The transcription factor AP4 is a critical regulator of epithelial–mesenchymal transition, migration, invasion, and metastasis in colorectal cancer cells.
Abstract
The basic helix-loop-helix transcription factor AP4/TFAP4/AP-4 is encoded by a c-MYC target gene and displays up-regulation concomitantly with c-MYC in colorectal cancer (CRC) and numerous other tumor types. Here a genome-wide characterization of AP4 DNA binding and mRNA expression was performed using a combination of microarray, genome-wide chromatin immunoprecipitation, next-generation sequencing, and bioinformatic analyses. Thereby, hundreds of induced and repressed AP4 target genes were identified. Besides many genes involved in the control of proliferation, the AP4 target genes included markers of stemness (LGR5 and CD44) and epithelial–mesenchymal transition (EMT) such as SNAIL, E-cadherin/CDH1, OCLN, VIM, FN1, and the Claudins 1, 4, and 7. Accordingly, activation of AP4 induced EMT and enhanced migration and invasion of CRC cells. Conversely, down-regulation of AP4 resulted in mesenchymal–epithelial transition and inhibited migration and invasion. In addition, AP4 induction was required for EMT, migration, and invasion caused by ectopic expression of c-MYC. Inhibition of AP4 in CRC cells resulted in decreased lung metastasis in mice. Elevated AP4 expression in primary CRC significantly correlated with liver metastasis and poor patient survival. These findings imply AP4 as a new regulator of EMT that contributes to metastatic processes in CRC and presumably other carcinomas.
The AP4 protein is a basic helix-loop-helix leucine-zipper (bHLH-LZ) transcription factor, which exclusively forms homodimers that bind to the E-box motif CAGCTG (Hu et al., 1990; Jung and Hermeking, 2009). So far, AP4 has only been shown to activate or repress the expression of a few genes (Mermod et al., 1988; Hu et al., 1990; Imai and Okamoto, 2006; Ku et al., 2009; Habib et al., 2010). We identified the AP4 gene as a direct transcriptional target of c-MYC, characterized p21 as a target for direct repression by AP4, and found that AP4 antagonizes cell cycle arrest induced by DNA damage and differentiation, at least in part, by down-regulating the cyclin-dependent kinase (CDK) inhibitor p21 (Jung et al., 2008). Up-regulation of c-MYC is a hallmark of colorectal cancer (CRC), which was recently confirmed by a genome-wide expression analysis of 276 CRC samples by the Cancer Genome Atlas (TCGA) consortium (Muzny et al., 2012). Consistent with being a c-MYC target gene, AP4 expression is up-regulated in colorectal tumors, which also display elevated c-MYC expression (Jung et al., 2008). However, given that AP4 binds to an E-box motif, which differs from the E-box motif occupied by c-MYC, it seemed likely that AP4 regulates a unique set of genes. Importantly, these may be integral components of the oncogenic program imposed on cells by constitutive activation of c-MYC.
Epithelial–mesenchymal transition (EMT) was initially discovered as a morphogenic program involved in the formation of several tissues and organs and in wound healing (Yang and Weinberg, 2008; Thiery et al., 2009). Epithelium-derived tumor cells undergo EMT, which facilitates the loss of cell adhesion and the gain of migratory as well as invasive properties (Fidler, 2003; Gupta and Massagué, 2006; Yilmaz and Christofori, 2009). Thereby the EMT of tumor cells contributes to metastasis, one of the hallmarks of cancer cells (Hanahan and Weinberg, 2011). In addition, cells that pass through an EMT acquire the self-renewing trait associated with stem cells and cancer stem cells (CSCs; Mani et al., 2008). A limited number of transcription factors are known to induce EMT such as SNAIL, SLUG, ZEB1, ZEB2, and TWIST (Peinado et al., 2007). Interestingly, these factors directly repress the cell adhesion mediator E-cadherin and also confer stemness to some extent. Only recently was it shown that c-MYC may induce EMT in human immortalized mammary epithelial cells (Cowling and Cole, 2007; Cho et al., 2010). However, the transcriptional program underlying c-MYC–induced EMT is still largely unknown.
Here we comprehensively identified AP4-regulated genes by mRNA profiling and DNA-binding analyses in a genome-wide manner. We show that AP4 directly regulates a set of genes reminiscent in size and complexity to the c-MYC–regulated transcriptome. Among these genes were EMT-associated factors and stemness markers. By further functional analysis we could show that AP4 is a direct inducer of EMT and mediates c-MYC–induced EMT in CRC cell lines. In addition, AP4 was necessary for metastasis formation of CRC lines injected into immunocompromised mice. Furthermore, the elevated AP4 expression in primary CRC samples correlated with distant metastases, which may be caused by the induction of EMT by AP4, and poor patient survival. Collectively, this study identified AP4 as a central regulator of EMT and metastasis. The detection of increased AP4 expression in primary CRC samples may therefore have prognostic value.
RESULTS
Characterization of the AP4-regulated transcriptome
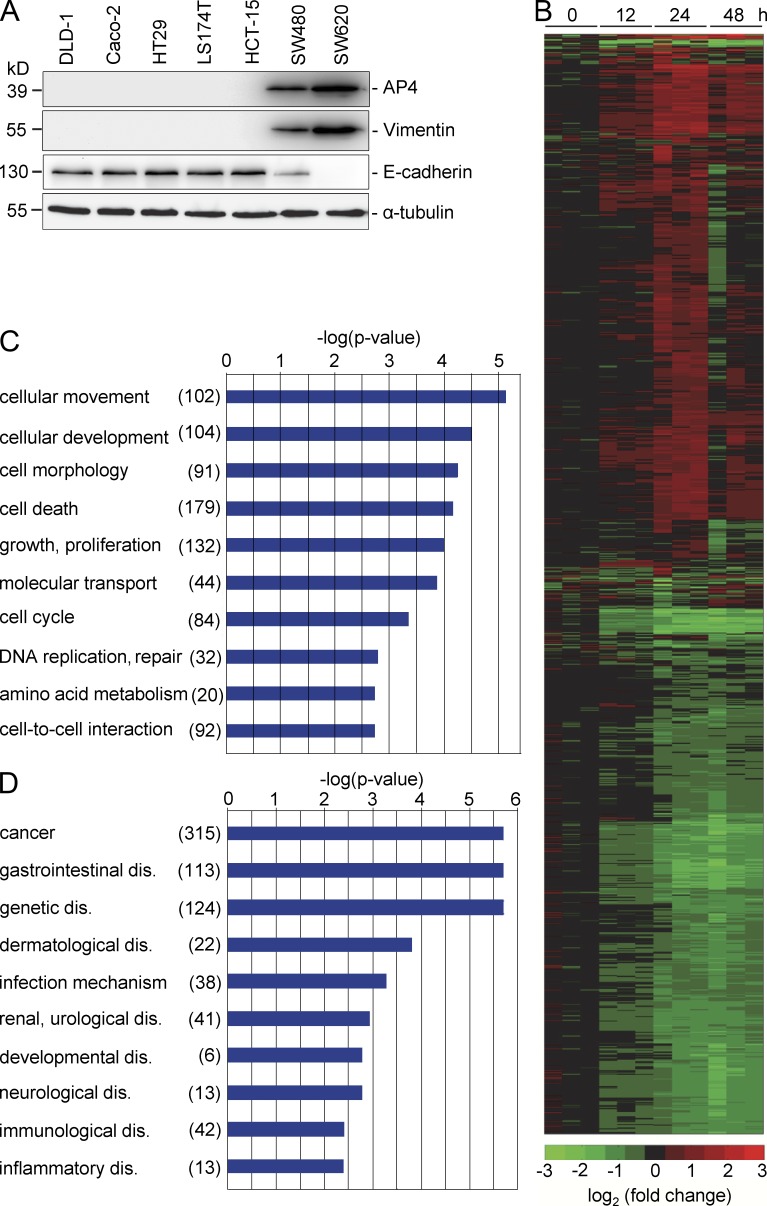
To identify AP4 target genes, we infected the human CRC cell line DLD-1 with adenoviruses driving the expression of AP4 and eGFP or, as control, only eGFP. DLD-1 cells were chosen because they express comparatively low levels of endogenous AP4 (Fig. 1 A). After adenoviral infection, >90% of the cells expressed eGFP (not depicted). The mRNA expression pattern was determined at the 0-, 12-, 24-, and 48-h time points after infection with either Ad-AP4 or Ad-eGFP using microarrays representing ∼27,000 genes. 1,458 genes were differentially expressed with ≥1.5-fold change 24 h after ectopic AP4 expression (Fig. 1 B and Table S10). The majority of the genes differentially regulated after ectopic AP4 expression reached the peak of their induction or repression at 24 h, which is presumably the time point at which ectopic expression of AP4 reaches its peak in most infected cells. A Gene Ontology analysis revealed that genes involved in cellular movement were highly enriched among the differentially expressed genes (Fig. 1 C). Furthermore, a disease-orientated Gene Ontology analysis implied an involvement of the AP4-regulated transcriptome in cancer and gastrointestinal disease (Fig. 1 D and Table S1). Interestingly, the AP4-regulated genes included many factors previously implicated in EMT, a process also involved in metastasis (Table S2). In addition, genes encoding proliferation and cell cycle regulators were overrepresented among the AP4-regulated genes (Table S2). Collectively, these results suggested that AP4 may play a role in cancer by coordinating the expression of a large set of genes, which ultimately might affect cellular processes like EMT and proliferation.
Figure 1.
Genome-wide analysis of transcriptional regulation by AP4. (A) The indicated human CRC cell lines were subjected to immunoblot analysis to detect expression of the indicated proteins. Detection of α-tubulin served as a loading control. (B) A heat map showing 1,458 differentially regulated mRNAs that displayed a change in expression ≥1.5-fold 0, 12, 24, and 48 h after Ad-AP4 versus Ad-GFP infection of DLD-1 cells is provided (see Table S10 for gene identities). A color scale is shown below. (C and D) Genes displaying >1.5-fold differential regulation after AP4 activation were subjected to Gene Ontology analyses. The results for the functional classes “molecular and cellular processes” (C) or “disease” (D) are shown. Numbers in parentheses indicate the number of genes assigned to the respective category.
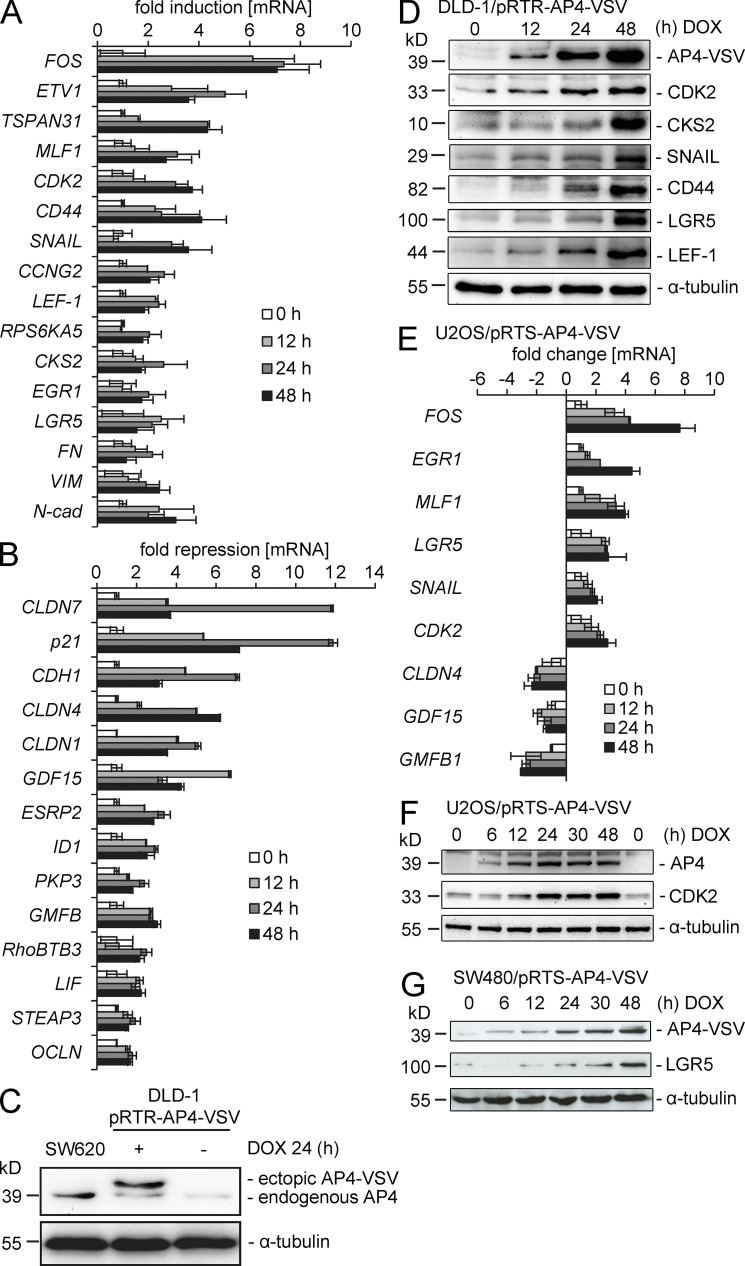
The AP4-mediated induction or repression of 30 selected genes was validated by quantitative PCR (qPCR), confirming the results obtained by microarray analysis (Fig. 2, A and B). In the following analyses, an improved episomal vector system (pRTR, see Materials and methods) that allowed the inducible expression of AP4 at levels similar to expression of endogenous AP4 in SW620 cells was used (Fig. 2 C). Although with varying kinetics, the mRNA inductions were translated into an induction of CDK2, CKS2, SNAIL, CD44, LGR5, and LEF-1 protein levels after activation of a conditional AP4 allele (Fig. 2 D). Furthermore, the levels of ectopic AP4 expression reached by the adenoviral and the pRTR expression vectors were similar (not depicted). Therefore, the ectopic AP4 expression studied here is likely to reproduce the effects of AP4 expression in cancer cells. AP4 also induced the expression of CDK2 mRNA and protein in the osteosarcoma cell line U2OS (Fig. 2, E and F). In addition, c-FOS, EGR1, MLF1, SNAIL, CLDN4, GDF15, and GMFB1 were differentially expressed. An induction of LGR5 protein expression by AP4 was also detected in the CRC cell line SW480 (Fig. 2 G). Therefore, the AP4-mediated changes in gene expression are not cell type specific.
