1, as well as expression of T-BET, GRANZYME B, and IFN-γ during in vitro polarization assays. We observed similar repression of these transcriptional regulators and effector proteins when we treated wild-type CD4+ and CD8+ T cells with a conventional GSI. Conversely, we did not see a similar reduction in protein expression when we used the NS-GSI JLK-6. Although we cannot rule out completely the contribution of additional γ-secretase substrates that may also be spared by JLK-6 treatment, the strong correlation of results obtained using GSIs and N1−/− T cells suggests that expression of proinflammatory molecules that are a hallmark of AA pathology are regulated downstream of NOTCH1 signaling. These findings support previous studies that showed GSI effectively targets the NOTCH signaling pathway (Adler et al., 2003; Palaga et al., 2003; Minter et al., 2005).
Aberrant expression of T-BET, the transcriptional regulator controlling Th1 cell differentiation, contributes significantly to the pathology of AA and has been found bound to the IFNG promoter in patients with AA (Solomou et al., 2006; Tang et al., 2010). Furthermore, in the Chinese population, polymorphisms in the TBX21 promoter may be associated with a predisposition to AA (Ge et al., 2012). In patients that respond fully to IST, T-BET levels decrease to those seen in healthy donor controls, with partial responders maintaining slightly elevated T-BET expression, compared with control samples. Interestingly, there is some indication that patients who do not respond to IST also fail to down-regulate T-BET at the protein level, and this correlates with disease progression (Solomou et al., 2006). These observations suggest that T-BET expression may not only provide a useful readout of therapeutic efficacy; reducing T-BET expression may also decrease disease severity because T-BET levels have been demonstrated to regulate Th1 versus Th2 cell fate determination, in part through its antagonistic actions on GATA3-regulated gene expression (Jenner et al., 2009; Zhu et al., 2012).
Although the downstream effects of T-BET expression are well described, the detailed mechanisms that regulate the TBX21 promoter remain ill defined. Experiments suggest that IL-12 and IFN-γ share redundancy in reinforcing T-BET expression in Th1-polarized cells, although individually, they are unable to induce Tbx21 expression in the absence of T cell stimulation (Placek et al., 2009). This is consistent with the notion that NOTCH1 is increased in activated but not naive T cells and thus may provide an initial signal to transcribe Tbx21 and with reports that NOTCH signaling influences the differentiation of Th1 T cells (Maekawa et al., 2003; Minter et al., 2005; Zhang et al., 2011; Tezgel et al., 2013).
In two of three randomly selected samples from patients with AA, we observed increased occupancy of NOTCH1 on the TBX21 promoter at two distinct binding sites within a 2-kb region upstream of the TBX21 start site. We identified these sites as putative CSL-binding sites. However, each of these sites contains nested NF-κB binding sites. Given the differential binding of CSL at these two different sites, it is possible and likely that NOTCH–CSL and/or NOTCH–NF-κB interaction at these sites is unique, although further experiments are required to verify this. This would be consistent with experiments using CSL conditional KO mice that showed T-BET expression in Th1 cells is robust in the absence of CSL (Osborne, B.A., personal communication) and suggests that, although NOTCH may bind to CSL to transcribe Tbx21, NOTCH–CSL interaction is not absolutely required. Importantly, after incubating in vitro Th1-polarized cells from healthy controls, or stimulated patient cells, with GSIs, NOTCH1 was lost from the TBX21 promoter. This correlated with a robust decrease in T-BET expression in CD4+ T cells, GRANZYME B expression in CD8+ T cells, and IFN-γ in both subsets, suggesting that targeting NOTCH signaling may represent an effective strategy for regulating T-BET in AA.
BM transplantation from an HLA-matched donor is the preferred treatment option for patients newly diagnosed with AA. This option carries a significant risk of graft versus host disease as immune-competent T cells carried within the BM graft react to disparate HLAs expressed on host tissues. Notably, graft versus host responses are abrogated in a BM transplant model that co-transfers T cells deficient in NOTCH signaling (Zhang et al., 2011). Extrapolating this finding, one could speculate that using GSIs in the setting of BM transplantation may prove advantageous, as it may reduce the high morbidity associated with acute graft versus host responses. In this context, our data showing extended GSI treatment has no adverse effects either on hematopoietic stem cell engraftment or long-term hematopoiesis are of utmost relevance. These data are also in agreement with previous studies suggesting NOTCH1 is not required to maintain adult hematopoiesis (Maillard et al., 2008; Varnum-Finney et al., 2011). Complete abrogation of NOTCH signaling during hematopoiesis has been studied using Pofut1-deficient mice, which lack the ability to fucosylate NOTCH receptors, a process necessary for regulating ligand binding (Yao et al., 2011). Although Pofut1−/− mice showed myeloid hyperplasia and defects in lymphopoiesis, we did not see such dysregulation in GSI-treated mice. Most likely, the relatively low dose of GSIs effective for treating disease permits a threshold level of NOTCH signaling in the BM that is sufficient to maintain hematopoiesis (Duncan et al., 2005). Alternatively, redundant signaling through noncanonical Wnt or hedgehog signaling pathways may compensate for decreased NOTCH signaling in the stem cell compartment (Fleming et al., 2008; Gao et al., 2009; Kokolus and Nemeth, 2010).
