Abstract
Background
Short‐term exposures to fine (<2.5 μm aerodynamic diameter) ambient particulate‐matter (PM) have been related with increased blood pressure (BP) in controlled‐human exposure and community‐based studies. However, whether coarse (2.5 to 10 μm) PM exposure increases BP is uncertain. Recent observational studies have linked PM exposures with blood DNA hypomethylation, an epigenetic alteration that activates inflammatory and vascular responses. No experimental evidence is available to confirm those observational data and demonstrate the relations between PM, hypomethylation, and BP.
Methods and Results
We conducted a cross‐over trial of controlled‐human exposure to concentrated ambient particles (CAPs). Fifteen healthy adult participants were exposed for 130 minutes to fine CAPs, coarse CAPs, or HEPA‐filtered medical air (control) in randomized order with ≥2‐week washout. Repetitive‐element (Alu, long interspersed nuclear element‐1 [LINE‐1]) and candidate‐gene (TLR4, IL‐12, IL‐6, iNOS) blood methylation, systolic and diastolic BP were measured pre‐ and postexposure. After adjustment for multiple comparisons, fine CAPs exposure lowered Alu methylation (β‐standardized=−0.74, adjusted‐P=0.03); coarse CAPs exposure lowered TLR4 methylation (β‐standardized=−0.27, adjusted‐P=0.04). Both fine and coarse CAPs determined significantly increased systolic BP (β=2.53 mm Hg, P=0.001; β=1.56 mm Hg, P=0.03, respectively) and nonsignificantly increased diastolic BP (β=0.98 mm Hg, P=0.12; β=0.82 mm Hg, P=0.11, respectively). Decreased Alu and TLR4 methylation was associated with higher postexposure DBP (β‐standardized=0.41, P=0.04; and β‐standardized=0.84, P=0.02; respectively). Decreased TLR4 methylation was associated with higher postexposure SBP (β‐standardized=1.45, P=0.01).
Conclusions
Our findings provide novel evidence of effects of coarse PM on BP and confirm effects of fine PM. Our results provide the first experimental evidence of PM‐induced DNA hypomethylation and its correlation to BP.
Keywords: air pollution, blood pressure, DNA methylation, epigenetics, mediation
Introduction
Air pollution is a major public health concern in the United States and worldwide, accounting for approximately 800 000 deaths annually.1 Historical episodes of accumulations of ambient particulate matter (PM), such as in London in December 1952 or in Donora Valley in the last 3 days of October 1948, have been associated with up to >10 times increased death rates, mostly from cardiovascular disease (CVD).2 In the past 30 years, massive efforts in controlling emissions have led to substantial lowering of air pollution levels. However, in the presence of stagnating weather, peaks more than 10 times higher than the background levels of PM pollution are still frequently recorded in U.S. cities3 and followed by increased CVD hospitalization and mortality within hours or days.4–5 A recent comparative study showed that PM and traffic pollution exposures, because they expose millions to unhealthy air, are among the most frequent triggers of myocardial infarction at the population level, with about twice as many events as heavy alcohol consumption and >10 times than cocaine abuse.6 As recently as in July 2012, warnings for unhealthy PM levels were issued for most large U.S. cities, as reported by the U.S. Environmental Protection Agency.3
Blood pressure (BP) can change rapidly in response to environmental stressors7–8 and has been proposed as a primary intermediate response for acute PM‐related cardiovascular events.4,7 Airborne PM ≤2.5 μm (PM2.5 or fine particles) in aerodynamic diameter, as well as larger particles between 2.5 and 10 μm (coarse particles), can be inhaled and deposited in the upper and lower airways. Because of their smaller size, fine particles can reach more deeply into the lungs and have been suggested to have more harmful effects on cardiovascular outcomes.4 Observational data and previous studies of controlled human exposures have reproducibly shown rapid adverse effects on BP as early as 2 hours during PM exposure.7,9 An increase as small as 1 mm Hg in usual systolic BP is estimated to increase the risk of CVD deaths by 2% to 4%,10–11 and transient increases have been linked to PM‐related triggering of cardiovascular events.4,12 The limited understanding of the mechanisms linking air pollution exposure to cardiovascular outcomes, including effects on BP, is identified as a critical research and clinical gap in a 2010 statement of the American Heart Association.4
DNA methylation, the most studied of the epigenetic mechanisms, is a natural process that suppresses gene expression via addition of methyl groups to the DNA. Loss of methylation in inflammatory genes has been shown in lymphocytes as early as 20 minutes after antigen presentation.13 Findings from human observational studies have suggested that PM exposure may determine loss of methylation in blood DNA, potentially reflecting activation of proinflammatory states in blood leukocytes.14 Specifically, PM‐related hypomethylation has been repeatedly observed in transposable repeated elements, including Alu15–17 and long interspersed nuclear element‐1 (LINE‐1),15–18 as well as in candidate proinflammatory genes.16 Consistent with these observations, reduced methylation of genomic DNA in blood has also been observed in patients with cardiovascular disease,14,19 or CVD‐related conditions and risk factors, including atherosclerosis,20 oxidative stress,21 and aging.22 However, current evidence on PM‐induced hypomethylation rests entirely on observational studies, and no human experimental data are yet available to demonstrate effects of air pollution on DNA methylation. In addition, whether PM‐induced hypomethylation mediates the effects of PM on cardiovascular outcomes, such as those on BP, has never been tested.
