Abstract
Patients with Alzheimer's disease show age-related cognitive decline. Postmortem autopsy of their brains shows the presence of large numbers of senile plaques, whose major component is the β-amyloid peptide. The β-amyloid peptide is a cleavage product of the amyloid precursor protein (APP). In addition to the neurodegeneration associated with β-amyloid aggregation in Alzheimer's disease patients, mutations in APP in mammalian model organisms have also been shown to disrupt several behaviors independent of visible amyloid plaque formation. However, the pathways in which APP function are unknown and difficult to unravel in mammals. Here we show that pan-neuronal expression of APL-1, the Caenorhabditis elegans ortholog of APP, disrupts several behaviors, such as olfactory and gustatory learning behavior and touch habituation. These behaviors are mediated by distinct neural circuits, suggesting a broad impact of APL-1 on sensory plasticity in C. elegans. Furthermore, we found that disruption of these three behaviors requires activity of the TGFβ pathway and reduced activity of the insulin pathway. These results suggest pathways and molecular components that may underlie behavioral plasticity in mammals and in patients with Alzheimer's disease.
Introduction
Alzheimer's disease (AD) is the most common form of dementia (Alzheimer's Association, 2010) and has been associated with type 2 diabetes (Ott et al., 1999; Luchsinger et al., 2004). Levels of insulin and IGF-1 receptors are lower in brains of AD patients (Steen et al., 2005). However, the underlying cellular mechanism that connects the association is unknown. Moreover, a hallmark in the diagnosis of AD is the deposition of amyloid plaques. The amyloid plaques contain aggregations of the β-amyloid peptide, which is a cleavage product of the amyloid precursor protein (APP) (Glenner and Wong, 1984; Masters et al., 1985; Kang et al., 1987). Families with an extra copy of the APP locus on chromosome 21 have been correlated with AD (Cabrejo et al., 2006; Rovelet-Lecrux et al., 2006; Sleegers et al., 2006), and the incidence of AD among Down syndrome patients, who have a trisomy of chromosome 21, is extremely high (Mann and Esiri, 1989; Schupf et al., 1998; Korbel et al., 2009). These findings suggest that higher levels of APP could contribute to the development of AD. Overexpression of APP in mice leads to learning defects and lethality independent of amyloid peptide aggregation (Hsiao et al., 1995; Simón et al., 2009). Similarly, overexpression of APL-1, the Caenorhabditis elegans ortholog of APP, leads to an incompletely penetrant lethality (Hornsten et al., 2007). Whether overexpression of APL-1 also leads to learning defects is unknown.
C. elegans shows olfactory and gustatory plasticity and touch habituation. These behaviors are generated by distinct neural circuits (Bargmann, 2006; Giles and Rankin, 2009). To navigate through the environment, C. elegans has to constantly monitor its surroundings to respond to novel stimuli and ignore nonthreatening persistent stimuli (Lee et al., 2010). For instance, C. elegans is attracted to chemicals that resemble food or its byproducts. However, after pre-exposing C. elegans for an hour to an attractive volatile or water-soluble chemical in the absence of food, animals will show associative plasticity by decreasing their response and avoiding the previously attractive chemical (Colbert and Bargmann, 1995; Tomioka et al., 2006).
Odorants are detected by chemosensory neurons that also control developmental decisions (Bargmann and Horvitz, 1991a). Under harsh environmental conditions, C. elegans enters an alternative larval stage called dauer (Riddle and Albert, 1997) by decreasing insulin signaling (Kimura et al., 1997; Henderson and Johnson, 2001). Reduction of daf-2/insulin/IGF-1 receptor function also impairs chemosensory associative plasticity (Tomioka et al., 2006; Lin et al., 2010). Because altered insulin signaling is associated with Alzheimer's disease, we determined whether overexpression of APL-1 could affect simple C. elegans learning behaviors and whether these effects are mediated via altered insulin signaling. Here, we show that varying levels of APL-1 expression affect the chemotaxis response and that neuronal expression of APL-1 disrupts associative and nonassociative learning. These APL-1-induced effects are mediated via TGFβ signaling and decreased insulin/IGF-1 signaling. APL-1 shares some motifs with the insulin-related peptides in C. elegans (see Fig. 1N) (Pierce et al., 2001) and could potentially bind or disrupt binding to the DAF-2 insulin/IGF-1 receptor.
Figure 1.
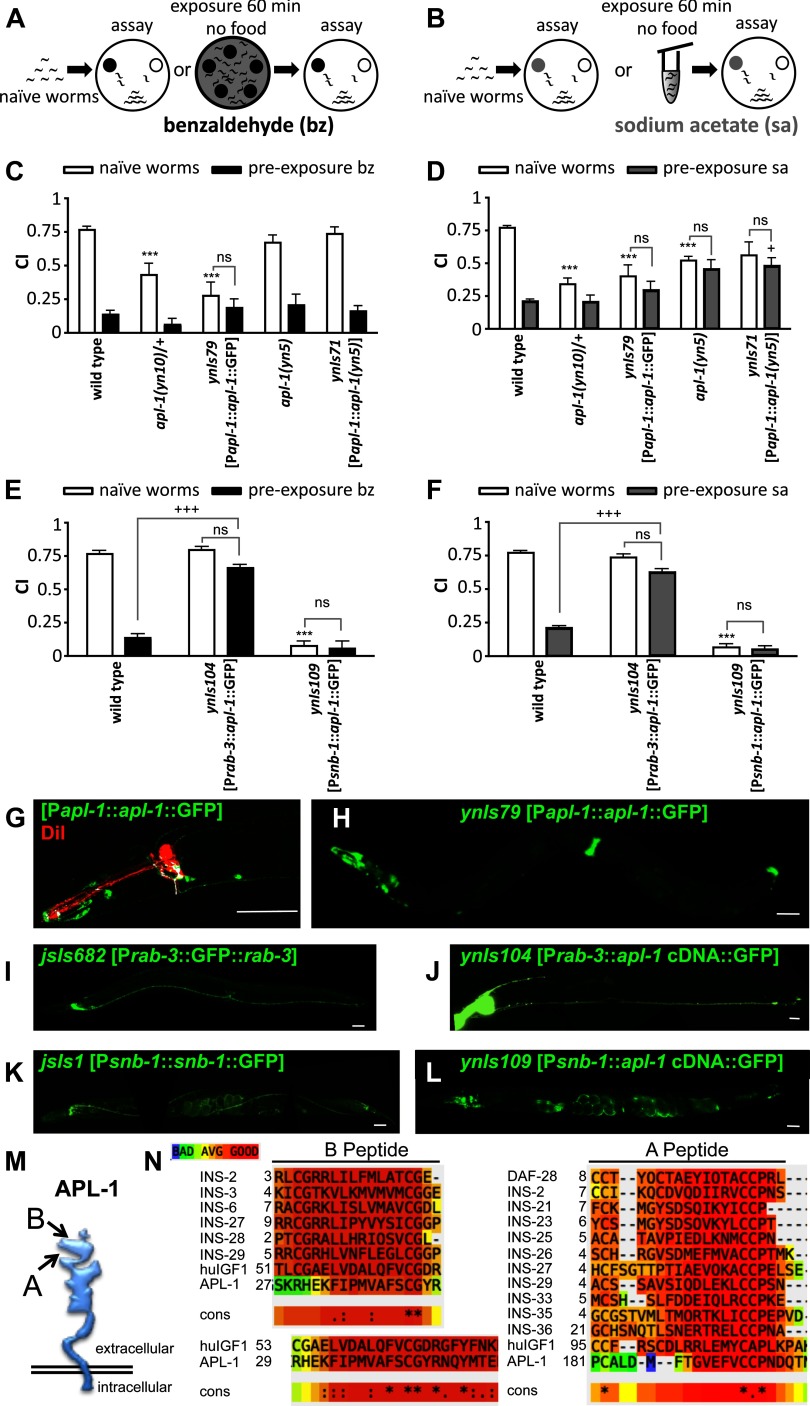
Increased APL-1 activity disrupts chemotaxis and chemosensory plasticity. A, B, Schematic of how naive worms or worms paired with starvation and a benzaldehyde (A) or sodium acetate (B) chemoattractant were assayed for chemotaxis. Chemotaxis index (CI) of naive worms (open bars) and conditioned worms (black or gray bars) of each strain is represented by bars. C, D, Heterozygous apl-1(yn10) (yn10/+) mutants, transgenic animals carrying an apl-1 genomic fragment with a GFP tag (ynIs79 [Papl-1::apl-1::GFP]), and homozygous apl-1(yn5) mutants showed diminished chemotaxis responses to benzaldehyde (C) and sodium acetate (D). Transgenic animals carrying an apl-1(yn5) genomic fragment (ynIs71 [Papl-1::apl-1(yn5)]) have slightly decreased chemotaxis responses to sodium acetate (D). Additional strains are shown in Table 1. E, F, C. elegans apl-1 driven by heterologous promoters. The rab-3 promoter drives expression pan-neuronally, while the snb-1 promoter drives expression not only pan-neuronally but also in non-neuronal cells types, such as the somatic gonad. Transgenic animals carrying the Prab-3::apl-1 cDNA::GFP transgene (ynIs104) had a wild-type chemotaxis response but showed no associative plasticity toward benzaldehyde or sodium acetate. Transgenic animals carrying the Psnb-1::apl-1 cDNA::GFP transgene (ynIs109) showed no chemoattraction to benzaldehyde or sodium acetate. Each trial consisted of 100–200 worms. Assay, Chemotaxis assay; bz, benzaldehyde; sa, sodium acetate. All data are represented as mean ± SEM. Additional strains and control strains are shown in Table 1. Statistical difference from wild-type chemotaxis response (***p < 0.001) and from wild-type associative plasticity response (+p < 0.05; +++p < 0.001) is shown by ANOVA with Bonferroni post hoc; for pairwise comparisons between no pre-exposure (naive) and pre-exposure treatments, only not significant (ns) is indicated. G–L, Anterior is to the left. G, H, apl-1 expression as monitored by a GFP-tagged APL-1 protein. Some of the chemosensory neurons can be visualized by DiI (red; G). Colocalizations are white. The ASJ chemosensory neuron appears to express apl-1. I, J, Pan-neuronal expression of a GFP-tagged synaptic protein RAB-3 GTPase (I) and APL-1 (J) using the rab-3 promoter. K, L, Expression of a GFP-tagged vesicular protein SNB-1 (K) and APL-1 (L) driven by the snb-1 promoter. Note staining in the somatic gonad. Scale bars, 50 μm. M, Schematic representation of the structure of APL-1: A and B indicate sequences where APL-1 shares similarity with the insulin A and B peptide domains, respectively. N, APL-1 shares sequence similarity with the B and A peptide domains of human IGF-1 (huIGF1) and the C. elegans insulin-related peptides (DAF-28 and INS peptides; only selected INS peptides shown); in addition, human IGF-1 has additional sequence similarities with APL-1. The degree of sequence similarity is indicated by the color scale (top left), as defined by the Expresso program; cons, strongly conserved amino acids indicated by dots; identical amino acids are indicated by asterisks.