An association between autoimmunity and hematological malignancies is well documented (Stern et al., 2007). Patients with AA are at significant risk of developing a clonal neoplasm within a decade of being diagnosed, with progression to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) occurring in 10–20% of patients (Bagby and Meyers, 2009). One theory postulates the inflammatory environment of the diseased BM places a selective pressure on hematopoietic stem cells, favoring those cells that have acquired random somatic mutations which endow them with a survival advantage. As unfit stem cells are lost via apoptotic signals, resistant clones survive and may develop into frank leukemias, such as AML. Of note, in patients with AML, increased expression of NOTCH1 and its ligands Jag1 and Dll-1 is associated with poor prognosis, especially in patients classified as an intermediate risk karyotype (Xu et al., 2011). These observations serve to broaden the rationale that targeting the NOTCH signaling pathway may have beneficial effects in patients with AA.
One early study examining the adverse effects of GSIs suggested that, at doses double that used in the present study, metaplasia was observed in intestinal goblet cells (Milano et al., 2004). Since that time, encouraging progress has been made in alleviating GSIs’ undesired effects on gut tissues, including using intermittent dosing schedules (Albain et al. 2011. Thirty-Fourth Annual CTRC-AACR San Antonio Breast Cancer Symposium. Abstr. S1-5.; Means-Powell et al. 2012. Thirty-Fifth Annual CTRC-AACR San Antonio Breast Cancer Symposium. Abstr. P2-14-04) or co-administering GSIs with dexamethasone, which has been shown to protect the intestinal crypt cells in the presence of GSIs (Real et al., 2009; Samon et al., 2012). Presently, there are more than 30 open clinical trials that include an investigational GSI in the treatment of cancer, denoting a substantial and acute interest in its use as a therapeutic modality. Our findings provide compelling evidence that NOTCH1 contributes to immune-mediated BMF and suggest that further investigation of GSIs as a therapeutic means of targeting NOTCH signaling in AA may be warranted.
MATERIALS AND METHODS
Animals.
All mouse protocols were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Amherst. F1 progeny were obtained by crossing BALB/c females with C57BL/6 males. Offspring between the ages of 9–12 wk were used in experiments. C57BL/6 GFP+/− mice, originally obtained from the Jackson Laboratory were maintained by crossing C57BL/6 wild-type females with GFP+/− males. All mice were maintained on acidified, antibiotic water.
Generating conditional Notch1 KO (N1KO) mice.
Notch1 conditional floxed mice were generated by crossing Notch1fl/fl (NOTCH1tm2Rko/GridJ) to Mx1Cre+/− (B6.Cg-Tg(Mx1-cre)1Cgn/J) from the Jackson Laboratory. To conditionally delete Notch1 from splenocytes, polyIC (GE Healthcare) was dissolved in PBS and given at a dose of 12–15 µg/g mouse weight to Notch1fl/fl × Mx1-Cre+/− (N1KO), Notch1fl/fl × Mx1-Cre−/− mice (cre controls), and Mx1-Cre+/− mice (cre + polyIC controls) via five i.p. injections, administered every other day. Some Notch1fl/fl × Mx1-Cre+/− mice were given an equivalent volume of PBS vehicle (polyIC controls). After the last injection, mice were rested for 3 wk before being used as a source of donor splenocytes in experiments.
BMF induction and analyses.
F1 progeny were conditioned with 3 Gy of total body irradiation using a 137Cs source. 4–6 h later, 5 × 107 bulk splenocytes from age- and gender-matched donors were given via i.p. injection. Mice were monitored daily for signs of disease and harvested on day 17. For survival studies, mice were considered lethally induced on the day they were no longer able to take food or water, at which time they were humanely euthanized. After CO2 asphyxiation, peripheral blood was obtained via cardiac puncture. Sterna were collected for histology. BM cells were recovered from the tibias and femurs of both legs by flushing the bones with 5% FBS/PBS. Splenocytes were isolated by manipulation through a 40-µM filter. Red blood cells were lysed in ACK lysis buffer, and the remaining white blood cells were enumerated using Trypan Blue exclusion. White and red cell counts were performed on peripheral blood using a HemaTrue Hematology Analyzer (Heska).
