Abstract
4D (3D spatial+time) echocardiography is gaining widespread acceptance at clinical institutions for its high temporal resolution and relatively low cost. We describe a novel method for computing dense 3D myocardial motion with high accuracy. The method is based on a classical variational optical flow technique, but exploits modern developments in optical flow research to utilize the full capabilities of 4D echocardiography. Using a variety of metrics, we present an in-depth performance evaluation of the method on synthetic, phantom, and intraoperative 4D Transesophageal Echocardiographic (TEE) data. When compared with state-of-the-art optical flow and speckle tracking techniques currently found in 4D echocardiography, the method we present shows notable improvements in error. We believe the performance improvements shown can have a positive impact when the method is used as input for various applications, such as strain computation, biomechanical modeling, or automated diagnostics.
Keywords: 4D transesophageal echocardiography, motion estimation, 3D optical flow
Introduction
Heart disease is one of the leading causes of death in the United States. In 2006, an estimated 26% of all deaths were caused by heart related conditions (Heron et al., 2009). Fortunately, many of these conditions can be treated or prevented if doctors have the right tools at their disposal and are able to diagnose pathologies early. The recovery of dense myocardial displacement from 4D echocardiography is an important endeavor in this regard, as it has many applications, including in diagnostics, modeling, simulation, and training.
Myocardial strain, which may be readily obtained from displacement vectors, can play a crucial role in the diagnosis of cardiovascular diseases (Abraham et al., 2002; Yip et al., 2003). An illustrative pathology related to strain-based diagnostics is Hypertrophic Cardiomyopathy (HCM) (Carasso et al., 2008; Serri et al., 2006; Afonso et al., 2008). HCM is an inherited condition that is manifested by an unhealthy enlargement of myocardial muscle. Such myocardial enlargement can cause cardiac arrhythmia and/or heart failure, and is an important cause of morbidity and sudden cardiac death in young adults (Maron et al., 1995). There are several functional and anatomical disorders associated with HCM, including severe cardiac hypertrophy, myocardial ischemia, outflow tract obstruction, and diastolic dysfunction. However, the mechanisms behind the aforementioned dysfunctions are still not fully understood. The availability of accurate strain information would allow clinicians to more deeply understand the mechanisms behind HCM, as well as possibly provide a fast and accurate screening technique for the disease.
Another important application of dense myocardial displacement recovery is in modeling and simulation. For example, most treatments for the mitral valve involve challenging time-constrained cardiothoracic surgical procedures; these procedures are often begun with limited information on the unique structures inside a patient’s heart. The outcome of such procedures could be improved if potential valve modifications are simulated beforehand, and if various reconstruction options are compared. However, reliable patient-specific simulations cannot be performed without first recovering each patient’s unique physiological characteristics.
Recent modeling and simulation efforts have included patient-specific information to improve simulation fidelity. For example, Votta et al. (2008) recovers and tracks the valve annulus from the patient’s 4D ultrasound (US) data. Similarly, Burlina et al. (2010); Sprouse et al. (2009) recover and exploit the annulus, leaflets, and atrial cavity from 2D and 3D US data. Obviously, any error in the observation of patient-specific parameters will propagate and could make the modeling and simulation stages unreliable. Thus, it is clear that a high accuracy method, like the one described in this paper, could improve patient-specific modeling and simulation fidelity.
Characterizing complex fast-moving heart structures also requires data with sufficiently high spatial and temporal resolution. 4D US represents one of the few viable options for this purpose. In contrast to other modalities such as fluoroscopy, X-ray CT, or MRI, 4D US involves minimal risks, the equipment required has a small form factor, and it is also relatively inexpensive. However, while great progress has been made in the development of piezoelectric elements and signal processing techniques, modern 4D US imaging (see Fig. 1) is still affected by noise an d artifacts that can make the data difficult to analyze. It is therefore also important to develop 4D US exploitation algorithms that can cope with these artifacts.
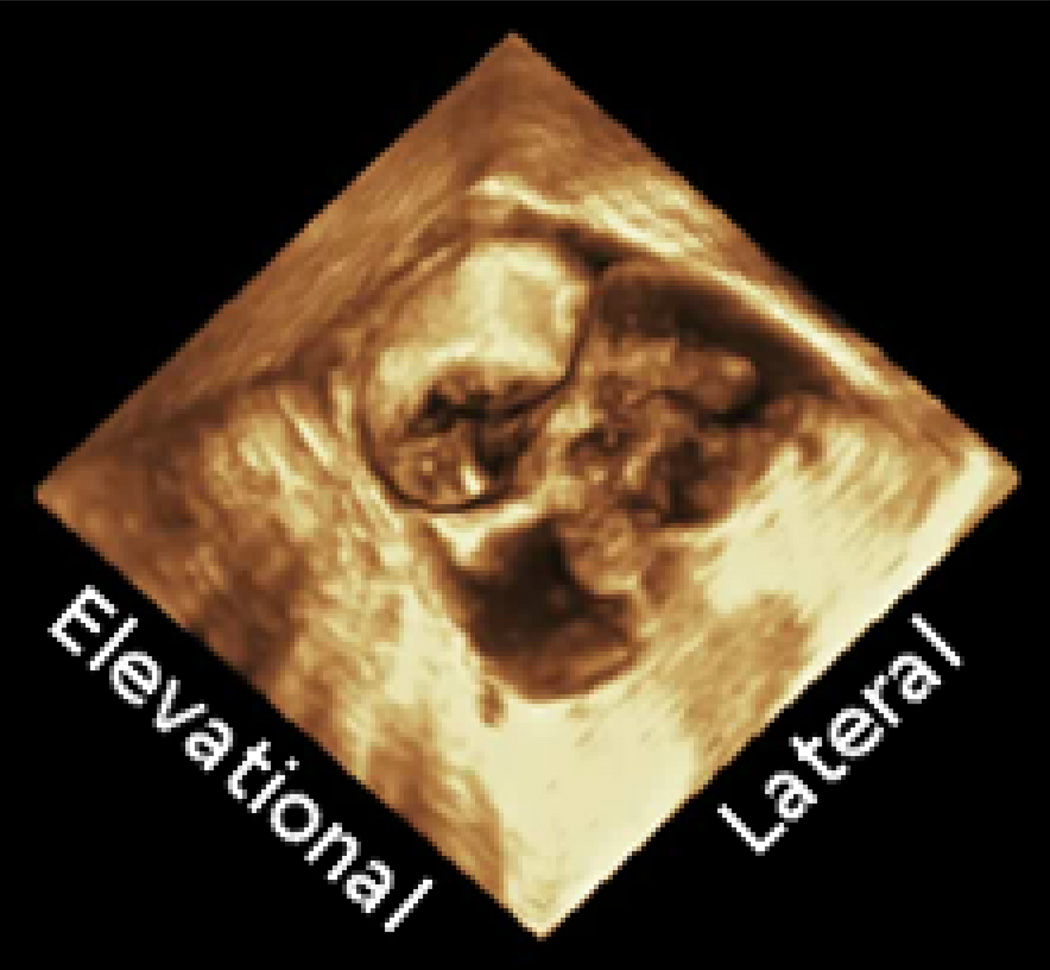
Figure 1.
A rendered 3D ultrasound image taken using a 4D TEE probe. The mitral valve, intra-atrial cavity, sections of the intraventricular cavity, and the aortic valve are all visible in the image
While the dense myocardial motion estimation method proposed in this paper may be applied to different types of 4D US data, our clinical data consists exclusively of sequences acquired using 4D Transesophageal Echocardiography (4D TEE), and our phantom experiments were performed using a 4D abdominal ultrasound probe. Differences between our data and data obtained from other 4D US modalities (such as 4D Transthoracic Echocardiography, or TTE) are mentioned in the discussion section.
Prior Work
In this paper, optical flow techniques are used to compute 3D velocity vectors for the myocardium and valves in 4D US imagery. Original approaches to optical flow computation date back to (Horn and Schunck, 1981; Lucas and Kanade, 1981). In the medical image analysis field, a substantial amount of work has been done on speckle tracking and optical flow using 2D+time US images (Cohen, 1993; O’Donnell et al., 1994; Mikic et al., 1998; Bohs et al., 2000; Burlina et al., 2011), however relatively little research has been performed using optical flow on 3D+time US images.
Recent research into correlation-based optical flow techniques applied to 4D US include works such as (Duan et al., 2005; Yu et al., 2006; Chen et al., 2005). Duan et al. implement a cross-correlation method using a 7×7×7 search window centered around every 5×5×5 pixel volume. They utilize this cross-correlation method to track an initial manual segmentation across frames, and report that the method produces s egmentation volumes within an acceptable margin of error.