Materials and Methods
Strains
Caenorhabditis elegans strains were grown and maintained on MYOB plates (Church et al., 1995) containing OP50 Escherichia coli bacteria at 20°C using methods as described previously (Brenner, 1974), unless noted. The wild-type strain used was N2 variant Bristol (Brenner, 1974), and hermaphrodite animals were used for all studies. All mutations used are described in WormBase (www.wormbase.org) and include the following: LGI, daf-16(mu86 and mgDf50); LGIII, daf-2(e1370), daf-7(e1372); LGX, daf-12(m20), lon-2(e678), apl-1(yn5 and yn10), and dpy-8(e130). Construction of the APL-1 transgenes and the resulting transgenic lines are described (Hornsten et al., 2007). Pmec-4::apl-1 cDNA::GFP was generated by PCR of the mec-4 promoter with primers Pmec4F-SbfI (5′-GTGTCCTGCAGGTCAGTCGGAGTTCCCGGTTC-3′) and Pmec4R (5′-TCCAAGCAAGGGTCCTCCTG-′) from the Pmec-4::CFP plasmid; this PCR fragment containing the mec-4 promoter was subcloned into the Psnb-1::apl-1 cDNA::GFP plasmid by replacing the snb-1 promoter; the promoter switch was confirmed by DNA sequencing. Pceh-36::apl-1 cDNA::GFP was generated by excising the ceh-36 promoter from ceh-36prom_TagRFPT_unc54UTR and inserting it into pPD49.26; the apl-1 cDNA fused to GFP was excised from Psnb-1::apl-1 cDNA::GFP and inserted into pPD49.26/Pceh-36. Nonintegrated transgenic lines used were as follows: dvEx371 [Psnb-1::humanAPP751 cDNA, Pmtl-2::GFP], dvEx372 [Psnb-1::humanAPP751 cDNA, Pmtl-2::GFP], ynEx1130 [Papl-1::apl-1, SUR-5::GFP], muEx169 [Punc-119::GFP::daf-16 cDNA, pRF4 rol-6(su1006gf)] (Libina et al., 2003), ynEx212 [Pmec-4::apl-1 cDNA::GFP, Pmyo-3::mCherry], ynEx213 [Pmec-4::apl-1 cDNA::GFP, Pmyo-3::mCherry], ynEx214 [Pceh-36::apl-1 cDNA::GFP, Pmyo-3::mCherry], ynEx215 [Pceh-36:: apl-1 cDNA::GFP, Pmyo-3::mCherry]. Integrated transgenic lines used were the following: ynIs14 [Phsp-16.2::apl-1 cDNA, lin-15(+)], ynIs71 [Papl-1::apl-1(yn5), SUR-5::GFP], ynIs112 [Prab-3:: apl-1 cDNA::GFP, Pmyo-3::RFP], ynIs113 [Pmec-4::apl-1 cDNA::GFP, Pmyo-3::mCherry], ynIs114 [Pmec-4::apl-1 cDNA::GFP, Pmyo-3::mCherry], ynIs115 [Pmyo-3::mCherry], jsIs1 [Psnb-1::snb-1::GFP, pRF4 rol-6(su1006gf)] (Nonet, 1999), lpIs14 [Pdaf-16::daf-16f::GFP, unc-119(+)] (Kwon et al., 2010); LGI: ynIs109 [Psnb-1::apl-1 cDNA::GFP]; LGIII: ynIs12 [Psnb-1::apl-1 cDNA, lin-15B(+)], jsIs682 [Prab-3::GFP::rab-3 cDNA; lin-15(+) (Mahoney et al., 2006); LGIV: vsIs13 [lin-11::pes-10::GFP, lin-15(+)] (Bany et al., 2003), ynIs104 [Prab-3::apl-1 cDNA::GFP, Pmyo-2::GFP], zIs356 [Pdaf-16::daf-16a/b::GFP, pRF4 rol-6(su1006gf)] (Henderson and Johnson, 2001); LGV: ynIs13 [Psnb-1:: apl-1 cDNA, lin-15B(+)], ynIs79 [Papl-1::apl-1::GFP], nuIs5 [Pglr-1::GFP; Pglr-1::Gαs(Q227L); lin-15(+)] (Berger et al., 1998); and LGX: dvIs62 [Psnb-1::humanTDP-43, Pmtl-2::GFP] (Ash et al., 2010), ynIs86 [Papl-1::apl-1, SUR-5::GFP], ynIs91 [Prab-3::apl-1 cDNA::GFP, pRF4 rol-6(su1006gf)], ynIs108 [Papl-1::apl-1(Δheparin ΔE2 domains)::GFP, SUR-5::GFP], and ynIs107 [Papl-1::apl-1(yn32/D342C/S362C)::GFP, Pmyo-2::GFP] (Hoopes et al., 2010).
Chemotaxis and conditioning learning assays
Chemotaxis and conditioning learning assays were performed at the same time and as described previously (Colbert and Bargmann, 1995; Tomioka et al., 2006). For each strain, worms were grown in duplicates, one set for chemotaxis and one for pre-exposure conditioning assays. To synchronize worm populations, ∼15 gravid adult worms were placed into a hypochlorite solution to release the eggs. Hatched worms were grown at 20°C (unless noted). Four days later, animals were washed off the growing plate with water into a 1.5 ml Microfuge tube and washed an additional three times with water.
Benzaldehyde assays.
For each strain, three 10 cm plates were poured with 10 ml of 2% agar; two were used for chemotaxis and one for pre-exposure to benzaldehyde without food (benzaldehyde conditioning). On the back of the chemotaxis plates, an equilateral triangle with 4 cm sides was drawn; the first corner was defined as starting point for the worms, on the second corner 1 μl of 1:200 benzaldehyde diluted in ethanol together with 1 μl of 1 m sodium azide was placed on the agar, and on the last corner 1 μl of ethanol together with 1 μl of 1 m sodium azide was placed on the agar as a control. The benzaldehyde dilution assays were performed the same way, except different concentrations of benzaldehyde (e.g., 1%, 0.1%, 0.05%, 0.01%) were used. For the conditioning plate, five molten plugs of 2% agar were placed on the lid; when solidified, 0.6 μl of undiluted benzaldehyde was placed on each plug (total of 3 μl of benzaldehyde). Of the two batches of washed worms per strain, one batch of worms (∼100–200 worms) was placed on the starting point of the chemotaxis plate, while the other batch (∼100–200 worms) was placed on an empty plate and covered with the benzaldehyde conditioning lid; the plate was sealed with Parafilm. Plates were maintained in a 20°C incubator for 60 min, after which animals were washed three times and assayed for chemotaxis (unless indicated). The chemotaxis index (CI) was determined after 60 min [CI = (number of worms on the test point − number of worms on control point)/(total number of worms on the plate)].
Salt assays.
The night before the assay, two 10 cm plates were poured for each strain with 10 ml of 2% agar. When the molten agar solidified, an equilateral triangle with 4 cm sides was drawn on the back of the chemotaxis plates. One corner was defined as a starting point for the worms. On a second corner a hole of 0.8 cm diameter was punched and filled with 2% agar containing 400 mm sodium acetate (NaAc), and on the third corner a hole of 0.8 cm diameter was punched and filled with 2% agar as a control. The sodium acetate was allowed to form a gradient overnight. Before the chemotaxis assay, 1 μl of 1 m sodium azide was placed on the test and control spot. Of the two batches of washed worms per strain, one batch of worms (∼100–200 worms) was placed on the starting point of the chemotaxis plate, and the other batch (∼100–200 worms) was incubated in 1 ml of 100 mm sodium acetate without food (sodium acetate conditioning) in a Microfuge tube. After 60 min at 20°C, the CI was scored for the chemotaxis plate. The worms conditioned to salt were washed three times with water and then placed on a chemotaxis plate. After 60 min at 20°C, the CI was scored. For each condition and strain, the experiment was conducted by at least two people who did not know the identities of the strains (either A.G., A.N., VC, L.T., and/or R.C); each experimental condition was repeated (T = trial) at least 3 times. For statistical analysis, repeated measures (mixed model) two-way ANOVAs with Bonferroni post-tests to compare trials and means (95% confidence intervals) were performed to assess similarity between groups using Prism 4.0a software (GraphPad).
Multiple sequence alignment
The alignment of multiple insulin-like protein sequences with the APL-1 protein sequence was performed with the program Expresso, which incorporates structural information with sequence information (Armougom et al., 2006).
Temperature shift assays
Worms carrying the temperature-sensitive daf-2(e1370) allele and corresponding control worms were grown, maintained, and synchronized at 15°C. Young adults were then placed at 25°C for 4 h and chemotaxis and conditioning learning assays were performed as described above.
Touch habituation assays
One day before the assay, fourth larval stage (L4) animals were placed onto a fresh plate with bacteria. One day adult animals were lightly touched with an eyebrow hair on the side of the head, which caused a backwards movement, and then at the side of the tail, which caused a forward movement. The touch assay was videotaped for wild-type animals (N2). The interstimulus interval (ISI) was determined by viewing the video recordings and measuring the time between head and tail touches. The ISI varied slightly from person to person. A.G.: ISI = 1.4 ± 0.02 s (N = 30); N2 mean habituation = 5.0 ± 0.3 (N = 31), A.N.: ISI = 1.7 ± 0.04 s (N = 30); N2 mean habituation = 5.4 ± 0.4 (N = 45), C.Y.E.: ISI = 1.3 ± 0.05 s (N = 30); N2 mean habituation = 5.9 ± 0.3 (N = 56), L.T.: ISI = 1.2 ± 0.03 s (N = 33); N2 mean habituation = 5.9 ± 0.4 (N = 60), and R.C: ISI = 2.1 ± 0.04 s (N = 36); N2 mean habituation = 9.9 ± 0.3 (N = 61). We found that the mean habituation not only depends on the ISI, but also on the force with which the animal was stroked with the eyebrow hair and on the properties of the eyebrow hair itself. Hence, data for all strains were acquired by C.Y.E., L.T., and R.C. equally with additional testing by A.G. and A.N.; strain identities were unknown to A.G., A.N., L.T., and R.C. during testing. For statistical analysis, a one-way ANOVA with post hoc Tukey test was used for comparison.
Touch recovery assays
One L4 animal was placed onto a fresh plate with bacteria. One day later, the animal was touched consecutively on the head and tail with an eyebrow hair until it no longer responded. This was repeated once after a pause of time X (X = 5 s, 15 s, 1 min, 3 min, 5 min, 10 min, or 20 min). Spontaneous touch recovery was calculated as follows: (number of head and tail touches until no response after time X)/(initial number of head and tail touches until no response).
Heat–shock assays
Heat–shock assays were performed as described previously (Tomioka et al., 2006). Animals were placed at 33°C for 2 h, then recovered for 2 h at 20°C and tested for either chemosensory plasticity toward benzaldehyde or sodium acetate or touch habituation.
High-osmolarity avoidance assays
High-osmolarity avoidance assays were performed as described previously (Fu et al., 2009). Quadrant Petri plates were prepared by filling opposite quadrants with either 2% agar containing 1 m sodium acetate or plain 2% agar. Worms were washed three times with water, placed at the intersection, and scored after 60 min. An osmolarity index (OI) was determined by calculating as follows: [(number of animals on NaAc quadrants − number of animals on plain agar quadrants)/(total number of animals)]. Because 1 m sodium acetate can be deadly for C. elegans, dead animals were omitted from the statistics. The number of dead ynIs104 [Prab-3::apl-1 cDNA::GFP] animals was similar to the number of dead wild-type animals on 1 m sodium acetate.
Egg-laying assays
L4 animals were transferred onto new plates. Forty-eight hours later, 10 animals were transferred to a new plate containing bacteria, left to lay eggs for 2 h, then transferred to a new plate without bacteria, and allowed to lay eggs for 2 h. The number of eggs laid in the presence and absence of food was scored. For statistical analysis, repeated measures (mixed model) two-way ANOVAs with Bonferroni post-tests to compare replicate means (95% confidence intervals) were performed to assess similarity between groups using Prism 4.0a software (GraphPad).
Dye fill assays
Worms were washed off plates with M9 and washed two additional times with M9 (M9 is defined by Brenner, 1974). Worms were incubated for 2 h in M9 containing 100 ng/μl DiI. Animals were mounted onto 2% agar pads containing a drop of 10 mm sodium azide, and amphidial neuronal morphology was scored at 400× magnification on a Zeiss Axioplan microscope. Images and z-stacks of the animals were taken at 400× magnification on a confocal microscope (Zeiss LSM 510 Confocal Laser Scanning System; slice thickness, 0.49 μm; for GFP: excitation wavelength, 488 nm; laser intensity, 5%; filter, BP505-530; pinhole, 80 μm; for DiI: excitation wavelength, 514 nm; laser intensity, 66%; filter, LP560; pinhole, 78 μm). The colocalization of the GFP and DiI fluorescence for z-stacks of each worm was determined by the Colocalization Analysis (test and highlighter) application in ImageJ.
