Abstract
The human genome comprises large chromosomes in the nucleus and mitochondrial DNA (mtDNA) housed in the dynamic mitochondrial network. Human cells contain up to thousands of copies of the double-stranded, circular mtDNA molecule that encodes essential subunits of the oxidative phosphorylation complexes and the rRNAs and tRNAs needed to translate these in the organelle matrix. Transcription of human mtDNA is directed by a single-subunit RNA polymerase, POLRMT, which requires two primary transcription factors, TFB2M and TFAM, to achieve basal regulation of the system. Here we review recent advances in understanding the structure and function of the primary human transcription machinery and the other factors that facilitate steps in transcription beyond initiation and provide more intricate control over the system.
Key Terms: Mitochondrial transcription, Human, Regulation, Transcription factor, RNA polymerase, TFAM
Organization of the mammalian mitochondrial genome
Mitochondria are dynamic double-membrane organelles that generate ATP via oxidative phosphorylation (OXPHOS) and are at the heart of other critical metabolic pathways. They are also involved in many other key cellular processes such calcium homeostasis, apoptosis, and signal transduction. A unique feature of these essential organelles is that they contain multiple copies of a circular, double-stranded DNA genome (mitochondrial DNA, mtDNA). Mammalian mtDNA is ~16.6 kb and encodes 13 mRNAs for key subunits of the OXPHOS pathway, 2 rRNAs of the mitochondrial ribosome, and 22 tRNAs necessary for translation by mitochondrial ribosomes in the matrix space (Figure 1) [1]. The remaining ~80 OXPHOS subunits and the ~1200–1500 other proteins present in mitochondria are encoded by the nuclear genome, translated by cytosolic ribosomes, and imported into mitochondria via specialized targeting and translocation mechanisms [2]. Because of the dual genomic origin of mitochondria components, mutations in mtDNA or nuclear genes can result in mitochondrial dysfunction that causes or contributes to inherited metabolic disorders, cancer, and neurodegenerative diseases, as well as the normal aging process and age-related pathology [3, 4].
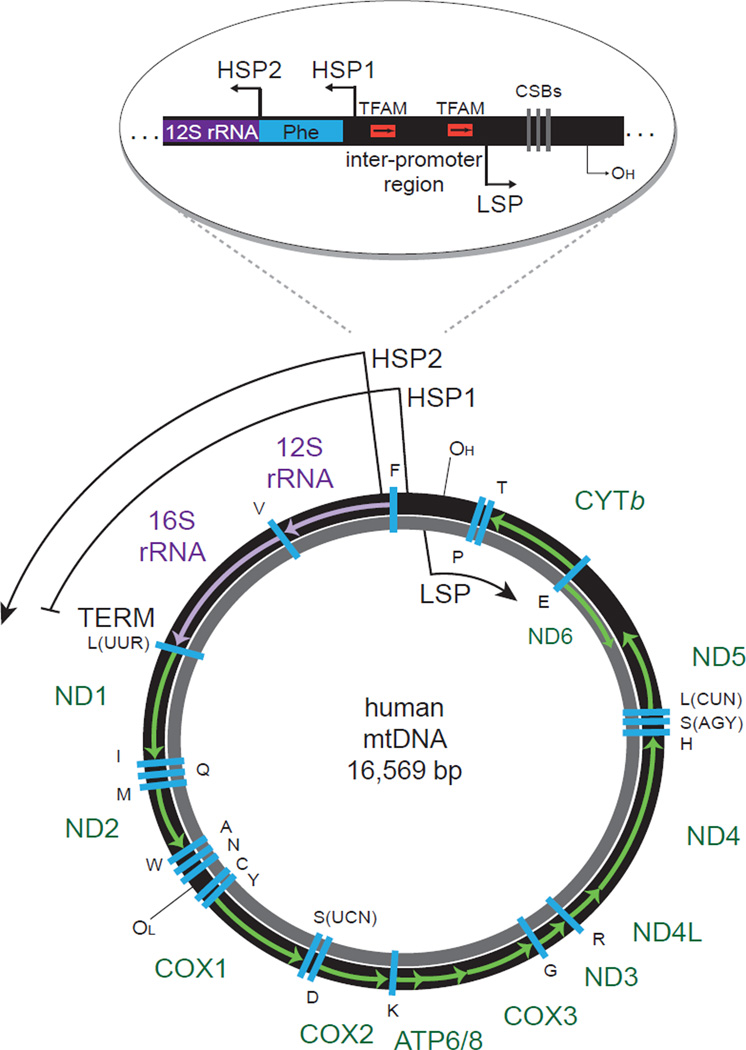
Figure 1. The human mitochondrial genome.
Human mtDNA is depicted with the heavy (H) strand in black and the light (L) strand in gray. The individual rRNAs (purple), mRNAs (green), and tRNAs (blue, letters represent cognate amino acids) are indicated with H-strand genes labeled outside and L-strand genes labeled inside the circle. H-strand transcription is initiated from two promoter sites, HSP1 and HSP2. HSP1 transcripts are terminated at the 22-bp termination sequence (TERM) within the tRNA-Leu(UUR) gene, where MTERF1 binds. HSP2 transcripts generate near full-length polycistronic transcripts that are then processed into the individual RNAs. L-strand transcription is initiated from a single promoter site, LSP, which also generates near full-length polycistronic messages that are processed. The primary replication origins of both the H- and L-strands are indicated (OH and OL, respectively). The D-loop control region, the major non-coding segment of human mtDNA, is shown expanded above. Indicated are HSP1, HSP2, and LSP, the TFAM binding sites at LSP and HSP1 (arrows indicate relative binding orientation of TFAM) within the inter-promoter region, conserved sequence blocks (CSBs I, II, III) important for proper RNA primer formation involved in H-strand replication, and the H-strand origin of replication (OH). mtDNA: mitochondrial DNA, HSP: heavy-strand promoter, LSP: light-strand promoter, MTERF1: mitochondrial termination factor 1, TFAM: transcription factor A mitochondrial.