In vivo administration of GSIs.
For experiments in which mice were pretreated with GSIs, mice were fed ad libitum either control chow or Harlan Teklad mouse/rat chow containing GSI (LY 411,575), formulated to deliver 5 mg/kg/day (GSI chow; gift of P. Das, Mayo Clinic, Jacksonville, FL). Mice were fed GSI chow beginning 2 wk before disease induction and were maintained on GSI chow through the disease course. For pretreatment or therapeutic treatment with GSIs, mice received GSI (LY 411,575) dissolved in DMSO at a dose of 5 mg/kg/day by i.p. injection beginning 3 d before disease induction or 5 d after disease induction and continued daily until mice were harvested. Control mice received an equivalent volume of DMSO vehicle. For extended GSI administration, C57BL/6 GFP+/− mice were fed control or GSI chow beginning at 6 wk of age and continuing for 6 mo, at which time mice were sacrificed and BM cells were harvested as above.
Serial BM transplantation and clonogenic assays.
Recipient C57BL/6 mice were preconditioned with 12 Gy of total body irradiation in split doses, using a 137Cs source. Mice were given 8 Gy of γ-irradiation, rested for 3 h, and then given an additional 4 Gy of γ-irradiation. Irradiated mice were reconstituted by i.v. injection with 2 × 106 total BM cells from mice maintained on GSI chow for 6 mo. Recipient mice were maintained on acidified, antibiotic water and monitored daily for signs of failed engraftment. Some recipient mice were placed on GSI chow for the duration of the reconstitution, which was allowed to proceed for 4 mo. At the end of the 4-mo period, mice were euthanized and 2 × 106 total BM cells were harvested and serially transplanted by i.v. injection into a second group of lethally irradiated mice, preconditioned as described above. Recipient mice were again maintained on acidified, antibiotic water and monitored daily for signs of failed engraftment. Mice were followed for an additional 2 mo. Some mice required treatment with silver nitrate cream because of complications from irradiation-induced dermatitis.
To determine the lineage-forming potential of BM cells from mice receiving extended GSI treatment, isolated BM cells were seeded in MethoCult media (STEMCELL Technologies) at a concentration of 2 × 104/ml/well, in duplicate wells. Plated samples were incubated for 12–14 d at 37°C in a humidified chamber. Colonies were counted and identified according to the manufacturer’s protocol (STEMCELL Technologies).
Histology.
Sterna were harvested on day 17 after BMF induction, fixed in 10% neutral buffered formalin (VWR) overnight, and decalcified in Cal-Rite (Richard Allen Scientific) for 48 h. Samples were preserved in 70% ethanol at 4°C until they were processed, paraffin-embedded, sectioned, and stained with hematoxylin and eosin.
T cell isolation and in vitro assays.
Spleens were isolated and manipulated through a 40-µM filter, and splenocytes were treated with ACK lysis buffer. CD4+ and CD8+ T cells were then isolated using the anti-mouse CD4 and CD8 magnetic particles (IMag; BD) and separated using the BD IMag system. Cells were plated at 2.25–3 × 106 cells/well in 12-well plates precoated with anti-CD3ε and anti-CD28, purified from 145-2c11 and 37N hybridoma cell lines, respectively. To polarize T cells toward a Th1 phenotype, 1 ng/ml mIL-12 (BD) and 10 µg/ml purified NA/LE anti–mIL-4 (BD) were added to culture medium at the time of plating. For some experiments, T cells were incubated with 50 µM of the GSI z-ILCHO or with 10 µM of the NS-GSI JLK-6 (Tocris Bioscience) for 30 min at 37°C before culture.
Validation of NOTCH1IC detection using flow cytometry.
To validate flow cytometric analysis, 40 µg of total protein lysates from DMSO- or z-ILCHO–treated, stimulated murine WT CD4+ and CD8+ T cells was resolved on an 8% SDS-PAGE, transferred to nitrocellulose, and probed with an anti-cleaved NOTCH1 mAb (mN1A; eBioscience). Membranes were stripped (Restore Western Blot Stripping Buffer; Thermo Fisher Scientific) and reprobed with an anti-cleaved NOTCH1 Val1744 mAb (D3B8; Cell Signaling Technology) and then probed a third time with an anti-actin mAb (AC-40; Sigma-Aldrich) to verify equal loading. The primary antibodies were detected with HRP-conjugated antibody (GE Healthcare) and ECL (Thermo Fisher Scientific). An aliquot of CD4+ and CD8+ T cells from the same experimental replicate was stained for the surface expression of CD4 or CD8 together with the expression of intracellular NOTCH1IC and analyzed using an LSRII flow cytometer (BD) according to the methods outlined below.