Controlled studies on human‐exposure to concentrated ambient particles (CAPs) provide a unique opportunity to simulate air pollution peaks, while allowing for experimental control of the exposures. Previous controlled human exposure experiments have shown increased BP after exposure to fine CAPs9,23—consistent with observational studies that have associated short‐term PM2.5 exposure with BP.4 To the best of our knowledge, changes in BP after coarse CAP exposure have not yet been documented. Albeit mostly deposited and cleared in the upper airways, coarse particles are enriched in organic components that activate innate inflammatory responses.24 Activation of specific innate immune responses in circulating leukocytes, such as those mediated through increased expression of the toll‐like receptor‐4 (TLR4), have been linked with BP and hypertension.25 Herein, we report the results of a double‐blind randomized cross‐over trial of controlled human exposures to fine and coarse CAPs. We experimentally determined effects on blood DNA methylation of LINE‐1 and Alu repetitive elements and candidate proinflammatory genes (TLR4, IL‐12, IL‐6, iNOS). In addition to evaluating CAPs effects on blood DNA methylation, we tested for CAPs‐induced effects on BP and conducted mediation analysis to estimate the proportion of the effects on BP mediated by DNA methylation.
Materials and Methods
Study Participants
The study included 15 healthy volunteers between 18 and 60 years of age. All participants were nonsmokers and free of CVD. All experiments were conducted between January 2008–March 2010 at the Gage Occupational and Environmental Health Unit, University of Toronto, Ontario, Canada. Exclusion criteria included a fasting total cholesterol >6.2 mmol/L, glucose >7 mmol/L, hypertension (resting BP >140/90 mm Hg), pregnancy/lactation, or ECG abnormalities. The study received institutional review board approval from St. Michael's Hospital and University of Toronto. All participants provided written informed consent before enrolling.
Study Design and Exposure Protocol
Using a double‐blind randomized placebo‐controlled cross‐over design, each participant underwent 3 exposures in random order: (i) fine CAPs; (ii) coarse CAPs; and (iii) High‐Efficiency‐Particulate‐Air (HEPA)‐filtered medical air (control), separated by a minimum 2‐week washout period. Volunteers and study personnel were blinded to the exposure order. Only the technologist who generated the exposure was aware of the exposure type. We utilized a controlled human exposure facility that concentrates fine or coarse particles under controlled conditions, using high‐flow (5000 L/min) Harvard ambient particle concentrators (see details in Data S1). Briefly, ambient particles were drawn into a 1.8 m high PM10 inlet located 10 m from a busy 4‐lane downtown Toronto street, with ≈2500 vehicles passing during the 130‐minute exposure. The CAP airstream was delivered directly to the volunteer who was seated inside a Lexan and steel tube frame enclosure (4.9 m3, see exposure apparatus during one of the experiments in Figure S1). Participants were at rest and breathing freely (no mouthpiece) via an “oxygen type” facemask covering their nose and mouth.26 The target levels for the fine and coarse CAP experiments were 250 and 200 μg/m3, respectively. The CAP/filtered air delivery system was designed so that there were no visual cues as to the exposure type while participants were seated in the exposure chamber.
BP Measures and Blood Sample Collection
All participants fasted (>8 hours) prior to their arrival at the facility at ≈7:30 am. Blood samples were collected at ≈9 am. Afterward, volunteers underwent the 130‐minute exposure, at rest. Seated BP was obtained 10 minutes before exposure and 5 minutes after completion of the exposure. Postexposure blood samples were collected ≈1 hour after the end of the exposure. A standardized protocol for BP measurements was used as recommended by the American Heart Association (see Data S1, Figure S2).27
DNA Methylation Analyses
Buffy coat was immediately obtained from blood in a preprocessing laboratory adjacent to the exposure facility, aliquoted, and frozen at −20°C until DNA isolation. All laboratory procedures on the buffy‐coat samples, from DNA isolation through DNA methylation analyses, were performed in a single batch. DNA was purified using Qiagen DNeasy Blood and Tissue Kit (Qiagen). All samples from each participant were placed on the same analytical plate to avoid plate effects. DNA methylation analyses were performed by bisulfite PCR‐Pyrosequencing. We performed DNA methylation analyses of Alu and LINE‐1 repeated sequences, as described previously,16 which allows for the amplification of a representative pool of repetitive elements. We developed assays for TLR4, IL6, IL12, and iNOS methylation by locating their promoters using the Genomatix Software (Genomatix Software Inc) and amplified the sequences shown in Tables S1 and S2. In each assay, we measured %5mC at multiple CpG dinucleotides (Table S1). Every sample was tested in duplicate to confirm reliability.
Statistical Analysis
In each blood sample, DNA methylation analysis produced 8 values each for LINE‐1 and TLR4 (methylation at 4 CpGs replicated in 2 runs), 6 values for Alu (methylation at 3 CpGs replicated in 2 runs), and 4 values each for IL‐6, IL‐12, and iNOS (methylation at 2 CpGs replicated in 2 runs). In addition, methylation was measured in each participant at 2 time points (pre‐ and postexposure) in each of the 3 randomized experiments. To account for this data structure and consider within‐individual effects, we used mixed‐effect models (PROC MIXED in SAS V9.2).16 We fitted mixed‐effect models with a random intercept for each subject—which captures the correlation among measurements within the same subject; a random intercept for each CpG—which captures the correlation among measurements within the same CpG position; and a random slope for each position—which captures potential different effects of the exposures across the different positions. To control for potential confounding due to period effect, we also included a numeric variable indicating the order of exposure.
For each DNA methylation marker we assumed the following:
| 1 |
where Yijkl is the value of methylation in subject i, CpG position j, time k (pre‐ or postexpsosure) and experiment l (fine CAPs, coarse CAPs, or medical air); β0 is the overall intercept, which indicates the average methylation in the control group (medical air) in preexposure samples; μi is the random intercept for the subject i; μ0j is the random intercept for each position; β1 and β2 are the main effects of the exposure to fine CAPs and to coarse CAPs, respectively, compared to the control exposure; β3 is the main effect of time (postexposure compared to preexposure); μ1j and μ2j are the random slopes of the different CpG positions for each exposure; β4 and β5 are the interaction effects between exposure (fine and coarse, respectively) and time; β6 represents the period effect.