Figure 2.
Validation of AP4-mediated differential gene expression. (A and B) The induction (A) or repression (B) of selected genes was confirmed by qPCR analysis with mRNA isolated at the indicated time points from DLD-1 cells infected with an adenovirus expressing AP4. Detection of β-actin was used for normalization. Results represent the mean ± SD (n = 3), and in the case of FOS mean ± SE (n = 2). (C) Expression of ectopic and endogenous AP4 was determined by immunoblot analysis using an AP4-specific antibody. Lysates were obtained from SW620 or DLD-1/pRTR-AP4-VSV cells treated with DOX for 24 h. (D) Lysates from DLD-1/pRTR-AP4-VSV cells treated with DOX for the indicated periods were subjected to immunoblot analysis for the indicated proteins. (E) Quantification of mRNA expression of several putative AP4 target genes after activation of a conditional AP4 allele in U2OS cells by addition of DOX for the indicated periods. Expression was normalized to β-actin expression. Experiments were performed in triplicates. Results represent the mean ± SD (n = 3). (F) Detection of the indicated protein expression levels by immunoblot analysis after activation of a conditional AP4 allele in U2OS cells by addition of DOX for the indicated periods. (G) Detection of the indicated protein expression levels by immunoblot analysis after activation of a conditional AP4 allele in SW480 cells by addition of DOX for the indicated periods. In C, D, F and G, the detection of α-tubulin served as a loading control.
Subsequently, we determined the in vivo expression of AP4 and proteins encoded by a selection of newly identified AP4 target genes in human colonic crypts using confocal laser-scanning microscopy. We had shown previously that AP4 expression is confined to the basal, highly proliferative part of the crypts, containing the transient amplifying unit including progenitor and stem cells. In contrast, p21, which is directly repressed by AP4, is exclusively expressed at the top of the crypts (Jung et al., 2008), where terminally differentiated cells are located, which eventually undergo cell death and are shed into the lumen of the colon (not depicted). As expected, AP4-positive colonocytes are devoid of the differentiation marker CDX2, whereas the differentiated, AP4-negative colonocytes in the upper crypt express CDX2 (not depicted; Du et al., 2008). AP4 expression largely overlapped with Ki67 expression in the transient amplifying unit of the lower crypt, containing highly proliferative cells (not depicted). Furthermore, the expression of SNAIL, CKS2, and CD44 overlapped with AP4 expression in the lower compartment of the crypts (not depicted). As described previously, expression of SNAIL was confined to the nucleus, CKS2 showed nucleolar expression, and CD44 was localized at the cell membranes (Roy et al., 2005; Lyng et al., 2006; Du et al., 2008). These results indicate that the AP4-regulated genes identified in a CRC cell line are presumably also regulated by AP4 in the colon and mediate the function of AP4 in vivo.
Genome-wide analysis of AP4 DNA binding
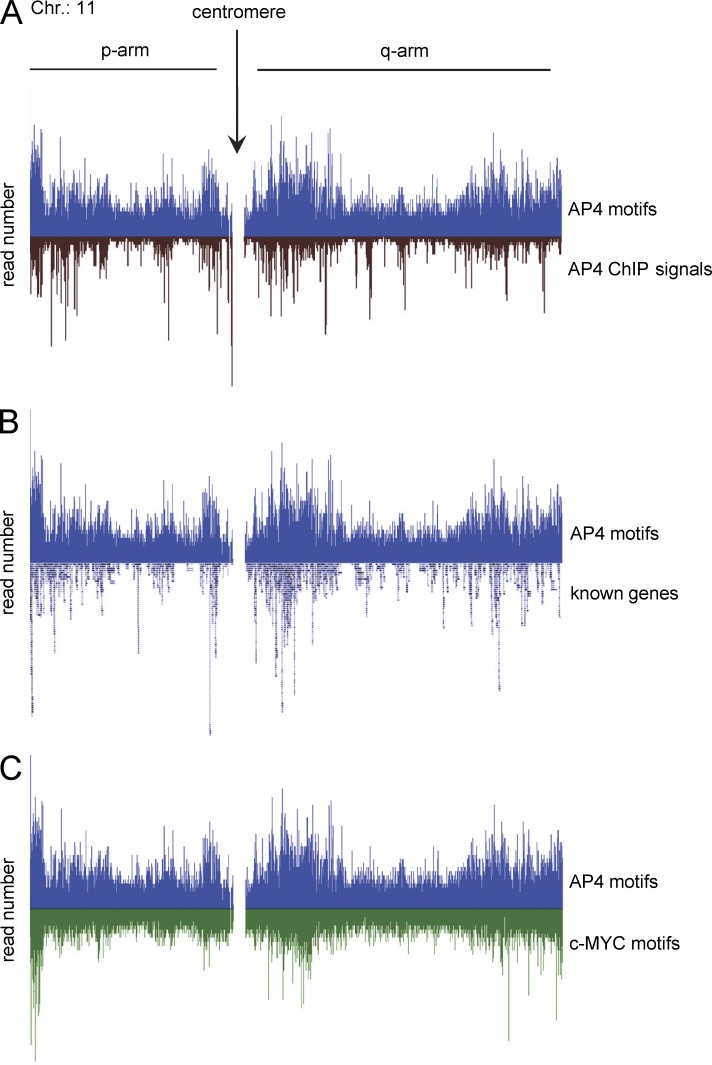
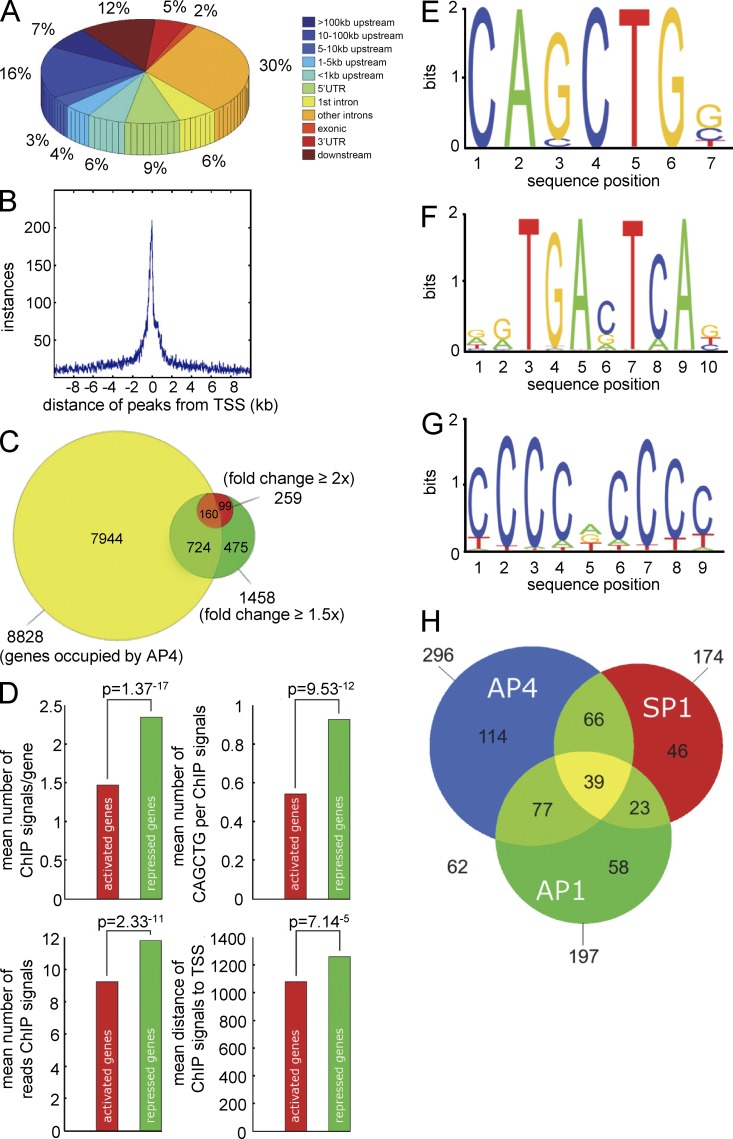
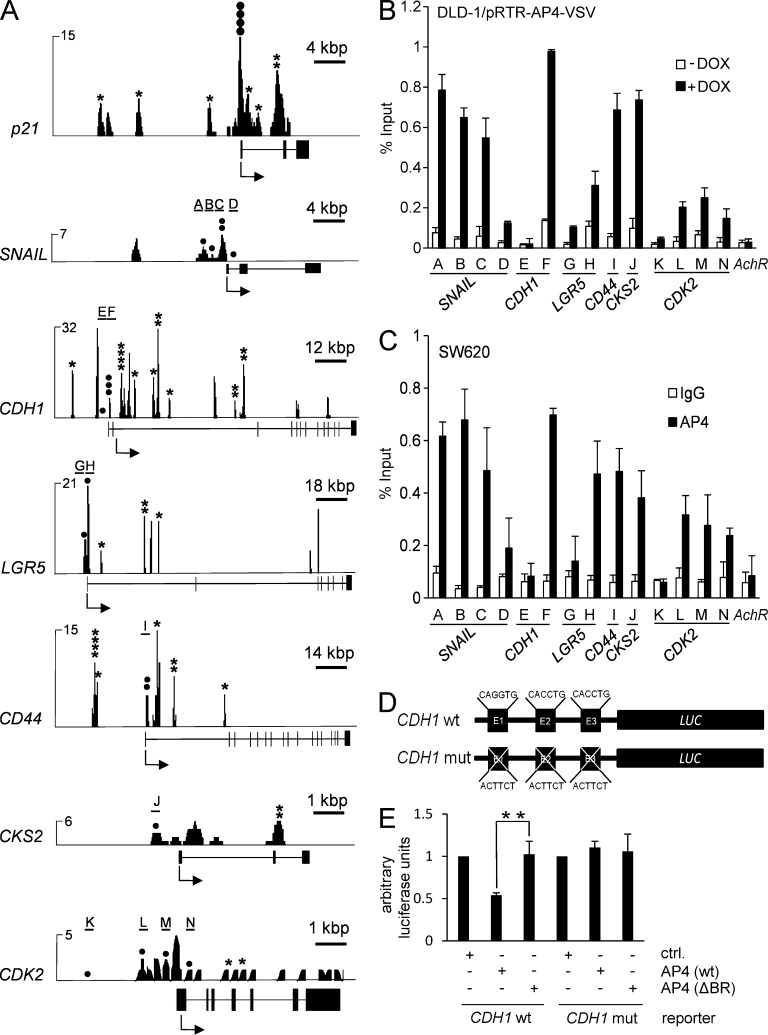
To comprehensively identify genes directly regulated by AP4, a genome-wide chromatin immunoprecipitation (ChIP) analysis followed by next generation sequencing (ChIP-seq) was performed after activation of a conditional AP4 allele in DLD-1 cells. AP4-associated DNA fragments obtained by ChIP were characterized by sequencing with a depth of ∼3.6 million reads. Genomic regions with a minimum ChIP signal height of six overlapping sequencing reads were considered as enriched. The genome-wide pattern of AP4-binding motifs and the distribution of ChIP-seq reads showed a substantial overlap (Fig. 3 A). AP4 motifs clustered in gene-dense regions, indicating that AP4 preferentially occupies transcriptionally active regions of the genome (Fig. 3 B). Interestingly, a similar distribution was found for AP4- and c-MYC–binding sites (Fig. 3 C). Binding sites occupied by AP4 were predominantly located in intragenic regions (52%) and regions <5 kbp upstream from transcriptional start sites (TSSs; 10%). 38% of the ChIP signals were detected >5 kbp up- or downstream from the TSS (Fig. 4 A). The ChIP signals located within a distance of 10 kbp up- and downstream of the TSS were centered on the TSS (Fig. 4 B). 8,828 genes displayed AP4 binding within 5 kbp up- and 3 kbp downstream of the TSS (Fig. 4 C). 5,674 (64.3%) of these genes contained at least one AP4-binding motif. Of the 1,458 genes (724 induced and 734 repressed) with ≥1.5-fold change 24 h after ectopic AP4 expression, 884 (60.6%; 354 induced and 530 repressed) were occupied by AP4 and therefore presumably direct AP4 target genes (listed in Table S10). Of the 259 genes regulated by AP4 with at least twofold change (128 induced and 131 repressed) as detected by microarray analysis, 160 genes (61.7%; 64 induced and 96 repressed) showed occupation of their promoters by AP4 (Table S10). Detailed analyses of 884 genes with >1.5-fold regulation by AP4 revealed significant differences between AP4-induced and -repressed genes with respect to the pattern of promoter occupation by AP4 (Fig. 4 D). On average, ∼2.3 ChIP signals were present in case of repression by AP4, whereas only ∼1.5 signals were detected at AP4-induced promoters. The ChIP signals derived from AP4-repressed genes also contained a larger number of CAGCTG motifs when compared with ChIP signals at AP4-activated genes. Furthermore, ChIP signals from AP4-repressed genes tend to be more pronounced than those of AP4-activated genes. On average, ChIP signals on AP4-activated promoters were located closer to the TSS than those present at AP4-repressed promoters. To characterize the transcription factor–binding motifs contained within AP4-derived ChIP signals, 485 sequences corresponding to ChIP signals with a minimum height of 25 reads from 5 kbp up- and 3 kbp downstream of TSSs were analyzed with MEME (multiple em for motif elicitation). In 61% of these promoter regions, the previously described AP4-binding motif CAGCTG (Mermod et al., 1988) was identified (Fig. 4 E). Interestingly, the MEME motif analysis revealed that a minor fraction of the AP4-regulated promoters contain an alternative AP4-binding motif (CACCTG), which is known to represent a binding motif for EMT-inducing factors such as SNAIL (Peinado et al., 2007). In the remaining promoter regions corresponding to prominent ChIP signals, no significant enrichment of other E-box motifs was detected. Therefore, an indirect DNA-binding mode involving AP4-associated proteins may exist. In addition, AP1 and SP1 motifs were detected within the regions covered by 41% and 36% of the AP4-derived ChIP signals, respectively (Fig. 4, F and G). The combinatorial distribution of AP4, SP1, and AP1 sites in the 485 promoters analyzed with MEME is depicted in Fig. 4 H. These results suggest that AP4 cooperates with SP1 and/or AP1 in the transcriptional regulation of common target genes.