Results
Modulating APL-1 levels disrupt olfactory chemotaxis
C. elegans is attracted to or repulsed from certain volatile and water-soluble chemicals that represent olfactory and gustatory responses, respectively (Bargmann and Horvitz, 1991b). This chemosensory behavior is mediated by 22 amphidial neurons (Ward et al., 1975). To determine whether a chemical is attractive or repulsive to C. elegans, animals are placed onto an agar plate that contains a spot with the test chemical and a control spot (Fig. 1A,B) (Colbert and Bargmann, 1995). Both spots also contain sodium azide to anesthetize the animal whenever it reaches either spot. A chemotaxis index is determined by calculating [(number of animals on the test spot − number of the animals on the control spot)/total number of animals on the plate]. A CI of 1 indicates a strong attractive test chemical, a CI of −1 a repulsive test chemical, and a CI of 0 no preference. As reported previously (Bargmann and Horvitz, 1991b), wild-type animals showed positive chemotaxis responses toward the volatile odorant benzaldehyde (CI = 0.77 ± 0.02; the number of independent times experiment was conducted or trials (T) = 30; Fig. 1C) and the water-soluble chemical sodium acetate (CI = 0.77 ± 0.01, T = 30; Fig. 1D). Hence, benzaldehyde and sodium acetate are considered strong chemoattractants. Benzaldehyde is sensed by two bilaterally symmetric AWC neurons (Bargmann et al., 1993), whereas sodium is mainly sensed by one of the ASE neurons, ASEL (Bargmann and Horvitz, 1991b; Pierce-Shimomura et al., 2001).
Loss of apl-1, such as with the yn10 null allele, leads to larval lethality (Hornsten et al., 2007); however, heterozygous apl-1(yn10) animals are viable and showed a reduced chemotaxis response toward both benzaldehyde (CI = 0.43 ± 0.09; T = 8; Fig. 1C) and sodium acetate (CI = 0.34 ± 0.05; T = 15; Fig. 1D), although both chemicals were still attractive to the heterozygous animals (Table 1). Hence, decreasing APL-1 levels lowers the chemosensory response without affecting chemical preference.
Table 1.
The defect in chemosensory plasticity in transgenic animals with pan-neuronal APL-1 expression requires daf-16 FOXO, daf-12 NHR, and daf-7 TGFβ activitya
| Genotype and/or transgene | Naive worms CI[bz] ± SEM | Pre-exposure CI[bz] ± SEM | T | Pc | Pa | Pnp | Naive worms CI[sa] ± SEM | Pre-exposure CI[sa] ± SEM | T | Pc | Pa | Pnp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type (N2) | 0.768 ± 0.021 | 0.135 ± 0.029 | 30 | °°° | 0.770 ± 0.014 | 0.210 ± 0.017 | 30 | °°° | ||||
| apl-1(yn10)/+ | 0.431 ± 0.085 | 0.063 ± 0.041 | 8 | *** | ns | °°° | 0.340 ± 0.046 | 0.205 ± 0.051 | 15 | *** | ns | ° |
| glr-1(ky176) glr-2(ak10); nmr-1(ak4) | 0.642 ± 0.042 | 0.039 ± 0.051 | 7 | ns | ns | °°° | 0.767 ± 0.089 | 0.597 ± 0.112 | 4 | ns | +++ | ns |
| Overexpression of APL-1 (and control strains) | ||||||||||||
| apl-1(yn10); ynEx1130 [Papl-1::apl-1]b | 0.516 ± 0.061 | 0.261 ± 0.098 | 4 | * | ns | °°° | 0.508 ± 0.110 | 0.309 ± 0.114 | 4 | * | ns | °° |
| ynIs86 [Papl-1::apl-1]b | 0.554 ± 0.097 | 0.322 ± 0.037 | 8 | ** | ns | °°° | 0.427 ± 0.060 | 0.362 ± 0.044 | 5 | *** | ns | ns |
| ynIs79 [Papl-1::apl-1::GFP] | 0.276 ± 0.100 | 0.184 ± 0.069 | 4 | *** | ns | ns | 0.401 ± 0.087 | 0.297 ± 0.065 | 4 | *** | ns | ns |
| apl-1(yn5) | 0.669 ± 0.056 | 0.205 ± 0.079 | 13 | ns | ns | °°° | 0.522 ± 0.030 | 0.456 ± 0.071 | 7 | *** | ns | ns |
| ynIs71 [Papl-1::apl-1(yn5)]b | 0.736 ± 0.051 | 0.161 ± 0.039 | 9 | ns | ns | °°° | 0.561 ± 0.099 | 0.480 ± 0.063 | 3 | ns | + | ns |
| ynIs107 [Papl-1::apl-1 (mut)::GFP]c,g | 0.818 ± 0.015 | 0.210 ± 0.084 | 3 | ns | ns | °°° | 0.746 ± 0.061 | 0.304 ± 0.026 | 4 | ns | ns | °°° |
| ynIs108 [Papl-1::apl-1 Δ::GFP]b,h | 0.755 ± 0.042 | 0.172 ± 0.142 | 3 | ns | ns | °°° | 0.735 ± 0.152 | 0.145 ± 0.090 | 3 | ns | ns | °°° |
| Pan-neuronal expression of APL-1 (and control strains) | ||||||||||||
| ynIs104 [Prab-3::apl-1 cDNA::GFP]c | 0.797 ± 0.023 | 0.661 ± 0.023 | 30 | ns | +++ | ns | 0.737 ± 0.025 | 0.624 ± 0.026 | 30 | ns | +++ | ns |
| ynIs112 [Prab-3::apl-1 cDNA::GFP]e | 0.747 ± 0.026 | 0.664 ± 0.035 | 23 | ns | +++ | ns | 0.705 ± 0.027 | 0.621 ± 0.027 | 22 | ns | +++ | ns |
| jsIs682 [Prab-3:: GFP::RAB-3]d | 0.682 ± 0.038 | 0.137 ± 0.033 | 6 | ns | ns | °°° | 0.722 ± 0.024 | 0.258 ± 0.030 | 3 | ns | ns | °°° |
| Ectopic expression of APL-1 (and control strains) | ||||||||||||
| ynIs12 [Psnb-1::apl-1 cDNA]d | 0.060 ± 0.022 | 0.031 ± 0.025 | 8 | *** | ns | ns | 0.291 ± 0.043 | 0.181 ± 0.060 | 6 | *** | ns | ° |
| ynIs13 [Psnb-1::apl-1 cDNA]d | 0.189 ± 0.062 | 0.099 ± 0.053 | 7 | *** | ns | ns | 0.309 ± 0.045 | 0.164 ± 0.075 | 5 | *** | ns | ° |
| ynIs109 [Psnb-1::apl-1::GFP] | 0.073 ± 0.036 | 0.054 ± 0.056 | 4 | *** | ns | ns | 0.065 ± 0.025 | 0.051 ± 0.024 | 5 | *** | ns | ns |
| vsIs13 [lin-15(+)]d | 0.764 ± 0.079 | 0.205 ± 0.038 | 3 | ns | ns | °°° | 0.697 ± 0.060 | 0.130 ± 0.106 | 3 | ns | ns | °°° |
| Expression of APL-1 in sensory neurons (and control strains) | ||||||||||||
| ynEx214 [Pceh-36:: apl-1 cDNA::GFP]e | 0.608 ± 0.045 | 0.293 ± 0.097 | 10 | ns | ns | °°° | ND | ND | ||||
| ynEx215 [Pceh-36:: apl-1 cDNA::GFP]e | 0.655 ± 0.065 | 0.386 ± 0.119 | 9 | ns | ns | °° | ND | ND | ||||
| ynEx212 [Pmec-4:: apl-1 cDNA::GFP]e | 0.701 ± 0.058 | 0.283 ± 0.080 | 9 | ns | ns | °°° | ND | ND | ||||
| ynEx213 [Pmec-4:: apl-1 cDNA::GFP]e | 0.517 ± 0.125 | 0.295 ± 0.081 | 7 | * | ns | °° | ND | ND | ||||
| ynIs115 [Pmyo-3::mCherry]e | 0.745 ± 0.063 | 0.038 ± 0.092 | 2 | ns | ns | °°° | ND | ND | ||||
| daf-2 mutant background | ||||||||||||
| daf-2(e1370) | 0.875 ± 0.035 | 0.575 ± 0.080 | 19 | ns | +++ | ° | 0.626 ± 0.053 | 0.475 ± 0.038 | 15 | ns | ++ | ns |
| daf-2(e1370); daf-16(mu86) | 0.655 ± 0.038 | 0.202 ± 0.024 | 16 | ns | ns | °°° | 0.563 ± 0.064 | 0.234 ± 0.035 | 11 | ** | ns | °°° |
| daf-2(e1370); daf-16(mgDf50) | 0.720 ± 0.023 | 0.259 ± 0.156 | 3 | ns | ns | °°° | ND | ND | ||||
| daf-2(e1370); daf-16(mgDf50); IpIs14 [DAF-16f::GFP] | 0.674 ± 0.068 | 0.556 ± 0.061 | 10 | ns | +++ | ns | 0.568 ± 0.038 | 0.365 ± 0.061 | 9 | * | + | ° |
| daf-2(e1370); ynIs86 [Papl-1::apl-1] | 0.795 ± 0.037 | 0.465 ± 0.069 | 12 | ns | +++ | ° | 0.559 ± 0.032 | 0.411 ± 0.153 | 3 | ** | ++ | ns |
| daf-2(e1370); ynIs79 [Papl-1::apl-1::GFP] | 0.734 ± 0.071 | 0.466 ± 0.063 | 3 | ns | +++ | ° | 0.461 ± 0.068 | 0.291 ± 0.065 | 6 | *** | ns | ° |
| daf-2(e1370); apl-1(yn5) | 0.716 ± 0.065 | 0.447 ± 0.063 | 11 | ns | +++ | ° | 0.549 ± 0.086 | 0.389 ± 0.097 | 5 | ** | + | ° |
| daf-2(e1370); ynIs104 [Prab-3:: apl-1 cDNA::GFP] | 0.735 ± 0.118 | 0.514 ± 0.086 | 6 | ns | +++ | ns | 0.435 ± 0.060 | 0.437 ± 0.098 | 4 | *** | + | ns |
| daf-2(e1370); ynIs12 [Psnb-1::apl-1 cDNA] | 0.539 ± 0062 | 0.372 ± 0.122 | 9 | * | + | ns | ND | ND | ||||
| daf-7 mutant background | ||||||||||||
| daf-7(e1372) | 0.588 ± 0.071 | 0.045 ± 0.029 | 9 | ns | ns | °°° | 0.713 ± 0.103 | 0.406 ± 0.063 | 6 | ns | ns | °° |
| daf-7(e1372); ynIs79 [Papl-1::apl-1::GFP] | 0.033 ± 0.014 | −0.034 ± 0.021 | 4 | *** | ns | ns | 0.212 ± 0.152 | 0.154 ± 0.069 | 5 | *** | ns | ns |
| daf-7(e1372); ynIs104 [Prab-3::apl-1 cDNA::GFP] | 0.733 ± 0.047 | 0.074 ± 0.046 | 9 | ns | ns | °°° | 0.610 ± 0.053 | 0.151 ± 0.051 | 6 | ns | ns | °°° |
| daf-7(e1372); ynIs12 [Psnb-1::apl-1 cDNA] | 0.585 ± 0.138 | 0.028 ± 0.028 | 5 | ns | ns | °°° | 0.691 ± 0.068 | 0.450 ± 0.112 | 4 | ns | + | °°° |
| daf-12 mutant background | ||||||||||||
| daf-12(m20) | 0.673 ± 0.054 | 0.321 ± 0.073 | 13 | ns | ++ | °° | 0.779 ± 0.035 | 0.269 ± 0.049 | 9 | ns | ns | °°° |
| daf-12(m20); ynIs104 [Prab-3::apl-1 cDNA::GFP] | 0.734 ± 0.049 | 0.152 ± 0.045 | 13 | ns | ns | °°° | 0.684 ± 0.074 | 0.404 ± 0.060 | 7 | ns | + | °°° |
| daf-12(m20); ynIs12 [Psnb-1::apl-1 cDNA] | 0.757 ± 0.028 | 0.363 ± 0.071 | 9 | ns | ++ | °° | 0.546 ± 0.038 | 0.323 ± 0.130 | 4 | * | ns | ° |
| daf-16 mutant background | ||||||||||||
| daf-16(mu86) | 0.737 ± 0.049 | 0.165 ± 0.039 | 12 | ns | ns | °°° | 0.477 ± 0.064 | 0.085 ± 0.029 | 12 | *** | ns | °°° |
| daf-16(mgDf50) | 0.653 ± 0.157 | 0.228 ± 0.037 | 10 | ns | ns | °°° | 0.527 ± 0.079 | 0.028 ± 0.090 | 4 | ** | ns | °°° |
| daf-16(mu86); ynIs79 [Papl-1::apl-1::GFP] | 0.267 ± 0.050 | 0.223 ± 0.056 | 6 | *** | ns | ns | ND | ND | ||||
| daf-16(mu86); ynIs104 [Prab-3::apl-1 cDNA::GFP] | 0.733 ± 0.050 | 0.165 ± 0.039 | 10 | ns | ns | °°° | 0.585 ± 0.053 | 0.161 ± 0.037 | 11 | ** | ns | °°° |
| daf-16(mgDf50); ynIs104 [Prab-3::apl-1 cDNA::GFP] | 0.759 ± 0.039 | 0.135 ± 0.057 | 10 | ns | ns | °°° | 0.477 ± 0.092 | 0.130 ± 0.077 | 4 | *** | ns | °°° |
| daf-16(mu86); ynIs12 [Psnb-1::apl-1 cDNA] | 0.125 ± 0.025 | −0.019 ± 0.052 | 12 | *** | ns | ° | 0.163 ± 0.062 | 0.121 ± 0.037 | 7 | *** | ns | ns |
| daf-16(mu86); ynIs13 [Psnb-1::apl-1 cDNA] | 0.194 ± 0.062 | 0.084 ± 0.031 | 7 | *** | ns | ° | ND | ND | ||||
| Ectopic expression of human APP | ||||||||||||
| dvEx371 [Psnb-1::humanAPP751]f | 0.539 ± 0.110 | −0.015 ± 0.053 | 11 | ns | ns | °°° | 0.574 ± 0.121 | 0.223 ± 0.061 | 4 | ns | ns | °°° |
| dvEx372 [Psnb-1::humanAPP751]f | 0.618 ± 0.056 | 0.049 ± 0.036 | 10 | ns | ns | °°° | 0.610 ± 0.055 | 0.234 ± 0.134 | 6 | ns | ns | °°° |
| Induction of neurodegeneration | ||||||||||||
| dvIs62 [Psnb-1::humanTDP-43]f | 0.000 ± 0.000 | 0.000 ± 0.000 | 6 | *** | ns | ns | 0.000 ± 0.000 | 0.000 ± 0.000 | 3 | *** | ns | ns |
| nuIs5 [Pglr-1::Gαs(Q227L)]d | 0.032 ± 0.023 | 0.181 ± 0.078 | 3 | *** | ns | ns | ND | ND | ||||
| nuIs5; ynIs104 [Prab-3::apl-1 cDNA::GFP] | 0.185 ± 0.094 | 0.070 ± 0.035 | 3 | *** | ns | ns | ND | ND | ||||
| Four hour up-shift to 25°C | ||||||||||||
| Wild type (N2) at 25°C | 0.734 ± 0.061 | 0.196 ± 0.106 | 9 | °°° | ND | ND | ||||||