The choice of the 6 methylation markers, and of BP as the clinical outcome, was made a priori. No other methylation markers or outcomes were examined. P values from the model described in equation 1 were adjusted for multiple comparisons using a permutation test that accounts for data correlation.28 Methylation data are expected to be correlated and statistical tests are likely to be not independent. In this situation, commonly used methods of multiple‐testing correction such as Bonferroni and FDR will overestimate the adjusted P value. Permutation test represents a straightforward—albeit computationally heavier—and accurate approach to correct for multiple comparisons.28 Briefly, we randomly permuted the exposures within subject and then regressed each of the 6 DNA methylation markers over the exposures on this permuted dataset. The permutation breaks the link in the data between the exposures and DNA methylation, thus the dataset generated will be under the null hypothesis. We repeated this process 1000 times. A total of 12 000 P values (6 genes×1000 datasets×2 exposures) were obtained. The adjusted permutation P value was equal to the number of simulations with P value smaller than the observed P value divided by 12 000. Adjusted permutation P values <0.05 were considered significant and reported alongside the nominal 95% CIs and P values.
We then examined the effects of the exposures on systolic or diastolic BP by using the following mixed‐effect models:
| 2 |
where Yikl is the measured value of either systolic or diastolic BP for subject i, time k, and experiment l; β0 is the overall intercept, which indicates the average value of BP in the control group (medical air) preexposure; β1 and β2 are the main effects of the exposure, respectively, to fine CAP and to coarse CAP compared to the control exposure; μi is the random intercept for the subject i; β3 is the main effect of time (postexposure compared to preexposure); β4 and β5 are the interaction effects between exposure (fine and coarse, respectively) and time; β6 represents the period effect.
From the regression coefficients of the models in equations 1 or 2, group means can be derived as the average values of the dependent variables for each combination of exposure and time. In our primary analysis, we present within‐subject mean differences between postexposure measurements (ie, fine versus medical air; and coarse versus medical air). For those outcomes showing a statistically significant difference, we are also reporting differences between pre‐ and postexposure means.
We finally evaluated the association of the DNA methylation markers with systolic or diastolic BP levels in postexposure measures. To reduce multiple testing, we prioritized and present in the paper only the results for the methylation markers that showed significant differences after either fine or coarse CAP exposure. For both systolic and diastolic BP, we assumed the following model:
| 3 |
where Yikl is the measured value of either systolic and diastolic BP for subject i, time k, and experiment l; β0 is the overall intercept; β1 is the regression coefficient for each the DNA methylation fitted as the mean of CpG positions and runs; μi is the random intercept for the subject. A nominal P value <0.05 was considered statistically significant.
Mediation Analysis
We performed mediation analysis to estimate the proportion of the exposure effects on BP mediated by DNA methylation. Based on the a priori assumption that a mediated effect is biologically plausible, this approach decomposes the total observed effect of exposure on BP into a direct effect of exposure and an indirect effect of exposure that acts via the mediator of interest29–30 (ie, DNA methylation). Mediation analysis usually requires a significant relation of the outcome to the exposure, a significant relation of the outcome to the mediator and a significant relation of the mediator to the exposure;31 as potential mediators, we therefore analyzed the methylation markers that satisfied all these assumptions. In order to establish mediation, a significant relation of the mediator to the outcome, when both the mediator and the exposure are predictors, is also required. To test the latter assumption we evaluated the potential mediators in the following model:
| 4 |
where Yikl is the measured value of BP (either systolic or diastolic) for subject i, time k, and experiment l; β0 is the overall intercept; μi is the random intercept for the subject; β1 is the regression coefficient for each CpG position; β2 and β3 are the main effects of the exposure to fine and coarse CAPs, respectively, relative to the medical air control exposure.
Once all these assumptions are verified, it is possible to evaluate the indirect effect, which estimates the size of the effect of CAPs exposure on BP that is mediated by DNA methylation.32 We estimated the indirect effect via a mixed‐effect mediation model using PROC MIXED in SAS 9.2.29–30
Results
Effects of Controlled Exposures on DNA Methylation
Table 1 shows the baseline characteristics of the study participants. The study included 8 male and 7 female participants (average age 27.7 years). The actual average levels of PM mass concentrations achieved during the experiments were 241.8 μg/m3, 210.6 μg/m3, and 0.6 μg/m3 during the fine CAP, coarse CAP, and control exposures, respectively.
Table 1.
Characteristics of the Study Participants at the Enrollment Visit*, n=15
| Characteristics | Mean±SD or n (%) |
|---|---|
| Age, y | 27.7±8.6 |
| Gender | |
| Male | 8 (53.3) |
| Female | 7 (46.7) |
| Race | |
| White | 10 (66.7) |
| Black | 1 (6.7) |
| Asian | 4 (26.6) |
| BMI, kg/m2 | 23.2±2.4 |
| Fasting glucose, mmol/L | 4.8±0.5 |
| Fasting cholesterol, mmol/L | 4.2±0.7 |
| Heart rate, beats/min | 67.9±12.6 |
| Systolic BP, mm Hg | 117.6±14.1 |
| Diastolic BP, mm Hg | 69.1±12.2 |
| White blood cell count, 109/L | 5.5±1.1 |
| DNA methylation, % 5mC | |
| Alu | 24.2±0.5 |
| LINE‐1 | 84.3±0.7 |
| TLR4 | 3.6±0.8 |
| IL‐6 | 45.5±7.4 |
| IL‐12 | 94.9±0.7 |
| iNOS | 62.9±2.8 |
BMI indicates body mass index; BP, blood pressure; LINE‐1, long interspersed nuclear element‐1; TLR4, toll‐like receptor‐4; IL‐6, interleukin 6; IL‐12, interleukin 12; iNOS, inducible nitric oxide synthase gene.