The gene organization of mammalian mtDNA is highly conserved [5]. Genes are present on both strands, which are designated the heavy (H)-strand or light (L)-strand. The H-strand encodes two rRNAs, twelve mRNAs and fourteen tRNAs, while the L-strand encodes one mRNA and eight tRNAs (Figure 1). There is one major non-coding region in human mtDNA called the D-loop regulatory region, so named for a stable three-stranded DNA structure that occurs here [5]. The D-loop region encompasses most of the known cis-acting regulatory elements necessary for mtDNA transcription and replication (Figure 1).
Transcription of human mtDNA initiates from three promoters and generates polycistronic transcripts [1]. As shown in Figure 1, there is one L-strand promoter (LSP) and two H-strand promoters (HSP1 and HSP2). Transcripts from HSP1 are preferentially terminated in a tRNA gene immediately after the rRNA genes, and transcripts from LSP and HSP2 are nearly full genome in length [6]. These primary transcripts are subsequently processed into individual mRNA, rRNA, and tRNA molecules by various RNases through a mechanism that was originally referred to as the “tRNA punctuation model” because excision of tRNAs that flank the mRNAs and rRNAs liberate most, but not all, of the 37 mature RNA species [3, 7]. Replication of mtDNA from the H-strand and the L-strand occurs from two primary origins of replication (OH and OL, Figure 1). While it is clear that mitochondrial transcription and replication are coupled as described below, alternate proposed mechanisms of mtDNA replication are not reviewed here (see [8]). Instead, this review will focus on the most recent developments in understanding human mitochondrial transcription.
The primary human mitochondrial transcription machinery
Transcription of mtDNA is dependent entirely on factors encoded by nuclear genes. There are three primary mitochondrial transcription components in humans, the human mitochondrial RNA polymerase itself (h-mtRNA polymerase or POLRMT), human mitochondrial transcription factor B2 (h-mtTFB2 or TFB2M), and human mitochondrial transcription factor A (h-mtTFA or TFAM) [3, 9]. There is an ongoing debate as to whether all three of these components or just POLRMT and TFB2M are required for basal, promoter-specific transcription initiation (i.e., a three-component versus a two-component system). We, and others, have provided evidence for a two-component system, with TFAM acting as a transcriptional activator or repressor, depending on the context [10–12], while others have argued for a three-component system [13–15]. We will not address this controversy head-on in this review, as we feel enough results are published at this point for those interested to scholarly ascertain and draw their own conclusions. Furthermore, arguments on both sides are based exclusively on analysis of data obtained in vitro using DNA templates that certainly do not recapitulate the situation in vivo and hence likely will not be resolved by this approach. Perhaps most importantly, there seems to be general agreement in the field that these three primary components are critical for the general operation and basic regulation of the system, with other factors that functionally interact with these key players being described at a fast rate. It is primarily from this perspective that we write this review, but acknowledge our thinking and coverage is biased by the tenets of the two-component model.
Mitochondrial RNA polymerase
Mitochondrial RNA polymerases, including human POLRMT, are single-subunit, DNA-dependent, RNA polymerases with extensive sequence homology to the RNA polymerases of the bacteriophages T3 and T7 [16, 17]. Although clearly evolutionarily related, an important functional difference between POLRMT and the bacteriophage enzymes is that it cannot initiate promoter-specific transcription by itself on double-stranded DNA [18–20]. Like bacteriophage T7 [21], POLRMT also generates RNA primers for initiation of DNA replication [5, 22, 23], thereby coupling transcription and mtDNA replication. However, it is noted that other modes of priming and replication have been proposed for mammalian mtDNA [24, 25].
The structure of human POLRMT has been solved by X-ray crystallography, confirming that it is structurally homologous to the bacteriophage T7 RNA polymerase family and comprises a C-terminal domain (CTD), an N-terminal domain (NTD), and an N-terminal extension (NTE) [26]. The CTD is the largest domain, and the subdomains are highly conserved from the catalytic domains of the bacteriophage enzymes. Enzymatic characterization of POLRMT in vitro indicates the mechanism of catalysis utilizes a series of conformational changes and small RNA-DNA scaffolds as substrates for nucleotide addition and elongation [27]. The NTD is most similar to bacteriophage promoter-binding domains, and contains elements important for promoter recognition such as the AT-rich recognition loop and intercalating hairpin [26]. These elements are apparently diverged enough from the bacteriophage RNA polymerase sequences such that POLRMT is unable to productively engage mtDNA promoters without the aid of other transcription factors. The NTE of POLRMT includes two pentatricopeptide repeats (PPRs, Box 1) and an uncharacterized flexible region [26]. The function of the POLRMT PPR domains remains unknown, but its interaction with the AT-rich recognition loop in the NTD suggests it may be involved in achieving promoter specificity. The unstructured/flexible region of the NTE is required for promoter-specific transcription, but not polymerase function, as a deletion mutant of this region retains catalytic activity [26]. Interestingly, the NTE of yeast mitochondrial RNA polymerase, Rpo41p, functions as a binding platform for factors involved in coupling transcription to RNA processing and translation [28]. Additionally, human POLRMT was recently shown to have a transcription-independent function in mitochondrial ribosome biogenesis [29]. That similar couplings of transcription to translation and/or ribosome biogenesis in human mitochondria might involve the NTE of POLRMT is an intriguing possibility.