| daf-16(mgDf50) at 25°C | 0.478 ± 0.150 | 0.119 ± 0.055 | 8 | ns | ns | °°° | ND | ND | ||||
| daf-2(e1370) at 25°C | 0.467 ± 0.061 | 0.466 ± 0.059 | 9 | ns | +++ | ns | ND | ND | ||||
| daf-2(e1370); daf-16(mgDf50) at 25°C | 0.410 ± 0.041 | 0.424 ± 0.077 | 8 | * | ++ | ns | ND | ND |
aEach trial consisted of 100–200 worms. All data are represented as mean ± SEM. A two-way ANOVA revealed a main effect of conditioning for benzaldehyde (F(1,56) = 822.9, p < 0.0001) and sodium acetate (F(1,51)=399.0, p < 0.0001) and a main effect of strain for benzaldehyde (F(1,56)=19.6, p < 0.0001) and sodium acetate (F(1,51) = 12.2, p < 0.0001. Statistically different from wild-type chemotaxis response (Pc = *p < 0.05; **p < 0.01; ***p < 0.001) and from wild-type associative plasticity response (Pa = +p < 0.05; ++p < 0.01; +++p < 0.001), two-way ANOVA with Bonferroni post hoc; for pair-wise comparisons between no pre-exposure (naive) and pre-exposure conditioning treatments are indicated (Pnp = °p < 0.05; °°p< 0.01; °°°p < 0.001). ns, Not significant; ND, not determined.
bCoinjection marker SUR-5::GFP.
cCoinjection marker Pmyo-2::GFP.
dCoinjection marker lin-15(+).
eCoinjection marker Pmyo-3::mCherry.
fCoinjection marker Pmtl-2::GFP.
g[apl-1(mut)::GFP]=[Papl-1::apl(yn32/D342C/S362C)::GFP].
h[apl-1Δ::GFP] = [apl-1(Δheparin ΔE2 domains)::GFP].
Transgenic animals carrying a genomic fragment of apl-1 express high levels of APL-1 (Hornsten et al., 2007) and were generally attracted to benzaldehyde and sodium acetate, but as with the heterozygous apl-1(yn10) animals, the response was much reduced compared to wild-type (Fig. 1C,D; Table 1). For example, homozygous apl-1(yn10) animals rescued by microinjection of a wild-type copy of an apl-1 fragment (apl-1(yn10); ynEx1130 [Papl-1::apl-1]) showed a decreased but positive chemotaxis response to benzaldehyde (CI = 0.52 ± 0.06; T = 4) and sodium acetate (CI = 0.5 ± 0.09; T = 4) (Table 1). Similarly, transgenic animals carrying an apl-1 genomic fragment in a wild-type background (ynIs86 [Papl-1::apl-1]) showed a positive chemotaxis response to benzaldehyde and sodium acetate, but the chemotaxis indices were significantly lower than those of wild-type; in particular, extremely high levels of APL-1 overexpression, such as in ynIs79 [Papl-1::apl-1::GFP] transgenic animals (Hornsten et al., 2007), resulted in a chemotaxis index that was only 40% of wild type (Fig. 1C,D; Table 1). Hence, high levels of APL-1 also disrupt the chemosensory response without affecting chemical preference. apl-1 is expressed in many cell types, including several amphidial chemosensory neurons such as ASJ, amphidial sheath cells, and several interneurons that are downstream of the chemosensory neurons (Fig. 1G,H) (Hornsten et al., 2007), suggesting that APL-1 exerts its effects on the chemosensory response indirectly.
Like mammalian APP, C. elegans APL-1 is cleaved by an α-secretase to release a large extracellular domain (sAPL-1) and a small cytoplasmic domain; no β-secretase has been found thus far that cleaves mammalian APP expressed in transgenic C. elegans (for review, see Ewald and Li, 2010). The apl-1(yn5) mutation deletes the region encoding the transmembrane and cytoplasmic domains. apl-1(yn5) mutants are viable and contain high levels of a fragment that contains the entire APL-1 extracellular domain (APL-1EXT); because APL-1EXT is not further cleaved by α-secretase, it is slightly larger than sAPL-1 (Hornsten et al., 2007). apl-1(yn5) mutants have a wild-type response to benzaldehyde but a reduced attractive response to sodium acetate (Fig. 1C,D; Table 1). Similarly, transgenic animals that carry an APL-1EXT transgene (ynIs71) and contain high levels of APL-1EXT (Hornsten et al., 2007) also showed a wild-type response to benzaldehyde but a reduced attractive response to sodium acetate (Table 1). Hence, high levels of APL-1EXT appear to differentially affect the chemosensory responses: the response to sodium acetate is partially compromised, whereas the response to benzaldehyde is unaffected. These results suggest that components mediating response to sodium acetate may be more sensitive to high levels of released sAPL-1 than those mediating the response to benzaldehyde. Control animals in which a deletion or point mutation was introduced into the transgene (ynIs108 [Papl-1::apl-1Δ::GFP] or ynIs107 [Papl-1::apl-1(mut)::GFP], respectively) showed wild-type responses (Table 1).
To determine whether the decreased chemotaxis response is due to defects in the chemosensory neurons, we examined the morphology of the chemosensory neurons by staining with a lipophilic dye, DiI. The ciliary endings of the chemosensory neurons are exposed to the extracellular milieu through the amphidial pore, through which DiI can be taken up by the ciliated dendritic endings of a subset of chemosensory neurons (Perkins et al., 1986). In apl-1(yn5) as well as the transgenic strains, the morphology of the labeled chemosensory neurons appeared wild type (Fig. 1G; N > 30 for each strain). Furthermore, when an RFP marker was expressed in AWC (Podr-1::dsRED), the morphology of AWC in these strains appeared wild type (data not shown). Hence, the chemosensory defects in apl-1(yn10 and yn5) mutants and the transgenic strains overexpressing APL-1 or APL-1EXT do not appear due to structural defects of the chemosensory neurons.
Ectopic expression of APL-1 disrupts chemotaxis
apl-1 is expressed in a subset of neurons that does not include the chemosensory neurons AWC and ASE that mediate the initial sensory responses to benzaldehyde and sodium acetate, respectively (Bargmann and Horvitz, 1991b; Bargmann et al., 1993); in addition, apl-1 is expressed in non-neuronal cells, such as somatic gonad, supporting cells, muscles, seam cells, and vulval cells (Fig. 1G,H) (Hornsten et al., 2007; Niwa et al., 2008). To determine whether ectopic apl-1 expression in AWC and ASE would affect the chemotaxis response, we used a pan-neuronal promoter, rab-3, to drive APL-1 expression in all neurons (Fig. 1J). These transgenic animals, ynIs104 [Prab-3::apl-1 cDNA::GFP] and ynIs112 [Prab-3::apl-1 cDNA::GFP], showed wild-type chemotaxis responses to benzaldehyde and sodium acetate, indicating that apl-1 overexpression in the chemosensory neurons does not disrupt their function (Fig. 1E,F; Table 1). By contrast, when apl-1 is driven by the snb-1 promoter, which drives expression not only pan-neuronally but also in numerous other cell types such as the somatic gonad, seam cells, hypodermis, and muscles (Fig. 1L), the transgenic ynIs12 [Psnb-1::apl-1 cDNA], ynIs13 [Psnb-1::apl-1 cDNA] and ynIs109 [Psnb-1::apl-1 cDNA::GFP] lines showed poor chemotaxis responses to benzaldehyde and only slightly better responses to sodium acetate (Fig. 1E,F; Table 1). Although APL-1 expression levels are higher in extracts from ynIs86 [Papl-1::apl-1] and ynIs79 [Papl-1::apl-1::GFP] animals than those from the ynIs12 and ynIs13 [Psnb-1::apl-1 cDNA] strains (Hornsten et al., 2007), the chemotaxis defects were more severe in the ynIs12 and ynIs13 strains (Fig. 1; Table 1), suggesting that the defect is more dependent on where APL-1 is expressed than on how much APL-1 is expressed. Again, the chemosensory neurons showed wild-type morphologies in ynIs12 and ynIs13 animals, as assayed by DiI staining or by driving RFP expression in AWC, and no neurodegenerative characteristics, such as swollen neuronal cells, vacuole-like structures in the head or nerve ring regions, or excitotoxicity during development, were seen (N>35 for each strain). As a further control, we tested dvEx371 [Psnb-1::humanAPP751 cDNA] and dvEx372 [Psnb-1::humanAPP751 cDNA] transgenic animals, in which human APP751 expression is driven by the snb-1 promoter, to exclude any nonspecific response caused by overexpression of any protein similar to APL-1; human APP751 does not rescue the apl-1 loss-of-function lethality (data not shown). These transgenic animals (dvEx371 and dvEx372) showed wild-type chemotaxis responses to benzaldehyde and sodium acetate (Table 1). As a second control, we tested dvIs62 [Psnb-1::humanTDP-43] animals, in which human TDP-43 expression is driven by the snb-1 promoter to induce neurotoxicity, presumably through protein aggregation or misfolding (Ash et al., 2010; Zhang et al., 2011). These dvIs62 animals showed a strong uncoordinated phenotype and did not show a chemotaxis response to benzaldehyde or sodium acetate (Table 1). Furthermore, inducing neurodegeneration in glutaminergic interneurons by expressing a mutated GTP-binding protein Gαs (nuIs5 [Pglr-1::GFP; Pglr-1::Gαs(Q227L); lin-15(+)]) (Berger et al., 1998) led to severe impairment of the chemotaxis response (Table 1), which is not rescued by pan-neuronal expression of APL-1 (ynIs104 [Prab-3::apl-1 cDNA::GFP]; Table 1). However, loss of glutaminergic receptors did not affect the chemotaxic response (glr-1(ky176) glr-2(ak10); nmr-1(ak4); Table 1). Collectively, these results indicate that the decreased chemotaxis responses seen in animals overexpressing APL-1 do not appear to be due to structural defects in the chemosensory neurons, but in signaling within the neurons or in cells outside the nervous system. Alternatively, the snb-1 and apl-1 promoters may drive higher relative levels of APL-1 expression in neurons than the rab-3 promoter, thereby disrupting neuronal function. However, transgenic animals in which APL-1 expression was driven by the snb-1 or apl-1 promoters responded normally on other sensory modalities and/or in different mutant backgrounds (see below, Impaired chemotaxis responses require the insulin/IGF-1 and TGF-β signaling pathways).