Figure 3.
Comparison of the distribution of AP4-binding motifs with ChIP signals, transcription units, and c-MYC motifs on chromosome 11. Gene distributions are shown according to UCSC annotation. Raw ChIP-seq reads are shown before peak calling. The transcription factor–binding motifs CAGCTG (AP4) and CACGTG (c-MYC) were mapped to the human genome (hg19). (A) AP4 motif distribution compared with AP4-derived ChIP signals. (B) Comparison of AP4-binding motif distribution with the location of known genes. (C) Comparison of the distribution of AP4- with c-MYC–binding motifs.
Figure 4.
Genome-wide analyses of AP4 occupancy. DLD-1/pRTR-AP4-VSV cells were treated with DOX for 24 h. After ChIP with an AP4-specific antibody, the coprecipitated DNA was subjected to next generation sequencing. For details of the ChIP-seq analysis, see Materials and methods. (A) Localization of called ChIP signals. ChIP signals were assigned using the closest TSS and mapped to specific genomic regions using UCSC annotations. (B) AP4-binding pattern in the vicinity of the TSS. ChIP signals located within a range of 10 kbp up- or downstream of the TSS of 10,636 genes were analyzed. (C) Proportion of differentially expressed genes with AP4 binding in the promoter region. Results of mRNA expression analysis (green and red) and AP4 occupancy were compared (yellow). (D) Comparison of AP4-derived ChIP signals at the promoters of AP4-induced and -repressed genes. Peaks located within 5 kbp up- and 3 kbp downstream of a TSS were considered. The p-values were calculated using a two-sample Kolmogorov-Smirnov test. (E) Refinement of the AP4-binding motif by MEME analysis of AP4 ChIP-seq results. 485 ChIP signals in close proximity to a TSS with at least 25 overlapping reads were included. (F and G) Specific enrichment of AP1 and SP1 sites in the vicinity of AP4 sites was determined by the MEME and Gibbs motif samplers. (H) Distribution of co-occurring motifs in proximity of 485 ChIP-seq peaks with at least 25 overlapping, AP4-derived reads. 62 ChIP signals did not contain any of the three motifs.
Binding of AP4 at selected promoters
When the AP4 occupation of seven selected AP4-regulated promoters was analyzed in detail, the pattern of sequence reads obtained by ChIP-seq analysis overlapped with the distribution of predicted AP4-binding motifs (Fig. 5 A). Similar results were obtained for 12 additional AP4 target genes (LEF-1, FOS, ETV1, RPS6KA5, EGR1, FN1, RHO-BTB3, CLDN4, PKP3, GMFB, STEAP3, and GDF15) selected because of their biological function (not depicted). However, as mentioned above, additional ChIP signals devoid of AP4 motifs were detected in these promoters. As expected, the ChIP-seq analysis also confirmed the previously reported characterization of AP4 occupation at the p21 promoter (Jung et al., 2008). The AP4-derived ChIP-seq results obtained for six AP4 target genes were confirmed by qPCR after induction of ectopic AP4 expression in DLD-1 cells (Fig. 5 B). Furthermore, the occupancy of these promoters by endogenous AP4 was confirmed in the CRC cell line SW620 (Fig. 5 C). Notably, the ChIP results obtained after immunoprecipitation of ectopically expressed and endogenous AP4 were highly similar. Collectively, these exemplary confirmations show that the ChIP-seq results presented here provide a representative, genome-wide overview of promoter occupancy by AP4.
Figure 5.
Analysis of AP4 occupancy at selected promoters. (A) Representative ChIP-seq results for seven selected promoters. All results besides those for CDK2 are shown after peak calling. Asterisks represent bioinformatically predicted CAGCTG motifs. Closed circles represent CAGCTG motifs (CACCTG in the CDH1 promoter) analyzed by qChIP. Letters and horizontal bars represent the qPCR amplicons used for qChIP analysis in B. (B) DLD-1/pRTR-AP4-VSV cells were treated with DOX for 24 h and subjected to qChIP analysis with an AP4-specific or, as a reference, IgG antibody. (C) SW620 cells were subjected to qChIP analysis with an AP4-specific or, as a reference, IgG antibody. In B and C, the acetylcholine receptor (AchR) promoter, which lacks AP4-binding motifs, served as a negative control, and results are depicted as mean ± SD (n = 3). (D and E) Scheme of CDH1/E-cadherin reporters (wt = wild-type; mut = three CACCTG motifs mutated) that were used for the dual-luciferase assay (E) 48 h after transfection of the indicated reporters and the indicated AP4 constructs (ΔBR = deleted DNA binding region) into DLD-1 cells. Experiments in E were performed in triplicates (n = 3); error bars indicate SD; significance level as indicated: **, P < 0.01.
The ChIP-seq analysis indicated AP4 binding at the promoter of CDH1/E-cadherin in the absence of a canonical AP4-binding motif (CAGCTG) in this region (amplicon F in Fig. 5, A–C). Interestingly, our MEME analysis of the AP4-bound DNA fragments revealed a low frequency of AP4 occupancy at CACCTG sequences (Fig. 4 E), which is the binding motif of known EMT-inducing transcription factors such as SNAIL. Notably, ectopic AP4 repressed a CDH1/E-cadherin reporter containing the three CACCTG motifs, whereas mutation of the three CACCTG motifs rendered the reporter resistant to repression by AP4 (Fig. 5, D and E). Furthermore, a mutant AP4 lacking the DNA-binding domain was not able to repress E-cadherin expression, excluding indirect repression by AP4 molecules, which are not bound to DNA. Therefore, the three E-boxes in the CDH1 promoter mediate the repressive effects of AP4.
Identification of AP4 as a regulator of EMT
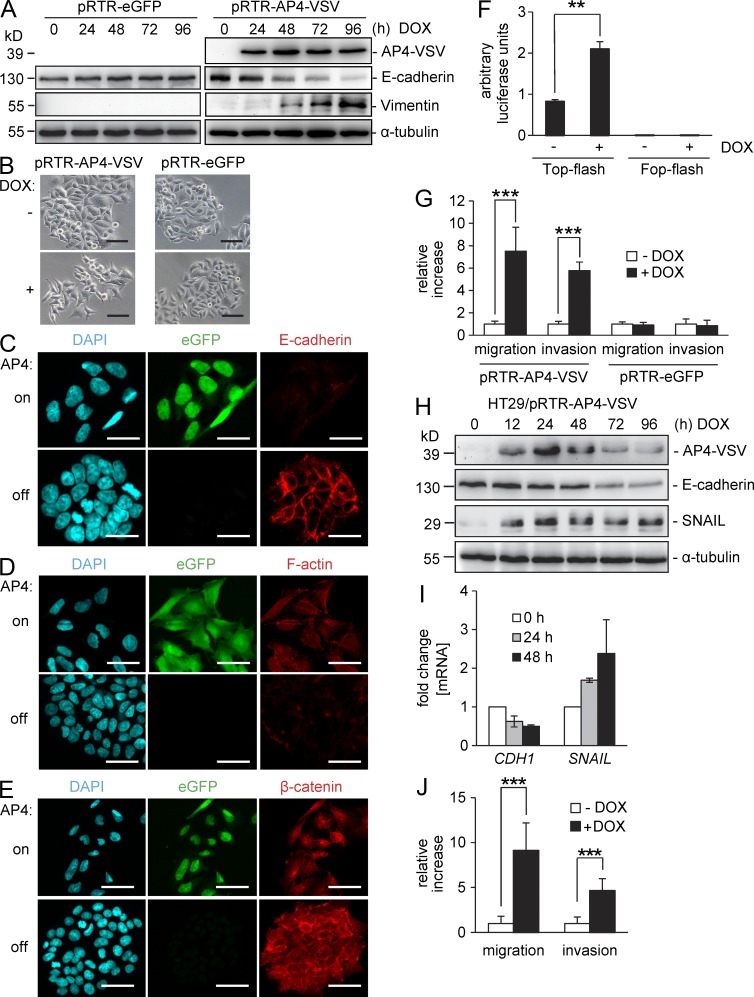
As mentioned above, numerous genes implicated in EMT were found to be differentially regulated by AP4 in our genome-wide analyses (Table S1). For example SNAIL, FN1, CDH2, VIM, TGF-α, TGF-β, LEF-1, TSPAN-3, MTA1, MMP14, MMP19, and FOS were induced after activation of AP4, whereas CDH1, CDH3, PKP3, STEAP3, OCLN, KRT19, JUP, VDR, several claudin genes (CLDNs), and integrins (ITGs) were repressed. qPCR analyses confirmed the regulation of the EMT-associated genes E-cadherin/CDH1, N-cadherin/CDH2, Fibronectin/FN1, Vimentin/VIM, Occludin/OCLN, LEF-1, SNAIL, FOS, and PKP3 by ectopic AP4 expression (Fig. 2, A and B). After activation of a conditional AP4 allele in DLD-1 cells, the E-cadherin protein was down-regulated and the mesenchymal marker protein Vimentin was induced, whereas treatment of DLD-1 cells harboring an empty pRTR vector with doxycycline (DOX) did not result in changes of E-cadherin or Vimentin expression (Fig. 6 A). As mentioned above, the SNAIL protein was up-regulated by ectopic AP4 expression (Fig. 2 C). Furthermore, the expression of endogenous AP4 protein was strictly associated with expression of Vimentin and inversely correlated with E-cadherin expression in seven CRC cell lines (Fig. 1 A). SW480 and SW620 cells showed the highest levels of AP4 protein and Vimentin and the lowest E-cadherin expression. In line with these results, SW480 and SW620 cells display a mesenchymal morphology and are more migratory, invasive, and metastatic than the other CRC cell lines analyzed here (Vécsey-Semjén et al., 2002; Malumbres et al., 2008; Mudduluru et al., 2011). Additionally, our analysis of mRNA expression profiles of seven CRC cell lines (COLO205, HCC2998, HCT116, HCT15, HT29, KM12, and SW620) belonging to the NCI-60 panel (Shoemaker, 2006) confirmed an inverse correlation between AP4 and E-cadherin expression (correlation coefficient = −0.821; P = 0.023; Table S3).
Figure 6.
Ectopic AP4 induces EMT in CRC cells. (A) A conditional AP4 allele was induced in DLD-1 cells by addition of DOX for the indicated periods. As a control, DLD-1 cells harboring a pRTR vector were treated with DOX. Protein lysates were subjected to immunoblot analysis for detection of the indicated proteins. (B) Representative phase-contrast pictures of DLD-1 cells 96 h after activation of a conditional AP4 allele or, as a control, of eGFP by addition of DOX. (C–E) Confocal microscopy of DLD-1 cells ectopically expressing AP4 after induction by DOX or the respective untreated control: E-cadherin and β-catenin were detected by indirect immunofluorescence. F-actin was visualized with Alexa Fluor 647–labeled Phalloidin. Cellular DNA was stained with DAPI. (B–E) Bars, 50 µm. (F) Dual-luciferase assay 24 h after transfection of the indicated TCF4 reporter constructs and simultaneous induction of conditional AP4 expression by addition of DOX. (G) Boyden chamber transwell assay of cellular migration or invasion in DLD-1 cells harboring a conditional AP4 allele or, as a control, the control vector only expressing eGFP. Cells were cultivated in the presence or absence of DOX for 96 h with serum reduction to 0.1% for the last 24 h. To analyze invasion, membranes were coated with Matrigel. After 48 h, cells were fixed and stained with DAPI. The mean number of cells in five fields per membrane was counted in three different inserts. Relative invasion or migration is expressed as the value of test cells to control cells. (H) A conditional AP4 allele was induced in HT29 cells by addition of DOX for the indicated periods. Protein lysates were subjected to immunoblot analysis for detection of the indicated proteins. (A and H) α-Tubulin detection served as a loading control. (I) qPCR analysis of RNA obtained at the indicated time points after AP4 activation. β-Actin was used for normalization. Results represent the mean ± SD (n = 3). (J) Boyden chamber transwell assay of cellular migration or invasion as in G in HT29 cells harboring a conditional AP4 allele. Experiments in F, G, and J were performed in triplicates (n = 3). Error bars indicate SD; significance level as indicated: **, P < 0.01; ***, P < 0.001.