Box 1: The pentatricopeptide (PPR) domain in proteins involved in mitochondrial gene expression.
The PPR domain family of proteins are a diverse set of proteins, originally discovered in plant plastids and mitochondria, involved in many aspects of RNA metabolism, including transcription, processing, stability, editing and translation [78]. The PPR motif is a 35 amino acid sequence that is usually present in multiple copies (ranging from 2 to 26) that are critical for RNA-binding and/or RNA-protein interactions [78, 79]. Seven PPR-domain proteins have been identified in mammals all of which are localized to mitochondria: mitochondria RNA polymerase (POLRMT), leucine-rich pentatricopeptide repeat containing protein (LRPPRC), PPR domain containing proteins 1, 2, and 3 (PTCD1-3), mitochondrial RNase P protein 3 (MRPP3) and mitochondrial ribosomal protein S27 (MRPS27) [78]. The mitochondrial RNA polymerase, POLRMT, has two tandem PPR domains in its N-terminal extension. These are the first PPR domains for which the structure has been solved, revealing that the domain forms a helix-turn-helix fold [26]. LRPPRC has 22 predicted PPR domains, and as described in the main text has reported roles in mitochondrial transcription, mRNA stability, and polyadenylation. PTCD1 has eight PPR domains and is part of an RNA-processing complex for mitochondrial tRNAs, PTCD2 has 5 PPR domains and is important for Cyt b RNA processing, and PTCD3 contains 15 PPR domains and associates with the 12S rRNA of the small mitochondrial ribosome subunit [78]. MRPP3 is part of a complex with mitochondrial RNase P activity and contains three PPR domains [78]. MRPS27 is a protein component of the small subunit of the mitochondrial ribosome, associates with the 12S rRNA in a non-regulatory way and has 6 PPR domains [78]. While it is clear that this family of proteins is critical for many aspects of mitochondrial gene expression, much remains to be learned about their overall functions in mitochondria, as well as the precise contributions of the individual PPR domains to the various protein-RNA transactions involved.
Mitochondrial transcription factor B2, TFB2M
The mitochondrial transcription machinery in yeast is a two-component system comprising Rpo41p and the mitochondrial transcription factor Mtf1p/sc-mtTFB1 [9]. There are two mammalian orthologs of this yeast transcription factor that likely resulted from a gene duplication event early in evolution [30]. The human proteins, h- mtTFB1/TFB1M and h-mtTFB2/TFB2M, are homologous to an ancestral bacterial rRNA dimethyltransferase and it appears that both proteins have retained this enzymatic activity, based on their ability to complement this function in Escherichia coli [15, 30–32]. However, multiple lines of evidence now suggest that TFB1M is the primary mitochondrial 12S rRNA methyltransferase important for small subunit ribosome biogenesis and TFB2M is the primary mitochondrial transcription initiation factor [15, 33, 34]. That is, TFB2M facilitates the transition from a closed to an open promoter conformation needed for initiation to occur (i.e. open complex formation) as well as other potentially rate-limiting steps, and acts transiently in the catalytic site for RNA polymerization [14, 18, 19]. Through its interaction with POLRMT, TFB2M facilitates open complex formation in part by inducing promoter melting [18–20], but does not appear to contribute significantly to promoter recognition, a function that resides in POLRMT [19, 20].
Mitochondrial transcription factor A, TFAM
TFAM was the first human mitochondrial transcription factor identified [35] and belongs to the high-mobility-group (HMG) family of DNA-binding proteins with characteristic DNA bending and wrapping abilities mediated by two HMG box domains [36]. The DNA bending and wrapping attributes of TFAM allow it to act both as a transcriptional activator and an mtDNA packaging protein [1]. The transcriptional activation role of TFAM has been linked with its ability to bind and bend DNA avidly at specific sites upstream of the mtDNA promoters (Figure 1, inset, and Figure 2A) [37, 38], and its role in packaging is associated with its ability to bind DNA non-specifically and bend it less dramatically throughout the mtDNA (Figure 2C) [39–42]. This latter function of TFAM in nucleoids (protein-mtDNA complexes) has been reviewed elsewhere [43], and is not discussed further.