Pan-neuronal APL-1 expression disrupts associative plasticity behavior
As with other organisms, C. elegans shows associative plasticity (Wen et al., 1997). Wild-type animals pre-exposed to the chemoattractant benzaldehyde (or sodium acetate) in the absence of food for 60 min (benzaldehyde conditioning) showed significantly reduced chemotaxis responses to benzaldehyde (or sodium acetate), such that the chemotaxis indices were now much lower than those of naive animals (Fig. 1C,D) (Colbert and Bargmann, 1995; Tomioka et al., 2006; Lin et al., 2010); this decreased response after benzaldehyde (or sodium acetate) conditioning will hereafter be referred to as chemosensory or associative plasticity. The chemosensory plasticity response was detected with as little as 30 min pairing of benzaldehyde (or sodium acetate) and starvation, and the chemosensory plasticity response was greater as the pairing time was increased from 30 to 150 min (Fig. 2C,D) (Colbert and Bargmann, 1995; Lin et al., 2010). Hence, pairing of a chemoattractant with starvation changes chemical preference, demonstrating that C. elegans displays behavioral plasticity toward benzaldehyde and sodium acetate (Colbert and Bargmann, 1995; Tomioka et al., 2006; Lin et al., 2010).
Figure 2.
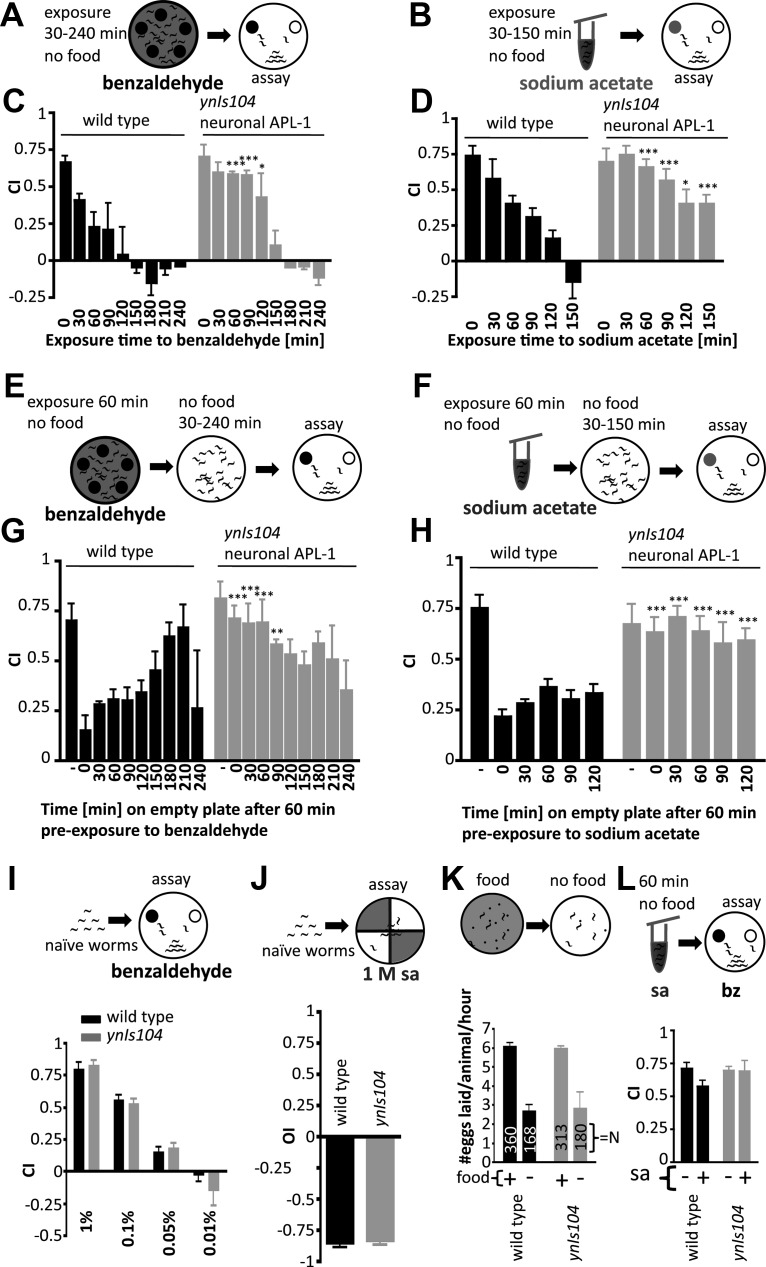
Wild-type animals show chemosensory plasticity, whereas transgenic animals expressing high levels of APL-1 only in neurons show little associative learning. A, B, Schematic representation of different pairing times of the chemoattractant benzaldehyde (A) or sodium acetate (B) with absence of food (30–240 min). C, D, Wild-type animals started to show associative plasticity after a 30 min pairing of chemoattractant and starvation; avoidance to the chemoattractants increased as the pairing time with starvation increased. After 150 min of pairing the chemoattractant with starvation, the chemoattractants became repulsive. By contrast, ynIs104 [Prab-3::apl-1 cDNA::GFP] transgenic animals, which overexpress APL-1 pan-neuronally, showed a strong associative plasticity response only after 150 min of pairing. C, Six trials; D, five trials. A two-way ANOVA revealed a main effect of pre-exposure time (benzaldehyde: F(8,1) = 29.5, p < 0.0001; sodium acetate: F(5,1) = 16.2, p < 0.0001) and a main effect for strain (benzaldehyde: F(8,1) = 10.8, p < 0.0001; sodium acetate: F(5,1) = 10.4, p = 0.0121). Statistical difference from wild-type associative plasticity response for the same pre-exposure time (*p < 0.05; ***p < 0.001) is shown by two-way ANOVA with Bonferroni post hoc. E, F, Animals were pre-exposed for 1 h to the chemoattractant, washed, and placed on empty agar plates for 30–240 min before their chemotaxis response was measured. G, H, Wild-type animals maintained the associative plasticity response for at least 2 h, whereas ynIs104 [Prab-3::apl-1 cDNA::GFP] animals did not show a strong associative plasticity response for the entire time. G, Three trials; H, four trials. A two-way ANOVA revealed a main effect of recovery time (on empty plates; benzaldehyde: F(9,1) = 3.1, p = 0.0077; sodium acetate: F(5,1) = 7.3, p = 0.0001) and a main effect for strain (benzaldehyde: F(9,1) = 6.3, p = 0.0066; sodium acetate, F(5,1) = 22.0, p = 0.0033). Statistical difference from wild-type associative plasticity response for the same recovery time (**p < 0.01; ***p < 0.001) is shown by two-way ANOVA with Bonferroni post hoc. I, Animals were exposed to serial dilutions of benzaldehyde (four trials). A two-way ANOVA revealed a main effect of dilutions (F(3,1) = 87.9, p < 0.0001) and no main effect for strain (F(3,1) = 0.509, p = 0.5023). J, Wild-type and ynIs104 [Prab-3::apl-1 cDNA::GFP] animals showed high-osmolarity avoidance behavior. OI, Osmolarity index; seven trials, not significant by two-tailed paired t test, t(6) = 0.37 p = 0.725. K, In the absence of food, wild-type animals lay fewer eggs on plates with no food compared to on a food source; ynIs104 [Prab-3::apl-1 cDNA1::GFP] animals also laid fewer eggs in the absence of food (three trials). A two-way ANOVA revealed a main effect of no food condition (F(1,1) = 33.6, p = 0.0044) and no main effect for strain (F(1,1) = 0.005, p = 0.9471). L, Wild-type or ynIs104 [Prab-3::apl-1 cDNA::GFP] animals were pre-exposed for 60 min in the absence of food to sodium acetate (sa) and their chemotaxic response to benzaldehyde (bz) was determined. Pre-exposure to sodium acetate does not affect benzaldehyde chemotaxis (five trials). A two-way ANOVA revealed no main effect of sa pre-exposure (F(1,1) = 2.2, p = 0.1724) and no main effect for strain (F(1,1) = 0.6, p = 0.4607). For C, D, G, H, I, J, L, each trial consisted of 100–200 worms per strain per condition tested.
To determine how long the association between the chemoattractant and starvation persists, wild-type animals were exposed for 60 min to benzaldehyde or sodium acetate under starvation conditions and transferred to plates without a food source or test chemical for 30 min intervals up to 240 min before assaying for chemotaxis (Fig. 2E,F). The animals continued to show the chemosensory plasticity response for 120 min, after which point the animals began to show an increased attraction toward the chemicals (Fig. 2G,H). Hence, this chemosensory plasticity response lasts about 2 h, suggesting a stable memory. Consistent with this observation, mutants [glr-1(ky176) glr-2(ak10); nmr-1(ak4)] with impaired AMPA or NMDA receptors that mediate long term potentiation did not show a compromised chemosensory plasticity response toward benzaldehyde (Table 1).
We examined the chemosensory plasticity response in transgenic APL-1 overexpression animals that showed a chemotaxis response. Transgenic animals carrying an apl-1 genomic fragment (ynIs86) showed a chemosensory plasticity response to benzaldehyde, but not to sodium acetate (Table 1). Similarly, animals that overexpressed only APL-1EXT, apl-1(yn5) mutants and ynIs71 [Papl-1::apl-1(yn5)] transgenic animals, showed a chemosensory plasticity response to benzaldehyde, but not to sodium acetate (Fig. 1C,D; Table 1). Hence, overexpression of APL-1 or APL-1EXT disrupts chemosensory plasticity to sodium acetate.
Surprisingly, after pairing benzaldehyde (or sodium acetate) to starvation, animals with pan-neuronal APL-1 expression (ynIs104 and ynIs112 [Prab-3::APL-1::GFP]) showed no chemosensory plasticity response to either benzaldehyde (or sodium acetate) for at least 120 min after exposure (Figs. 1E,F, 2C,D; Table 1). Indeed, only when pairing times of benzaldehyde (or sodium acetate) with starvation was increased to 150 min did ynIs104 [Prab-3::apl-1 cDNA::GFP] animals start to show associative plasticity (Fig. 2C,D). This requirement for an increased pairing time for ynIs104 [Prab-3::apl-1 cDNA::GFP] animals to show associative plasticity was not due to a decreased sensitivity to benzaldehyde, as the animals showed wild-type sensitivity to dilutions of benzaldehyde (Fig. 2I); furthermore, the transgenic animals showed wild-type avoidance to exposure to high-osmolar concentrations of sodium acetate (Fig. 2J) and wild-type responses to the absence of food (Fig. 2K). As a control to exclude the possibility that any protein overexpressed pan-neuronally affects the chemosensory plasticity response, we assayed jsIs682 [Prab-3::GFP::rab-3] transgenic animals in which a GFP-tagged RAB-3 GTPase is expressed under the rab-3 promoter; these animals showed wild-type chemosensory plasticity responses to benzaldehyde and sodium acetate (Table 1).
To confirm that the chemosensory plasticity toward benzaldehyde and sodium acetate was due to pairing of the chemoattractant with starvation, we exposed wild-type animals to sodium acetate in the absence of food for 60 min and assayed their response to benzaldehyde. Wild-type animals were attracted to benzaldehyde (Fig. 2L), indicating that the starvation state of the animal does not affect chemotaxis responses. Hence, although pan-neuronal APL-1 expression does not disrupt chemotaxis, this expression disrupts the associative plasticity response to both benzaldehyde and sodium acetate.