DLD-1 cells have a cuboidal, epithelial morphology and form cobblestone-like islands with extensive cell–cell contacts. After ectopic expression of AP4, DLD-1 cells adopted a spindle-shaped, mesenchymal morphology and showed a scattered distribution with decreased cell–cell contacts, indicating an EMT, whereas treatment of DLD-1 cells harboring an empty pRTR vector with DOX did not result in morphological changes (Fig. 6 B). We had previously observed these phenotypic changes in DLD-1 cells after ectopic expression of SNAIL, a known inducer of EMT (Siemens et al., 2011). Furthermore, the membrane-associated expression of E-cadherin, which is characteristic for epithelial cells, was decreased after activation of AP4 in DLD-1 cells (Fig. 6 C). AP4 activation also caused a rearrangement of F-actin stress fibers in the cytoplasm (Fig. 6 D), which is another characteristic feature of EMT (Moreno-Bueno et al., 2009). In addition, ectopic expression of AP4 in DLD-1 cells resulted in loss of β-catenin from the cell membrane and a staining centered around the nucleus (Fig. 6 E). Ectopic AP4 also enhanced the activity of β-catenin/TCF4 reporter (Fig. 6 F). Because the activation of β-catenin/TCF4 activity is another hallmark of EMT (Brabletz et al., 2005), the combination of these findings suggests that AP4 represents an inducer of EMT. As EMT contributes to enhanced migration and invasion of tumor cells, we determined the effect of AP4 expression on these processes. Indeed, ectopic AP4 expression increased invasion and migration of DLD-1 cells in a transwell Boyden chamber assay, whereas treatment of DLD-1 cells harboring an empty pRTR vector with DOX did not affect migration or invasion (Fig. 6 G). Furthermore, ectopic expression of AP4 in the CRC cell line HT29 also resulted in the induction of SNAIL and repression of E-cadherin at the protein and mRNA levels (Fig. 6, H and I), and an increased invasion and migration in a transwell Boyden chamber assay (Fig. 6 J). Therefore, the ability of AP4 to induce EMT and related functions is not restricted to DLD-1 cells.
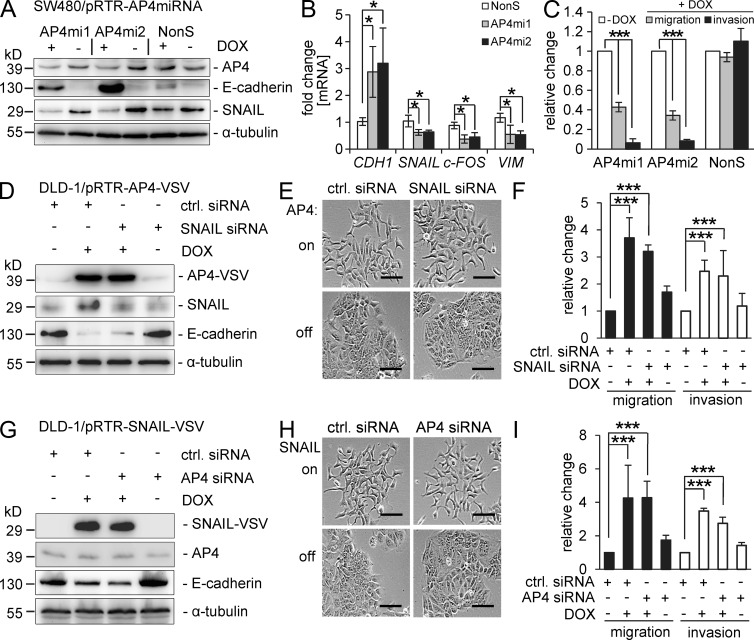
Subsequently, we used SW480 cells, which display comparatively high AP4 expression (see also Fig. 1 A), to determine whether AP4 expression is required to maintain mesenchymal features of CRC cells, such as migration and invasion. Conditional expression of two different, recombinant microRNAs (miRNAs) directed against the AP4 mRNA led to a significant accumulation of E-cadherin and a decrease in the expression of SNAIL at the protein and mRNA levels (Fig. 7, A and B). Furthermore, down-regulation of AP4 decreased migration and invasion of SW480 cells (Fig. 7 C). Therefore, elevated AP4 expression is necessary to maintain the EMT-associated state, which allows increased migration and invasion of CRC cells. In addition to repressing E-cadherin, AP4 may contribute to the phenotypic changes associated with EMT by regulating additional genes encoding components of cell junction complexes. For example, the expression of mRNAs encoding proteins involved in tight junctions (OCLN and CLDNs), adherens junctions (CDH3), and desmosomes (PKP3 and JUP) was repressed by AP4 as determined by microarray analysis (Table S1).
Figure 7.
Analyses of the requirement of AP4 and SNAIL for EMT-associated traits. (A and B) Immunoblot (A) and qPCR analysis (B) of SW480 cells harboring episomal plasmids for conditional expression of two different miRNAs directed against AP4 (AP4mi1 or AP4mi2) or a nonsilencing miRNA (NonS). For generation of the miRNAs, see Materials and methods. Cells were treated with DOX for 48 h. (C) Transwell Boyden chamber assay of cellular migration or invasion after conditional down-regulation of AP4 in SW480 cells. (D and E) 96 h after AP4 activation by addition of DOX and concomitant SNAIL-specific siRNA transfection of DLD-1 cells, lysates were subjected to immunoblot analysis of the indicated proteins (D) or phase-contrast pictures were obtained (E). (F) Transwell Boyden chamber assay of cellular migration and invasion in DLD-1 cells with and without AP4 activation by addition of DOX and/or transfection of SNAIL-specific siRNAs for 96 h. For the last 24 h, the serum concentration was reduced to 0.1%. To analyze invasion, membranes were coated with Matrigel. After 48 h, cells were fixed and stained with DAPI. The mean number of cells in five fields per membrane was counted in three different inserts. Relative invasion or migration is expressed as the cell number in the test versus the control (ctrl. siRNA or without DOX) samples. (G and H) 96 h after SNAIL activation by DOX and concomitant AP4-specific siRNA transfection of DLD-1 cells, lysates were subjected to immunoblot analysis of the indicated proteins (G) or phase-contrast pictures were obtained (H). (E and H) Bars, 50 µm. (I) Transwell Boyden chamber assay as in F but after SNAIL activation and AP4 down-regulation as indicated. For siRNA transfections (D–I), identical total siRNA concentrations (20 nM) were achieved by reconstitution with control siRNA. Experiments in B, C, F, and I were performed in triplicates (n = 3); error bars indicate SD; significance level as indicated: *, P < 0.05; ***, P < 0.001.
Because we had identified SNAIL, which encodes an EMT-inducing transcription factor, as a direct AP4 target and EMT transcription factors often form redundant, regulatory networks (Peinado et al., 2007), we determined whether AP4 and SNAIL are functionally interdependent. When ectopic AP4 expression was induced in DLD-1 cells, concomitant down-regulation of SNAIL by RNA interference did not prevent the down-regulation of E-cadherin (Fig. 7 D). In combination with the results shown in Fig. 4 E, these results demonstrate that AP4 directly regulates the expression of E-cadherin via CACCTG motifs. Furthermore, AP4 activation in the absence of SNAIL induction resulted in morphological changes indicative of EMT (Fig. 7 E) and an enhancement of migration and invasion (Fig. 7 F). Therefore, AP4 exerts these functions via mechanisms that do not require enhanced expression of SNAIL. Similarly, when SNAIL was activated in the presence of siRNA-mediated inhibition of AP4 repression of E-cadherin, EMT-associated morphological changes as well as enhanced migration and invasion were observed (Fig. 7, G–I). Therefore, AP4 and SNAIL seem to function largely independent from each other. Furthermore, AP4 itself harbors the capacities of an EMT-inducing transcription factor.
Role of AP4 in c-MYC–induced EMT
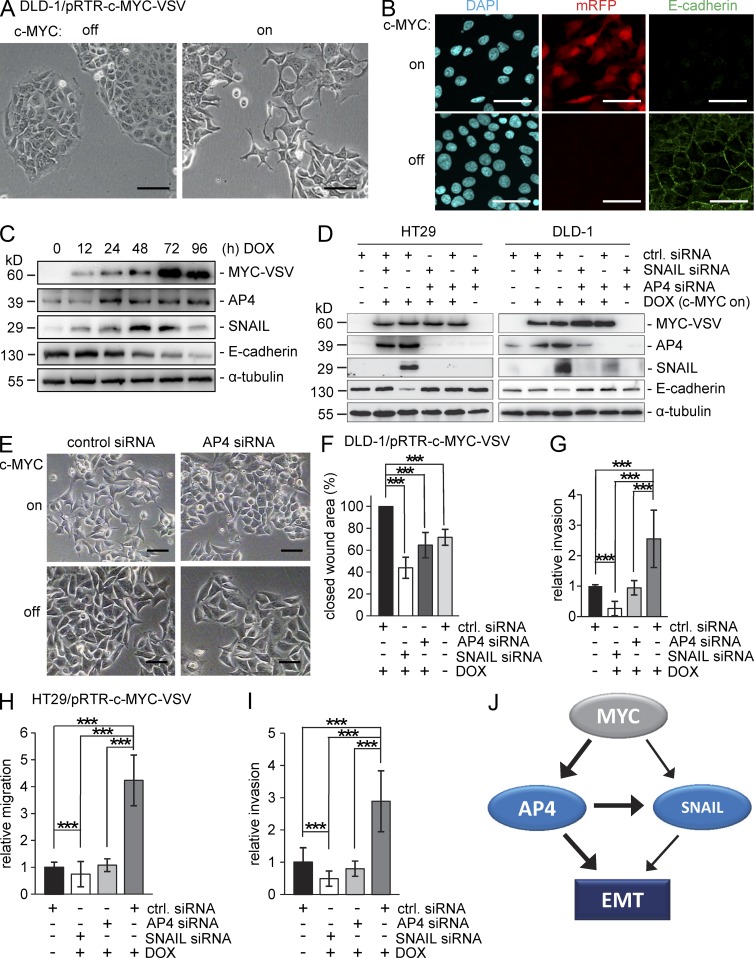
Because AP4 is as a known c-MYC target gene and, as we show here, represents an inducer of EMT, we addressed the question of whether c-MYC induces EMT in CRC cells and which role AP4 may have in this process. As mentioned above, DLD-1 cells display an epithelial morphology and grow in cobblestone-like islands. After ectopic expression of c-MYC in DLD-1, the cells adopted a mesenchymal shape and a scattered growth pattern (Fig. 8 A). Indicative of an EMT, the membrane-associated expression of E-cadherin was decreased after activation of c-MYC (Fig. 8 B). A similar decrease in membrane-associated localization of E-cadherin was detected after ectopic c-MYC expression in HT29 cells (not depicted). Furthermore, c-MYC activation resulted in repression of E-cadherin and in the induction of AP4 and SNAIL expression (Fig. 8 C). The induction of SNAIL protein expression 12 h after c-MYC seemingly precedes the induction of AP4. However, these differences may be caused by different sensitivities of the antibodies used for SNAIL and AP4. Alternatively, c-MYC may directly induce SNAIL expression (Smith et al., 2009). When c-MYC was activated in the presence of concomitant siRNA-mediated down-regulation of AP4, the repression of E-cadherin was abolished, and induction of SNAIL was prevented in HT29 and strongly decreased in DLD-1 cells (Fig. 8 D). These results demonstrate an important role of AP4 in mediating the induction of SNAIL expression by c-MYC. After concomitant c-MYC activation and siRNA-mediated down-regulation of SNAIL, the repression of E-cadherin was also prevented, although AP4 was still induced (Fig. 8 D). In addition, the morphological changes provoked by c-MYC activation were prevented by siRNA-mediated down-regulation of AP4 in DLD-1 cells (Fig. 8 E). Furthermore, the activation of c-MYC enhanced wound closure and invasion of DLD-1 cells, which was prevented by siRNA-mediated down-regulation of AP4 or SNAIL (Fig. 8, F and G). Also in HT29 cells, the enhancing effects of c-MYC on migration and invasion were dependent on the induction of either AP4 or SNAIL (Fig. 8, H and I). In summary, these analyses show that c-MYC mediates EMT in CRC cells by inducing AP4, which induces SNAIL (Fig. 8 J). Furthermore, c-MYC–induced EMT requires both SNAIL and AP4 expression. Because both factors have the same targets, e.g., E-cadherin, these results suggest a synergism between AP4 and SNAIL. The induction of SNAIL by c-MYC is largely mediated through up-regulation of AP4 but also seems to have an AP4-independent, albeit less prominent, component, which may be mediated by direct binding of c-MYC to the SNAIL promoter reported previously (Smith et al., 2009).