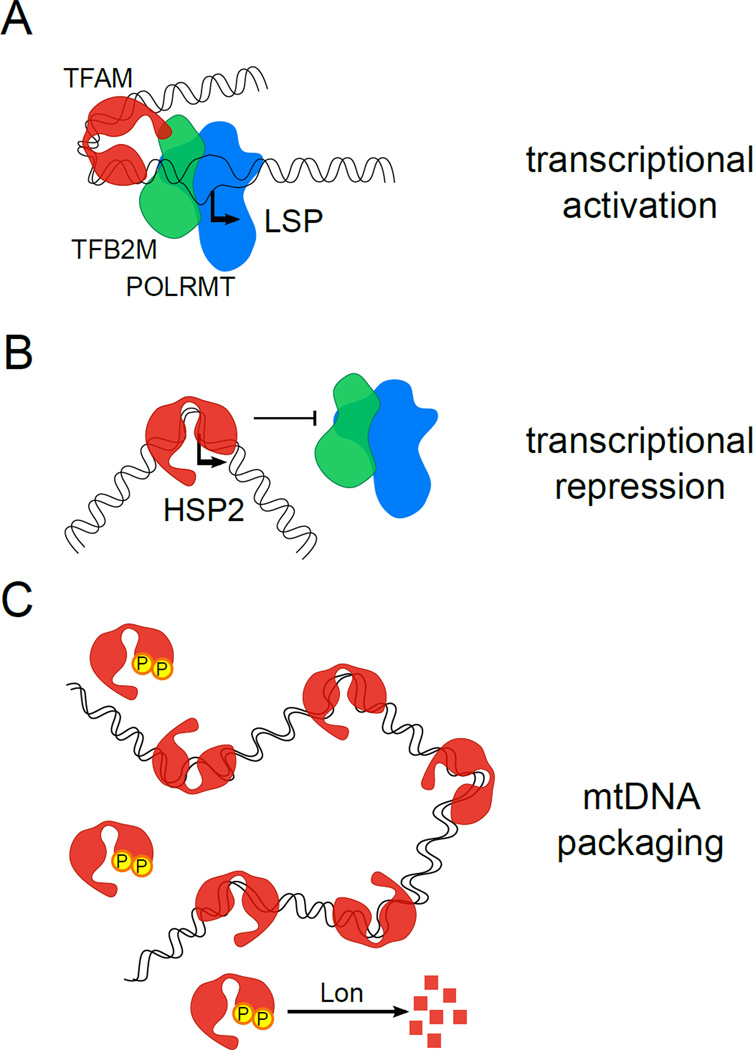
Figure 2. TFAM differentially regulates mtDNA promoters and packages mtDNA.
TFAM is a multi-functional protein. (A) TFAM activation of transcription at the LSP is shown. TFAM, via its C-terminal tail that interacts with TFB2M (green) and unique DNA-bending capacity, promotes high levels of specific initiation by POLRMT (blue) and TFB2M. (B) TFAM can also inhibit transcription at HSP2 in vitro at concentrations that activate LSP and HSP1. It is postulated that this is due to a unique binding mode at this site that competitively inhibits promoter binding by POLRMT and TFB2M. C) In addition to its role in transcription, TFAM also packages mtDNA to facilitate nucleoid formation. This is accomplished by its ability to bind many sites on mtDNA in a more or less nonspecific manner and bend the DNA, albeit to a lesser degree than promoter DNA. TFAM is phosphorylated at sites within the HMG-box domains (denoted as round circles with a “P”), which reduces DNA binding and promotes its degradation by Lon protease. This may allow dynamic remodeling of nucleoids to achieve specific outcomes (e.g. to relieve inhibition of HSP2 transcription). TFAM: transcription factor A mitochondrial, TFB2M: transcription factor B2 mitochondrial, POLRMT: mitochondrial RNA polymerase, LSP: light-strand promoter, HSP: heavy-strand promoter, mtDNA: mitochondrial DNA, HMG box: high-mobility-group box.
The X-ray crystal structures of TFAM bound to the LSP promoter site demonstrate that it imposes a “U-turn” in the promoter DNA [41, 44]. Specifically, each HMG box facilitates a 90-degree turn that bends the promoter DNA back on itself. The C-terminal tail of TFAM is important for transcriptional activation and interacts with TFB2M (and TFB1M) (Figure 2A) [38, 45]. It is likely that the dramatic bend in promoter DNA mediated by TFAM is needed to enhance the interaction of the C-terminal tail with TFB2M to increase the rate of transcription initiation at the LSP [41]. TFAM binding at non-specific mtDNA sites also imparts DNA bending but to a lesser degree than observed at the HSP1 and LSP promoter sites (Figure 2C) [39–41].
It is now becoming clear that the interaction of TFAM with mtDNA is regulated both by post-translational modification and targeted degradation. TFAM is degraded by the Lon protease [46, 47] and this is mediated by phosphorylation at multiple serine residues in the HMG1 and HMG2 domains by protein kinase A (PKA) [46]. These results support a novel mode of regulation whereby PKA-mediated phosphorylation of TFAM inhibits DNA binding and results in degradation of the “free” (not bound to DNA) pool of the protein. This mode of regulation may allow a dynamic remodeling of when and where TFAM is bound to mtDNA that allows precise modulation of transcription output and nucleoid structure (via altering the packaging mode) under different physiological circumstances (Figure 2C). TFAM may also be acetylated [48], thus it may turn out to be the case that TFAM activity is modulated by multiple inputs and modifications akin to histones in the nucleus. Breaking this “mitochondrial code” promises to be an exciting area of future study.
Basal regulation of transcription by POLRMT, TFB2M and TFAM
With the structural and functional properties of the three components (POLRMT, TFB2M and TFAM) that provide basal regulation of human mitochondrial transcription introduced above, we now turn to how they interface with the mtDNA promoters. It has long been known that the LSP is preferentially activated by TFAM in vitro and has the strongest binding site upstream of the transcription initiation site [35, 49]. The HSP1 binding site for TFAM is significantly weaker, hence its activation profile is much different that that of LSP [15, 35, 49]. Shutt et al. [12] quantified this in dose-response experiments with TFAM using templates that contained both promoters. In the absence of TFAM, POLRMT and TFB2M direct significant promoter-specific initiation from both promoters in vitro, but more efficiently at HSP1 [12]. The addition of TFAM stimulates transcription at LSP and HSP1 (Figure 2A), but the activation profiles are markedly different. That is, significantly less TFAM is needed to activate and attain maximal transcriptional stimulation at LSP compared to HSP1. Certainly the relative degree of DNA bending by TFAM is important for its differential transcriptional activation at the HSP1 and LSP promoters [39–41, 44], but other parameters may play a role, such as the influence of the inter-promoter sequences [12] and/or the fact that the TFAM binding sites for these two promoters are in reverse orientation relative to the transcription initiation site (Figure 1, inset) [49].