Impaired chemotaxis responses require the insulin/IGF-1 and TGF-β signaling pathways
In C. elegans the absence of food or overpopulation induces animals to undergo an alternative life cycle and form dauer larva. At least three pathways act in parallel to integrate sensory information, such as starvation, with developmental decisions: (1) DAF-2 insulin/IGF-1 receptor signaling, which promotes the phosphorylation of a FOXO transcription factor DAF-16 to prevent movement of DAF-16 into the nucleus to activate target genes that regulate dauer formation, stress resistance, and longevity (Fig. 3A); (2) DAF-7 TGFβ signaling (Fig. 3A); and (3) DAF-11 cyclic GMP signaling. The three pathways converge on the DAF-12 nuclear hormone receptor (NHR), which acts as a switch between reproductive growth and dauer formation (Daniels et al., 2000). Reducing the activity of any one of the three pathways can induce dauer formation (Daniels et al., 2000). To determine whether the impaired chemotaxis response in transgenic animals carrying an apl-1 genomic fragment or Psnb-1::apl-1 cDNA transgene requires signaling from one of these pathways, we first characterized the chemotaxis and associative plasticity responses of mutants from two of the pathways; mutants in the daf-11 cGMP branch are completely chemotaxis defective and could not be assayed. Although daf-12(m20) NHR mutants show an impaired chemotaxis response toward butanone (Daniels et al., 2000), daf-12(m20) mutants showed wild-type chemoattractive responses to benzaldehyde and sodium acetate (Fig. 3C,D; Table 1). Similarly, daf-2(e1370) reduction of function mutants and daf-16(mu86) or daf-16(mgDf50) null mutants showed wild-type chemoattractive responses, while daf-7(e1372) mutants showed slightly lowered chemoattractive responses to benzaldehyde (Fig. 3B–D; Table 1). For sodium acetate, daf-2(e1370), daf-16(mu86), or daf-16(mgDf50) mutants showed decreased chemoattraction, whereas daf-7(e1372) mutants showed wild-type chemoattraction (Table 1). Collectively, these results suggest that daf-12, daf-2, daf-16, and daf-7 are not required for chemoattraction to benzaldehyde or sodium acetate.
Figure 3.
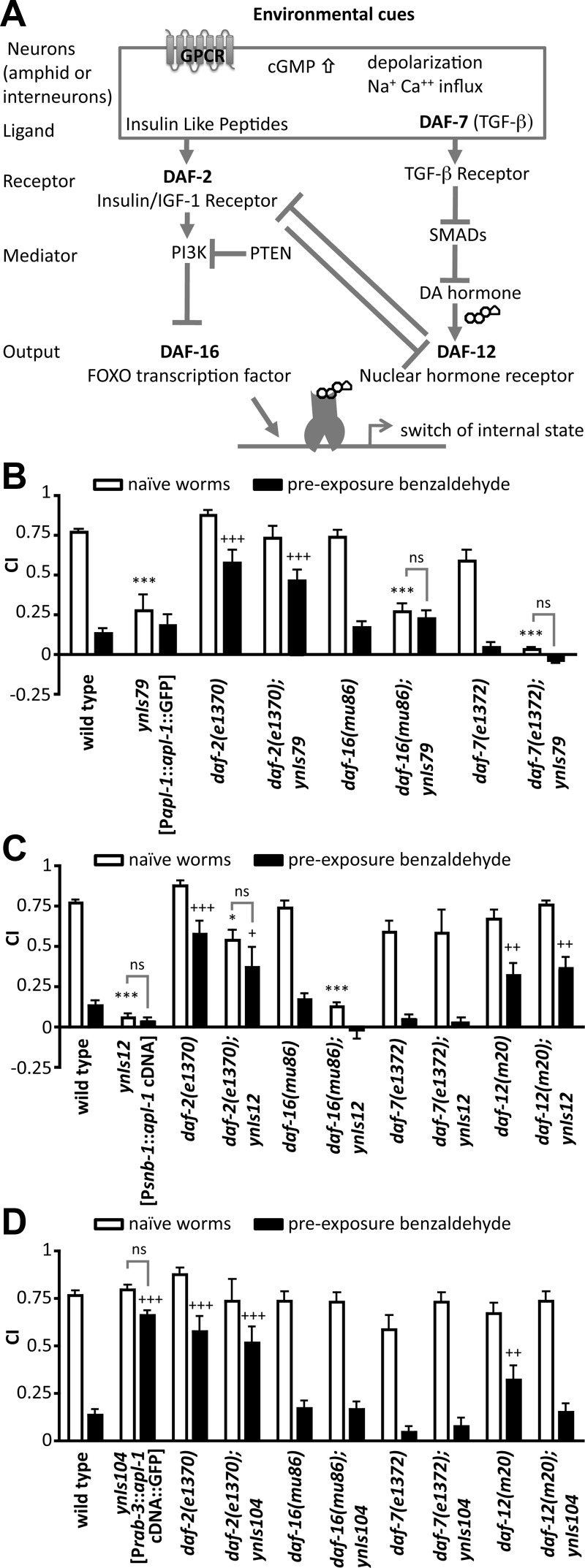
The impaired chemosensory plasticity response in transgenic animals with pan-neuronal APL-1 expression requires daf-16 FOXO, daf-12 NHR, and daf-7 TGFβ activity. A, Schematic showing how environmental conditions are translated by the TGFβ and insulin/IGF-1 pathways for dauer formation; these pathways also regulate olfactory plasticity (Daniels et al., 2000; Fielenbach and Antebi, 2008). B, Transgenic animals carrying an apl-1 genomic fragment fused to GFP (ynIs79 [Papl-1::apl-1::GFP]) showed poor chemotaxis to benzaldehyde. The chemotaxis response was rescued with decreased daf-2 insulin/IGF-1 receptor activity. C, Transgenic animals with ectopic apl-1 expression (ynIs12 [Psnb-1::apl-1]) showed no chemotaxis to benzaldehyde, but the chemotaxis response was rescued with decreased activity of daf-2 insulin/IGF-1 receptor, daf-7 TGFβ, and daf-12 NHR activity. D, Pan-neuronal overexpression of apl-1 (ynIs104 [Prab-3::apl-1 cDNA::GFP]) disrupted chemosensory plasticity. The chemosensory plasticity response was restored when daf-16 FOXO, daf-7 TGFβ, and daf-12 NHR activity were decreased. Each trial consisted of about 100–200 worms. Assay, Chemotaxis assay; bz, benzaldehyde; sa, sodium acetate. B–D, More than five trials each. All data are represented as mean ± SEM. For statistical details, please see Figure 1 legend. Statistical difference from wild-type chemotaxis response (*p < 0.05; ***p < 0.001) and from wild-type avoidance response (++p < 0.01; +++p < 0.001) to wild-type, two-way ANOVA with Bonferroni post hoc; for pairwise comparisons between no pre-exposure (naive) and pre-exposure treatments, only not significant (ns) are indicated.
Transgenic animals carrying an apl-1 genomic fragment fused to GFP (i.e., ynIs79 [Papl-1::apl-1::GFP] transgenic animals) showed a poor chemotaxis response to benzaldehyde (Figs. 1C, 3B; Table 1). However, when daf-2 insulin/IGF-1 receptor activity was decreased, ynIs79 [Papl-1::apl-1 cDNA::GFP] transgenic animals showed a robust chemosensory response (Fig. 3B; Table 1). This rescued chemosensory response by decreasing daf-2 activity may be due to increased daf-16 FOXO activity. Consistent with this possibility, decreasing the activity of daf-16 FOXO did not restore the chemosensory response in ynIs79 [Papl-1::apl-1 cDNA::GFP] transgenic animals (Fig. 3B; Table 1). Hence, the impaired ability to chemotax in ynIs79 [Papl-1::apl-1 cDNA::GFP] transgenic animals requires activation of the insulin pathway to decrease daf-16 FOXO activity. Transgenic animals carrying the Psnb-1::apl-1 cDNA transgene showed no chemotaxis response in a wild-type background (Fig. 1E,F; Table 1); these animals (ynIs12 [Psnb-1::apl-1 cDNA]) showed a robust chemotaxis response to benzaldehyde and sodium acetate when daf-2 insulin/IGF-1 receptor, daf-12 NHR activity, and daf-7 TGFβ signaling were decreased (Fig. 3C; Table 1), but not when daf-16 FOXO activity was decreased (Fig. 3C; Table 1). The impaired ability to chemotax when APL-1 is overexpressed with the snb-1 promoter, therefore, requires activation of the TGFβ and DAF-12 NHR pathways. Hence, depending on the cells in which apl-1 is expressed, apl-1 signaling modulates different metabolic pathways to affect the chemotaxis response.
The impaired associative plasticity response in transgenic animals with pan-neuronal APL-1 expression requires daf-16 FOXO and daf-12 NHR activity
daf-16(mu86 and mgDf50) and daf-7(e1372) mutants showed wild-type associative plasticity responses to benzaldehyde and sodium acetate, whereas daf-2(e1370) and daf-12(m20) mutants showed impaired or slightly impaired associative plasticity responses, respectively (Fig. 3; Table 1) (Tomioka et al., 2006; Lin et al., 2010). DAF-2 insulin/IGF-1 receptor activity negatively regulates daf-16 FOXO; in addition, the longevity and dauer phenotypes of daf-2 mutants require daf-16 FOXO activity (Lin et al., 1997; Ogg et al., 1997). Similarly, daf-2(e1370); daf-16(mu86) or daf-2(e1370); daf-16(mgDf50) double mutants showed a wild-type or daf-16 chemosensory plasticity behavior at 20°C (Table 1), suggesting that daf-16 FOXO activity is required for the impaired associative plasticity response of daf-2 insulin/IGF-1 receptor mutants. These findings are in contrast to observations from Tomioka et al. (2006), who reported that salt chemotaxis learning defects in daf-2 mutants was mediated independently of daf-16. These differences can be due to use of different salts (sodium acetate, which mainly activates the ASEL neuron, versus sodium chloride, which activates both the ASEL and ASER neurons) and different temperatures used during the assays and conditioning exposures (20°C versus 25°C). daf-2(e1370) is a temperature-sensitive allele, whereby 25°C is the restrictive temperature and 20°C is a partially permissive temperature (Gems et al., 1998). When we performed a temperature up-shift to 25°C for 4 h before the assays and benzaldehyde conditioning, the daf-2(e1370) impairment in plasticity were not rescued in a daf-16(mgDf50) mutant background (Table 1). These results suggest that under conditions of partial daf-2 activity at 20°C, the defect in associative plasticity in daf-2(e1370) mutants is mediated through daf-16/FOXO activity, whereas under conditions of complete daf-2 functional impairment (i.e., at 25°C), the defect in associative plasticity in daf-2(e1370) mutants is mediated independently of daf-16 activity.
To investigate whether signaling in the dauer and stress pathways also affects the neuronal APL-1 associative plasticity response, we examined transgenic animals with pan-neuronal APL-1 expression; ynIs104 [Prab-3::apl-1 cDNA::GFP] transgenic animals showed a wild-type chemotaxis response to benzaldehyde but did not show associative plasticity (Fig. 1E). In a daf-16(mu86) or daf-16(mgDf50) background, however, ynIs104 [Prab-3::apl-1 cDNA::GFP] transgenic animals showed wild-type associative plasticity responses toward benzaldehyde and sodium acetate (Fig. 3D; Table 1). Similarly, in daf-12(m20) NHR and daf-7(e1372) TGFβ mutant backgrounds, ynIs104 [Prab-3::apl-1 cDNA::GFP] transgenic animals showed associative plasticity responses to benzaldehyde and sodium acetate similar to or more robustly than those of daf-12(mu20) or daf-7(e1372) single mutants (Fig. 3D; Table 1). These results suggest that the impaired chemosensory plasticity responses in transgenic animals with pan-neuronal APL-1 expression require the TGFβ and/or DAF-12 NHR pathways and decreased insulin signaling.