Figure 8.
A circuitry involving c-MYC, AP4, and SNAIL regulates EMT and invasion. (A) Representative phase-contrast pictures of DLD-1/pRTR–c-MYC–VSV cells after addition of DOX for 96 h. (B) Confocal microscopy analysis 96 h after DOX treatment as in Fig. 5 (C–E). (C) Protein lysates obtained after DOX treatment for the indicated periods were subjected to immunoblot analysis. Detection of α-tubulin served as a loading control. (D and E) Immunoblot analysis of the indicated cell lines 96 h after c-MYC activation by DOX and concomitant siRNA transfection (D) and phase-contrast microscopy of DLD-1/pRTR–c-MYC–VSV cells 96 h after addition of DOX and/or transfection with the indicated siRNAs (E). (A, B, and E) Bars, 50 µm. (F) 72 h after c-MYC activation and/or transfection with the indicated siRNAs, cells were subjected to a wound healing assay. Wound closure was determined 48 h after scratching. (G) Cellular invasion was determined in a transwell Boyden chamber assay 96 h after addition of DOX and/or transfection with the indicated siRNAs. After 48 h, cells were fixed and stained with DAPI. The mean number of cells in five fields per membrane was counted in three different inserts. (H and I) Cellular migration (H) and invasion (I) of HT29 ectopically expressing c-MYC as described in G. Relative invasion or migration is expressed as the value of test cells to control cells. (J) Schematic model of the c-MYC/AP4/SNAIL circuitry. (D–G) For siRNA transfections, identical total siRNA concentrations (20 nM) were achieved by reconstitution with control siRNA. (F–I) Experiments were performed in triplicates (n = 3); error bars indicate SD; significance level as indicated: ***, P < 0.001.
Role of AP4 in metastasis
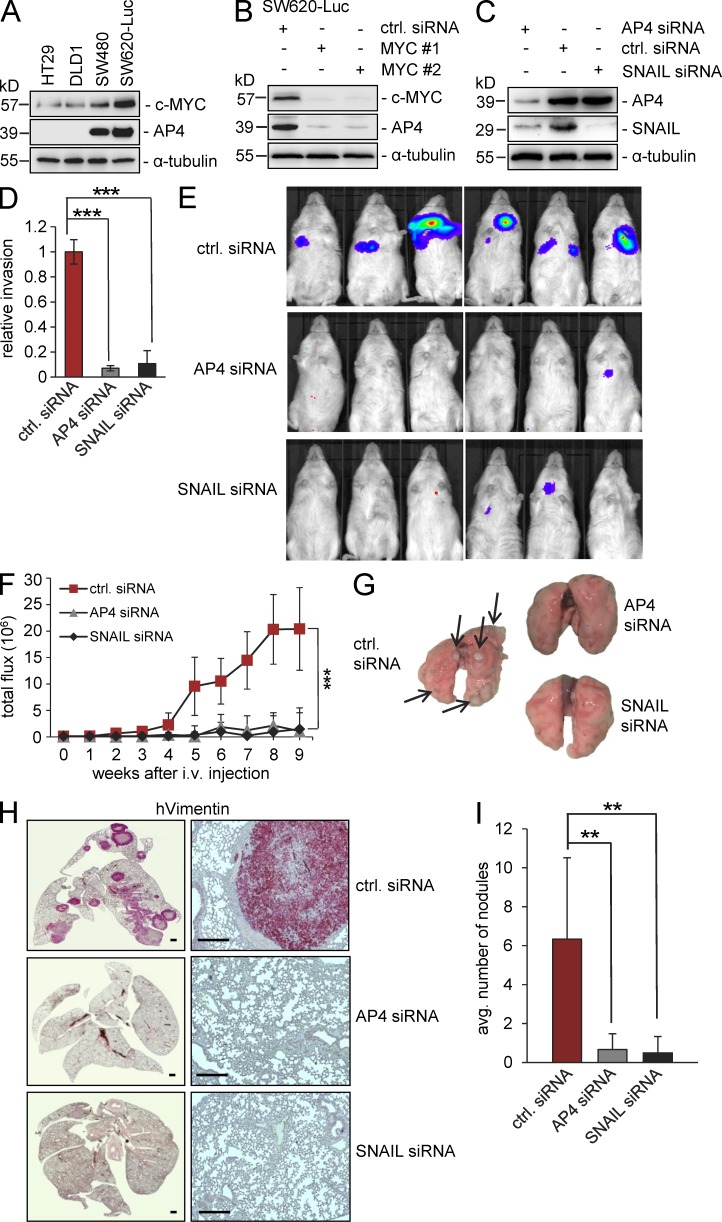
Because EMT has been implicated in the formation of metastases, we asked whether AP4 function is required during metastases formation in a xenograft mouse model. Among the four CRC cell lines used for functional analysis in this study, those lines with the most pronounced mesenchymal phenotype (SW480 and SW620) expressed the highest levels of AP4 and c-MYC, whereas the more epithelial lines (DLD-1 and HT29) showed less c-MYC expression and almost undetectable expression of AP4 (Fig. 9 A). In line with AP4 being a c-MYC target in CRC, c-MYC and AP4 expression correlated in the four CRC lines (Fig. 9 A). Furthermore, the down-regulation of c-MYC by two specific siRNAs led to a pronounced decrease in AP4 expression in SW620 cells (Fig. 9 B) and also in SW480 cells (not depicted). Therefore, AP4 is downstream of c-MYC in SW480 and SW620 cells, which is in line with AP4 being a direct target of c-MYC (Jung et al., 2008). To determine the role of AP4 and its target SNAIL during metastases formation, AP4 expression was suppressed by specific siRNAs in SW620 CRC cells stably expressing luciferase (Fig. 9 C). As shown above, SW620-Luc cells express comparatively high levels of AP4 (Fig. 9 A). Down-regulation of AP4 also resulted in a decrease of the SNAIL protein, in line with SNAIL being a target gene of AP4, whereas inhibition of SNAIL did not affect AP4 expression. Furthermore, the down-regulation of AP4 or SNAIL in SW620–Luc cells resulted in decreased invasion in a Boyden chamber assay (Fig. 9 D). Subsequently, these cells were injected in the tail vein of immunocompromised NOD/SCID mice, and their spreading and colonization were monitored over time in a noninvasive manner, as described previously (Malumbres et al., 2008). 9 wk after xenografting, the luminescence signals were ∼20-fold higher for SW620-Luc cells treated with control oligonucleotides than for SW620-Luc cells transfected with siRNAs directed against SNAIL or AP4 (Fig. 9, E and F). Metastatic growth was detected within 4 wk after injection of SW620-Luc cells transfected with control siRNAs (Fig. 9 F). However, cells treated with AP4- or SNAIL-specific siRNAs were not detected earlier than 6 wk after injection and generally gave rise to relatively weak luminescence signals. The lungs of the control mice displayed macroscopically visible metastases, whereas lungs of mice treated with SW620-Luc cells transfected with AP4- or SNAIL-specific siRNAs did not display macroscopically visible metastases (Fig. 9 G). Histochemical detection of human Vimentin expression confirmed the presence of SW620-Luc cells treated with control siRNAs in the lung of the mice, whereas the knockdown of AP4 or SNAIL had largely prevented colonization of the lung with SW620-Luc cells (Fig. 9 H). Furthermore, quantification of detailed histological examinations of the lung revealed a significant decrease in the total number of metastatic nodules when AP4 or SNAIL was inhibited by siRNAs (Fig. 9 I). Collectively, these results demonstrate that the function of AP4 and SNAIL is required for the metastatic growth of CRC tumor cells in this mouse model.
Figure 9.
Requirement of AP4 for metastasis in a xenograft mouse model. (A) Detection of c-MYC and AP4 protein in the indicated CRC cell lines by immunoblot analysis. (B and C) SW620 cells stably expressing luciferase were transfected with the indicated siRNAs. 72 h later, protein lysates were subjected to immunoblot analysis of the indicated proteins. (A–C) Detection of α-tubulin served as loading control. (D) Transwell Boyden chamber assay of cellular invasion. SW620-Luc cells were transfected with the indicated siRNAs. After 72 h, cells were seeded into a transwell chamber and analyzed as in Fig. 7 G. (E) SW620-Luc cells were transfected with the indicated siRNAs. After 72 h, cells were injected into the tail veins of NOD/SCID mice (n = 6 for each siRNA). 9 wk later, representative bioluminescence images were obtained. (F) Bioluminescence signals recorded at the indicated time points are represented as total flux. (G) Lungs were resected 9 wk after injection of SW620-Luc cells. Arrows indicate metastatic tumor nodules. (H) Detection of human Vimentin (red) in the lungs of mice injected with SW620-Luc cells transfected with the indicated siRNAs. Bars, 500 µm. (I) Quantification of metastatic nodules in the lung per mouse 9 wk after injection of SW620-Luc cells transfected with the indicated siRNAs. For the analysis in D (n = 3) and F and I (n = 6), error bars represent SD; significance level as indicated: **, P < 0.01; ***, P < 0.001.
Clinical relevance of AP4 expression in CRC
The results described above suggested that elevated expression of AP4 may be associated with an increased metastatic capacity of primary, human CRC. To test this hypothesis, we analyzed the expression of AP4 in a cohort of 55 primary colon tumors with synchronous liver metastasis and compared it with 55 colon cancer specimens derived from patients showing no distant spread within a follow-up period of 5 yr using a case-control study design. For this analysis, we used a score consisting of four degrees of nuclear AP4 expression intensity (Fig. 10 A). Indeed, elevated AP4 expression showed a significant correlation with distant metastasis (P ≤ 0.05) and lymph node metastasis (P ≤ 0.05; Fig. 10 B). In addition, a significant correlation between AP4 expression and the tumor grade was found (P ≤ 0.03), whereas no correlation with gender and tumor size was detected. The detection of AP4 expression in primary CRC may therefore have prognostic value for predicting the risk of metastases.
Figure 10.
Association of AP4 with metastasis and poor patient survival. (A) The intensity of nuclear AP4 staining was assigned the following scores: none = −, weak = +, moderate = ++, and strong = +++ expression. Examples of representative immunohistochemistry results are shown. Bars, 100 µm. (B) AP4 expression in primary colon cancer samples of 110 patients who underwent surgical tumor resection at the Ludwig-Maximilians University of Munich between 1994 and 2005. Percentage values are given in parentheses. 55 control cases had colon cancers without distant metastases at the time of diagnosis and disease-free survival of at least 5 yr after primary surgical resection. Data were analyzed using the χ2 test. (C) Kaplan–Meier plot of CRC specimens (n = 227) with weak (1+), moderate (2+), and strong (3+) AP4 expression. A log-rank test indicated statistical significance (P = 0.016). (D) The expression of AP4 and c-MYC mRNA was analyzed in the TCGA CRC dataset (http://cancergenome.nih.gov/). 197 of 276 CRC samples were informative. The linear regression was determined using a Pearson correlation analyses. Dashed lines indicate the confidence interval. (E) Model summarizing the main findings of this study. c-MYC induces expression of AP4, which either induces or represses target genes, of which selected examples are depicted. The biological outcomes of the AP4-mediated regulations with relevance to cancer are indicated in gray boxes. Dotted lines indicate inferred biological functions of AP4 that remain to be proven experimentally. The gene or genes linking AP4 to the regulation of stemness remain to be characterized.
Because metastatic potential generally affects the long-term survival of patients after curative resection of the primary tumor, we analyzed AP4 expression in a cohort of 227 CRC patients with a median follow-up of 6 yr (range 0.8–13.4 yr). Only colorectal adenocarcinomas with moderate or poor differentiation (G2 or G3 according to WHO) and T-category T2 or T3 having neither nodal (N0) nor distant metastasis (M0) at the time of diagnosis (UICC stage I and IIA) were considered. Indeed, tumors with the highest AP4 expression were associated with the shortest overall patient survival, whereas patients with tumors displaying intermediate- or low-grade AP4 expression showed a better clinical outcome (P = 0.016; Fig. 10 C). 5 yr after the resection of the primary tumor, ∼50% of the patients with tumors showing high AP4 expression had died from CRC, whereas ∼70% with intermediate and ∼80% with low AP4 expression levels had survived. Analyses of SNAIL expression in this cohort revealed a significant, positive correlation between AP4 and SNAIL expression (Table S4). Furthermore, a positive correlation of AP4 mRNA expression and c-MYC mRNA expression was detected by analysis of published expression data obtained from 276 CRC samples provided by the TCGA (Muzny et al., 2012) project (Fig. 10 D). These correlations suggest that the regulatory circuits identified in this study in CRC cell lines are also present in clinical CRC samples and may critically contribute to tumor progression and metastasis. Collectively, these results imply that increased AP4 expression contributes to decreased patient survival, presumably by enforcing an EMT program, which facilitates metastatic spread of the primary tumor (see summarizing model in Fig. 10 E).