Yu et al. attempt to quantitatively measure and compare the reliability of tracking features across ultrasound frames. Using echocardiographic imaging models derived from the work of Meunier and Bertrand (see Meunier, 1998; Meunier and Bertrand, 1995), Yu validates that speckle decorrelation appears when tissue undergoes lateral rotation (rotation about an axis orthogonal to the axial axis) or deformation along any axis. The authors also hypothesize that when using an imaging system with frame rates between 20 to 28 Hz, frame-to-frame cardiac strain variation may be limited to between 1.5% and 2%.
The sensitivity to noise of early differential methods is often cited as a reason why correlation-based methods should be used. However, modern 2D performance comparisons, which utilize datasets with a variety of noise levels (see Barron et al., 1994; Baker et al., 2007), have consistently ranked variational methods, rather than region-based methods, among the best performers in terms of accuracy and reliability recovering flow from both real and synthetic image sequences, and these comparisons take into account many of the improvements that have been made to optical flow techniques over their original implementations.
The method we develop in this paper is inspired by the recent approaches of Brox et al. (2004); Liu et al. (2008); Sun et al. (2010). The method uses a variational technique that takes into account 4D TEE brightness intensity constraints as well as spatiotemporal smoothness regularization constraints. We use intraoperative 4D TEE frames to qualitatively compare the results of our method with the known behavior of the ventricle and mitral valve during both diastolic and systolic phases. To quantitatively evaluate the method, synthetic transformations are applied to single intraoperative 4D TEE images, creating pairs of frames for which ground-truth motion is known. A multipurpose ultrasound phantom is also used with an external 4D probe to generate real ultrasound data with known ground-truth motion.
Method
The 3D optical flow method presented in this paper is inspired by recent works reported by Brox et al. (2004); Liu et al. (2008); Sun et al. (2010). The first assumption to computing optical flow is based on the idea that brightness intensity should remain constant when visually following a feature point within a bounded temporal window. The applicability of this idea to ultrasound has been validated in several 2D+time US applications and forms the basis for speckle tracking, strain computation, as well as recent work related to elastography (Luo et al., 2007; Ophir et al., 2000; Pellot-Barakat et al., 2004).
Consider two 3D frames taken at times t and t + 1. The brightness intensity constraint states that the intensity value of a voxel in the first frame of an image at time t and at location x ≡ (x, y, z), must remain constant and match the intensity value of a voxel in our second frame taken at time t + 1, at position x + u, where u ≡ (u, v, w), i.e.:
| (1) |
One method for applying this constraint to all of the voxels in our images is to form an energy function that penalizes differences in voxel value between pairs of frames:
| (2) |
The function constitutes an L2 norm, which allows the application of quadratic optimization techniques, while behaving as a robust L1 norm when ε → 0.
Because the use of this sole constraint leads to an ill-posed problem (one constraint for three unknowns u), an additional spatiotemporal smoothness constraint is utilized and is based on the assumption that the flow between two successive frames and two neighboring voxels does not involve sudden local changes. This constraint is incorporated by forming an energy function that penalizes large gradients in our flow vectors at each voxel:
| (3) |
The final objective function is formed by summing together the two previously discussed energy components. To allow for the amount of flow vector smoothness to be adjusted as desired, a weight α > 0 is used, as in:
| (4) |
To minimize the objective function, which involves an unknown under an integral operator, the Euler-Lagrange equation must be used.
The Euler-Lagrange equations for (4) are as follows:
| (5a) |
| (5b) |
| (5c) |
where
is the Lagrangian density and
| (6a) |
| (6b) |
| (6c) |
Here ux ≡ ∂u/∂x. Expanding (5a) through (5c) results in the following equations:
| (7a) |
| (7b) |
| (7c) |
Due to the nonlinearity of the preceding equations, an iterative numerical approximation technique is used as in Liu et al. (2008). Incorporating inner and outer fixed point iterations, combined with a coarse-to-fine multiscale image warping approach (as in Brox et al., 2004; Lucas and Kanade, 1981), we obtain three equations whose solutions make up the displacement vector for a given slice of the coarse-to-fine pyramid on which we iterate.
Defining we write (7a) through (7c) at level k + 1 as
| (8a) |
| (8b) |
| (8c) |
where k represents the current level of the coarse-to-fine pyramid. Using Taylor expansions of the following form
we obtain
| (9a) |
| (9b) |
| (9c) |
where
and l represents the current step of the fixed point iteration loop. Immediately following each warping step in the coarse-to-fine pyramid, a 5×5×5 median filter is applied to the flow vectors (as in Sun et al., 2010). More specifically, we independently apply a 5×5×5 median filter to each of our three 3D cubes representing the values u, v, and w for each displacement vector u. Note that as the displacement vectors are 3D, the computation of uk for each level of the coarse-to-fine pyramid involves trilinear interpolation.
Lastly, to prevent the computation of flow vectors in areas where flow is not well defined (e.g., regions of uniform intensity), we remove empty and/or blood-filled cavities from our data and process only the myocardium. To remove all but the myocardium, we use k-means clustering, after compensating for attenuation, to group all the voxels of our cube into two clusters, and subsequently run the above variational optical flow algorithm on the brighter intensity cluster.
Experiments
Intraoperative Data
To qualitatively evaluate the proposed method and discover any challenges associated with its clinical application, we performed tests on real human TEE data. We collected intraoperative real-time 4D TEE data from patients at the Johns Hopkins University’s School of Medicine. Coronary artery bypass surgery was performed on a majority of the patients, while the remaining few underwent interventions for valvular dysfunction. Acquisition was performed under a protocol approved by the JHU Institutional Review Board, and all patients gave informed consent.
4D TEE acquisition was performed using an iE33 Philips console fitted with an X7-2t TEE probe (Philips Medical Systems, Bothell, WA). The resulting TEE cube sizes were 224×208×208 along the elevational, lateral, and axial directions. These directions denote the 3D ultrasound canonical directions: the axial direction is along the acoustic path and corresponds here to the z-axis, and the lateral-elevational plane, which contains the array of transducer elements, corresponds to the x-y plane (see labels in Fig. 1). The 4D TEE probe was run at frequencies ranging from 3 to 5 MHzl. The spatia resolutions ranged approximately from 0.4 mm to 1 mm. Data was collected intraoperatively using ECG-gating and a 7 breath-hold protocol leading to frame rates varying from 24 to 56 Hz. More specific parameters for each sequence used are detailed in Table 1.
Table 1.
Cardiologist evaluation: Vector Motion ranges from 1 - plausible, to 5 - not plausible; Image Quality ranges from 1 - high quality/no artifacts, to 5 - poor quality/many artifacts
| 4D TEE Sequence | Voxel Resolution (mm) | Frame Rate (Hz) | Ultrasound Depth (cm) | Vector Motion | Image Quality |
|---|---|---|---|---|---|
| 1 | 0.41×0.4×0.44 | 52 | 9 | 1 | 2 |
| 2 | 0.37×0.36×0.39 | 56 | 8 | 1 | 2 |
| 3 | 0.61×0.6×0.53 | 44 | 11 | 1 | 3 |
| 4 | 0.61×0.6×0.53 | 44 | 11 | 2 | 2 |
| 5 | 0.61×0.6×0.53 | 44 | 11 | 1 | 2 |
| 6 | 0.41×0.4×0.44 | 52 | 9 | 1 | 1 |
| 7 | 0.67×0.66×0.58 | 41 | 12 | 1 | 1 |
| 8 | 0.67×0.66×0.58 | 41 | 12 | 1 | 1 |
| 9 | 0.72×0.71×0.63 | 39 | 13 | 1 | 2 |
| 10 | 0.72×0.71×0.63 | 39 | 13 | 1 | 2 |
| 11 | 0.72×0.71×0.63 | 39 | 13 | 1 | 2 |
| 12 | 0.67×0.66×0.58 | 41 | 12 | 1 | 2 |
| 13 | 0.67×0.66×0.58 | 41 | 12 | 1 | 1 |
| 14 | 0.5×0.49×0.44 | 52 | 9 | 1 | 2 |
| 15 | 0.5×0.49×0.44 | 52 | 9 | 1 | 2 |
| 16 | 0.78×0.77×0.68 | 36 | 14 | 1 | 2 |
| 17 | 0.78×0.77×0.68 | 36 | 14 | 1 | 2 |
| 18 | 0.78×0.77×0.68 | 36 | 14 | 1 | 2 |
| 19 | 0.78×0.77×0.68 | 36 | 14 | 1 | 1 |
| 20 | 0.56×0.55×0.48 | 48 | 10 | 1 | 2 |
| 21 | 0.56×0.55×0.48 | 48 | 10 | 1 | 2 |
| 22 | 0.56×0.55×0.48 | 48 | 10 | 1 | 2 |
| 23 | 0.56×0.55×0.48 | 48 | 10 | 1 | 2 |
| 24 | 0.67×0.66×0.58 | 41 | 12 | 1 | 3 |
| 25 | 0.67×0.66×0.58 | 41 | 12 | 1 | 3 |
| 26 | 0.67×0.66×0.58 | 41 | 12 | 1 | 3 |
| 27 | 0.5×0.49×0.44 | 30 | 9 | 1 | 2 |
| 28 | 0.67×0.66×0.58 | 24 | 12 | 1 | 2 |
The breath-hold procedure used to collect data is intended to minimize artifacts caused by the patient’s breathing apparatus. Before acquiring a TEE volume, the anesthesiologist temporarily deactivates the patient’s respirator and allows the patient’s chest to steady. However, the patient is still breathing during acquisition, and this may result in unintended probe motion that can overwhelm meaningful flow. Possible solutions include examining relative flow within the heart, or registering the myocardium between frame pairs to eliminate the effects of probe motion or global heart movement.