Variables assessed at a preliminary screening visit conducted before the beginning of the experiments except DNA methylation—methylation values were measured on blood samples collected before the first exposure experiments.
Figure 1 shows the average postexposure differences in methylation in blood samples collected after exposures to CAPs (fine or coarse) relative to the medical air control exposure, taken as reference (ie, mean within‐subject differences between postexposure measurements obtained from model [1]); because methylation showed large between‐marker differences in mean and range of values, to allow for comparability we report standardized βs expressing the difference between exposures as a fraction of the SD of DNA methylation. Alu methylation was significantly lower after fine‐CAPs exposure (standardized β=‐0.74, P=0.0006), compared to postmedical air (control) exposure. TLR4 methylation was significantly lower after both fine‐ (standardized β=‐0.21, P=0.02) and coarse CAPs (standardized β=‐0.27, P=0.01) exposures, relative to the postcontrol exposure (Figure 1). After P value adjustment for multiple comparisons, the postexposure difference in Alu methylation between fine CAPs and medical filtered air remained significant (permutation‐adjusted P=0.03). The effect on TLR4 methylation remained significant for coarse CAPs (permutation‐adjusted P=0.04), but not for fine CAPs (permutation‐adjusted P=0.08). No significant differences were observed in postexposure LINE‐1, IL‐6, IL‐12, or iNOS methylation. Postexposure comparisons for each of the 6 methylation markers are reported in Table S5.
Figure 1.
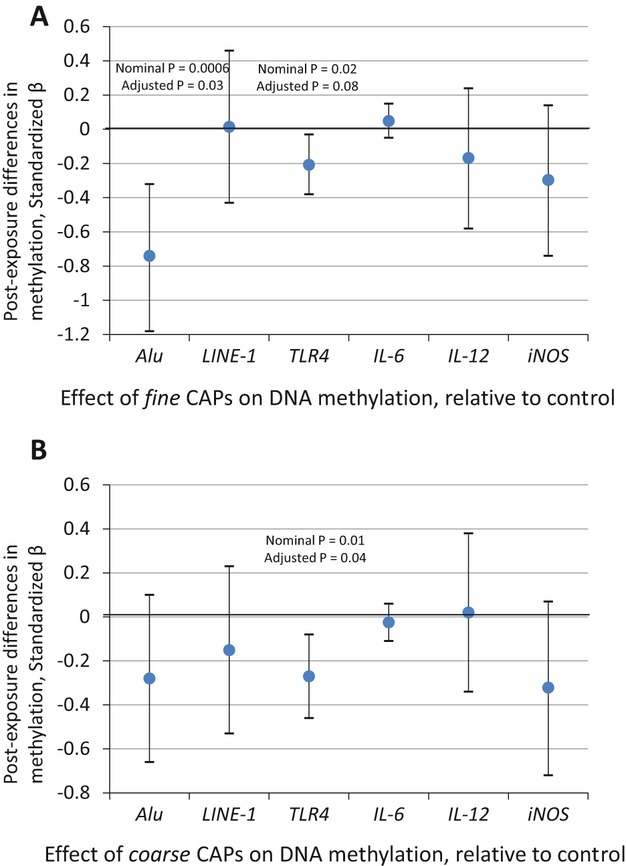
Effect of controlled exposures to fine CAPs (A) and coarse CAPs (B) on blood DNA methylation. Differences in CAPs vs medical air (control) exposures of DNA methylation in postexposure samples reported as standardized βs and 95% confidence intervals. βs indicate the differences between CAPs exposure and control exposure (HEPA‐filtered medical air) in the postexposure blood samples. βs are standardized to express the effect of CAPs exposure on DNA methylation as a fraction of the standard deviation of DNA methylation. Nominal P values (P), as well as P values adjusted for multiple testing (adjusted P) are shown for the significant effects. CAP indicates concentrated ambient particle; HEPA, high‐efficiency particulate air; LINE‐1, long interspersed nuclear element‐1; TLR4, toll‐like receptor‐4; IL‐6, interleukin 6; IL‐12, interleukin 12; iNOS, inducible nitric oxide synthase gene.
We confirmed these findings by evaluating the average within‐participant change in DNA methylation in postexposure relative to preexposure blood samples in each of the 3 exposures (fine CAPs, coarse CAPs, or control). Alu methylation showed a significant average within‐participant decrease in postexposure samples after fine CAPs exposure (standardized β=−0.40, P=0.05), and no change after coarse CAPs (standardized β=0.02, P=0.86) or control exposures (standardized β=0.20, P=0.26). TLR4 methylation showed a significant average within‐participant decrease after fine (standardized β=−0.21, P=0.02) and coarse (standardized β=−0.16, P=0.05) CAPs exposures, and no significant changes after control exposure (standardized β=0.11, P=0.28).
Effects of Controlled Exposures on BP
Figure 2 shows the average BP differences after exposures to CAPs (fine or coarse) relative to the control exposure, taken as reference (ie, mean within‐subject differences between postexposure measurements obtained from model [2]);. Systolic BP postexposure was significantly higher after exposure to fine (β=2.53 mm Hg, P=0.001) and coarse (β=1.56 mm Hg, P=0.03) CAPs relative to measurements after the control exposure. Postexposure differences in diastolic BP were not statistically significant (Figure 2).
Figure 2.