Patient samples and healthy controls.
PBMCs from nine patients with severe AA who had not received IST were obtained from the National Marrow Donor Program Research Sample Repository. PBMCs from six healthy donors (STEMCELL Technologies) were included as controls. PBMCs were plated at 106 cells/ml in RPMI 1640 medium supplemented with 10% FBS (Gibco), 2 mM l-glutamine, 1 mM Na pyruvate, 0.1 mM nonessential amino acids, 100 U/ml penicillin, 100 µg/ml streptomycin, and 50 µM 2-mercaptoethanol (Gibco) at 37°C in a humidified atmosphere with 5% CO2. Media and supplements were purchased from Lonza unless otherwise specified. PBMCs from healthy donors were incubated with the IMag T lymphocyte enrichment set (BD) to isolate T cells by negative selection. Control T cells and AA patient PBMCs were preincubated with 0.1% DMSO or the GSI z-ILCHO (40 µM in DMSO) for 30 min at 37°C before being stimulated with 5 µg/ml of plate-bound anti-CD3ε (UCHT1) and 2.5 µg/ml anti-CD28 (clone 37407) for 48–72 h. Th1-polarized control T cells were generated by adding 10 ng/ml hIL-12 and 10 µg/ml anti–hIL-4 (clone 3007) at the time of plating. All antibodies and cytokines were purchased from R&D Systems.
Surface and intracellular flow cytometry of murine and human samples.
Murine samples were surface stained with fluorescent-conjugated antibodies. For intracellular staining, cells were fixed and permeabilized using the Foxp3 staining buffer set (eBioscience) according to the manufacturer’s protocol. For IFN-γ staining, cells were harvested and cultured in fresh media on anti-CD3ε–coated plates for 5 h in the presence of Brefeldin A (GolgiPlug; BD). A complete list of anti–mouse antibodies is found in Table S1.
Human samples were surface-stained with FITC- and PE-Cy7–conjugated anti-CD4 (RPA-T4) and PerCP-Cy5.5–conjugated anti-CD8 (RPA-T8) antibodies. For intracellular staining, cells were fixed and permeabilized using the Foxp3 staining buffer set (eBioscience) and stained with PE-conjugated anti-NOTCH1 (mN1A), eFluor 660–conjugated anti–T-BET (4B10), and PE-conjugated anti–GRANZYME B (GB11). To detect IFN-γ production, 72-h-stimulated GSI- or DMSO-treated cells were restimulated with plate-bound anti-CD3ε and anti-CD28 for 6 h, and Brefeldin A (GolgiPlug) was added after 2 h of stimulation. Then the cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD) and stained with APC-conjugated anti–IFN-γ (clone 45–15; Miltenyi Biotec) antibody. All human antibodies were purchased from eBioscience unless otherwise indicated. Samples were acquired on an LSRII flow cytometer and analyzed using the acquisition software FACSDiva (BD). Analysis of FACS data was performed using FACSDiva or FlowJo (Tree Star) software.
Cytometric bead array.
Cytokine levels were determined in plasma using either the Th1/Th2 or Th1/Th2/Th17 cytometric bead array kit (BD) according to the manufacturer’s protocol. Sample data were acquired on an LSRII flow cytometer and analyzed using FCAP array software (BD).
RNA isolation and real-time PCR of murine and human samples.
Total RNA was isolated from murine and human samples with the RNAqueous kit (Ambion) and concentrated with the RNeasy MiniElute Cleanup kit (QIAGEN) according to the manufacturer’s protocol. 1 µg of total RNA was reverse transcribed to cDNA using dNTPs (Roche), M-MuLV reverse transcription buffer (New England Biolabs), oligo-DT12–18 (Invitrogen), RNase inhibitor (Promega), and M-MuLV reverse transcription (New England Biolabs, Inc.) on a Mastercycler gradient Thermal Cycler (Eppendorf). Quantitative real-time PCR primers were designed with PrimerBank and are listed in Tables S2 (mouse) and S3 (human). Quantitative real-time PCR was performed in duplicate with SYBR Premix Ex Taq (Takara Bio Inc.) using the Mx3000P system (Agilent Technologies). Quantitative real-time PCR conditions were as follows: 95°C for 1 min, 95°C for 25 s, 62°C for 25 s (35 cycles), 95°C for 1 min, 62°C for 1 min, and 95°C for 30 s. Relative gene expression was determined using the 2−ΔΔCt method. The results are presented as the fold change in gene expression normalized to the housekeeping gene β-actin for mouse cells and ribosomal protein RP9 for human cells and relative to unstimulated controls.