3D flow was computed on a sequence of frames spanning one full heart cycle. A total of 28 sequences from 12 separate patients were used. Each sequence contained anywhere between 30 and 50 frames. After computing 3D optical flow, movies were generated for each sequence displaying flow vector and heat map overlays (see Fig. 7). As shown in Table 1, the generated movies and 3D flow results were evaluated by a cardiologist and all but one sequence obtained the highest marks for physiological plausibility in both direction and magnitude.
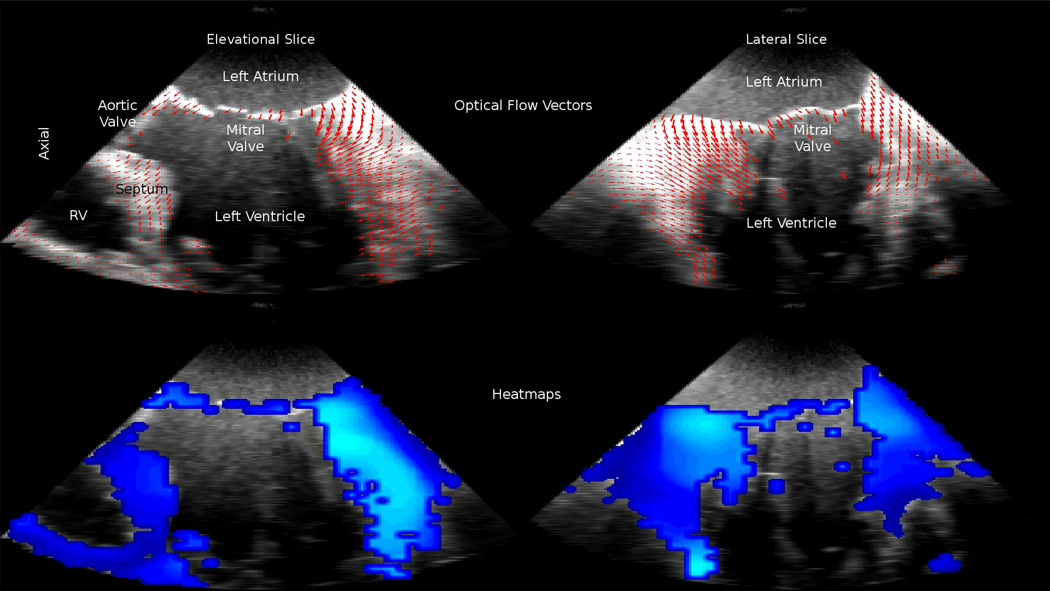
Figure 7.
3D optical flow computed on intraoperative data. Top row: down-sampled 3D displacement vectors overlayed in 2D, Bottom row: heatmap of 3D displacement vectors, Left side: long-axis 4-chamber view, Right side: long-axis 2-chamber view
Intraoperative Data with Synthetic Motion
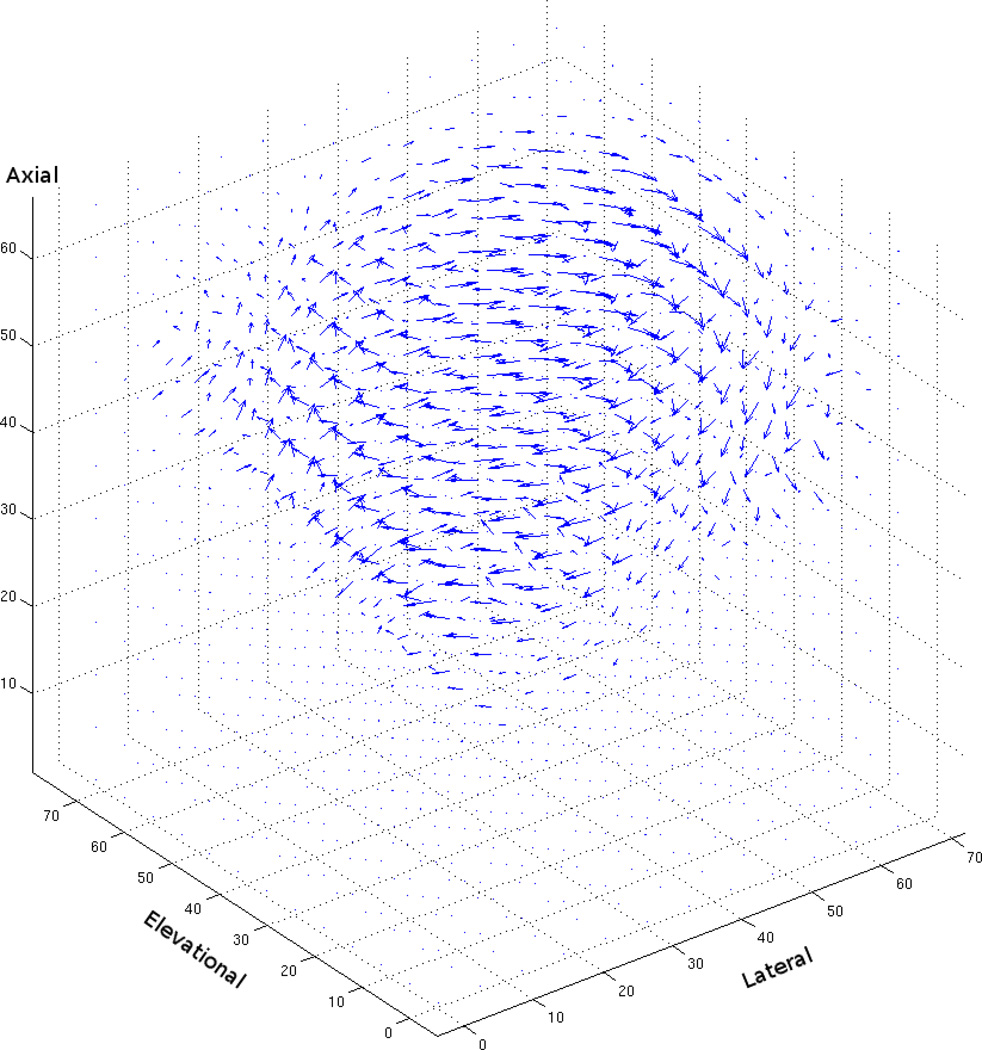
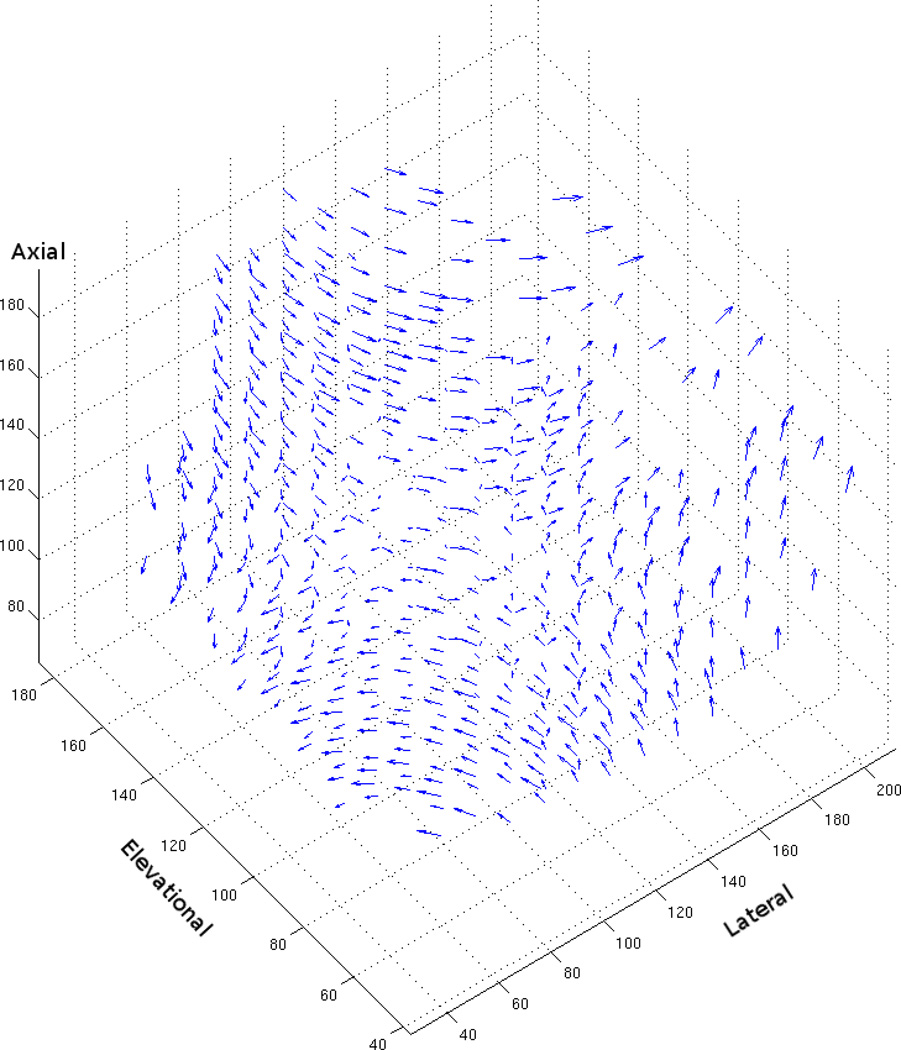
To quantitatively assess algorithm performance, we synthetically generated sequences by subjecting clinical 3D volumes to known motion transformations. Because the motion transformations were known, ground-truth flow vectors for each voxel could be obtained. Using these ground-truth vectors, angular and endpoint error, popular metrics for comparing the performance of optical flow algorithms, were computed. Both error metrics were computed as in Baker et al. (2007) with a simple extension to handle 3D flow vectors. Figures 2 and 3 are examples of the 3D flow vectors computed by the variational technique after lateral rotation of 6° and deformation of 6%.