Despite evidence from early RNA mapping studies indicating the existence of a second H-strand promoter (HSP2) [6], initiation from this promoter in vitro proved elusive for many years. Transcription from HSP2 was eventually observed in vitro using mitochondrial lysates and reported to be dependent on mitochondrial termination factor 1 (MTERF1) [50]. However, more recent studies revealed that promoter-specific transcription from HSP2 is achievable in vitro with just POLRMT and TFB2M (i.e., the core two-component system) [10, 11]. Furthermore, not only did transcription initiation from HSP2 not require TFAM, it was actually inhibited by concentrations of TFAM that activate HSP1 and LSP. Compared to HSP1 and LSP, TFAM binds much closer to the initiation site of HSP2, and increasing the amount of POLRMT and TFB2M overcomes inhibition of transcription initiation from HSP2 (Figure 2B), suggesting that TFAM binding competitively inhibits the core machinery from binding HSP2 in vitro [10, 11].
The ability of TFAM to differentially activate HSP1 and LSP at various protein/DNA ratios in vitro, and to repress HSP2 at concentrations that activate the other two promoters, invites speculation that TFAM activity can differentially activate mtDNA promoters to achieve specific outcomes. For example, transcription from LSP not only is needed for expression of genes on the L-strand, but also primes mtDNA for replication [1, 23]. Furthermore, Attardi and colleagues proposed many years ago that transcription initiated at HSP1 is preferentially terminated after the rRNA genes, while initiation from HSP2 leads to expression of almost the full complement of H-strand genes [6, 51]. Thus, it may be important to alter TFAM activity at the promoters to balance the needs for wholesale gene expression, ribosome biogenesis, and mtDNA replication in different tissues or in response to physiological or environmental changes. One can envision many ways this could be achieved, but three that come to mind are: 1) having different steady-state amounts of TFAM and/or mtDNA in cells or tissues so that different TFAM/mtDNA ratios are achieved [52]; 2) modulating the amount of TFAM bound to mtDNA within nucleoids (e.g. via phosphorylation and/or Lon-mediated degradation; Fig. 2C); and 3) influence of other factors that interact with TFAM or the core transcription machinery. The remainder of this review is devoted to the last of these possibilities, as multiple accessory factors have been described recently that modulate mitochondrial transcription in humans.
Mitochondrial transcriptional regulatory factors
Although the primary transcriptional machinery is relatively simple in nature, it is becoming clear that the regulation of mitochondrial transcription is much more complex, involving additional factors acting at multiple stages of the process (Figure 3). These regulatory factors are described in more detail in this section and in Box 2.
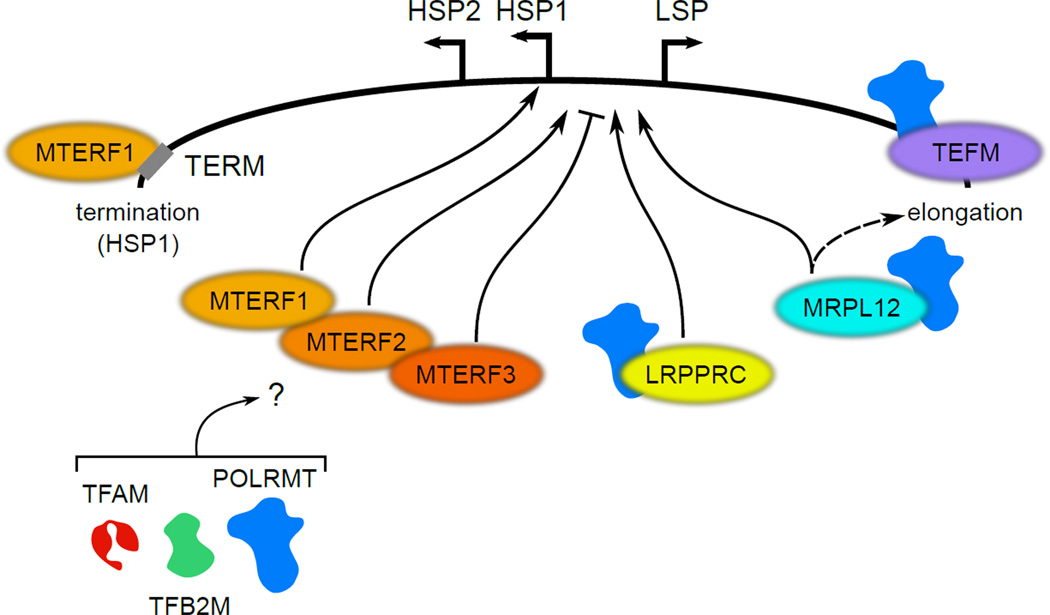
Figure 3. Mitochondrial transcription accessory factors.