DISCUSSION
Here we characterized the transcriptome regulated by AP4 and the genome-wide DNA-binding pattern of AP4. This allowed us to uncover several processes regulated by AP4, which may critically contribute to its role in cancer. Our results revealed a large number of genes regulated by AP4 that are likely to connect c-MYC to additional processes previously not regarded as being regulated by c-MYC. The regulation of EMT by AP4 exemplifies how this knowledge may help to identify new networks of regulation exerted by c-MYC via induction of AP4.
When only genes containing AP4-binding motifs were considered, gene repression was found to occur more frequently than induction by AP4. Nonetheless, both types of regulation are mediated via the binding of AP4 to E-boxes with the sequence CAGCTG or, at a lower frequency, CACCTG. This property clearly distinguishes AP4 from c-MYC, which is known to transactivate genes via E-box binding, whereas c-MYC–mediated repression occurs via association with MIZ-1 and binding to initiator sequences (Staller et al., 2001). Differences in the chromatin composition and/or AP4-associated proteins may determine whether a gene is induced or repressed by AP4. Another plausible determinant could be the presence of other transcription factor binding sites in the vicinity of motifs occupied by AP4. Similar to c-MYC, AP4 regulates a large number of genes and occupies an even larger number of genomic binding sites, which are present in intergenic regions and therefore presumably not connected to the direct regulation of specific target genes. c-MYC has been shown to function as a global regulator of chromatin accessibility and organization in addition to directly regulating single target genes (Knoepfler et al., 2006). c-MYC achieves this by binding to a large number of E-box motifs in intergenic regions and recruiting histone-modifying enzymes as well as chromatin-remodeling complexes. Interestingly, AP4 was found to be associated with histone-modifying enzymes such as HDAC1 and HDAC3 to repress expression of HIV1 and the PAXH-AP1 gene, respectively (Imai and Okamoto, 2006). Furthermore, recent evidence suggests that c-MYC activates DNA replication by nontranscriptional mechanisms (Dominguez-Sola et al., 2007). Similar to c-MYC (Classon et al., 1987), AP4 binds the origin of the simian virus SV40 gene (Mermod et al., 1988). Therefore, AP4 may also influence replication and chromatin organization in a transcription-independent manner.
Here we found that AP1 and SP1 motifs are frequently located proximal to occupied AP4 motifs. Interestingly, AP1 and SP1 have both been implicated in proliferation and cancer (Li and Davie, 2010; Lopez-Bergami et al., 2010). SP1 sites are known to mediate transcriptional activation after mitogenic as well as antimitogenic signals (Li and Davie, 2010). In line with our results, a cooperation of AP4 and SP1 has already been described for the dopamine β-hydroxylase DBH promoter (Hong et al., 2008). This may represent an example of a context-dependent regulation by AP4 involving SP1.
The AP1 motif represented the second most prevalent transcription factor motif in proximity to AP4-binding sites. The transcription factor AP1, which is composed of a FOS/JUN heterodimer, is involved in diverse cellular functions ranging from proliferation and development to metastasis and angiogenesis (Lopez-Bergami et al., 2010). An example of a coordinated gene regulation mediated by AP1 and AP4 is the late transcription of SV40 (Mermod et al., 1988). In the future, it will be necessary to characterize the AP4-associated proteins and the chromatin state of AP4-regulated genes in detail to deduce the mechanisms by which AP4 exerts opposing effects via the same DNA-binding motif.
Our results show that AP4 represents a new member of the small group of transcription factors able to induce EMT. Furthermore, the results suggest that AP4 may be an important mediator of c-MYC–induced EMT. Among many gene regulations characteristic for EMT, AP4 directly repressed E-cadherin via a noncanonical AP4-binding motif and induced N-cadherin, which represents a hallmark of EMT (Gupta and Massagué, 2006). Additional indirect contributions to the repression of E-cadherin by AP4 may result from the AP4-mediated induction of Vitamin D receptor (VDR), SNAIL, FOS, and LEF-1, as these factors are known to repress E-cadherin (Palmer et al., 2001; Kim et al., 2002; Medici et al., 2006). Interestingly, SNAIL was shown to induce EMT after TGF-β stimulation in concert with LEF-1 (Medici et al., 2006). Ectopic expression of FOS promotes EMT in cooperation with TGF-β by activating β-catenin/LEF-1 (Eger et al., 2004). Ectopic AP4 expression in DLD-1 cells resulted in TCF4 reporter activation, which could be mediated by the induction of LEF-1 and/or the translocation of β-catenin to the nucleus, which is presumably caused by repression of E-cadherin. Interestingly, elevated LEF-1 expression enhanced metastasis, which could be caused by induction of EMT (Nguyen et al., 2009). Furthermore, experimental down-regulation of AP4 diminished migration, invasiveness, and metastasis of CRC cell lines. These effects were presumably mediated by reversion of EMT-associated features; e.g., the expression of E-cadherin was regained after AP4 down-regulation. The parallel regulation of multiple EMT regulators and effectors by AP4 may serve to stabilize the signaling circuitries necessary to establish EMT. In the future, it will be interesting to determine whether AP4 is also relevant for EMT during embryonic development.
c-MYC has been implicated in metastasis and invasion (Wolfer and Ramaswamy, 2011). For example, c-MYC was found to coordinate gene expression signatures in breast cancer, which is associated with metastasis and poor survival (Wolfer et al., 2010). Furthermore, c-MYC amplification correlates with distant metastasis and poor survival in CRC (Kozma et al., 1994). Recently, it was shown that c-MYC may induce EMT through the GSK3β/SNAIL axis or repression of the Wnt inhibitors DKK1 and SFRP1 in mammary epithelial cells (Cowling et al., 2007; Cho et al., 2010). Furthermore, induction of SNAIL by TGF-β was abrogated by knockdown of c-MYC (Smith et al., 2009). We could show that the AP4-mediated repression of E-cadherin and induction of EMT is independent of SNAIL, whereas the repression of E-cadherin and induction of migration and invasion caused by c-MYC were dependent on both SNAIL and AP4. Therefore, SNAIL and AP4 are presumably important downstream mediators of c-MYC–induced EMT, which may cooperate at the level of target gene promoters. These findings were supported by the positive correlation of AP4 and c-MYC expression in the CRC dataset of the TCGA and the association of AP4 and SNAIL expression detected in our survival cohort of 227 patients. A similar redundant mode of regulation has been shown for other EMT-inducing transcription factors, such as ZEB1/2 and SNAIL1/2, which induce each other and simultaneously regulate genes encoding EMT effectors, such as E-cadherin or Vimentin (Taube et al., 2010). These parallel regulatory circuits may serve to confer robustness to the cellular states, which are enforced and maintained by the EMT-inducing transcription factors.
Additionally, it was shown that the c-MYC–induced nucleosome remodeling and deacetylating complex MTA1 can induce EMT (Zhang et al., 2005). As we identified MTA1 as an AP4 target, it may represent an alternative route of AP4-mediated EMT. Interestingly, it was shown that c-MYC–induced migration and EMT are attenuated by p21 in immortalized mammary epithelial cells (Liu et al., 2009). Furthermore, the EMT-inducing transcription factors TWIST1/2 were shown to override oncogene-induced premature senescence by the transcriptional repression of p21 and p16 (Ansieau et al., 2008). Because AP4 directly down-regulates p21 (Jung et al., 2008), it is conceivable that AP4 may also suppress senescence by repressing p21 in the context of inducing EMT and metastasis. Collectively, our results imply that AP4 represents an important mediator of c-MYC–induced EMT and associated metastatic processes in CRC. This may also be the case in other tumor types exhibiting deregulation of c-MYC. In the future, it will be interesting to determine whether these properties of AP4 are conserved in other organs/tissues, tumor entities, and species.
Here we found that AP4 regulates the expression of CD44 and LGR5, which represent two markers of colorectal CSCs. LGR5 was described as a stem cell marker in the mouse intestine and colon, which is also induced by Wnt signaling (Barker et al., 2007; Sato et al., 2009). In the mouse intestine, we observed expression of AP4 in cycling columnar cells at the base of the crypt (unpublished data), which are known to express LGR5, indicating that AP4 may be involved in the in vivo regulation of LGR5. CRCs display increased expression of CD44 and LGR5 (Wielenga et al., 1993; Takahashi et al., 2011), which may be caused by elevated AP4 expression. In support of this conclusion, deletion of one c-Myc allele in mice, and therefore reduced AP4 expression, leads to reduced expression of LGR5 and CD44 (Athineos and Sansom, 2010). However, further studies will be necessary to address a potential role of AP4 in CSCs.
We could show that the down-regulation of AP4 or SNAIL in highly metastatic CRC cells prevents lung metastasis of CRC cells in a xenograft mouse model. Other metastasis-regulating factors have been characterized in similar analyses (Png et al., 2012). A decrease in metastases formation was also observed after knockdown of SNAIL in breast cancer cells (Olmeda et al., 2007). Furthermore, immunohistochemical analysis of human tumor samples revealed a positive correlation between AP4 protein expression and distant metastases in right-sided colon cancer. Because distant spreading in CRC is driven by EMT of tumor cells, AP4 may play an important role in the development of distant metastases in human CRC. By analysis of two independent cohorts of CRC patients we could demonstrate that elevated AP4 expression in the primary tumor is associated with distant metastasis and poor survival. In summary, these results suggest that detection of AP4 protein expression may serve as a prognostic marker for predicting the risk of developing distant metastases.
MATERIALS AND METHODS
Cell lines and reagents.
SW480, SW620, Caco-2, HCT-15, HT29, and LS174T CRC cell lines and the U2OS osteosarcoma cell line were maintained in DMEM (Invitrogen) containing 10% FBS (Invitrogen). The CRC cell line DLD-1 was maintained in McCoy’s 5A Medium (Invitrogen) containing 10% FBS. All cells were cultivated in the presence of 100 U/ml penicillin and 0.1 mg/ml streptomycin. DOX (Sigma-Aldrich) was dissolved in water (100 µg/ml stock solution) and always used at a final concentration of 100 ng/ml unless otherwise indicated. Luciferase-expressing SW620 cells were provided by X.-F. Wang (Duke University Medical Center, Durham, NC).
Adenoviral infections.
Adenovirus amplification and purification were performed as previously described (He et al., 1998). The minimal amount of virus needed to reach >90% infection efficiency was determined by monitoring eGFP signals with fluorescence microscopy. DLD-1 cells were infected in serum-free medium with adenovirus for 3 h, and an equal amount of medium containing 20% FBS was added subsequently.
Boyden chamber assays of migration and invasion.
DLD-1 and HT29 cells harboring a DOX-inducible AP4 or c-MYC allele were analyzed as described in Siemens et al. (2011). The mean number of cells in five fields per membrane was counted in triplicate inserts. The relative invasion/migration was expressed as the number of treated cells to control cells. The p-value was calculated with a Student’s t test.
Cell-based reporter assays.
DLD-1 CRC cells harboring a conditional AP4 allele were transfected using FuGENE Reagent (Roche) in 24-well plates with 10 ng Renilla luciferase control reporter plasmid and 100 ng Topflash or Fopflash luciferase reporter constructs containing either wild-type or mutant TCF-binding sites (Veeman et al., 2003). Simultaneously, the conditional AP4 allele was activated by addition of DOX. Firefly and Renilla luciferase activities were determined 24 h after transfection using a Dual-luciferase assay (Promega). Generation of pcDNA3-AP4-VSV and pcDNA3-AP4ΔBR-VSV was previously described in Jung et al. (2008), and the generation of pXP2–E-cadherin and pXP2–E-cadherin (mut-E1E2E3) in Yang et al. (2010). The reporter constructs were cotransfected into DLD-1 cells using FuGENE Reagent in 24-well plates with 10 ng Renilla luciferase control reporter plasmid, 100 ng pXP2–E-cadherin and pXP2–E-cadherin (mut-E1E2E3) reporter, and 100 ng pcDNA3-AP4-VSV and pcDNA3-AP4ΔBR, or an equimolar amount of pcDNA3-VSV backbone. Firefly luciferase activity was normalized to Renilla luciferase activity.
ChIP analysis.
Before cross-linking, DLD-1 cells were treated with DOX for 24 h to induce ectopic expression of AP4. Cross-linking and harvesting of cells was performed as described in Frank et al. (2001). Chromatin was sheered by 8 sonication cycles (HTU SONI 130; G. Heinemann) to generate DNA fragments with a mean size of 700 bp for qChIP and 30 sonication cycles to generate DNA fragments with a mean size of 300 bp for ChIP-seq analyses. Preclearing and incubation with polyclonal AP4 antibody (HPA001912; Sigma-Aldrich) or IgG control (M-7023; Sigma-Aldrich) for 16 h was performed as previously described (Menssen et al., 2007). The sequences of oligonucleotides used as qChIP primers are listed in Table S6.
ChIP-seq analysis.
DLD-1 cells were treated and harvested as described in the section above. ChIP was performed as described in the section above with DNA being fragmented by sonication to a mean size of 300 bp. After immunoprecipitation with the AP4-specific antibody HPA001912, the DNA fragments were quantified using a Bioanalyzer (Agilent Technologies). Libraries were generated with a ChIP-Seq Sample Prep kit (Illumina), sequenced with a read length of 36 bases using a Genome Analyzer IIx (Illumina), and aligned to the hg19 genome assembly using Eland. The library comprised 3,617,696 reads, which could be mapped to the genome. The ChIP-seq data can be accessed in the GEO database using accession no. GSE46935.