Neuronal overexpression of APL-1 diminishes touch habituation
The lack of associative learning in the chemosensory plasticity response in animals with pan-neuronal APL-1 expression may be restricted to chemosensation or may represent a general impairment with learning of other behaviors as well. To distinguish between these alternatives, we examined a different sensory modality, gentle body touch, which is mediated by six mechanosensory or touch neurons (Chalfie et al., 1985). A gentle touch to the animal's head causes the animal to move backwards, whereas a gentle touch to the animal's tail causes it to move forward; responses to anterior and posterior touch are mediated through anterior and posterior mechanosensory neural circuits, respectively (Chalfie and Sulston, 1981). When wild-type animals are touched repeatedly on the head or tail, they no longer respond but habituate to the touch stimulus (Chalfie and Sulston, 1981). When the interstimulus interval is 10 s between head touches, we found that the animals no longer responded after 10.6 ± 0.2 touches (N = 39), similar to what was previously reported (Xu et al., 2002). To better characterize this response, we modified the touch paradigm whereby animals were touched on the head, allowed to move backwards, touched on the tail, allowed to move forward, and then touched on the head again to restart the cycle; one cycle, therefore, is composed of one head touch followed by a tail touch. When the ISI between the head and tail touches averaged 1.6 ± 0.2 s (N = 129), wild-type animals no longer responded, i.e., became touch unresponsive after six touch cycles (6.2 ± 0.2 touches to the head and tail; N = 244) (Fig. 4C). To determine the rate of spontaneous recovery, we calculated an ht/h0 index, where h0 is the initial number of consecutive head and tail touch cycles until no touch response for one animal, and ht is the number of consecutive head and tail touches until no touch response at given time points after the animal became unresponsive to touch; an ht/h0 value of 1 indicates full recovery. Wild-type animals took between 10 to 20 min to show full recovery (Fig. 4A). To distinguish whether this decreased response corresponds to habituation or sensory fatigue, we examined whether increasing the ISI would increase the spontaneous recovery time, as would be expected in habituation (Rankin et al., 2009). When the ISI was increased to 10 s, wild-type animals became touch unresponsive after 13 cycles of head–tail touches (13.1 ± 0.2 head touches; 12.6 ± 0.3 tail touches; N = 40); full recovery of the touch response occurred after 30 min (Fig. 4B), suggesting that the decreased response to repeated touch stimuli corresponds to habituation. We also examined the effect of increasing the intercycle interval between head–tail touch cycles to 10 s (i.e., head–tail interval was 1.6 s and intercycle interval was 10 s); wild-type animals became touch unresponsive after 15.2 ± 0.2 cycles (N = 42), again suggesting that the decreased response to repeated touch stimuli is due to habituation rather than sensory fatigue.
Figure 4.
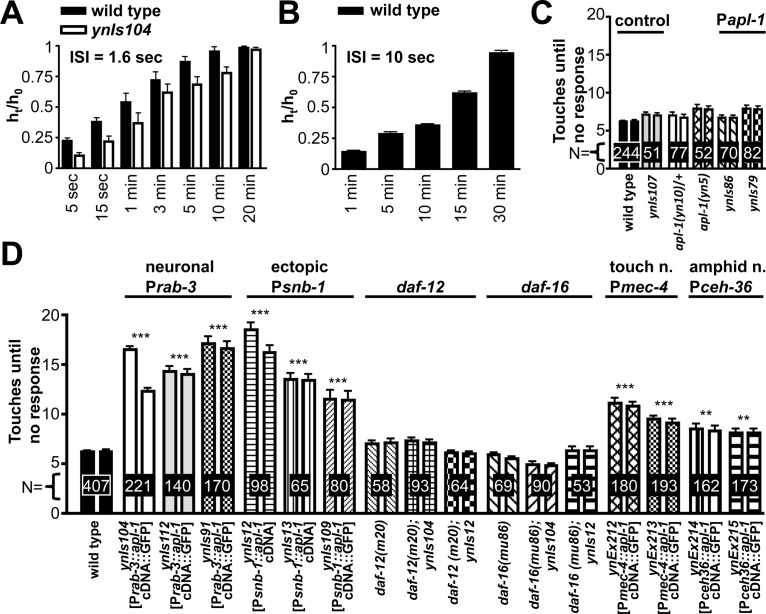
APL-1 expression in sensory neurons impairs touch habituation. A, B, Animals were touched with an eyebrow hair sequentially on the head and tail with an average interstimulus interval of 1.6 s (A) or 10 s (B) until the animal became touch unresponsive. At different time points, the animals were tested again by sequential head–tail touches until they no longer responded. h0 = initial number of consecutive head and tail touches until no response for one animal; ht = number of consecutive head and tail touches until no response at given time points after the animal became touch insensitive. C, D, Animals were touched with an eyebrow hair sequentially on the head and tail with an average interstimulus interval of 1.6 s. Transgenic strains are as follows (see Materials and Methods for more details about strains, including the coinjection markers; transgene is indicated in brackets): ynIs107 [Papl-1::apl-1(yn32/D342C/S362C)::GFP] (the yn32 mutation is an apl-1 null mutation), ynIs86 [Papl-1::apl-1], ynIs79 [Papl-1::apl-1::GFP], ynIs104 [Prab-3::apl-1 cDNA::GFP], ynIs112 [Prab-3::apl-1 cDNA::GFP], ynIs91 [Prab-3::apl-1 cDNA::GFP], ynIs12 [Psnb-1::apl-1 cDNA], ynIs13 [Psnb-1::apl-1 cDNA], ynIs109 [Psnb-1::apl-1 cDNA], ynEx212 [Pmec-4::apl-1 cDNA::GFP], ynEx213 [Pmec-4::apl-1 cDNA::GFP], ynEx214 [Pceh-36::apl-1 cDNA::GFP], ynEx215 [Pceh-36::apl-1 cDNA::GFP]. All data are represented as mean ± SEM. The promoters used to drive expression or the mutant background of the animals is indicated above the lines. N, Number of animals, trials, >3. Additional strains tested: transgenic strains containing integrated arrays of Pmec-4::apl-1 cDNA::GFP: ynIs113: head 8.1 ± 0.4, tail 8.1 ± 0.4, N = 155, p < 0.01; ynIs114: head 8.1 ± 0.4, tail 7.7 ± 0.4, N = 131, p < 0.01. Control strains were as follows: jsIs1 [Psnb-1::snb-1::GFP] head 6.9 ± 0.2, tail 6.5 ± 0.2, N = 139, p > 0.05; jsIs682 [Prab-3::GFP::rab-3] head 6.1 ± 0.2, tail 6.0 ± 0.2, N = 130, p > 0.05; dvEx371 [Psnb-1::humanAPP751] head 4.7 ± 0.2, tail 4.7 ± 0.2, N = 84, p > 0.05; dvEx372 [Psnb-1::humanAPP751] head 4.1 ± 0.2, tail 4.0 ± 0.2, N = 83, p > 0.05; dvIs62 [Psnb-1::humanTDP-43] head 1.2 ± 0.2, tail 1.2 ± 0.2, N = 71, p > 0.05; daf-2(e1370) head 7.3 ± 0.2, tail 7.6 ± 0.2, N = 329, p > 0.05; daf-16(mu86); daf-2(e1370); muEx169 [Punc-119::GFP::DAF-16] head 13.2 ± 0.7, tail 12.68 ± 0.5, N = 125, p < 0.001; daf-16(mgDf50); daf-2(e1370); IpIs14 [Pdaf-16::DAF-16f::GFP] head 11.1 ± 0.5, tail 10.5 ± 0.4, N = 136, p < 0.001; N2 at 25°C head 6.5 ± 0.3, tail 6.2 ± 0.3, N = 46, p > 0.05; daf-2(e1370) at 25°C head 8.3 ± 0.4, tail 8.3 ± 0.4, N = 35, p < 0.05. A: N > 30, a two-way ANOVA revealed a main effect of recovery time (F(6,1) = 38.95, p < 0.0001) and no main effect for strain (F(6,1) = 1.4 p = 0.2440). B: N > 30, one-way ANOVA with Tukey post hoc (F(4) = 888.1, p < 0.0001). C, D: ***p < 0.001 to wild-type, one-way ANOVA with Tukey post hoc (F(96) = 73.12 p < 0.0001).
We tested different apl-1 overexpression strains for habituation to repeated head–tail touch cycles with an average ISI of 1.6 s. Heterozygous apl-1(yn10) and homozygous apl-1(yn5) mutants and transgenic animals carrying an apl-1 genomic fragment or a mutated apl-1 fragment showed wild-type habituation (Fig. 4C), indicating that modulating APL-1 levels does not disrupt this sensory modality, presumably because apl-1 is not expressed in the six mechanosensory neurons or other neurons in the mechanosensory neural circuit. By contrast, transgenic animals with APL-1 expression driven by the pan-neuronal rab-3 promoter (ynIs91, ynIs104, ynIs112) or ectopic (including pan-neuronal) expression driven by the snb-1 promoter (ynIs12, ynIs13, ynIs109) needed significantly more head-tail stimuli before they habituated (Fig. 4D); the recovery rate of the animals, however, was similar to that of wild type (data not shown). Again, overexpression of a human APP751 cDNA driven by the snb-1 promoter (dvEx371, dvEx372) did not affect touch habituation (Fig. 4D legend). These results indicate that pan-neuronal APL-1 expression disrupts learning responses to multiple behaviors.
daf-12(m20) and daf-16(mu86) mutants showed wild-type touch habituation (Fig. 4D). The impaired habituation response of transgenic animals with pan-neuronal APL-1 expression is suppressed in a daf-12(m20) or daf-16(mu86) mutant background (Fig. 4D), suggesting that daf-12 NHR and daf-16 FOXO activity are required downstream of APL-1 signaling to inhibit touch habituation.
Expression of APL-1 in chemosensory or touch neurons is sufficient to disrupt the touch habituation response
Because transgenic animals in which APL-1 is pan-neuronally expressed showed touch habituation defects, whereas animals in which APL-1 is expressed with the apl-1 promoter did not, APL-1 disruption of the habituation response could lie within the touch cells. To determine the site of apl-1 action in disrupting touch habituation, we generated transgenic animals (ynEx212 and ynEx213) in which APL-1 expression was driven by the touch cell-specific promoter mec-4 (Lai et al., 1996). Like the transgenic animals in which APL-1 is pan-neuronally expressed, transgenic animals carrying the Pmec-4::apl-1 cDNA::GFP construct needed significantly more touch stimuli before they habituated (Fig. 4D). Hence, APL-1 expression within the touch cells is sufficient to disrupt the touch habituation response. We also examined whether APL-1 expression in the ASE and AWC chemosensory neurons affected the touch habituation response. Surprisingly, when apl-1 was expressed in the ASE and AWC chemosensory neurons, which mediate sodium and benzaldehyde chemoattraction, respectively, using the ceh-36 promoter (Lanjuin et al., 2003), the transgenic animals also showed defective touch habituation (Fig. 4D). These results suggest that apl-1 signaling does not have to occur specifically in the touch cells to disrupt touch habituation; for instance, release of sAPL-1 from other neurons can modulate touch responsiveness in the touch cells or other downstream neurons in the touch circuit.
Similar experiments were performed to determine the site of action of the chemosensory plasticity response. Naive and benzaldehyde-conditioned transgenic animals expressing apl-1 in the ASE and AWC chemosensory neurons or the touch cells were examined for chemotaxis and associative plasticity responses. All transgenic animals showed a robust chemotaxis response to benzaldehyde, and all showed chemosensory plasticity when conditioned to benzaldehyde (Table 1). Hence, the site of action for the defects in chemosensory plasticity appears to lie downstream of the ASE and AWC chemosensory neurons.
Induction of APL-1 overexpression during adulthood is sufficient to impair chemotaxis responses and to diminish touch habituation
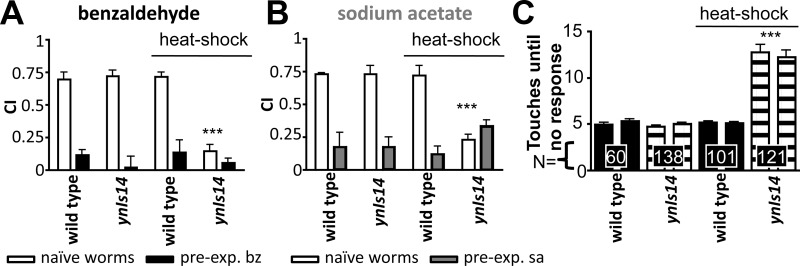
Since apl-1 is essential for development and overexpression of APL-1 causes an incompletely penetrant early larval lethality, we considered whether APL-1 overexpression affected the development of neurons or other cell types. Adult animals carrying an apl-1 cDNA under the control of a heat shock promoter (ynIs14 [Phsp-16.2::apl-1 cDNA]) showed wild-type chemotaxis and chemosensory plasticity responses toward benzaldehyde and sodium acetate and wild-type touch habituation at 20°C (Fig. 5). However, after a short heat shock to induce ubiquitous APL-1 expression (Hornsten et al., 2007), ynIs14 [Phsp-16.2::apl-1 cDNA] animals showed defective chemotaxis responses toward benzaldehyde and sodium acetate and diminished touch habituation (Fig. 5). These results indicate that the APL-1 overexpression behavioral phenotypes are not caused by developmental changes, but rather by APL-1 signaling that activates downstream pathways to interfere with behavioral plasticity.