Figure 2.
3D flow vectors computing using the variational method on ultrasound data that has been synthetically rotated by 6° (i.e., α, β, and γ are all set equal to 1, and θ is set to 6°)
Figure 3.
3D flow vectors computed using the variational method on ultrasound data that has been synthetically laterally deformed by 6% (i.e., α = 1.06, , and γ = 1)
For further comparison, we implemented one of the most commonly used correlation-based optical flow approaches and evaluated it using our data. The method computes the cross-correlation coefficient r across a given search window centered around every 5×5×5 volume (as in Duan et al., 2009):
| (10) |
Translation, rotation, and deformation transformations were applied to intraoperative data and errors were calculated for both methods. Translation was performed by shifting a sub-cube of the data by a known number of voxels in either the lateral, elevational, or axial directions. Translation can be expressed by the following equation:
| (11) |
Tables 2 and 3 compare angular and endpoint error values between the variational method presented in this paper and the correlation-based method as computed on our data. The correlation-based method returned zero angular and endpoint error across all translations tested, while the variational method recorded near-zero angular and endpoint error values, with angular error values decreasing as the amount of translation increased.
Table 2.
Synthetic translation angular error comparison
| Trans. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1 voxel | 0.09° | 0.73° | 0° | 0° |
| 2 voxels | 0.06° | 0.46° | 0° | 0° |
| 3 voxels | 0.04° | 0.33° | 0° | 0° |
| 4 voxels | 0.03° | 0.26° | 0° | 0° |
| 5 voxels | 0.03° | 0.20° | 0° | 0° |
| 6 voxels | 0.02° | 0.16° | 0° | 0° |
Table 3.
Synthetic translation endpoint error comparison (voxels)
| Trans. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1 voxel | 0 | 0.02 | 0 | 0 |
| 2 voxels | 0 | 0.02 | 0 | 0 |
| 3 voxels | 0 | 0.02 | 0 | 0 |
| 4 voxels | 0 | 0.02 | 0 | 0 |
| 5 voxels | 0 | 0.02 | 0 | 0 |
| 6 voxels | 0 | 0.02 | 0 | 0 |
Rotation and deformation transformations we re computed using the following equation (as in Meunier and Bertrand, 1995):
| (12) |
where α, β, and γ, restricted to αβγ = 1 by an incompressibility constraint, represent the amount of lateral, elevational, and axial deformation and θ represents the amount of rotation about the z axis (i.e. the 3D ultrasound axial direction). Tables 4 and 5 compare flow error values for transformations where α, β, and γ are all set equal to 1 (i.e., no deformation), but θ is varied according to the number of degrees rotation desired. Tables 6 and 7 compare error values for lateral deformation, which is obtained by setting θ to 0, γ to 1, and adjusting α (and consequently β) according to the percentage deformation desired. Note that 1% lateral deformation is achieved when α = 1.01 and . Tables 8 and 9 compare error values for lateral-axial deformation, which is obtained by varying γ together with α as in the lateral deformation case. Lateral-axial deformation involves deformation in all three axes, as the elevational axis is forced to expand due to the incompressibility constraint when the lateral and axial axes are compressed. Finally, Tables 10 and 11 compare error values for lateral-axial deformation, as described above, combined with rotation.
Table 4.
Synthetic rotation angular error comparison
| Rot. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1° | 0.73° | 2.64° | 1.43° | 7.95° |
| 2° | 0.78° | 2.83° | 2.22° | 10.03° |
| 3° | 1.01° | 3.91° | 3.55° | 12.32° |
| 4° | 1.13° | 5.26° | 9.43° | 21.34° |
| 5° | 1.28° | 6.40° | 21.79° | 34.22° |
| 6° | 1.52° | 7.86° | 35.43° | 41.54° |
Table 5.
Synthetic rotation endpoint error comparison (voxels)
| Rot. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1° | 0.02 | 0.09 | 0.04 | 0.22 |
| 2° | 0.04 | 0.14 | 0.09 | 0.41 |
| 3° | 0.07 | 0.20 | 0.26 | 0.75 |
| 4° | 0.10 | 0.31 | 0.93 | 1.60 |
| 5° | 0.14 | 0.43 | 2.14 | 2.68 |
| 6° | 0.18 | 0.61 | 3.55 | 3.42 |
Table 6.
Synthetic lateral deformation angular error comparison
| Axial Def. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% | 1.16° | 0.94° | 20.77° | 7.75° |
| 2% | 1.15° | 0.71° | 14.05° | 8.58° |
| 3% | 1.39° | 0.67° | 10.98° | 7.60° |
| 4% | 1.71° | 0.72° | 9.36° | 8.54° |
| 5% | 2.07° | 0.81° | 8.78° | 10.22° |
| 6% | 2.44° | 0.92° | 9.20° | 12.31° |
Table 7.
Synthetic lateral deformation endpoint error comparison (voxels)
| Axial Def. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% | 0.02 | 0.02 | 0.46 | 0.24 |
| 2% | 0.03 | 0.02 | 0.41 | 0.29 |
| 3% | 0.05 | 0.03 | 0.41 | 0.25 |
| 4% | 0.09 | 0.04 | 0.44 | 0.37 |
| 5% | 0.13 | 0.06 | 0.51 | 0.58 |
| 6% | 0.18 | 0.08 | 0.68 | 0.85 |
Table 8.
Synthetic axial-lateral deformation angular error comparison
| Axial-Lateral Deformatio | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% | 1.47° | 1.15° | 20.69° | 9.78° |
| 2% | 1.61° | 0.79° | 22.82° | 12.71° |
| 3% | 2.05° | 0.82° | 33.14° | 21.75° |
| 4% | 2.60° | 0.95° | 44.49° | 28.14° |
| 5% | 3.16° | 1.12° | 54.81° | 31.40° |
| 6% | 3.71° | 1.29° | 62.82° | 32.33° |
Table 9.
Synthetic axial-lateral deformation endpoint error comparison (voxels)
| Axial-Lateral Deformation | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% | 0.04 | 0.03 | 0.54 | 0.24 |
| 2% | 0.08 | 0.04 | 0.96 | 0.60 |
| 3% | 0.15 | 0.07 | 1.85 | 1.20 |
| 4% | 0.26 | 0.12 | 2.92 | 1.69 |
| 5% | 0.39 | 0.19 | 4.04 | 2.08 |
| 6% | 0.55 | 0.26 | 5.12 | 2.38 |
Table 10.
Synthetic axial-lateral deformation and rotation angular error comparison
| Deformation and Rotation | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% + 1° | 1.32° | 1.08° | 18.69° | 10.35° |
| 2% + 2° | 1.38° | 1.16° | 26.51° | 21.24° |
| 3% + 3° | 1.67° | 1.44° | 41.96° | 32.16° |
| 4% + 4° | 2.17° | 1.97° | 53.67° | 35.29° |
| 5% + 5° | 2.70° | 2.45° | 61.86° | 35.65° |
| 6% + 6° | 3.24° | 2.87° | 67.63° | 35.12° |
Table 11.