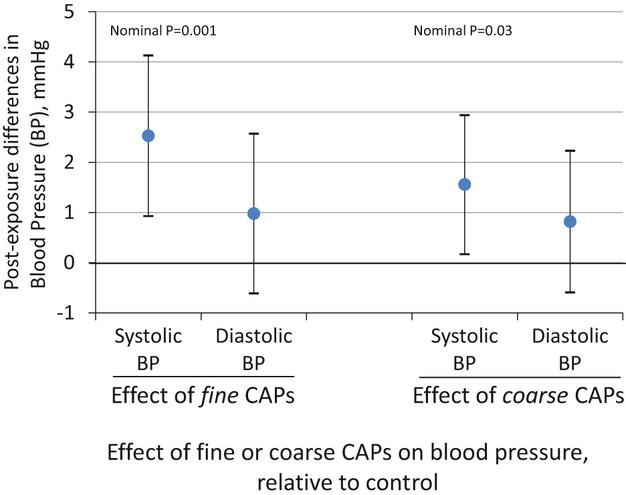
Effect of controlled exposures to fine concentrated ambient particles (CAPs) and coarse CAPs on systolic and diastolic blood pressure (BP). Differences in CAPs vs medical air (control) exposures of systolic and diastolic BP in postexposure measurements. βs and 95% confidence intervals expressing the difference in BP (mm Hg) between CAPs exposures and control exposure (HEPA‐filtered medical air) in postexposures measurements. HEPA indicates high‐efficiency particulate air.
To confirm these findings, we also estimated the average within‐participant BP change in postexposure measurements relative to preexposure for each of the 3 exposures. Both systolic and diastolic BP showed significant average within‐participant postexposure increases after fine CAPs (β=2.97 mm Hg, P=0.002 for systolic BP; and β=1.87 mm Hg, P=0.005 for diastolic BP), and coarse CAPs (β=2.11 mm Hg, P=0.0008 for systolic BP; and β=2.36 mm Hg, P=0.0007 for diastolic BP). However, as expected due to normal circadian variations in BP, we also found in the control experiments a moderate postexposure increase in systolic BP (β=0.25 mm Hg, P=0.74) and a stronger increase in diastolic BP (β=1.83 mm Hg, P=0.006), albeit both were less pronounced than those found after fine or coarse CAPs exposures.
Association of DNA Methylation With Blood Pressure
To reduce false positive findings from multiple testing, we prioritized and present only the results on the association between DNA methylation and BP for the 2 markers that showed effects from CAP exposures, that is, Alu and TLR4. Decreased Alu methylation was associated with significantly increased diastolic BP (β=0.41, P=0.04) and nonsignificantly increased systolic BP (β=0.40, P=0.15). Decreased TLR4 methylation was associated with significant increases of both diastolic (β=0.84, P=0.02) and systolic BP (β=1.45, P=0.01). Analyses stratified by exposure type showed correlations between DNA methylation and BP substantially homogeneous across the different exposures (data not shown) (Figure 3).
Figure 3.
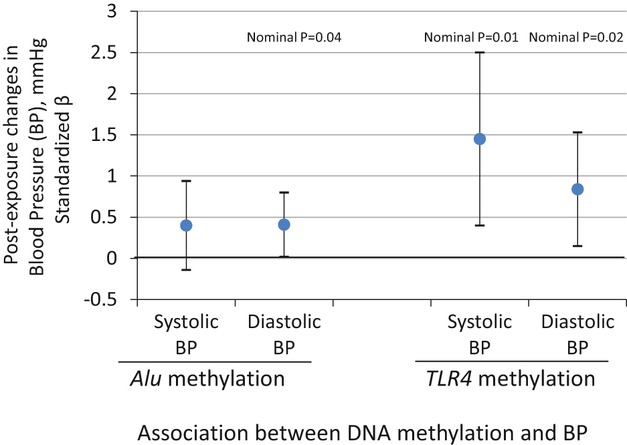
Associations of Alu and TLR4 methylation with systolic and diastolic blood pressure (BP). Standardized βs and 95% confidence intervals are shown in the figure. Standardize βs express the changes in BP as fractions of the BP standard deviation associated with a decrease in DNA methylation equal to its standard deviation. TLR4 indicates toll‐like receptor‐4.
Sensitivity Analyses
By study design, exposure‐order randomization is expected to balance potential confounders across exposure groups. We adjusted in our primary analysis for experiment order to limit confounding from random order imbalances. Moreover, by fitting a random intercept for each participant, our modeling approach allowed for controlling of participant characteristics that do not vary over time. A source of residual confounding may be represented by carryover effect. To control for this potential confounder we performed a sensitivity analysis adjusting models (1) and (2) for the previous exposure (binary variable indicating if the previous exposure was either CAPs or medical air). No major differences in the results were observed (Tables S6 and S7). As additional sensitivity analyses, multiple regression models were fitted to control for potential time‐varying confounders such as white blood cell counts and proportions of blood leukocyte cell types in differential blood counts (neutrophils, lymphocytes, monocytes, eosinophils, and basophils). We first checked if there was any difference in blood cell proportions between pre‐ and postexposure samples or when comparing exposure types in postexposure blood samples, and found no statistically significant differences (Table S8 and S9). Nonetheless, we further adjusted model (1) for white blood cell counts and proportions of blood leukocyte cell to exclude potential influences on DNA methylation. Results from these adjusted models (Table S10) were similar to those from our primary analysis.
Mediation Analysis
We performed mediation analysis to estimate the proportion of the effects of the exposures on BP that were mediated by postexposure changes in DNA methylation. Alu methylation satisfied the underlying assumptions for mediation analysis, described in the methods section, as it showed significant associations with fine CAPs exposures (β=−0.74, adjusted P value=0.03), as well as with systolic BP in the model including both Alu methylation and fine CAPs exposure as independent predictors (β=−0.37, P=0.03). These results fulfill the assumptions for mediation analysis; therefore, we considered the potential pathway of fine CAPs exposure, Alu methylation and systolic BP and estimated indirect effect and proportion of mediation.
In the mediation analysis models, estimates of the proportion of mediation indicated that Alu hypomethylation mediated 10% of the positive association between fine CAP exposure and systolic BP (indirect effect: 0.40).