ChIP.
Cells were suspended at 106 cells/ml in RPMI 1640 medium (Lonza), treated for 10 min at room temperature with 1% formaldehyde (Thermo Fisher Scientific) to cross-link proteins to DNA, and treated with 125 mM glycine to quench unreacted formaldehyde. Then cells were lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.1) and sonicated using a Bioruptor sonicator (Diagenode, high setting, 30 s on/30 s off for 30 min) to shear the DNA. Sonicated cell lysates were diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.1, and 167 mM NaCl) and precleared with ChIP-grade protein G agarose beads (Cell Signaling Technology) before immunoprecipitation with 5 µg anti-CSL (RBP-Jκ; clone H-50), anti-NOTCH1 (clone C-20), or normal rabbit IgG overnight at 4°C (all antibodies from Santa Cruz Biotechnology, Inc.). Next, protein–DNA complexes were recovered by adding protein G agarose beads and washed as follows: low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% Na deoxycholate, 1 mM EDTA, and 10 mM Tris, pH 8.1), and TE buffer (1 mM EDTA and 10 mM Tris, pH 8.1). After wash and elution steps, cross-linking was reversed overnight at 65°C, and the DNA was purified by proteinase K digestion followed by extraction with the QIAEX II gel extraction kit (QIAGEN). Two regions of the TBX21 promoter containing putative CSL-binding sites were amplified by PCR. Primer set I (product size 361 bp) forward, 5′-ATGCTGCCCCACTTTGAACATCAG-3′; and reverse, 5′-TCTCTCTACCTTCACCCCAGTCC-3′; and primer set II (product size 297 bp) forward, 5′-CTTTAGGGGGTGGAATTTGGGGTG-3′; and reverse 5′-CTGCTTTGAACAGATTCAGAGCCG-3′. PCR conditions were as follows: 95°C for 2 min, 95°C for 30 s, 56°C for 30 s, 72°C for 1 min (35 cycles), and 72°C for 5 min.
Statistics.
The results are expressed as mean ± SEM; all in vitro experimental replicates were repeated at least three times. Unpaired, two-tailed Student’s t test (Prism 5; GraphPad Software) was used for statistical comparison of two groups, with Welch’s correction applied when variances were significantly different. For in vivo treatment experiments, survival benefit was determined using Kaplan–Meier estimates with an applied log-rank test. P-values of ≤0.05 were considered significant.
Online supplemental material.
Table S1 lists antibodies used for surface and intracellular staining of murine samples. Tables S2 and S3 list primers used for real-time PCR of murine and human samples, respectively. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112615/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Michael Katz (Hampshire Veterinary Clinic, Amherst, MA) for use of his hematology analyzer, Brooke Bentley and Theresa Stec for excellent technical assistance in histology and clonogenic assays, respectively, and Pritam Das for GSIs. We are indebted to Juan Anguita and Dominique Alfandari for thoughtful discussions and critical reading of the manuscript, Barbara A. Osborne for continued support and commitment, and Stephanie L. Phakos for endless inspiration.
This work was supported by grants from the National Institutes of Health (PO1 AG025531), Aplastic Anemia & MDS International Foundation, Inc., Charles H. Hood Foundation for Child Health Research, and American Heart Association.
The authors declare they have no competing financial interests.
Author contributions: J.E. Roderick, G. Gonzalez-Perez, C.A. Kuksin, A. Dongre, E.R. Roberts, and J. Srinivasan contributed to study design, performed experiments, and analyzed data. A.H. Fauq synthesized GSIs and C. Andrzejewski Jr., L. Miele, and T.E. Golde consulted on experimental design and data interpretation and offered critical review of the manuscript. L.M. Minter conceived of this project, supervised the experimental design and collection, interpretation, and analyses of data, and wrote the manuscript with help from J.E. Roderick., G. Gonzalez-Perez., C.A. Kuksin, E.R. Roberts, and L. Miele.