Synthetic axial-lateral deformation and rotation endpoint error comparison (voxels)
| Deformation and Rotation | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1% + 1° | 0.04 | 0.04 | 0.61 | 0.39 |
| 2% + 2° | 0.09 | 0.08 | 1.57 | 1.42 |
| 3% + 3° | 0.16 | 0.15 | 3.02 | 2.43 |
| 4% + 4° | 0.29 | 0.27 | 4.48 | 3.18 |
| 5% + 5° | 0.46 | 0.42 | 5.88 | 3.86 |
| 6% + 6° | 0.66 | 0.61 | 7.24 | 4.53 |
In each rotation and/or deformation experiment, lower angular and end-point error values are observed for the variational method when compared with the correlation-based method. Across all the synthetic experiment tables, the variational method’s error values increase steadily as the amount of rotation and/or deformation increases. In some cases (e.g., Tables 4 and 5), the correlation-based method experiences a sudden increase in error values after a certain amount of rotation and/or deformation. Also, in the case of lateral deformation (i.e., Tables 6 and 7), the angular and endpoint error values of the correlation-based method improve as deformation increases from 1–3%, and subsequently deteriorate as deformation further increases from 4–6%.
Phantom Data
To account for the various challenges of ultrasound imagery (e.g., attenuation, shadowing, reverberation, speckle decorrelation, and polar-to-cartesian conversion artifacts) in our quantitative assessment, we performed a series of experiments on data obtained using a multipurpose tissue-equivalent ultrasound phantom (Nuclear Associates, Carle Place, NY, Model#: 84–317). The phantom is composed of Zerdine, a solid-elastic water-based polymer, and is designed to emulate liver tissue. Embedded within the phantom are numerous nylon wires and cyst-like structures that, for our purposes, add a degree of heterogeneity to the data. Still, a majority of the phantom’s volume is relatively uniform and featureless, and this uniformity makes the computation of flow even more difficult.
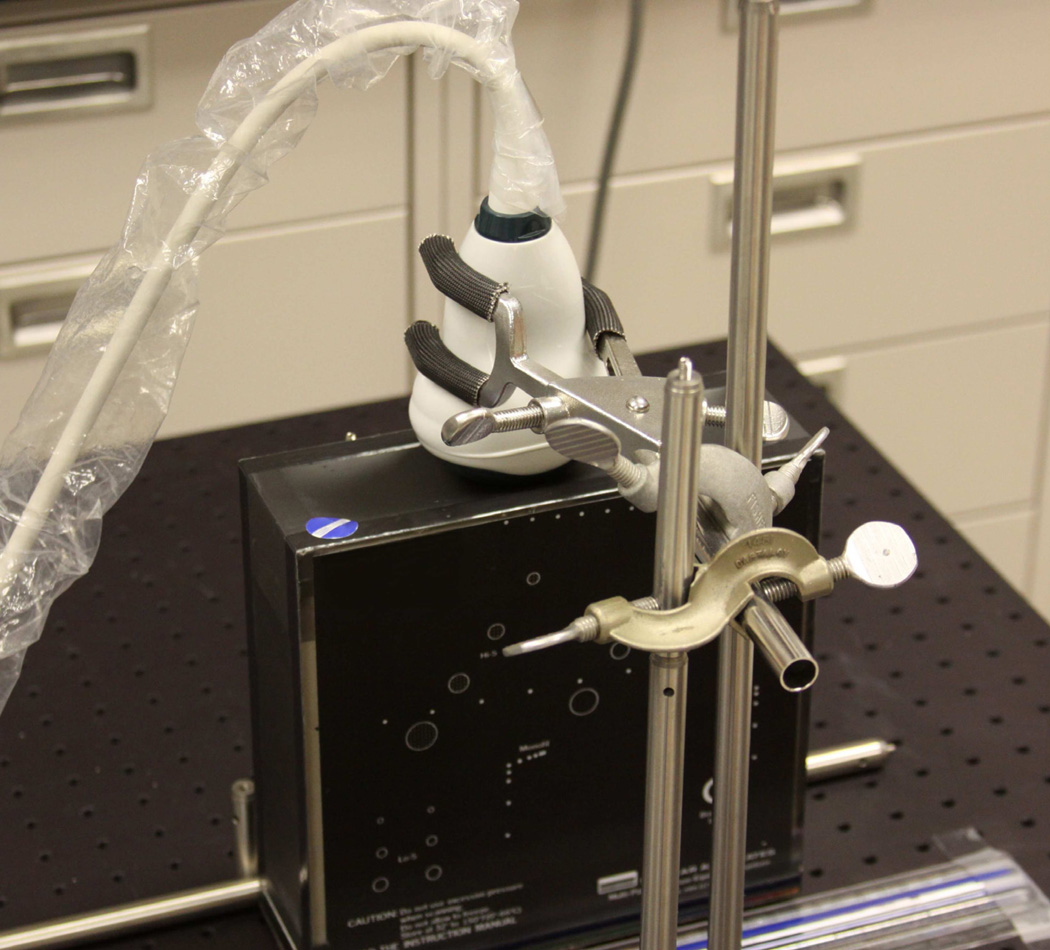
Ultrasound acquisition was performed using an Ultrasonix SonixTouch Research console (Vancouver, BC) and 4DC7-3/40 probe. The probe contains 128 elements in a curved linear array that is panned back and forth by a motor with 410 steps, where each step tilts the array by 0.183°. Volumes with a resolution of 484×364×90 were collected using a depth of 15 cm, transmit frequency of 3.3 MHz, and focus depth of 7.5 cm, resulting in an approximate spatial resolution of 0.485 mm. The ultrasound probe and phantom were placed inside an apparatus that maintained their positions, contacting the probe with the phantom, and allowed for controlled and measurable movement (see Fig. 4).
Figure 4.
Phantom apparatus. The 4D abdominal probe is held in place, contacting the top of the multipurpose tissue-equivalent ultra-sound phantom (black object with white markings)
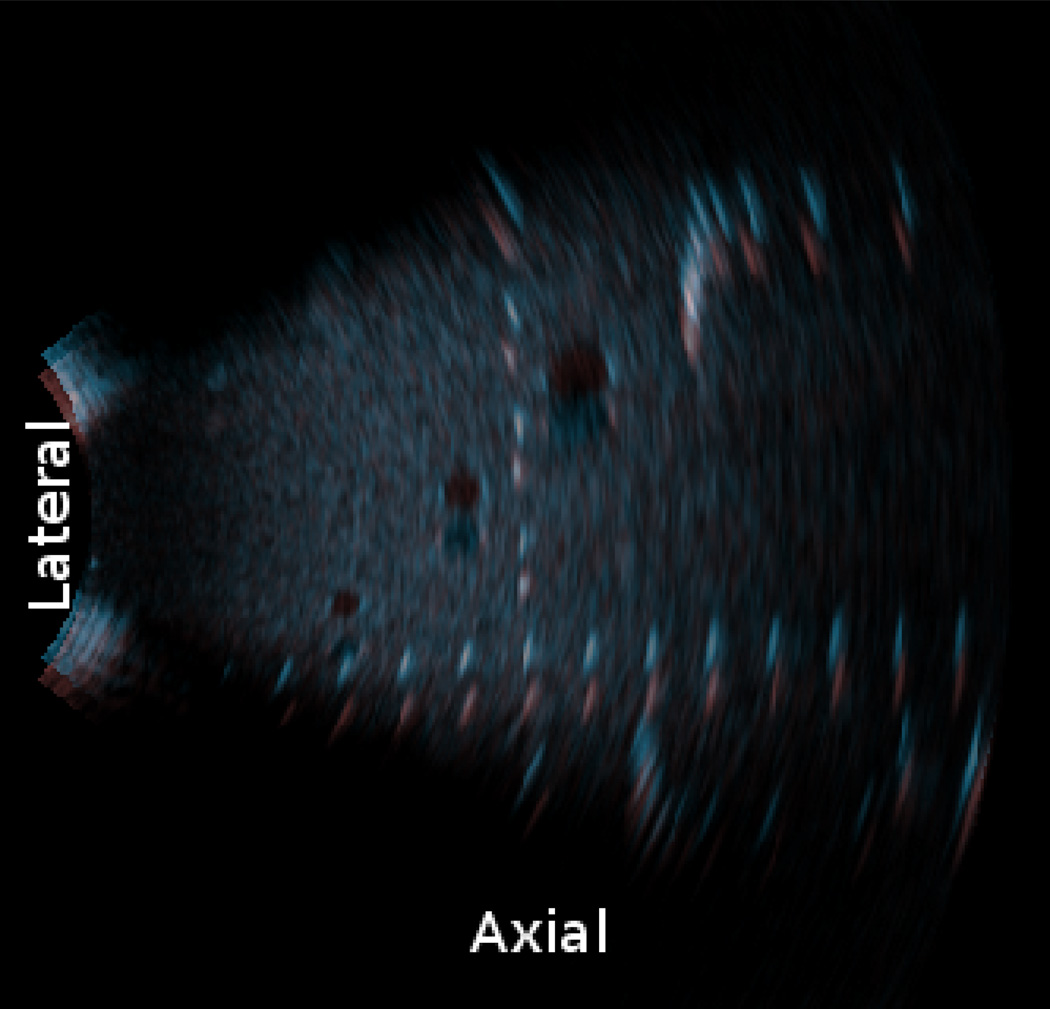
An initial series of experiments were performed by translating the phantom a known distance (example frame pair shown in Fig. 5). As the probe’s surface is slightly larger and more curved than the surface of the phantom, a portion of each volume collected required masking to prevent the computation of flow outside the boundaries of the phantom.