Discussion
In the present study of controlled human exposures, fine and coarse CAPs induced blood hypomethylation of the Alu repetitive elements and TLR4 gene. Hypomethylation of both Alu and TLR4 was found to be associated with increased systolic BP after the exposures. A wealth of epidemiological studies have reported associations between peaks of ambient particulate matter exposure and cardiovascular disease and deaths.4–5 Several biological mechanisms that may in part explain these findings have been reported through the use of controlled human exposures to CAPs.4 Blood pressure, a well‐known predictor of cardiovascular risk, has been shown to exhibit small, but potentially critical acute increases in response to fine CAPs exposures.9 Decreased global and gene‐specific DNA methylation has been proposed as a novel mechanism mediating the effect of air pollution exposure to CVD‐related events.19 Castro et al20 found lower DNA methylation content in peripheral blood leukocytes from patients with atherosclerotic cardiovascular disease. Furthermore, recent findings from the Normative Aging Study have shown that lower blood LINE‐1 methylation predicts incidence and mortality from ischemic heart disease and stroke.14 Processes related to cardiovascular disease, such as oxidative stress,21 atherosclerosis,20 and aging22 have been associated with lower DNA methylation content in blood. In vascular tissues, hypomethylated DNA has been shown to be prone to mutations or aberrant gene expression patterns leading to the transition from a normal phenotype to vascular fibrocellular lesions by increasing proliferation of vascular smooth cells and lipid deposition.20
Previous in vitro experiments have shown that repetitive‐element and gene‐specific hypomethylation is induced by biological processes, such as oxidative stress,21 which are generated by PM in exposed individuals.4 Oxidative DNA damage can interfere with the ability of methyltransferases to interact with DNA,21 thus resulting in hypomethylation of cytosine residues at CpG sites. The association between ambient particle exposure and DNA methylation has been observed in repetitive elements, such as Alu and LINE‐1,15 in innate immune and proinflammatory pathways (TLR4, IL‐12, IL‐6),17–18 and in the inducible nitric oxide synthase gene (iNOS).16
In our study, Alu and TLR4 methylation were found to be significantly lower after CAPs exposures, providing for the first time—to the best of our knowledge—direct experimental evidence that PM exposure induces DNA hypomethylation in humans. We also observed a significant increase in systolic BP after CAPs exposures, confirming previous results on fine particle exposure4 and providing novel experimental evidence indicating that coarse exposure induces cardiovascular responses. Our findings also provide results consistent with the hypothesis that rapid hypomethylation of Alu and TLR4 is an epigenetic mechanism that mediates the effects of particle exposure on BP. Activation of Alu repetitive elements, which is associated with hypomethylation of genomic DNA, increases in older individuals and has been suggested to contribute to biological aging and age‐related chronic disease.33 Nearly 1 million copies of Alu sequences are dispersed throughout the genome.34 Alu methylation states have been shown to control DNA compaction and alter the expression of nearby genes.34 Previous studies have linked the presence of Alu sequences in a number of genes related with hypertension—including the serine/threonine protein kinase family member WNK1,35 angiotensin I converting enzyme,36–37 tissue‐type plasminogen activator,38 and 11beta‐hydroxysteroid dehydrogenase type 2.39 Whether the association found in our study between global Alu hypomethylation and BP is driven by hypomethylation of specific Alu sequences in these genes, or in other genomic regions, remains to be determined.
Recent investigations have consistently indicated that expression of TLR4 on circulating leukocytes may act as a primary communication between the innate immune system and systemic vascular functions. Experimental models have shown that TLR4, which is activated by endotoxin contained in PM, contributes to induce PM‐related inflammation, oxidative stress, and CVD‐related responses.40–41 Kampfrath et al40 showed that TLR4 deficiency prevented the increased microvascular vasoconstriction induced in mice by PM2.5 exposure, as well as PM‐related inflammatory and oxidative‐stress responses. Bonfim et al41 showed that TLR4 inactivation with anti‐TLR4 antibodies reduced the mean arterial pressure in spontaneously hypertensive rats. Recent molecular studies have shown that TLR4 binds a wide range of endogenous ligands related to BP regulation and cardiovascular function, including angiotensin II (AT‐II), heat shock protein, fibrinogen, and fibronectin.41 Downstream products of TLR4 signaling include thromboxane A2, a potent vasoconstrictor that can induce rapid increases in BP.42 TLR4 expression is increased in both peripheral mononuclear leukocytes and cardiac tissues of hypertensive patients.25,43 Our results are consistent with these animal models that indicated PM‐induced activation of TLR4 pathways as integral to PM‐related cardiovascular effects.