A portion of the human mtDNA with salient cis-acting transcriptional regulatory elements (LSP, HSP1, HSP2, and TERM) is shown at the top. Accessory factors discussed in the main text that have been implicated in human mitochondrial transcriptional regulation are depicted as colored ovals, with stimulatory (arrows) or inhibitory (“T”) functions indicated. Known interactions with POLRMT are shown for MRPL12, LRPPRC and TEFM. That it is currently unknown if or how MTERF1-3 interact with the primary mitochondrial transcription machinery (POLRMT, TFB2M and TFAM) is indicated by the question mark. As indicated, MTERF1, 2 and 3 have been shown to interact in a complex. MTERF1 is shown bound to the TERM site, where it facilitates transcription termination. mtDNA: mitochondrial DNA, LSP: light-strand promoter, HSP: heavy-strand promoter, TERM: termination sequence, POLRMT: mitochondrial RNA polymerase, TFB2M: transcription factor B2 mitochondrial, TFAM: transcription factor A mitochondrial, MTERF: mitochondrial termination factor, LRPPRC: leucine-rich pentatricopeptide repeat containing, MRPL12: mitochondrial ribosomal protein L12, TEFM: transcription elongation factor mitochondrial.
Box 2: Nuclear transcription factors implicated in mitochondrial transcriptional regulation.
In addition to dedicated mitochondrial proteins, there is now an extensive list of nuclear transcription factors that have been reported to also localize to mitochondria. Some appear to regulate mitochondrial transcription, while others likely have non-transcription related functions in mitochondria. For example, alternative forms of several nuclear hormone receptors have been reported in mitochondria, where they have been postulated to bind to their cognate response elements found in mtDNA [80]. However, in no case has a direct interaction with the primary mitochondrial transcription machinery been shown, nor has activation from direct binding to mtDNA sites in vivo been unequivocally demonstrated. A direct interaction of p53 with TFAM has been reported [81, 82], which may indicate that mitochondrial localized forms of p53 regulate transcription, mtDNA packaging and/or repair. Likewise, Cockayne syndrome B protein has been localized to mitochondria and implicated as a transcriptional activator that is modulated by TFAM levels and may be involved in elongation [83]. Finally, additional nuclear transcription factors have been reported in mitochondria, including NF-κB (nuclear factor kappa-light-chain enhancer of activated B cells), BRCA1 (breast cancer 1), STAT3 (signal transducer and activator of transcription 3), HMGA1 (high-mobility-group protein 1), and others [80]. While, in most of these cases, the jury is still out as to whether these factors regulate mitochondrial transcription directly or have other functions in the organelle, they may indicate that a rich mode of tissue- and context-dependent regulation of mitochondrial gene expression exists that awaits better definition.
The MTERF family
The MTERF family of proteins (MTERF1-4) is a diverse set of mitochondrial regulatory factors defined by sequence homology to the founding member, MTERF1 [53]. Transcripts initiated at HSP1 in vivo are preferentially terminated after the rRNA genes via MTERF1 binding to a 22-bp termination site located within the tRNA-Leu(UUR) gene, immediately downstream of the 16S rRNA gene (Figure 1 and Figure 3) [51, 53, 54]. Structural studies show that base flipping and DNA unwinding promote site-specific termination by MTERF1 [54, 55]. By contrast, transcripts initiated at HSP2 are seemingly immune to this termination event, leading to expression of virtually all of the genes on the H-strand. Furthermore, MTERF1 has been reported to simultaneously bind near HSP1 and its distant termination site, suggesting a DNA-looping mechanism that facilitates recycling of the core transcription machinery to this promoter after termination [50]. This combination of events is thought to achieve the observed, and apparently important, greater steady-state level of rRNAs relative to mRNAs [50].
The other members of the MTERF family are not involved in termination, but do have reported functions in transcription and gene expression. For example, MTERF2 is present in nucleoids and can bind mtDNA non-specifically or at specific sites within the HSP1 promoter region [56, 57]. Loss of MTERF2 in primary fibroblasts and certain tissues of mice reduced OXPHOS activity and the steady-state levels of mtDNA-encoded mRNAs [56]. In this same study, MTERF2 was shown to interact with both MTERF1 and MTERF3 in an mtDNA-dependent manner [56]. These results are consistent with a role for MTERF2 in the activation of mitochondrial transcription (Figure 3). Interestingly, loss of MTERF3 also results in OXPHOS deficiency, but unlike loss of MTERF2, this is accompanied by increased and aberrant mtDNA gene expression [58], perhaps consistent with disruption of its repressive function at HSP1 (Figure 3). Similar to MTERF1, both MTERF2 and MTERF3 likely play regulatory roles in mtDNA replication [59]. Thus, it appears MTERF1-3 somehow modulate mitochondrial transcription and replication, but it remains unclear what the exact relationships are and how their activities are coordinated. It also remains to be determined if and how these proteins are interfacing with the primary transcriptional machinery to exert their proposed regulatory effects (Figure 3). The last member of the MTERF family, MTERF4, plays a unique role in ribosome biogenesis by interacting with NSUN4, a mitochondrial 16S rRNA methyltransferase [60].
Mitochondrial LRPPRC/LRP130
The leucine-rich pentatricopeptide repeat containing protein (LRPPRC or LRP130) has 22 PPR domains (Box 1) and is implicated in a range of cellular functions involving various protein complexes [61–67]. Although reported to be in the cytoplasm and nucleus, its primary localization is in mitochondria [68], where it has multiple roles in RNA metabolism. Accordingly, mutations in the LRPPRC gene cause the inherited French Canadian variant of Leigh Syndrome (LSFC), a rare neurodegenerative disorder characterized by deficiency of mitochondrial OXPHOS complex IV (Cytochrome cOxidase, or COX) [64, 65, 67, 69].