Footnotes
Abbreviations used:
- AA
- aplastic anemia
- AML
- acute myeloid leukemia
- BMF
- BM failure
- ChIP
- chromatin immunoprecipitation
- GSI
- γ-secretase inhibitor
- IST
- immunosuppressive therapy
- MFI
- median fluorescence intensity
- MS
- multiple sclerosis
- NS-GSI
- NOTCH-sparing GSI
- polyIC
- polyinosinic:polycytidylic acid
References
- Adler S.H., Chiffoleau E., Xu L., Dalton N.M., Burg J.M., Wells A.D., Wolfe M.S., Turka L.A., Pear W.S. 2003. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 171:2896–2903 [DOI] [PubMed] [Google Scholar]
- Auderset F., Schuster S., Coutaz M., Koch U., Desgranges F., Merck E., MacDonald H.R., Radtke F., Tacchini-Cottier F. 2012. Redundant Notch1 and Notch2 signaling is necessary for IFNγ secretion by T helper 1 cells during infection with Leishmania major. PLoS Pathog. 8:e1002560 10.1371/journal.ppat.1002560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagby G.C., Meyers G. 2009. Myelodysplasia and acute leukemia as late complications of marrow failure: future prospects for leukemia prevention. Hematol. Oncol. Clin. North Am. 23:361–376 10.1016/j.hoc.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom M.L., Wolk A.G., Simon-Stoos K.L., Bard J.S., Chen J., Young N.S. 2004. A mouse model of lymphocyte infusion-induced bone marrow failure. Exp. Hematol. 32:1163–1172 10.1016/j.exphem.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Chen J. 2005. Animal models for acquired bone marrow failure syndromes. Clin. Med. Res. 3:102–108 10.3121/cmr.3.2.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Brandt J.S., Ellison F.M., Calado R.T., Young N.S. 2005. Defective stromal cell function in a mouse model of infusion-induced bone marrow failure. Exp. Hematol. 33:901–908 10.1016/j.exphem.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Chen J., Ellison F.M., Eckhaus M.A., Smith A.L., Keyvanfar K., Calado R.T., Young N.S. 2007. Minor antigen h60-mediated aplastic anemia is ameliorated by immunosuppression and the infusion of regulatory T cells. J. Immunol. 178:4159–4168 [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J.S., Schroeter E.H., Schrijvers V., Wolfe M.S., Ray W.J., et al. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 398:518–522 10.1038/19083 [DOI] [PubMed] [Google Scholar]
- Deftos M.L., He Y.-W., Ojala E.W., Bevan M.J. 1998. Correlating notch signaling with thymocyte maturation. Immunity. 9:777–786 10.1016/S1074-7613(00)80643-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deftos M.L., Huang E., Ojala E.W., Forbush K.A., Bevan M.J. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 13:73–84 10.1016/S1074-7613(00)00009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezern A.E., Brodsky R.A. 2011. Clinical management of aplastic anemia. Expert Rev Hematol. 4:221–230 10.1586/ehm.11.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A., Durham A.D., Di Piazza M., Vauclair S., Koch U., Ferrand G., Ferrero I., Demehri S., Song L.L., Farr A.G., et al. 2010. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS ONE. 5:e9258 10.1371/journal.pone.0009258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Rattis F.M., DiMascio L.N., Congdon K.L., Pazianos G., Zhao C., Yoon K., Cook J.M., Willert K., Gaiano N., Reya T. 2005. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 6:314–322 10.1038/ni1164 [DOI] [PubMed] [Google Scholar]
- Fauq A.H., Simpson K., Maharvi G.M., Golde T., Das P. 2007. A multigram chemical synthesis of the γ-secretase inhibitor LY411575 and its diastereoisomers. Bioorg. Med. Chem. Lett. 17:6392–6395 10.1016/j.bmcl.2007.07.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming H.E., Janzen V., Lo Celso C., Guo J., Leahy K.M., Kronenberg H.M., Scadden D.T. 2008. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2:274–283 10.1016/j.stem.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A., Geffers R., Hunger J.K., Pförtner S., Piao W., Ivanyi P., Grosse J., Probst-Kepper M., Ganser A., Buer J. 2006. Identification of novel regulators in T-cell differentiation of aplastic anemia patients. BMC Genomics. 7:263–273 10.1186/1471-2164-7-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Graves S., Koch U., Liu S., Jankovic V., Buonamici S., El Andaloussi A., Nimer S.D., Kee B.L., Taichman R., et al. 2009. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell. 4:548–558 10.1016/j.stem.2009.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M., Zheng Y., Li X., Shi J., Ge J., Li H., Feng S. 2012. The polymorphisms of T cell-specific TBX21 and STAT4 genes may contribute to the susceptibility of Chinese individuals to aplastic anemia. Hum. Immunol. 73:118–121 10.1016/j.humimm.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Grabher C., von Boehmer H., Look A.T. 2006. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6:347–359 10.1038/nrc1880 [DOI] [PubMed] [Google Scholar]