Results from regression models in the present study were reported as standardized beta that indicated the effects of the exposures as a fraction of a SD of the DNA methylation markers. This presentation allowed to compare the size of CAPs effects across different markers and also showed that the effects of CAPs on DNA methylation were sizable relative to the methylation SDs, as—for instance—they amounted to as much as 74% of the SD of Alu methylation. However, the SDs of both Alu and TLR4 were small, thus indicating that Alu and TLR4 methylation is tightly regulated. These findings suggest that even small changes in absolute DNA methylation, potentially corresponding to profound demethylation in a subset of blood leukocytes, may lead to effects on BP. In our study, CAP exposures did not show consistent effects on LINE‐1 methylation. Methylation measures in LINE‐1 and Alu repetitive elements have been proposed as markers of genomic DNA methylation content based on results in cancer cells.34 However, the 2 repetitive elements are controlled through different mechanisms and have been shown to have different transcription patterns in response to environmental stressors and other conditions.33 For instance, Alu—but not LINE‐1—methylation has been shown to decrease through aging,33 potentially reflecting differential susceptibility to cumulative environmental insults over time. Our data provide further evidence that Alu and LINE‐1 methylation have different sensitivities to environmental stressors,15 and that they can show different associations with cardiovascular outcomes, such as increased BP. In contrast to previous evidence from in vitro and observational investigations,16–18 CAPs exposures did not affect IL‐6, IL‐12, or iNOS methylation in our study. Despite the advantage of using high‐precision quantitative pyrosequencing measures, the lower number of CpG sites analyzed in these genes, compared to those measured in Alu and TLR4, may have limited our capability to detect potential effects. Blood DNA is derived from a mixed cell population of different types of circulating white blood cells. Therefore, our findings on DNA methylation may have resulted from a CAP‐induced shift in cell populations. However, CAPs exposure had no significant effects on the proportions of the major leukocyte cell types and sensitivity analysis showed no substantial differences in the results from models adjusted for leukocyte total count and proportions of neutrophils, lymphocytes, monocytes, basophils, and eosinophils. Although these findings limit the chances that shifts on major cell types underlie CAP‐induced effects on DNA methylation, it is still possible that our results may be driven by changes in other smaller subpopulations, such as the different types of circulating lymphocytes. TLR4 has been found to be expressed in a variety of hematopoietic cells, including lymphocytes, monocytes, and myeloid cells.24 Future studies are needed to confirm our findings and determine the cell compartment responsible for the observed changes. Finally, the volunteers enrolled in the present study were healthy and not necessarily representative of the general population. Our results may not be generalizable to different population strata and, particularly, should be replicated in a higher risk population. Moreover, the relatively small sample size might have limited our capability to detect significant effects.
In conclusion, our results demonstrate for the first time in a human experimental study that PM exposure induces rapid DNA hypomethylation. Alu and TLR4 hypomethylation may represent a novel mechanism that mediates environmental effects on BP.
Sources of Funding
The present study was supported from grants from Health Canada, Environment Canada, AllerGen NCE, US EPA (RD‐83241601; RD‐83479801) and National Institute of Environmental Health Science (ES000002, ES020268, ES009825, and ES019773). The contents of this publication are solely the responsibility of the grantee and do not necessarily represent the official views of the funding agencies. Further, the funding agencies do not endorse the purchase of any commercial products or services mentioned in the publication.
Disclosures
None.
References
- 1.WHO Air Quality and Health. 2009Geneva, Switzerland: WHO [Google Scholar]
- 2.Bell ML, Davis DL. Reassessment of the lethal London fog of 1952: novel indicators of acute and chronic consequences of acute exposure to air pollution. Environ Health Perspect. 2001; 109suppl 3:389-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Office of Air Quality Planning and Standards Air Now: Action Days. 2011U.S. Environmental Protection Agency [Google Scholar]
- 4.Brook RD, Rajagopalan S, Pope CA, III, Brook JR, Bhatnagar A, Diez‐Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, Peters A, Siscovick D, Smith SC, Jr, Whitsel L, Kaufman JD. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation. 2010; 121:2331-2378 [DOI] [PubMed] [Google Scholar]
- 5.O'Toole TE, Conklin DJ, Bhatnagar A. Environmental risk factors for heart disease. Rev Environ Health. 2008; 23:167-202 [DOI] [PubMed] [Google Scholar]
- 6.Nawrot TS, Perez L, Kunzli N, Munters E, Nemery B. Public health importance of triggers of myocardial infarction: a comparative risk assessment. Lancet. 2011; 377:732-740 [DOI] [PubMed] [Google Scholar]
- 7.Brook RD, Rajagopalan S. Particulate matter, air pollution, and blood pressure. J Am Soc Hypertens. 2009; 3:332-350 [DOI] [PubMed] [Google Scholar]
- 8.Zanobetti A, Canner MJ, Stone PH, Schwartz J, Sher D, Eagan‐Bengston E, Gates KA, Hartley LH, Suh H, Gold DR. Ambient pollution and blood pressure in cardiac rehabilitation patients. Circulation. 2004; 110:2184-2189 [DOI] [PubMed] [Google Scholar]
- 9.Urch B, Silverman F, Corey P, Brook JR, Lukic KZ, Rajagopalan S, Brook RD. Acute blood pressure responses in healthy adults during controlled air pollution exposures. Environ Health Perspect. 2005; 113:1052-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamler J, Stamler R, Neaton JD. Blood pressure, systolic and diastolic, and cardiovascular risks. US population data. Arch Intern Med. 1993; 153:598-615 [DOI] [PubMed] [Google Scholar]
- 11.Van den Hoogen PC, Feskens EJ, Nagelkerke NJ, Menotti A, Nissinen A, Kromhout D. The relation between blood pressure and mortality due to coronary heart disease among men in different parts of the world. Seven countries study research group. N Engl J Med. 2000; 342:1-8 [DOI] [PubMed] [Google Scholar]