Figure 5.
Example phantom data: two long-axis slices (one highlighted blue, the other highlighted red) showing 6 mm translational phantom movement
Angular and endpoint error values for both the variational and correlation-based methods are presented in Tables 12 and 13. Both the variational and correlation-based methods exhibit a significant increase in angular and end-point error values when compared with the synthetic translation experiments. Also, the variational method shows consistently lower angular and endpoint error values than the correlation-based method, and both methods display an increase in error values as the amount of phantom translation increases.
Table 12.
Phantom translation angular error comparison
| Trans. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1 mm | 15.68° | 9.27° | 26.10° | 24.07° |
| 2 mm | 14.98° | 15.46° | 27.67° | 33.92° |
| 3 mm | 13.55° | 18.97° | 46.77° | 43.37° |
| 4 mm | 13.87° | 20.39° | 71.93° | 42.29° |
| 5 mm | 16.88° | 25.42° | 81.02° | 39.40° |
| 6 mm | 20.10° | 29.27° | 83.68° | 38.84° |
Table 13.
Phantom translation endpoint error comparison
| Trans. | Variational | Cross-Correlation | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| 1 mm | 0.36 | 0.19 | 0.95 | 1.01 |
| 2 mm | 0.60 | 0.48 | 1.37 | 1.35 |
| 3 mm | 0.97 | 1.01 | 2.86 | 2.21 |
| 4 mm | 1.47 | 1.63 | 5.48 | 2.24 |
| 5 mm | 2.35 | 2.51 | 7.68 | 2.19 |
| 6 mm | 3.49 | 3.44 | 9.61 | 2.21 |
Discussion
This paper describes a novel 3D motion flow computation method for estimating myocardial motion between pairs of volumetric ultrasound frames. The method extends to 3D and incorporates features of state-of-the-art 2D techniques by Brox et al. (2004); Liu et al. (2008); Sun et al. (2010). The salient features of the method are as follows: using first order (brightness constancy) and second order (smoothness) constraints; exploiting coarse-to-fine pyramid computation; applying median filtering at each intermediate computation stage of the pyramid; and using iterative numerical techniques to solve non-linear equations.
Qualitative evaluations of intraoperative data were performed by a cardiologist to determine if the variational method returned physiologically plausible flow vectors. Also, to better quantify algorithm performance, an evaluation is presented using several metrics, operating both on synthetically transformed intraoperative data, as well as phantom data. Additionally, comparisons are shown between the variational method presented in this paper and a commonly used correlation-based approach, such as the method presented in Duan et al. (2009).
While many of the metrics that we utilize to compare methods are currently used by researchers in the field, the results shown in our comparisons need to be qualified with a word of caution, as there are many factors that can reduce the meaningfulness of such a comparison. Perhaps the most important factor is that we are simply not processing the same data. Tables 14 and 15, containing error values for 2D versions of the variational and correlation-based methods, as run on the Middlebury public dataset (Baker et al., 2007), are included to demonstrate the large variation in error values across different test data. In the case of 4D TEE data, we believe that large variations in error values can be explained by differences in intensity uniformity between sets of test data (size and location of regions where optical flow is difficult to compute), as well as motion magnitude. To clarify what we mean by motion magnitude, especially as it may pertain to our synthetic experiments where motion magnitude is seemingly controlled, let us use the rotation transformation as an example. To generate Tables 4 and 5, we rotated intraoperative data about the center of each 224×208×208 cube. As a result, voxels further from the center of rotation underwent larger displacements. But, because each intraoperative data set contains varying levels of contrast and different tissue distributions, and because we do not compute flow in empty and/or blood-filled cavities, the amount of motion present in a pair of synthetically rotated frames may vary. Thus, to present a proper comparison of methods using identical data, we implemented one of the most commonly used correlation-based approaches.
Table 14.
Error in degrees and pixels, using the variational method in 2D, for images in the Middlebury public dataset (Baker et al., 2007)
| Image Pair | Angular Error | Endpoint Error | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| Dimetrodon | 3.21° | 5.71° | 0.21 | 0.33 |
| Grove2 | 2.87° | 8.07° | 0.20 | 0.47 |
| Grove3 | 7.36° | 18.71° | 0.81 | 1.54 |
| Hydrangea | 2.67° | 7.59° | 0.26 | 0.51 |
| RubberWhale | 10.14° | 27.05° | 0.23 | 0.49 |
| Urban2 | 4.40° | 13.38° | 0.48 | 1.49 |
| Urban3 | 6.57° | 24.03° | 0.80 | 1.96 |
| Venus | 12.41° | 34.53° | 0.65 | 1.01 |
Table 15.
Error in degrees and pixels, using the correlation-based method in 2D, for images in the Middlebury public dataset (Baker et al., 2007)
| Image Pair | Angular Error | Endpoint Error | ||
|---|---|---|---|---|
| Mean | Stdev. | Mean | Stdev. | |
| Dimetrodon | 17.71° | 26.84° | 0.79 | 1.13 |
| Grove2 | 13.77° | 24.87° | 0.83 | 1.14 |
| Grove3 | 39.42° | 43.41° | 2.90 | 3.18 |
| Hydrangea | 25.70° | 39.18° | 1.88 | 2.11 |
| RubberWhale | 11.92° | 17.17° | 0.42 | 0.62 |
| Urban2 | 54.06° | 46.70° | 7.97 | 8.50 |
| Urban3 | 68.43° | 51.45° | 7.01 | 5.21 |
| Venus | 42.46° | 45.48° | 2.90 | 2.80 |
Based on the comparisons performed, the variational method shows promising improvements in error values that may have a positive impact when the method is applied to a broader algorithmic pipeline. As shown in Tables 2 and 3, both the variational and correlation-based methods exhibit low angular and endpoint error for synthetic translation experiments. Because synthetic translation does not result in any decorrelation, the correlation-based flow perfectly matched the ground-truth, mimicking results reported by Duan et al. (2007). On the other hand, the smoothness constraint and coarse-to-fine approach used by the variational method caused it to return flow with very small amounts of error, as it must contend with handling the sudden flow changes at the translated subcube’s boundaries. Error values for rotation and deformation transformations, presented in Tables 4 – 11, show a more sizable performance improvement for the variational method over the correlation-based method. For both methods, angular and endpoint error rises steadily as the number of degrees rotation or percent deformation increase. However, the variational method’s average endpoint and angular error values are consistently lower than the correlation-based method’s error values, at times recording up to a twenty-fold improvement in the case of rotation, and four-fold improvement in the case of deformation. The standard deviation of endpoint and angular error values for flow returned by the variational method are also consistently and significantly lower than for the correlation-based method. Of note is the fact that the endpoint and angular error of flow returned by the correlation-based method suddenly increases after 4° rotation. We believe this sudden increase to be due to the magnitude of flow vectors extending beyond the range of the 7×7×7 search window used by the correlation-based method.