- Haapasalo A., Kovacs D.M. 2011. The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 25:3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellström M., Phng L.K., Hofmann J.J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A.K., Karlsson L., Gaiano N., et al. 2007. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 445:776–780 10.1038/nature05571 [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Townsend M.J., Jackson I., Sun K., Bouwman R.D., Young R.A., Glimcher L.H., Lord G.M. 2009. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. USA. 106:17876–17881 10.1073/pnas.0909357106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I., Minter L.M., Telfer J., Demarest R.M., Capobianco A.J., Aster J.C., Sicinski P., Fauq A., Golde T.E., Osborne B.A. 2009. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 113:1689–1698 10.1182/blood-2008-03-147967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokolus K., Nemeth M.J. 2010. Non-canonical Wnt signaling pathways in hematopoiesis. Immunol. Res. 46:155–164 10.1007/s12026-009-8116-7 [DOI] [PubMed] [Google Scholar]
- Kook H., Zeng W., Guibin C., Kirby M., Young N.S., Maciejewski J.P. 2001. Increased cytotoxic T cells with effector phenotype in aplastic anemia and myelodysplasia. Exp. Hematol. 29:1270–1277 10.1016/S0301-472X(01)00736-6 [DOI] [PubMed] [Google Scholar]
- Lee S.-Y., Kumano K., Masuda S., Hangaishi A., Takita J., Nakazaki K., Kurokawa M., Hayashi Y., Ogawa S., Chiba S. 2005. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 19:1841–1843 10.1038/sj.leu.2403896 [DOI] [PubMed] [Google Scholar]
- Ma D., Dai J., Zhu X., Yan S., Zhao P., Zhang J., Zhu Y., Sun J., Peng J., Ji C., Hou M. 2010. Aberrant expression of Notch signaling molecules in patients with immune thrombocytopenic purpura. Ann. Hematol. 89:155–161 10.1007/s00277-009-0790-y [DOI] [PubMed] [Google Scholar]
- Maekawa Y., Tsukumo S.-I., Chiba S., Hirai H., Hayashi Y., Okada H., Kishihara K., Yasutomo K. 2003. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 19:549–559 10.1016/S1074-7613(03)00270-X [DOI] [PubMed] [Google Scholar]
- Maillard I., Koch U., Dumortier A., Shestova O., Xu L., Sai H., Pross S.E., Aster J.C., Bhandoola A., Radtke F., Pear W.S. 2008. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2:356–366 10.1016/j.stem.2008.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesic L.E., Haines D.C., Copeland N.G., Jenkins N.A. 2006. Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum. Mol. Genet. 15:3485–3497 10.1093/hmg/ddl425 [DOI] [PubMed] [Google Scholar]
- Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R.T., Zacco A., Greenberg B., Ciaccio P.J. 2004. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82:341–358 10.1093/toxsci/kfh254 [DOI] [PubMed] [Google Scholar]
- Minter L.M., Osborne B.A. 2012. Canonical and non-canonical Notch signaling in CD4+ T cells. Curr. Top. Microbiol. Immunol. 360:99–114 10.1007/82_2012_233 [DOI] [PubMed] [Google Scholar]
- Minter L.M., Turley D.M., Das P., Shin H.M., Joshi I., Lawlor R.G., Cho O.H., Palaga T.P., Gottipati S., Telfer J.C., et al. 2005. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 6:680–688 10.1038/ni1209 [DOI] [PubMed] [Google Scholar]
- Osborne B.A., Minter L.M. 2007. Notch signalling during peripheral T-cell activation and differentiation. Nat. Rev. Immunol. 7:64–75 10.1038/nri1998 [DOI] [PubMed] [Google Scholar]
- Palaga T., Minter L.M. 2012. NOTCH signaling and its emerging role in autoimmunity. Front. Biol. 10.1007/s11515-012-1209-z [DOI] [Google Scholar]
- Palaga T., Miele L., Golde T.E., Osborne B.A. 2003. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J. Immunol. 171:3019–3024 [DOI] [PubMed] [Google Scholar]
- Petit A., Bihel F., Alvès da Costa C., Pourquié O., Checler F., Kraus J.L. 2001. New protease inhibitors prevent gamma-secretase-mediated production of Abeta40/42 without affecting Notch cleavage. Nat. Cell Biol. 3:507–511 10.1038/35074581 [DOI] [PubMed] [Google Scholar]
- Petit A., Pasini A., Alves Da Costa C., Ayral E., Hernandez J.F., Dumanchin-Njock C., Phiel C.J., Marambaud P., Wilk S., Farzan M., et al. 2003. JLK isocoumarin inhibitors: selective γ-secretase inhibitors that do not interfere with notch pathway in vitro or in vivo. J. Neurosci. Res. 74:370–377 10.1002/jnr.10747 [DOI] [PubMed] [Google Scholar]