Clinical samples (metastasis cohort).
AP4 expression was evaluated using formalin-fixed, paraffin-embedded (FFPE) colon cancer samples of 110 patients who underwent surgical tumor resection at the Ludwig-Maximilians University of Munich (LMU) between 1994 and 2005. Follow-up data were recorded by the tumor registry Munich. All tumors were located on the right side of the colon. Half of the patients had colon cancers with synchronous liver metastases, where metastasis was diagnosed by clinical imaging or liver biopsy. Controls consisted of colon cancer patients without distant metastases at the time of diagnosis and with a disease-free survival of at least 5 yr after primary surgical resection. The samples of cases and controls were matched with respect to tumor grade (according to WHO 2000), T-classification (according to TNM Classification of Malignant Tumors 2009), and tumor localization, resulting in 55 matched pairs. Frequencies were analyzed using the χ2 test. Statistical procedures were performed using PASW Statistics 18.0 (SPSS, Inc). The study was performed according to the recommendations of the ethics committee of the Medical Faculty of the LMU.
Clinical samples (survival cohort).
CRC specimens from patients that underwent curative surgical resection between 1994 and 2005 at the LMU were drawn from the Institute’s archives, assuring that a 6-yr follow-up could be surveyed. Follow-up data were recorded by the tumor registry Munich. Only colorectal adenocarcinomas with moderate or poor differentiation (G2 or G3 according to WHO 2000) and T-categories T2 and T3 having neither nodal (N0) nor distant metastasis (M0) at the time of diagnosis were considered (UICC stage I or IIA). This resulted in a collection of tissues from 227 patients, of whom 65 (29%) died from CRC within the follow-up period. The survival data of 162 cases (71%) was censored when case follow-up was discontinued or patients died of reasons other than CRC. Overall survival was estimated by the Kaplan–Meier method and tested for significance by the log-rank procedure. Statistics were calculated using PASW Statistics 18.0. The study complied with the requirements of the ethics committee of the Medical Faculty of the LMU.
De novo motif discovery.
For motif discovery, a subset of ChIP signals that were located within a range of 5 kbp up- or 3 kbp downstream of a TSS and had a minimum height of 25 reads after peak calling were used. These criteria were met by 485 ChIP signals. To increase accuracy of the motif discovery algorithm, we restricted the sequence to a 300-bp radius around the summit point of the ChIP signal. For this the software MEME (Bailey et al., 2009) was used. The program was applied twice, and the best resulting motifs corresponding to TFBS were mapped back to the sequences through position weight matrices using the R statistics tool.
Generation of cell pools stably expressing conditional alleles.
The generation of pools of DLD-1 or SW480 cells harboring episomal vectors has been described previously in Jung et al. (2008) and Siemens et al. (2011).
Genome-wide analyses.
Microarray analyses were performed using Human Exon ST arrays (Affymetrix) according to the instruction of the manufacturer. Analyses were performed using Expression Console software (Affymetrix). ChIP-seq analysis was performed using a Genome Analyzer IIx and aligned to the hg19 genome assembly using Eland. Peak calling was performed using FindPeaks (Fejes et al., 2008), applying a cut-off set at a minimum of six overlapping reads considering bidirectional enrichment. This threshold was chosen for two reasons. First, the distribution of ChIP signals (peaks) relative to the regions of the closest genes (intronic, exonic, promoter region, and intergenic) was stabilized starting at six-read height. Second, previously validated targets of AP4 showed ChIP signal heights of around six as well. In addition to the peak calling, a peak splitting was applied to better estimate the amount of single binding sites and enhance the accuracy of de novo motif prediction. Applied parameters for the peak splitting were the factor 0.6, which multiplied with the maximum signal heights represents the threshold to initiate splitting and a minimum signal height of five. The algorithm was provided by PeakAnalyzer (Salmon-Divon et al., 2010), which was also used to assign the ChIP signals to the gene with the closest TSS. A subset of these ChIP signals was further analyzed. By comparison with the array data, the functionally relevant ChIP signals (at least 1.5-fold change, 5 kbp up- and 3 kbp downstream of a TSS) were identified and subdivided according to their activating or repressing effect. This resulted in 520 ChIP signals corresponding to 354 activated genes and 1,243 ChIP signals mapping to 530 repressed genes. The two groups were compared in terms of their mean height, absolute distance to the TSS, number of AP4 sites they contain, and their co-appearance in promoter regions. For peak splitting and assigning the ChIP signals to the gene with the closest TSS, the software PeakAnalyzer was used (Salmon-Divon et al., 2010). The MEME software (Bailey et al., 2009) was used for de novo motif discovery.
Immunohistochemistry.
Immunohistochemical staining was performed using 5-µm sections of whole standard tissue sections of the M0/M1 cohort blocks and tissue microarray blocks of the survival collection, respectively. As primary antibody, prediluted anti-TFAP4 monoclonal mouse antibody (HPA001912; Sigma-Aldrich) was used. SNAIL immunohistochemistry and evaluation of immunostaining were described in Siemens et al. (2013). Sections were pretreated for antigen retrieval by boiling in a microwave oven twice for 15 min at 750 W in Target Retrieval Solution (RE7113; Novocastra). Signals were generated using ImmPress Reagent kit rabbit Ig (Vector Laboratories) together with AEC (Dako) as the chromogen. Finally, slides were counterstained with hematoxylin (Vector Laboratories). Evaluation of immunostaining for AP4 was performed double-blinded by two investigators (R. Jackstadt and J. Neumann), both not aware of the clinical outcome of the respective samples. In discrepant cases, a consensus agreement was reached. The intensity of nuclear AP4 staining was categorized into none = −, weak = +, moderate = ++, and strong = +++ expression (Fig. 9 A). AP4 detection was considered as positive when staining was detected in the nucleus of tumor cells, consistent with its previously described localization (Jung et al., 2008).
Metastasis formation in a xenograft mouse model.
6–8-wk-old age-matched male immune-compromised NOD/SCID mice were used for lung metastasis assays after tail vein injection. NOD/SCID mice were purchased from the Jackson Laboratory. 72 h after transfection of siRNAs, 0.2 ml of suspension of 4 × 106/0.2 ml SW620-Luc cells (Ma et al., 2008) was injected into the lateral tail vein of the mice using 25-gauge needles. In weekly intervals, anesthetized mice were injected intraperitoneally with 150 mg/kg d-luciferin and imaged with the IVIS Illumina System (Caliper Life Sciences) 10 min after injection. The acquisition time was 2 min. After 9 wk, whole lungs were resected and photographed. For the immunohistochemical staining of human Vimentin (Dako), lungs were fixed with 4% paraformaldehyde and stained as described above. The number of metastases positive for Vimentin was determined microscopically. Pictures were obtained with an Axioplan 2 microscope equipped with an AxioCam 2 camera and AxioVision software (Carl Zeiss). All experiments involving mice were conducted with approval by the local animal experimentation committee (Regierung of Oberbayern).
Microarray analysis.
cDNA was generated from total RNA and amplified using the GeneChip WT cDNA Amplification kit (Affymetrix). cDNA was labeled using the GeneChip WT terminal labeling kit and hybridized to Human Exon ST arrays (Affymetrix) according to the manufacturer’s instructions. Arrays were hybridized, washed, stained, and scanned according to the manufacturer’s recommendations. Expression values were derived from CEL files generated from GeneChip Exon 1.0 ST arrays using the Expression Console implemented by Affymetrix. The normalization method of choice was the robust multichip average provided by the software. In addition, a filtering step was attached to the normalization. To reduce noise, the sum of the expression values of all replicates was calculated for each gene, and the 10% lowest values were excluded from the set. To calculate the p-value, a Student’s t test was applied to the 0- and 24-h measurements. From these values, the q-value was computed. The microarray data can be accessed in the GEO database using accession no. GSE46578.
Microscopic and indirect immunofluorescence analyses.
Phase-contrast images of cells in culture were captured on an Axiovert 25 microscope (Carl Zeiss) equipped with a Sony Digital Hyper HAD camera (software: Kappa Image Base; Kappa Opto-electronics) or an Axiovert Observer Z.1 microscope connected to an AxioCam MRm camera equipped with AxioVision software (Carl Zeiss). Immunohistochemical pictures were captured on an Axioplan 2 microscope equipped with an AxioCam 2 camera and AxioVision software. Indirect immunofluorescence analysis was performed as described previously (Siemens et al., 2011) and documented with an Axiovert Observer Z.1 microscope connected to an AxioCam MRm camera equipped with AxioVision software. The antibodies used for these analyses are listed in Table S9. LSM (laser-scanning microscopy) images were captured with a confocal microscope (LSM 700; Carl Zeiss) using a Plan Apochromat 20×/0.8 M27 objective, ZEN 2009 software (Carl Zeiss), and the following settings: image size of 2048 × 2048 and 16 bit, pixel/dwell of 25.2 µs, pixel size of 0.31 µm, laser power of 2%, and master gain of 600–1,000. After image capturing, the original LSM files were converted into TIFF files.
Plasmids and siRNAs.
pRTR was generated by replacing the KRAB repressor domain containing Tet trans-silencer of the pRTS (Bornkamm et al., 2005) vector with an alternative trans-silencer containing a CtBP-recruiting PLDLS repression motif (Molloy et al., 2001) and a relaxed effector specificity (Krueger et al., 2006), together with an IRES-coupled Puromycin-resistance gene. To generate the episomal pRTR-AP4-VSV vector, the AP4-VSV ORF was isolated from pcDNA3-AP4-VSV (Jung et al., 2008) and ligated into the pUC19-SfiI shuttle vector via BamHI and NotI restriction sites, released with SfiI, and the resulting fragment of interest was ligated into pRTR. All plasmids were verified by sequencing.
For the generation of miRNA-encoding plasmids directed against AP4, a two-step PCR was used. In the first step, the miRNA-AP4 coding sequences were amplified by primers listed in Table S7. The targeted AP4 sequence is indicated in Table S7. The first PCR product was used as a template and amplified by the universal miR30-XhoI/EcoRI primers and cut with XhoI and EcoRI and inserted into pSHUMI vector (Epanchintsev et al., 2006), which contains the flanking miR30 regions. A SfiI fragment containing the miRNA cassette was inserted into pRTR. The nonsilencing miRNA was isolated from a commercial pSM2c vector (Expression Arrest; Open Biosystems).
CDH1/E-cadherin reporter constructs were provided by M.-H. Yang (National Yang-Ming University, Taipei, Taiwan). siRNAs were transfected at 10 nM final concentration using HiPerFect reagent (QIAGEN). siRNA target sequences for AP4, SNAIL, and c-MYC are listed in Table S8. AP4 and SNAIL siRNAs and the negative control Silencer siRNA #1 were obtained from Ambion. c-MYC siRNAs and negative control AllStars #1 were obtained from QIAGEN.
Real-time qPCR.
The quantification of mRNA expression and DNA fragments obtained by ChIP by qPCR was performed as described in Menssen and Hermeking (2002) and Jung et al. (2008). The sequences of oligonucleotides used as primers for qPCR or qChIP analyses are listed in Tables S5 and S6, respectively.
Tissue microarray analysis.
Colorectal tissue microarrays were constructed as described previously (Kononen et al., 1998). In brief, hematoxylin and eosin–stained sections were used to define representative areas of viable tumor tissue. 1.0-mm needle core biopsies were taken from corresponding areas on the FFPE tumor blocks using a tissue arraying instrument (Beecher Instruments) and then placed in recipient paraffin array blocks at defined coordinates. To ensure the presence of representative parts of the tumors, six probes of each tumor were included. The cores in the paraffin block were incubated for 30 min at 37°C to improve adhesion between cores and paraffin of the recipient block.
Western blot analysis.
Western blot analyses were performed as described previously (Siemens et al., 2011). A list of antibodies used for this purpose is provided in Table S9.
Wound healing assay.
The wound healing assay was performed as described previously (Siemens et al., 2011).
Online supplemental material.
Table S1 lists results obtained by Gene Ontology analysis of AP4-regulated genes. Table S2 lists EMT- and proliferation-associated genes regulated by AP4. Table S3 shows AP4 and E-cadherin expression in seven CRC cell lines from the NCI-60 panel. Table S4 shows correlations of AP4 and SNAIL expression in the survival cohort. Tables S5 and S6 list oligonucleotides used for qPCR and qChIP analyses, respectively. Table S7 lists oligonucleotides used for miRNA construct generation. Table S8 lists siRNA target sequences. Table S9 depicts antibodies used for Western blot, immunofluorescence, and ChIP analyses. Table S10 lists microarray and ChIP-seq results. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120812/DC1.
Supplementary Material
Acknowledgments
We wish to thank Andrea Sendelhofert and Anja Heier for assistance with immunohistochemistry, Andreas Jung and Silvio Scheel for discussions, Sybille Liebmann for technical advice, and Muh-Hwa Yang and Xiao-Fan Wang for providing materials.