While it is clear that a loss of LRPPRC leads to impaired OXPHOS and COX activity and its over-expression increases OXPHOS [61–64, 67, 70, 71], the precise mechanisms involved have not been resolved fully. Several groups have demonstrated LRPPRC binds multiple mtDNA-encoded mRNAs directly to increase their stability and, in one case, specific PPR domains were shown to interact with the COX1 mRNA [64–66, 70]. LRPPRC also interacts with a mitochondrial stem-loop RNA-binding protein (SLIRP) in a high molecular weight complex involved in maintaining the polyadenylation and stability of mature non-translating mRNAs and inhibiting the polynucleotide phosphorylase (PNPase) [61, 62, 64, 66]. However, reports differ on whether this complex is dependent on RNA and which mitochondrial mRNAs it specifically binds. Finally, LRPPRC also stimulates mitochondrial transcription in vitro using either mitochondrial extracts or purified, recombinant transcription components (Figure 3) [63, 71]. This is likely through a direct interaction with POLRMT and/or other transcriptional regulatory proteins [63] (Fig. 3). Labeling studies in organello revealed that over-expression of LRPPRC increases the synthesis of newly synthesized mtDNA-encoded transcripts, including mRNA, rRNAs and tRNAs [63]. In an attempt to synthesize the multiple studies on the mitochondrial function of LRPPRC, we propose that it may act as an RNA chaperone that ferries transcripts from the early stages of initiation of their synthesis to their eventual translation, and facilitates their transfer between the multiple complexes that are involved. In this regard, it may have some functions similar to the distantly related Pet309p and other factors that interact with mitochondrial RNA polymerase to couple transcription to RNA processing and translation in budding yeast [28].
Mitochondrial ribosomal protein L12 (MRPL12)
MRPL12 is a component of the large subunit of the mitochondrial ribosome, but in its “free” (non-ribosome associated) form interacts directly with POLRMT to stimulate mitochondrial transcription [72, 73]. MRPL12 stimulates both promoter-dependent and promoter-independent transcription in vitro, and exists in some non-ribosomal complexes in vivo that are separate from POLRMT-TFB2M initiation complexes [72]. Based on these results, we have proposed that MRPL12 activates transcription at multiple steps, perhaps including the transition from initiation to elongation (Figure 3).
The connections of MRPL12, in particular, to mitochondrial transcription are intriguing given how it was discovered and the other cellular functions in which it has been implicated. MRPL12, the first human mitochondrial ribosomal protein identified, was found in a screen for genes that are up-regulated when serum-starved cells reenter the cell cycle [74]. Mutations in MRPL12 were also found in a screen for genes that suppress pro-growth pathways in Drosophila [75]. Finally, the acetylation state of MRPL12 may be dependent on the mitochondrial protein deacetylase sirtuin 3 [48]. Taken together, these observations indicate to us that MRPL12 may be exquisitely regulated at multiple levels in response to cellular growth cues and could relay such information directly to the mitochondrial transcription machinery to alter mitochondrial gene expression and function in response.
Mitochondrial transcription elongation factor (TEFM)
Mitochondrial transcripts are generated as polycistronic messages that are processed into individual rRNAs, tRNAs and mRNAs. Enzymatic studies in vitro suggest that POLRMT alone is likely insufficient to generate these long transcripts [27], and that elongation factors are required. TEFM is a mitochondrial protein that enhances POLRMT processivity both in vitro and in vivo [76]. It interacts with the CTD of POLRMT in an RNA-independent manner (Figure 3), but not TFB2M or TFAM, consistent with a role in elongation. Accordingly, depletion of TEFM results in reduced levels of promoter-distal H- and L-strand mitochondrial transcripts that may underlie the observed OXPHOS defects [76]. The mechanism by which TEFM interacts with POLRMT and facilitates transcriptional elongation has yet to be determined, but it is part of a large complex that may also be involved in coupling transcription elongation complexes to the translation machinery [76].
Concluding Remarks
We have seen exciting recent advances in the field of mitochondrial transcription. An explosion of structural and functional studies has provided key new insights. And, while POLRMT, TFB2M and TFAM provide the basal regulation of mitochondrial transcription, additional accessory factors that provide more intricate control of the system at multiple steps in the process (e.g. initiation, elongation and termination) are being uncovered at an unprecedented rate. It will be critically important to now move toward understanding how these new factors interact with the primary transcription machinery to exert their specific effects. While in vitro mitochondrial transcription systems have and will continue to provide important basic insights, it is clear that what is needed going forward is to better ascertain how mitochondrial transcription is regulated in vivo. In this regard, understanding how post-translational modifications regulate the system and the potential for epigenetic regulation (e.g. mtDNA methylation [77]) are important to consider. Finally, the mitochondrial transcriptional response to metabolic variations, changing energy demands and environmental factors in vivo remains in its infancy, especially in terms of tissue-specific responses. This is an area of great interest that may involve the increasing number of nuclear transcriptional regulators that are also localized to mitochondria (Box 2), and will no doubt be important to decipher if we are to fully comprehend how mitochondria are involved in normal cell physiology and go awry in human disease and aging.
Highlights.