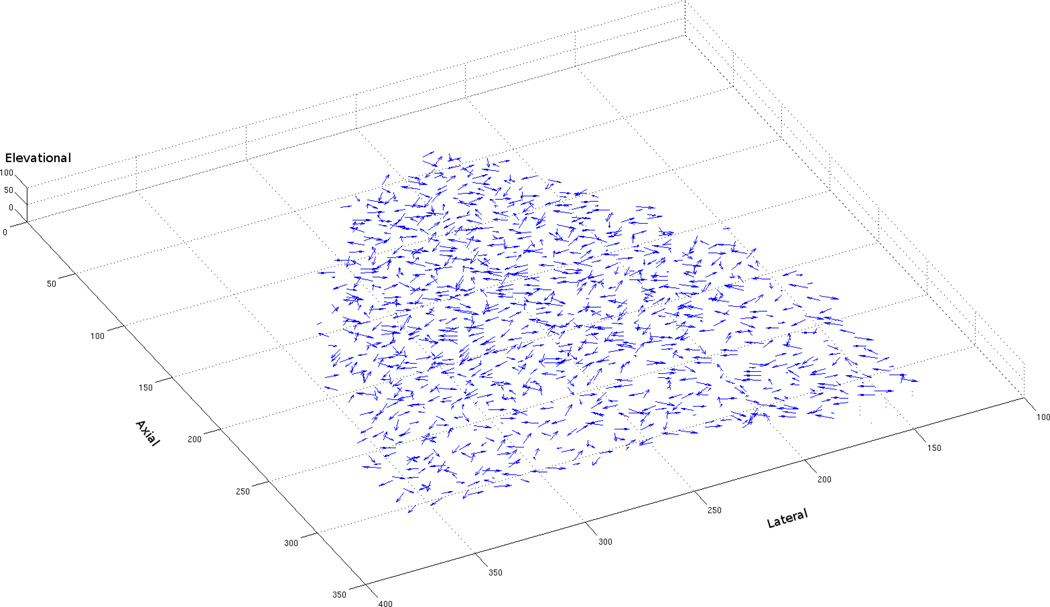
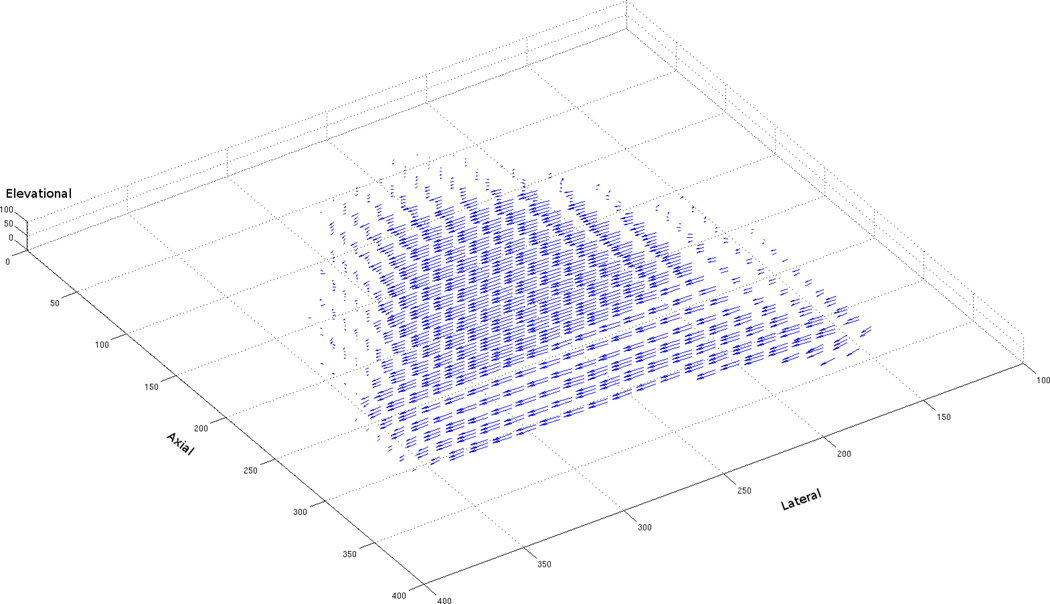
Phantom flow comparisons were also performed. The phantom data used contains noise, artifacts (which are discussed later), and a limited number of features for optical flow algorithms to easily track, thus making it a more realistic and difficult problem to solve. As previously mentioned, the sensitivity to noise of early differential methods is a common rationale for why correlation-based methods should be used. Citing modern 2D performance comparisons, which utilize datasets with a variety of noise levels (see Barron et al., 1994; Baker et al., 2007), we argued that modern variational methods can outperform region-based methods on noisy imagery. Our findings from the phantom flow experiments confirm this hypothesis. As can be seen from Tables 12 and 13, the variational approach performs significantly better than the correlation-based approach on translational phantom data. While the correlation-based approach is able to successfully match some regions within the phantom, the high amount of noise and lack of features cause it to produce flow vectors with very high error values (see Fig. 6(a)). On the other hand, the variational approach, with the aid of a smoothness constraint and coarse-to-fine technique, is able to match the sparse nylon features and produce relatively consistent flow vectors across the entire volume of the phantom (see Fig. 6(b)).
Figure 6.
Computed 3D flow vectors for a pair of phantom images containing a translation of 6 mm (a) Correlation-Based Method, (b) Variational Method
As previously mentioned, optical flow results generated from real intraoperative sequences using the variational method described in this paper were also evaluated by a cardiologist. This evaluation provides qualitative insight into the performance of the algorithm when applied to real clinical data. While the synthetic and phantom experiments previously discussed help characterize algorithmic performance operating on images of varying motion magnitude and noise, they do not incorporate all the complexity of physiological heart motion. For example, the synthetic rotation experiments test unidirectional rotation only. For healthy individuals, the ventricle will exhibit a twisting motion as it contracts and expands, resulting in opposite rotation at the basal and apical regions of the ventricle. The cardiologists qualitative evaluation of intraoperative flow results provides confirmation of satisfactory algorithmic performance on imagery containing more complex motion, and supplements the quantitative synthetic and phantom evaluations.
The intraoperative data evaluated consisted of 4D TEE imagery. TEE images are characterized by having the left atrial cavity closest to the probe, separated from the left ventricle (LV) chamber (seen at the bottom of the frustum) by the mitral annulus and the mitral valve apparatus (valve leaflets). The computed flow movement in each sequence appears complex, but the overall assessment can be broadly summarized as follows: During the first phase of the heart cycle, the mitral valve leaflets open up and the flow vectors on the leaflets correctly point downwards to wards the apex of the left ventricle. At the same time, the myocardial wall relaxes and the computed flow vectors on the myocardium are seen moving up and outwards towards the peripheral part of the field. These two movements happen together and correctly represent the diastolic portion of cardiac cycle. After the LV cavty fills with blood, opposite computed flow vectors on both structures are observed. Flow vectors lying on the mitral valve leaflets point up towards the left atrium, and, as the myocardium contracts and shortens its length, the myocardial flow vectors point inwards toward the apex and center of the LV cavity drawing a curve with an inside-cavity direction. At the end of the systolic phase, during isovolumic relaxation, as the atrium fills up and starts a descending motion, the vectors on the mitral valve reverse direction and point downward.
There are several intricacies to clinical data that can affect optical flow results: For example, signal attenuation can degrade contrast, and as a result, make it more difficult to match voxel intensities and compute optical flow for objects further away from the ultrasound probe; Shadowing and reverberation can obscure texture or create spurious texture that detracts from the true motion present in a cube; Polar-to-Cartesian conversions can artificially stretch or compress features; Stitching artifacts resulting from the 7 breath-hold protocol can result in chunks of a cube being artificially translated; And finally, limited acquisition frame rates may mean that fast moving tissue (such as the mitral valve) will have large displacements between pairs of frames, which can result in increased error (as shown in the synthetic experiments). Despite all these factors, after running the variational method across sequences of varying image quality and frame rates, all but one sequence obtained the highest marks from a cardiologist for following physiological motion, as shown in Table 1.
Another issue worth considering is the applicability of presented performance results to transthoracic echocardiography (TTE). Compared to TEE, TTE generally entails additional signal attenuation because it must traverse skin and fat layers. TTE imagery may also contain more significant shadowing due to the ribs, and additional reverberation caused by the lungs and pleural interface. In addition, patient specific conditions such as emphysema, obesity, and rib cage deformities can worsen the aforementioned artifacts. However, despite the increased prevalence of artifacts in TTE data, the clinical TEE and phantom data used still contain shadowing, attenuation, and reverberation effects, albeit to a lesser degree. Thus, we believe our performance comparisons provide some insight into the method’s performance on TTE data, but more investigation into the performance of variational methods on TTE data is required.
Finally, it is worth noting that when these methods are applied to more complex algorithmic pipelines, to track tissue for example, flow estimation errors will compound over time. In other words, without correction, residual flow estimation errors in each frame will accumulate, resulting in increased error as an object is tracked over a sequence of frames. Also, as can be seen from the presented comparisons, both optical flow methods perform better on images with less movement (fewer degrees rotation, less deformation, etc.). One way to achieve less motion between two frames in a sequence is to capture data with a higher frame rate, but most systems used clinically today operate at frame rates significantly lower than necessary for correlation-based methods to maintain reasonable errors (especially given the rapid motion of heart structures). As such, we believe that variational methods will show significant improvements over correlation-based methods when used in real-world applications.
Conclusion and Future Work
We present a novel method for the estimation of dense 3D myocardial displacement from 4D TEE. Tests were performed on synthetic, phantom, and intraoperative TEE data, and demonstrate a considerable reduction in error. This method may be applied to biomechanical simulations, myocardial strain computation, elastography and automated diagnostics of pathologies, such as HCM.
Our future work will involve applying flow estimation to the characterization and clinical validation of pathologies such as HCM or mitral valve disease (MVD). As seen in Figure 7, optical flow is also computed on the mitral valve. Our preliminary analysis of mitral valve flow has shown promising results. More work on quantitatively evaluating the ability of optical flow techniques to characterize the complex and rapid motion of the mitral valve is required. We view the work reported in this manuscript, especially the performance evaluation of the method against the state of the art in speckle tracking, as a necessary first step towards the application of the variational motion flow method to clinical diagnostics.