- 12.Baccarelli A, Benjamin EJ. Triggers of MI for the individual and in the community. Lancet. 2011; 377:694-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin‐2 gene enhances transcription by an active process. Nat Immunol. 2003; 4:235-240 [DOI] [PubMed] [Google Scholar]
- 14.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010; 21:819-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009; 179:572-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009; 117:217-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011; 119:977-982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low‐dose benzene. Cancer Res. 2007; 67:876-880 [DOI] [PubMed] [Google Scholar]
- 19.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010; 3:567-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and S‐adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003; 49:1292-1296 [DOI] [PubMed] [Google Scholar]
- 21.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl‐CpG sequences inhibits the binding of the methyl‐cpg binding domain (MBD) of methyl‐CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004; 32:4100-4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5‐methylcytosine content in human peripheral leukocytes and placentas: an HPLC‐based study. Ann Hum Genet. 2004; 68:196-204 [DOI] [PubMed] [Google Scholar]
- 23.Brook RD, Urch B, Dvonch JT, Bard RL, Speck M, Keeler G, Morishita M, Marsik FJ, Kamal AS, Kaciroti N, Harkema J, Corey P, Silverman F, Gold DR, Wellenius G, Mittleman MA, Rajagopalan S, Brook JR. Insights into the mechanisms and mediators of the effects of air pollution exposure on blood pressure and vascular function in healthy humans. Hypertension. 2009; 54:659-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehli M, Poltorak A, Schwarzfischer L, Krause SW, Andreesen R, Beutler B. PU.1 and interferon consensus sequence‐binding protein regulate the myeloid expression of the human toll‐like receptor 4 gene. J Biol Chem. 2000; 275:9773-9781 [DOI] [PubMed] [Google Scholar]
- 25.Marketou ME, Kontaraki JE, Zacharis EA, Kochiadakis GE, Giaouzaki A, Chlouverakis G, Vardas PE. TLR2 and TLR4 gene expression in peripheral monocytes in nondiabetic hypertensive patients: the effect of intensive blood pressure‐lowering. J Clin Hypertens (Greenwich). 2012; 14:330-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovic B, Urch J, Brook J, Datema J, Purdham L, Liu Z, Lukic B, Zimmerman G, Tofler E, Downar P, Corey S, Tarlo I, Broder R, Dales F, Silverman S. Cardiorespiratory effects of concentrated ambient PM2. 5: a pilot study using controlled human exposures. Inhalation Toxicol. 2000; 12:173-188 [Google Scholar]
- 27.Alpert B, McCrindle B, Daniels S, Dennison B, Hayman L, Jacobson M, Mahoney L, Rocchini A, Steinberger J, Urbina E, Williams R. Recommendations for blood pressure measurement in human and experimental animals; Part 1: blood pressure measurement in humans. Hypertension. 2006; 48:e3. [DOI] [PubMed] [Google Scholar]
- 28.Wilker EH, Alexeeff SE, Suh H, Vokonas PS, Baccarelli A, Schwartz J. Ambient pollutants, polymorphisms associated with microRNA processing and adhesion molecules: the normative aging study. Environ Health. 2011; 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanderWeele TJ. Marginal structural models for the estimation of direct and indirect effects. Epidemiology. 2009; 20:18-26 [DOI] [PubMed] [Google Scholar]
- 30.Bauer DJ, Preacher KJ, Gil KM. Conceptualizing and testing random indirect effects and moderated mediation in multilevel models: new procedures and recommendations. Psychol Methods. 2006; 11:142-163 [DOI] [PubMed] [Google Scholar]
- 31.Vanderweele TJ, Valeri L, Ogburn EL. The role of measurement error and misclassification in mediation analysis: mediation and measurement error. Epidemiology. 2012; 23:561-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron RM, Kenny DA. The moderator‐mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986; 51:1173-1182 [DOI] [PubMed] [Google Scholar]
- 33.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009; 130:234-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009; 21:243-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Putku M, Kepp K, Org E, Sober S, Comas D, Viigimaa M, Veldre G, Juhanson P, Hallast P, Tonisson N, Shaw‐Hawkins S, Caulfield MJ, Khusnutdinova E, Kozich V, Munroe PB, Laan M. Novel polymorphic AluYb8 insertion in the WNK1 gene is associated with blood pressure variation in Europeans. Hum Mutat. 2011; 32:806-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agirbasli M, Guney AI, Ozturhan HS, Agirbasli D, Ulucan K, Sevinc D, Kirac D, Ryckman KK, Williams SM. Multifactor dimensionality reduction analysis of MTHFR, PAI‐1, ACE, PON1, and eNOS gene polymorphisms in patients with early onset coronary artery disease. Eur J Cardiovasc Prev Rehabil. 2011; 18:803-809 [DOI] [PubMed] [Google Scholar]
- 37.Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999; 22:59-62 [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Zhou X, Dang A, Liu G, He R. Alu‐repeat polymorphism in the gene coding for tissue‐type plasminogen activator and the risk of hypertension in a Chinese han population. Hypertens Res. 2002; 25:949-953 [DOI] [PubMed] [Google Scholar]
- 39.Lovati E, Ferrari P, Dick B, Jostarndt K, Frey BM, Frey FJ, Schorr U, Sharma AM. Molecular basis of human salt sensitivity: the role of the 11beta‐hydroxysteroid dehydrogenase type 2. J Clin Endocrinol Metab. 1999; 84:3745-3749 [DOI] [PubMed] [Google Scholar]
- 40.Kampfrath T, Maiseyeu A, Ying Z, Shah Z, Deiuliis JA, Xu X, Kherada N, Brook RD, Reddy KM, Padture NP, Parthasarathy S, Chen LC, Moffatt‐Bruce S, Sun Q, Morawietz H, Rajagopalan S. Chronic fine particulate matter exposure induces systemic vascular dysfunction via NADPH oxidase and TLR4 pathways. Circ Res. 2011; 108:716-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bomfim GF, Dos Santos RA, Oliveira MA, Giachini FR, Akamine EH, Tostes RC, Fortes ZB, Webb RC, Carvalho MH. Toll‐like receptor 4 contributes to blood pressure regulation and vascular contraction in spontaneously hypertensive rats. Clin Sci (Lond). 2012; 122:535-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez Y, Briones AM, Balfagon G, Alonso MJ, Salaices M. Hypertension increases the participation of vasoconstrictor prostanoids from cyclooxygenase‐2 in phenylephrine responses. J Hypertens. 2005; 23:767-777 [DOI] [PubMed] [Google Scholar]
- 43.Eissler R, Schmaderer C, Rusai K, Kuhne L, Sollinger D, Lahmer T, Witzke O, Lutz J, Heemann U, Baumann M. Hypertension augments cardiac toll‐like receptor 4 expression and activity. Hypertens Res. 2011; 34:551-558 [DOI] [PubMed] [Google Scholar]