POLRMT, TFB2M and TFAM provide basal regulation of human mitochondrial transcription
Structures of key mitochondrial transcription components have provided new insights
The need to understand regulation of mitochondrial transcription in vivo is eminent
Accessory factors provide additional layers of mitochondrial transcriptional regulation
Acknowledgements
This work was supported by the U.S. NIH via an NRSA Postdoctoral Fellowship F32DK091042 to M.L.B. and grant HL-059655 to G.S.S. Due to imposed referencing limitations, we certainly were not able to cite all of the relevant literature in this area and in many cases used review articles to cover broader topics in lieu of the primary literature. We apologize for any inadvertent oversights in this regard.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENES
- 1.Bonawitz ND, et al. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Becker T, et al. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci. 2012;37:85–91. doi: 10.1016/j.tibs.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 6.Montoya J, et al. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 7.Ojala D, et al. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 8.Wanrooij S, Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim Biophys Acta. 2010;1797:1378–1388. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Shadel GS, Clayton DA. Mitochondrial transcription initiation. Variation and conservation. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 10.Lodeiro MF, et al. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc Natl Acad Sci U S A. 2012;109:6513–6518. doi: 10.1073/pnas.1118710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zollo O, et al. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc Natl Acad Sci U S A. 2012;109:6508–6512. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shutt TE, et al. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci U S A. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, et al. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci U S A. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litonin D, et al. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 16.Tiranti V, et al. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 17.Masters BS, et al. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 18.Lodeiro MF, et al. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sologub M, et al. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaspari M, et al. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato M, et al. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 22.Fuste JM, et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 23.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt IJ. Mitochondrial DNA replication and repair: all a flap. Trends Biochem Sci. 2009;34:358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 26.Ringel R, et al. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 27.Smidansky ED, et al. Human mitochondrial RNA polymerase: evaluation of the single-nucleotide-addition cycle on synthetic RNA/DNA scaffolds. Biochemistry. 2011;50:5016–5032. doi: 10.1021/bi200350d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Surovtseva YV, Shadel GS. Transcription-independent role for human mitochondrial RNA polymerase in mitochondrial ribosome biogenesis. Nucleic Acids Res. 2013;41:2479–2488. doi: 10.1093/nar/gks1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J Mol Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 31.Seidel-Rogol BL, et al. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 32.McCulloch V, et al. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Cotney J, et al. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavyand light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 36.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 37.Fisher RP, et al. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 38.Dairaghi DJ, et al. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 39.Farge G, et al. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat Commun. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
- 40.Malarkey CS, et al. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngo HB, et al. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Rubio-Cosials A, et al. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 45.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu B, et al. Phosphorylation of Human TFAM in Mitochondria Impairs DNA Binding and Promotes Degradation by the AAA(+) Lon Protease. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsushima Y, et al. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc Natl Acad Sci U S A. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hebert AS, et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher RP, et al. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 50.Martin M, et al. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 51.Kruse B, et al. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 52.Shutt TE, et al. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription. 2011;2:55–59. doi: 10.4161/trns.2.2.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberti M, et al. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta. 2009;1787:303–311. doi: 10.1016/j.bbabio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Yakubovskaya E, et al. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jimenez-Menendez N, et al. Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nat Struct Mol Biol. 2010;17:891–893. doi: 10.1038/nsmb.1859. [DOI] [PubMed] [Google Scholar]
- 56.Wenz T, et al. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Pellegrini M, et al. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 58.Park CB, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 59.Hyvarinen AK, et al. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol Biol Rep. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 60.Camara Y, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Chujo T, et al. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruzzenente B, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. [Google Scholar]
- 63.Liu L, et al. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J Biol Chem. 2011;286:41253–41264. doi: 10.1074/jbc.M111.276121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sasarman F, et al. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu F, et al. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sterky FH, et al. LRPPRC is a mitochondrial matrix protein that is conserved in metazoans. Biochem Biophys Res Commun. 2010;398:759–764. doi: 10.1016/j.bbrc.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 69.Debray FG, et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J Med Genet. 2011;48:183–189. doi: 10.1136/jmg.2010.081976. [DOI] [PubMed] [Google Scholar]
- 70.Xu F, et al. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem J. 2012;441:275–283. doi: 10.1042/BJ20110985. [DOI] [PubMed] [Google Scholar]
- 71.Sondheimer N, et al. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Surovtseva YV, et al. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci U S A. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, et al. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marty L, Fort P. A delayed-early response nuclear gene encoding MRPL12, the mitochondrial homologue to the bacterial translational regulator L7/L12 protein. J Biol Chem. 1996;271:11468–11476. doi: 10.1074/jbc.271.19.11468. [DOI] [PubMed] [Google Scholar]
- 75.Frei C, et al. The Drosophila mitochondrial ribosomal protein mRpL12 is required for Cyclin D/Cdk4-driven growth. EMBO J. 2005;24:623–634. doi: 10.1038/sj.emboj.7600523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minczuk M, et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shock LS, et al. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rackham O, Filipovska A. The role of mammalian PPR domain proteins in the regulation of mitochondrial gene expression. Biochim Biophys Acta. 2012;1819:1008–1016. doi: 10.1016/j.bbagrm.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 79.Lightowlers RN, Chrzanowska-Lightowlers ZM. PPR (pentatricopeptide repeat) proteins in mammals: important aids to mitochondrial gene expression. Biochem J. 2008;416:e5–e6. doi: 10.1042/BJ20081942. [DOI] [PubMed] [Google Scholar]
- 80.Szczepanek K, et al. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 2012;22:429–437. doi: 10.1016/j.tcb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park JY, et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105:705–712. doi: 10.1161/CIRCRESAHA.109.205310. 711 p following 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshida Y, et al. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 83.Berquist BR, et al. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012;40:8392–8405. doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]