This work was supported by a grant from the Deutsche Krebshilfe (grant number 108243) to H. Hermeking. H. Hermeking also received support from the German Cancer Consortium (DKTK) and the Rudolf-Bartling Foundation.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CRC
- colorectal cancer
- CSC
- cancer stem cell
- DOX
- doxycycline
- EMT
- epithelial–mesenchymal transition
- miRNA
- microRNA
- TSS
- transcriptional start site
References
- Ansieau S., Bastid J., Doreau A., Morel A.P., Bouchet B.P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., et al. 2008. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 14:79–89 10.1016/j.ccr.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Athineos D., Sansom O.J. 2010. Myc heterozygosity attenuates the phenotypes of APC deficiency in the small intestine. Oncogene. 29:2585–2590 10.1038/onc.2010.5 [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Bornkamm G.W., Berens C., Kuklik-Roos C., Bechet J.M., Laux G., Bachl J., Korndoerfer M., Schlee M., Hölzel M., Malamoussi A., et al. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33:e137 10.1093/nar/gni137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T., Hlubek F., Spaderna S., Schmalhofer O., Hiendlmeyer E., Jung A., Kirchner T. 2005. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs (Print). 179:56–65 10.1159/000084509 [DOI] [PubMed] [Google Scholar]
- Cho K.B., Cho M.K., Lee W.Y., Kang K.W. 2010. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 293:230–239 10.1016/j.canlet.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Classon M., Henriksson M., Sümegi J., Klein G., Hammarskjöld M.L. 1987. Elevated c-myc expression facilitates the replication of SV40 DNA in human lymphoma cells. Nature. 330:272–274 (published erratum appears in Nature 2001. 412:748) 10.1038/330272a0 [DOI] [PubMed] [Google Scholar]
- Cowling V.H., Cole M.D. 2007. E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene. 26:3582–3586 10.1038/sj.onc.1210132 [DOI] [PubMed] [Google Scholar]
- Cowling V.H., D’Cruz C.M., Chodosh L.A., Cole M.D. 2007. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol. Cell. Biol. 27:5135–5146 10.1128/MCB.02282-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D.A., Gu W., Gautier J., Dalla-Favera R. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature. 448:445–451 10.1038/nature05953 [DOI] [PubMed] [Google Scholar]
- Du L., Wang H., He L., Zhang J., Ni B., Wang X., Jin H., Cahuzac N., Mehrpour M., Lu Y., Chen Q. 2008. CD44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. 14:6751–6760 10.1158/1078-0432.CCR-08-1034 [DOI] [PubMed] [Google Scholar]
- Eger A., Stockinger A., Park J., Langkopf E., Mikula M., Gotzmann J., Mikulits W., Beug H., Foisner R. 2004. beta-Catenin and TGFbeta signalling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene. 23:2672–2680 10.1038/sj.onc.1207416 [DOI] [PubMed] [Google Scholar]
- Epanchintsev A., Jung P., Menssen A., Hermeking H. 2006. Inducible microRNA expression by an all-in-one episomal vector system. Nucleic Acids Res. 34:e119 10.1093/nar/gkl624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes A.P., Robertson G., Bilenky M., Varhol R., Bainbridge M., Jones S.J. 2008. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 24:1729–1730 10.1093/bioinformatics/btn305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler I.J. 2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 3:453–458 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069–2082 10.1101/gad.906601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.P., Massagué J. 2006. Cancer metastasis: building a framework. Cell. 127:679–695 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Habib S.L., Bhandari B.K., Sadek N., Abboud-Werner S.L., Abboud H.E. 2010. Novel mechanism of regulation of the DNA repair enzyme OGG1 in tuberin-deficient cells. Carcinogenesis. 31:2022–2030 10.1093/carcin/bgq189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 95:2509–2514 10.1073/pnas.95.5.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.J., Choi H.J., Hong S., Huh Y., Chae H., Kim K.S. 2008. Transcription factor GATA-3 regulates the transcriptional activity of dopamine beta-hydroxylase by interacting with Sp1 and AP4. Neurochem. Res. 33:1821–1831 10.1007/s11064-008-9639-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.F., Lüscher B., Admon A., Mermod N., Tjian R. 1990. Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev. 4:1741–1752 10.1101/gad.4.10.1741 [DOI] [PubMed] [Google Scholar]
- Imai K., Okamoto T. 2006. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J. Biol. Chem. 281:12495–12505 10.1074/jbc.M511773200 [DOI] [PubMed] [Google Scholar]
- Jung P., Hermeking H. 2009. The c-MYC-AP4-p21 cascade. Cell Cycle. 8:982–989 10.4161/cc.8.7.7949 [DOI] [PubMed] [Google Scholar]
- Jung P., Menssen A., Mayr D., Hermeking H. 2008. AP4 encodes a c-MYC-inducible repressor of p21. Proc. Natl. Acad. Sci. USA. 105:15046–15051 10.1073/pnas.0801773105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lu Z., Hay E.D. 2002. Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol. Int. 26:463–476 10.1006/cbir.2002.0901 [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Zhang X.Y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N. 2006. Myc influences global chromatin structure. EMBO J. 25:2723–2734 10.1038/sj.emboj.7601152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen J., Bubendorf L., Kallioniemi A., Bärlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M.J., Sauter G., Kallioniemi O.P. 1998. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4:844–847 10.1038/nm0798-844 [DOI] [PubMed] [Google Scholar]
- Kozma L., Kiss I., Szakáll S., Ember I. 1994. Investigation of c-myc oncogene amplification in colorectal cancer. Cancer Lett. 81:165–169 10.1016/0304-3835(94)90198-8 [DOI] [PubMed] [Google Scholar]
- Krueger C., Danke C., Pfleiderer K., Schuh W., Jäck H.M., Lochner S., Gmeiner P., Hillen W., Berens C. 2006. A gene regulation system with four distinct expression levels. J. Gene Med. 8:1037–1047 10.1002/jgm.932 [DOI] [PubMed] [Google Scholar]
- Ku W.C., Chiu S.K., Chen Y.J., Huang H.H., Wu W.G., Chen Y.J. 2009. Complementary quantitative proteomics reveals that transcription factor AP-4 mediates E-box-dependent complex formation for transcriptional repression of HDM2. Mol. Cell. Proteomics. 8:2034–2050 10.1074/mcp.M900013-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Davie J.R. 2010. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat. 192:275–283 10.1016/j.aanat.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Liu M., Casimiro M.C., Wang C., Shirley L.A., Jiao X., Katiyar S., Ju X., Li Z., Yu Z., Zhou J., et al. 2009. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. USA. 106:19035–19039 10.1073/pnas.0910009106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bergami P., Lau E., Ronai Z. 2010. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer. 10:65–76 10.1038/nrc2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyng H., Brøvig R.S., Svendsrud D.H., Holm R., Kaalhus O., Knutstad K., Oksefjell H., Sundfør K., Kristensen G.B., Stokke T. 2006. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. 7:268 10.1186/1471-2164-7-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Rong Y., Radiloff D.R., Datto M.B., Centeno B., Bao S., Cheng A.W., Lin F., Jiang S., Yeatman T.J., Wang X.F. 2008. Extracellular matrix protein betaig-h3/TGFBI promotes metastasis of colon cancer by enhancing cell extravasation. Genes Dev. 22:308–321 10.1101/gad.1632008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Pevarello P., Barbacid M., Bischoff J.R. 2008. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol. Sci. 29:16–21 10.1016/j.tips.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici D., Hay E.D., Goodenough D.A. 2006. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol. Biol. Cell. 17:1871–1879 10.1091/mbc.E05-08-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A., Hermeking H. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA. 99:6274–6279 10.1073/pnas.082005599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A., Epanchintsev A., Lodygin D., Rezaei N., Jung P., Verdoodt B., Diebold J., Hermeking H. 2007. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle. 6:339–352 10.4161/cc.6.3.3808 [DOI] [PubMed] [Google Scholar]
- Mermod N., Williams T.J., Tjian R. 1988. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 332:557–561 10.1038/332557a0 [DOI] [PubMed] [Google Scholar]
- Molloy D.P., Barral P.M., Bremner K.H., Gallimore P.H., Grand R.J. 2001. Structural determinants outside the PXDLS sequence affect the interaction of adenovirus E1A, C-terminal interacting protein and Drosophila repressors with C-terminal binding protein. Biochim. Biophys. Acta. 1546:55–70 10.1016/S0167-4838(00)00071-6 [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G., Peinado H., Molina P., Olmeda D., Cubillo E., Santos V., Palacios J., Portillo F., Cano A. 2009. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat. Protoc. 4:1591–1613 10.1038/nprot.2009.152 [DOI] [PubMed] [Google Scholar]
- Mudduluru G., Ceppi P., Kumarswamy R., Scagliotti G.V., Papotti M., Allgayer H. 2011. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 30:2888–2899 10.1038/onc.2011.13 [DOI] [PubMed] [Google Scholar]
- Muzny D.M., Bainbridge M.N., Chang K., Dinh H.H., Drummond J.A., Fowler G., Kovar C.L., Lewis L., Morgan M.B., Newsham I.F.; Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.X., Chiang A.C., Zhang X.H., Kim J.Y., Kris M.G., Ladanyi M., Gerald W.L., Massagué J. 2009. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 138:51–62 10.1016/j.cell.2009.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D., Moreno-Bueno G., Flores J.M., Fabra A., Portillo F., Cano A. 2007. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 67:11721–11731 10.1158/0008-5472.CAN-07-2318 [DOI] [PubMed] [Google Scholar]
- Pálmer H.G., González-Sancho J.M., Espada J., Berciano M.T., Puig I., Baulida J., Quintanilla M., Cano A., de Herreros A.G., Lafarga M., Muñoz A. 2001. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. J. Cell Biol. 154:369–387 10.1083/jcb.200102028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7:415–428 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Png K.J., Halberg N., Yoshida M., Tavazoie S.F. 2012. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 481:190–194 10.1038/nature10661 [DOI] [PubMed] [Google Scholar]
- Roy H.K., Smyrk T.C., Koetsier J., Victor T.A., Wali R.K. 2005. The transcriptional repressor SNAIL is overexpressed in human colon cancer. Dig. Dis. Sci. 50:42–46 10.1007/s10620-005-1275-z [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P. 2010. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 11:415 10.1186/1471-2105-11-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Shoemaker R.H. 2006. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 6:813–823 10.1038/nrc1951 [DOI] [PubMed] [Google Scholar]
- Siemens H., Jackstadt R., Hünten S., Kaller M., Menssen A., Götz U., Hermeking H. 2011. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 10:4256–4271 10.4161/cc.10.24.18552 [DOI] [PubMed] [Google Scholar]
- Siemens H., Neumann J., Jackstadt R., Mansmann U., Horst D., Kirchner T., Hermeking H. 2013. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 19:710–720 10.1158/1078-0432.CCR-12-1703 [DOI] [PubMed] [Google Scholar]
- Smith A.P., Verrecchia A., Fagà G., Doni M., Perna D., Martinato F., Guccione E., Amati B. 2009. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 28:422–430 10.1038/onc.2008.395 [DOI] [PubMed] [Google Scholar]
- Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392–399 10.1038/35070076 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Ishii H., Nishida N., Takemasa I., Mizushima T., Ikeda M., Yokobori T., Mimori K., Yamamoto H., Sekimoto M., et al. 2011. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann. Surg. Oncol. 18:1166–1174 10.1245/s10434-010-1373-9 [DOI] [PubMed] [Google Scholar]
- Taube J.H., Herschkowitz J.I., Komurov K., Zhou A.Y., Gupta S., Yang J., Hartwell K., Onder T.T., Gupta P.B., Evans K.W., et al. 2010. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA. 107:15449–15454 10.1073/pnas.1004900107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Vécsey-Semjén B., Becker K.F., Sinski A., Blennow E., Vietor I., Zatloukal K., Beug H., Wagner E., Huber L.A. 2002. Novel colon cancer cell lines leading to better understanding of the diversity of respective primary cancers. Oncogene. 21:4646–4662 10.1038/sj.onc.1205577 [DOI] [PubMed] [Google Scholar]
- Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., Moon R.T. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13:680–685 10.1016/S0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Wielenga V.J., Heider K.H., Offerhaus G.J., Adolf G.R., van den Berg F.M., Ponta H., Herrlich P., Pals S.T. 1993. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 53:4754–4756 [PubMed] [Google Scholar]
- Wolfer A., Ramaswamy S. 2011. MYC and metastasis. Cancer Res. 71:2034–2037 10.1158/0008-5472.CAN-10-3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A., Wittner B.S., Irimia D., Flavin R.J., Lupien M., Gunawardane R.N., Meyer C.A., Lightcap E.S., Tamayo P., Mesirov J.P., et al. 2010. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc. Natl. Acad. Sci. USA. 107:3698–3703 10.1073/pnas.0914203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Weinberg R.A. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14:818–829 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang M.H., Hsu D.S., Wang H.W., Wang H.J., Lan H.Y., Yang W.H., Huang C.H., Kao S.Y., Tzeng C.H., Tai S.K., et al. 2010. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat. Cell Biol. 12:982–992 10.1038/ncb2099 [DOI] [PubMed] [Google Scholar]
- Yilmaz M., Christofori G. 2009. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 28:15–33 10.1007/s10555-008-9169-0 [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., DeSalle L.M., Patel J.H., Capobianco A.J., Yu D., Thomas-Tikhonenko A., McMahon S.B. 2005. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc. Natl. Acad. Sci. USA. 102:13968–13973 10.1073/pnas.0502330102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.