- Placek K., Gasparian S., Coffre M., Maiella S., Sechet E., Bianchi E., Rogge L. 2009. Integration of distinct intracellular signaling pathways at distal regulatory elements directs T-bet expression in human CD4+ T cells. J. Immunol. 183:7743–7751 10.4049/jimmunol.0803812 [DOI] [PubMed] [Google Scholar]
- Real P.J., Tosello V., Palomero T., Castillo M., Hernando E., de Stanchina E., Sulis M.L., Barnes K., Sawai C., Homminga I., et al. 2009. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 15:50–58 10.1038/nm.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samon J.B., Castillo-Martin M., Hadler M., Ambesi-Impiobato A., Paietta E., Racevskis J., Wiernik P.H., Rowe J.M., Jakubczak J., Randolph S., et al. 2012. Preclinical analysis of the γ-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol. Cancer Ther. 11:1565–1575 10.1158/1535-7163.MCT-11-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih IeM., Wang T.L. 2007. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 67:1879–1882 10.1158/0008-5472.CAN-06-3958 [DOI] [PubMed] [Google Scholar]
- Skokos D., Nussenzweig M.C. 2007. CD8− DCs induce IL-12–independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J. Exp. Med. 204:1525–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomou E.E., Keyvanfar K., Young N.S. 2006. T-bet, a Th1 transcription factor, is up-regulated in T cells from patients with aplastic anemia. Blood. 107:3983–3991 10.1182/blood-2005-10-4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M., Buser A.S., Lohri A., Tichelli A., Nissen-Druey C. 2007. Autoimmunity and malignancy in hematology—more than an association. Crit. Rev. Oncol. Hematol. 63:100–110 10.1016/j.critrevonc.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Talora C., Campese A.F., Bellavia D., Felli M.P., Vacca A., Gulino A., Screpanti I. 2008. Notch signaling and diseases: an evolutionary journey from a simple beginning to complex outcomes. Biochim. Biophys. Acta. 1782:489–497 10.1016/j.bbadis.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Tang Y., Desierto M.J., Chen J., Young N.S. 2010. The role of the Th1 transcription factor T-bet in a mouse model of immune-mediated bone-marrow failure. Blood. 115:541–548 10.1182/blood-2009-03-211383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teachey D.T., Seif A.E., Brown V.I., Bruno M., Bunte R.M., Chang Y.J., Choi J.K., Fish J.D., Hall J., Reid G.S., et al. 2008. Targeting Notch signaling in autoimmune and lymphoproliferative disease. Blood. 111:705–714 10.1182/blood-2007-05-087353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezgel A.O., Gonzalez-Perez G., Telfer J.C., Osborne B.A., Minter L.M., Tew G.N. 2013. Novel protein transduction domain mimics as nonviral delivery vectors for siRNA targeting NOTCH1 in primary human T cells. Mol. Ther. 21:201–209 10.1038/mt.2012.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum-Finney B., Halasz L.M., Sun M., Gridley T., Radtke F., Bernstein I.D. 2011. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J. Clin. Invest. 121:1207–1216 10.1172/JCI43868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Walls M., Qiu M., Ding R., Denlinger R.H., Wong A., Tsaparikos K., Jani J.P., Hosea N., Sands M., et al. 2010. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol. Cancer Ther. 9:1618–1628 10.1158/1535-7163.MCT-10-0034 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Wilson A., Trumpp A. 2006. Bone-marrow haematopoietic-stem-cell niches. Nat. Rev. Immunol. 6:93–106 10.1038/nri1779 [DOI] [PubMed] [Google Scholar]
- Xu X., Zhao Y., Xu M., Dai Q., Meng W., Yang J., Qin R. 2011. Activation of Notch signal pathway is associated with a poorer prognosis in acute myeloid leukemia. Med. Oncol. 28:S483–S489 10.1007/s12032-010-9667-0 [DOI] [PubMed] [Google Scholar]
- Yada S., Takamura N., Inagaki-Ohara K., O’leary M.K., Wasem C., Brunner T., Green D.R., Lin T., Pinkoski M.J. 2005. The role of p53 and Fas in a model of acute murine graft-versus-host disease. J. Immunol. 174:1291–1297 [DOI] [PubMed] [Google Scholar]
- Yao D., Huang Y., Huang X., Wang W., Yan Q., Wei L., Xin W., Gerson S., Stanley P., Lowe J.B., Zhou L. 2011. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 117:5652–5662 10.1182/blood-2010-12-326074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.S., Scheinberg P., Calado R.T. 2008. Aplastic anemia. Curr. Opin. Hematol. 15:162–168 10.1097/MOH.0b013e3282fa7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sandy A.R., Wang J., Radojcic V., Shan G.T., Tran I.T., Friedman A., Kato K., He S., Cui S., et al. 2011. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood. 117:299–308 10.1182/blood-2010-03-271940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 37:660–673 10.1016/j.immuni.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.