Also, the experiments presented in this paper are applied primarily to 4D TEE data. However, the variational method we present is not limited to one modality. More investigation is required to evaluate the performance of our method on TTE data. We will also investigate performance of the method against motion computed using tagged MRI.
Supplementary Material
Acknowledgments
Support from the Johns Hopkins University Applied Physics Laboratory Science and Technology, Research and Development fund and the NIH NHLBI R21HL098765 fund are gratefully acknowledged. The views expressed in this document reflect those of the authors and not those of the NIH, NIHLB, or its representatives. We would also like to acknowledge Drs. Yuh, McVeigh (Johns Hopkins University), and Francisco Contijoch (University of Pennsylvania) for very useful discussions and help in data acquisition.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Exact center frequencies for the sequences were not available, as they were set automatically by the platform as a byproduct of selecting other parameters and were not stored. However, the general frequency range has three settings (Penetration, General, and Resolution) that can be adjusted using the “2D Opt” control.
References
- Abraham T, Nishimura R, Holmes D, Jr, Belohlavek M, Seward J. Strain rate imaging for assessment of regional myocardial function: results from a clinical model of septal ablation. Circulation. 2002;10512:11403. doi: 10.1161/01.cir.0000013423.33806.77. [DOI] [PubMed] [Google Scholar]
- Afonso L, Bernal J, Bax J, Abraham T. Echocardiography in hypertrophic cardiomyopathy: The role of conventional and emerging technologies. JACC Cardiovascular Imaging. 2008;16:787–800. doi: 10.1016/j.jcmg.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Baker S, Scharstein D, Lewis J, Roth S, Black M, Szeliski R. A database and evaluation methodology for optical flow; Proc. IEEE Int. Conf. on Computer Vision; 2007. [Google Scholar]
- Barron J, Fleet D, Beauchemin S. Performance of optical flow techniques. Int. Journal of Computer Vision. 1994;121:43–77. [Google Scholar]
- Bohs L, Geiman B, Anderson M, Gebhart S, Trahey G. Speckle tracking for multi-dimensional flow estimation. Ultrasonics. 2000;381:369–375. doi: 10.1016/s0041-624x(99)00182-1. [DOI] [PubMed] [Google Scholar]
- Brox T, Bruhn A, Papenberg N, Weickert J. High accuracy optical flow estimation based on a theory for warping; Proc. European Conf. Computer Vision; 2004. pp. 25–36. [Google Scholar]
- Burlina P, Hoffmann B, Abraham T. Life Science Systems and Applications Workshop (LiSSA), 2011. IEEE/NIH; 2011. Computing left ventricular hemodynamics from echographic optical flow of ceus microspheres; pp. 67–72. [Google Scholar]
- Burlina P, Sprouse C, DeMenthon D, Jorstad A, Juang R, Contijoch F, Abraham T, Yuh D, McVeigh E. Patient-specific modeling and analysis of the mitral valve using 3D-TEE. Information Processing in Computer-Assisted Interventions. 2010:135–146. [Google Scholar]
- Carasso S, Yang H, Woo A, Vannan M, Jamorski M, Wigle E, Rakowski H. Systolic myocardial mechanics in hypertrophic cardiomyopathy: novel concepts and implications for clinical status. Journal of the American Society of Echocardiography. 2008;216:675–683. doi: 10.1016/j.echo.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Chen X, Xie H, Erkamp R, Kim K, Jia C, Rubin J, O’Donnell M. 3-D correlation-based speckle tracking. Ultrasonic Imaging. 2005;271:21–36. doi: 10.1177/016173460502700102. [DOI] [PubMed] [Google Scholar]
- Cohen I. Nonlinear variational method for optical flow computation. Proceedings of the Scandanavian Conference on Image Analysis. 1993;Vol. 1:523–523. [Google Scholar]
- Duan Q, Angelini E, Herz S, Gerard O, Allain P, Ingrassia C, Costa K, Holmes J, Homma S, Laine A. Tracking of LV endocardial surface on real-time three-dimensional ultrasound with optical flow. Functional Imaging and Modeling of the Heart. 2005:434–445. [Google Scholar]
- Duan Q, Angelini E, Herz S, Ingrassia C, Costa K, Holmes J, Homma S, Laine A. Region-based endocardium tracking on real-time three-dimensional ultrasound. Ultrasound in Medicine & Biology. 2009;352:256–265. doi: 10.1016/j.ultrasmedbio.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Angelini E, Homma S, Laine A. Validation of optical-flow for quantification of myocardial deformations on simulated RT3D ultrasound; Proc. IEEE Int. Symp. Biomedical Imaging; 2007. pp. 944–947. [Google Scholar]
- Heron M, Hoyert D, Murphy S, Xu J, Kochanek K, Tejada-Vera B. Deaths: final data for 2006. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2009;5714:1. [PubMed] [Google Scholar]
- Horn B, Schunck B. Determining optical flow. Artificial Intelligence. 1981;171-3:185–203. [Google Scholar]
- Liu C, Freeman W, Adelson E, Weiss Y. Human-assisted motion annotation; Proc. IEEE Int. Conf. Computer Vision and Pattern Recognition; 2008. pp. 1–8. [Google Scholar]
- Lucas B, Kanade T. An iterative image registration technique with an application to stereo vision. Int. Joint Conf. on Artificial Intelligence. 1981;Vol.3:674–679. [Google Scholar]
- Luo J, Fujikura K, Homma S, Konofagou E. Myocardial elastography at both high temporal and spatial resolution for the detection of infarcts. Ultrasound in medicine & biology. 2007;338:1206–1223. doi: 10.1016/j.ultrasmedbio.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Maron B, Gardin J, Flack J, Gidding S, Kurosaki T, Bild D. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the cardia study. Circulation. 1995;924:785. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- Meunier J. Tissue motion assessment from 3D echographic speckle tracking. Physics in Medicine and biology. 1998;43:1241. doi: 10.1088/0031-9155/43/5/014. [DOI] [PubMed] [Google Scholar]
- Meunier J, Bertrand M. Ultrasonic texture motion analysis: theory and simulation. IEEE Transactions on Medical Imaging. 1995;142:293–300. doi: 10.1109/42.387711. [DOI] [PubMed] [Google Scholar]
- Mikic I, Krucinski S, Thomas J. Segmentation and tracking in echocardiographic sequences: Active contours guided by optical flow estimates. Medical Imaging, IEEE Transactions on. 1998;172:274–284. doi: 10.1109/42.700739. [DOI] [PubMed] [Google Scholar]
- O’Donnell M, Skovoroda A, Shapo B, Emelianov S. Internal displacement and strain imaging using ultrasonic speckle tracking. Ultrasonics, Ferroelectrics and Frequency Control, IEEE Transactions on. 1994;413:314–325. [Google Scholar]
- Ophir J, Garra B, Kallel F, Konofagou E, Krouskop T, Righetti R, Varghese T. Elastographic imaging. Ultrasound in Medicine and Biology. 2000;261:23. doi: 10.1016/s0301-5629(00)00156-3. [DOI] [PubMed] [Google Scholar]
- Pellot-Barakat C, Frouin F, Insana M, Herment A. Ultrasound elastography based on multiscale estimations of regularized displacement fields. IEEE Transactions on Medical Imaging. 2004;232:153–163. doi: 10.1109/TMI.2003.822825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serri K, Reant P, Lafitte M, Berhouet M, Le Bouffos V, Roudaut R, Lafitte S. Global and regional myocardial function quantification by two-dimensional strain: application in hypertrophic cardiomyopathy. Journal of the American College of Cardiology. 2006:1175–1181. doi: 10.1016/j.jacc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- Sprouse C, Yuh D, Abraham T, Burlina P. Computational hemodynamic modeling based on transesophageal echocardiographic imaging. Proc. Int. Conf. Engineering in Medicine and Biology Society. 2009;2009:3649–3652. doi: 10.1109/IEMBS.2009.5332519. [DOI] [PubMed] [Google Scholar]
- Sun D, Roth S, Black M. Secrets of optical flow estimation and their principles; Proc. IEEE Conf. Computer Vision and Pattern Recognition; 2010. pp. 2432–2439. [Google Scholar]
- Votta E, Caiani E, Veronesi F, Soncini M, Montevecchi F, Redaelli A. Mitral valve finite-element modelling from ultrasound data: a pilot study for a new approach to understand mitral function and clinical scenarios. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 2008;3661879:3411. doi: 10.1098/rsta.2008.0095. [DOI] [PubMed] [Google Scholar]
- Yip G, Abraham T, Belohlavek M, Khandheria B. Clinical applications of strain rate imaging. journal of the american society of echocardiography. 2003;1612:1334–1342. doi: 10.1067/j.echo.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Yu W, Yan P, Sinusas A, Thiele K, Duncan J. Towards pointwise motion tracking in echocardiographic image sequences-Comparing the reliability of different features for speckle tracking. Medical Image Analysis. 2006;104:495–508. doi: 10.1016/j.media.2005.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.