Figure 5.
Short induction of APL-1 overexpression is sufficient to disrupt the chemotaxis response and to diminish touch habituation. At 20°C, ynIs14 [Phsp-16.2::apl-1 cDNA] animals showed wild-type chemosensory plasticity toward benzaldehyde (bz; A) and sodium acetate (sa; B) and also wild-type habituation to light touch (C). By contrast, when wild-type and ynIs14 [Phsp-16.2::apl-1 cDNA] animals were heat-shocked for 2 h at 33°C, recovered at 20°C for 2 h, and then assayed, ynIs14 [Phsp-16.2::apl-1 cDNA] animals showed poor chemotaxis behavior toward bz (A) and sa (B) and took longer to habituate to light touch (C) compared to wild-type animals. Each trial consisted of 100–200 worms. All data are represented as mean ± SEM. A, B, Trials, >5. ***p < 0.001 to wild-type, two-way ANOVA with Bonferroni post hoc (for more detail please see Fig. 1 legend). C, N, Number of animals, trials, >3, ***p < 0.001 to heat-shocked wild-type, one-way ANOVA with Tukey post hoc (F(96) = 73.12, p < 0.0001).
Discussion
APL-1 overexpression using either the apl-1 or the snb-1 promoter, but not the pan-neuronal rab-3 promoter, caused a diminished chemotaxis response to both sodium acetate and benzaldehyde. Similarly, a short heat shock during adulthood to induce ubiquitous APL-1 expression was sufficient to cause an impaired chemotaxis response. These results suggest that APL-1 expression in non-neuronal cells decreases or disrupts the chemotaxis response. For instance, apl-1 signaling may interfere with the feedback from downstream neurons or non-neuronal tissue to the sensory neurons. Strikingly, the chemotaxis impairments of these transgenic animals were fully or partially restored by decreased activity of daf-2 insulin-IGF-1 receptor, daf-7 TGFβ, or daf-12 NHR signaling. Hence, the effects of APL-1 levels on chemotaxis are unlikely to be due to disrupted neuronal structure or neurodegeneration, but rather to modulation of the insulin/IGF-1 and/or DAF-12 NHR signaling pathways. Similarly, disruption of apl-1 by the apl-1(yn5) mutation reduces the activity of the insulin pathway to delay development (Ewald et al., 2012).
Although transgenic animals with pan-neuronal APL-1 [Prab-3::apl-1 cDNA::GFP] expression show wild-type chemotaxis and touch responses, these animals needed longer pairing times to an olfactory or gustatory stimulus with starvation for associative learning to occur and more repeated stimuli for touch habituation, suggesting that neuronal APL-1 expression inhibits processes for associative and nonassociative learning. This inhibition requires daf-16 FOXO and daf-12 NHR activity. Although the neural circuitry mediating the chemotaxis and touch responses are distinct, the signaling pathways underlying the two learning responses share common molecular elements. We propose that APL-1 signaling interferes with insulin signaling, thereby increasing DAF-16 FOXO or DAF-12 NHR activity to inhibit associative and nonassociative learning pathways, perhaps at the memory acquisition step. Whether APL-1 signaling directly affects daf-2 insulin/IGF-1 receptor activity or acts downstream of daf-2 to affect learning is unclear. Several mechanisms can account for how APL-1 signaling affects the physiological state of the neurons. APL-1 expression in the touch cells or ASE and AWC chemosensory neurons is sufficient to disrupt touch habituation. Release of sAPL-1 from neurons may inhibit insulin signaling in touch neurons, thereby maintaining their receptive state to touch and inhibiting pathways to decrease touch responsiveness. Curiously, sAPL-1 shares some motifs, including a putative zinc-binding domain (for review see Dunn, 2005) with the DAF-28 and INS peptides, which are putative ligands of the DAF-2 insulin/IGF-1 receptor (Fig. 1N) (Pierce et al., 2001), suggesting that sAPL-1 could potentially bind DAF-2 or disrupt DAF-2 ligand binding. In addition, apl-1 genetically interacts with a receptor protein tyrosine phosphatase (Ewald et al., 2012), whose activity could modulate the insulin pathway. Alternatively, release of sAPL-1 from neurons could affect insulin signaling in downstream cells of the touch circuit to prevent a decreased response to touch and keep a high level of responsiveness. apl-1 knockdown by RNA interference results in hypersensitivity to aldicarb, which is rescued by the expression of the extracellular domain of APL-1, an indication that sAPL-1 affects acetylcholine neurotransmission (Wiese et al., 2010), which could affect its touch response. Taken together, excess APL-1 signaling maintains activity of the touch circuit, preventing animals from undergoing behavioral plasticity.
By contrast, APL-1 expression in the ASE and AWC chemosensory neurons or touch cells is not sufficient to disrupt the chemosensory plasticity response. We propose that release of sAPL-1 from unidentified neurons, perhaps neighboring amphidial neurons or the AIA interneuron, which is required for salt chemotaxis learning (Tomioka et al., 2006), results in a change in the metabolic state of the ASE and AWC chemosensory neurons. This metabolic reset decreases daf-2 insulin/IGF-1 receptor activity, which has multiple effects, including extending lifespan (Kenyon, 2005) and impairing chemosensory plasticity toward sodium and benzaldehyde (Table 1) (Tomioka et al., 2006; Lin et al., 2010), and which affects daf-16 FOXO-dependent and independent pathways (Table 1). In other behaviors that show plasticity, such as thermal or butanone associative plasticity, mutations in daf-2 insulin/IGF-1 receptor also resulted in learning impairments that required daf-16 activity (Murakami et al., 2005; Kauffman et al., 2010). Olfactory plasticity is also regulated by pheromone levels (Yamada et al., 2010). Food resources and pheromone levels from crowded conditions are sensed by amphidial neurons; this external information is integrated via the insulin and/or daf-7 TGFβ pathways (Fielenbach and Antebi, 2008) to modulate daf-12 NHR activity to reprogram the internal state of the animal and adjust behavior accordingly.
In summary, our results suggest novel signaling interactions between APL-1 and the insulin/daf-16 FOXO and daf-12 NHR pathways. In three behavioral assays, pan-neuronal expression of APL-1 may delay or inhibit memory acquisition, since larger stimuli will allow the animals to show behavioral plasticity. In mammals, overexpression of APP shows impairments of several behaviors independent of plaque formation (Hsiao et al., 1995; Simón et al., 2009). Our results suggest that APP activity may not be directly involved in forming those behaviors, but may indirectly affect them via the insulin/IGF-1 pathway.
Footnotes
This work was supported by grants from the Alzheimer's Association, National Institutes of Health (R21AG033912 and R01AG32042), and National Science Foundation (IOS08207) (C.L.) and a National Institutes of Health RCMI grant (G12-RR03060) to City College. We thank Chris Link for kindly providing the dvEx371, dvEx372, and dvIs62 strains, Scott Clark and Oliver Hobert for kindly providing plasmids, Catharine Rankin and Piali Sengupta for comments on the manuscript, lab members for helpful discussions, Sarah Tichelli for help with statistical analysis, and Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources, for providing strains. The following work was performed by the authors listed. Benzaldehyde assays: R.C., L.T., V.S., A.G., A.N., and C.Y.E.; benzaldehyde dilution assays: A.N., R.C., and C.Y.E.; sodium acetate assays: L.T., R.C., V.S., and C.Y.E.; high-osmolarity avoidance assays: L.T., R.C., and C.Y.E.; touch habituation: C.Y.E., R.C., L.T., A.N., and A.G.; egg-laying assays: C.Y.E.; heat-shock assays: L.T., R.C., and C.Y.E.; temperature up-shift assays: R.C., L.T., and C.Y.E.; generation of transgenic strains: C.Y.E. and C.L.
The authors declare no competing financial interests.
References
- Alzheimer's Association. 2010 Alzheimer's disease facts and figures. Alzheimers Dement. 2010;6:158–194. doi: 10.1016/j.jalz.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bany IA, Dong MQ, Koelle MR. Genetic and cellular basis for acetylcholine inhibition of Caenorhabditis elegans egg-laying behavior. J Neurosci. 2003;23:8060–8069. doi: 10.1523/JNEUROSCI.23-22-08060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI The C. elegans Research Community, editor. Chemosensation in C. elegans. WormBook. 2006. doi: 10.1895/wormbook.1.123.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991a;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991b;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Berger AJ, Hart AC, Kaplan JM. G alphas-induced neurodegeneration in Caenorhabditis elegans. J Neurosci. 1998;18:2871–2880. doi: 10.1523/JNEUROSCI.18-08-02871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrejo L, Guyant-Maréchal L, Laquerrière A, Vercelletto M, De la Fournière F, Thomas-Antérion C, Verny C, Letournel F, Pasquier F, Vital A, Checler F, Frebourg T, Campion D, Hannequin D. Phenotype associated with APP duplication in five families. Brain. 2006;129:2966–2976. doi: 10.1093/brain/awl237. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- Daniels SA, Ailion M, Thomas JH, Sengupta P. egl-4 acts through a transforming growth factor-beta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn MF. Zinc-ligand interations mediate assembly and stability of the insulin hexamer-a review. BioMetals. 2005;18:295–303. doi: 10.1007/s10534-005-3685-y. [DOI] [PubMed] [Google Scholar]
- Ewald CY, Li C. Understanding the molecular basis of Alzheimer's disease using a Caenorhabditis elegans model system. Brain Struct Funct. 2010;214:263–283. doi: 10.1007/s00429-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald CY, Raps DA, Li C. APL-1, the Alzheimer's amyloid precursor protein in Caenorhabditis elegans, modulates multiple metabolic pathways throughout development. Genetics. 2012;191:493–507. doi: 10.1534/genetics.112.138768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ren M, Feng H, Chen L, Altun ZF, Rubin CS. Neuronal and intestinal protein kinase d isoforms mediate Na+ (salt taste)-induced learning. Sci Signal. 2009;2:ra42. doi: 10.1126/scisignal.2000224. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles AC, Rankin CH. Behavioral and genetic characterization of habituation using Caenorhabditis elegans. Neurobiol Learn Mem. 2009;92:139–146. doi: 10.1016/j.nlm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer's disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hoopes JT, Liu X, Xu X, Demeler B, Folta-Stogniew E, Li C, Ha Y. Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site. J Biol Chem. 2010;285:2165–2173. doi: 10.1074/jbc.M109.018432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O'Connor G, Plasterk R, Li C. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci U S A. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KK, Borchelt DR, Olson K, Johannsdottir R, Kitt C, Yunis W, Xu S, Eckman C, Younkin S, Price D. Age-related CNS disorder and early death in transgenic FVB/N mice overexpressing Alzheimer amyloid precursor proteins. Neuron. 1995;15:1203–1218. doi: 10.1016/0896-6273(95)90107-8. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Müller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Tirosh-Wagner T, Urban AE, Chen XN, Kasowski M, Dai L, Grubert F, Erdman C, Gao MC, Lange K, Sobel EM, Barlow GM, Aylsworth AS, Carpenter NJ, Clark RD, Cohen MY, Doran E, Falik-Zaccai T, Lewin So, Lott IT, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A. 2009;106:12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Hong K, Kinnell M, Chalfie M, Driscoll M. Sequence and transmembrane topology of MEC-4, an ion channel subunit required for mechanotransduction in Caenorhabditis elegans. J Cell Biol. 1996;133:1071–1081. doi: 10.1083/jcb.133.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lee JI, O'Halloran DM, Eastham-Anderson J, Juang BT, Kaye JA, Scott Hamilton O, Lesch B, Goga A, L'Etoile ND. Nuclear entry of a cGMP-dependent kinase converts transient into long-lasting olfactory adaptation. Proc Natl Acad Sci U S A. 2010;107:6016–6021. doi: 10.1073/pnas.1000866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin CH, Tomioka M, Pereira S, Sellings L, Iino Y, van der Kooy D. Insulin signaling plays a dual role in Caenorhabditis elegans memory acquisition and memory retrieval. J Neurosci. 2010;30:8001–8011. doi: 10.1523/JNEUROSCI.4636-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Luchsinger JA, Tang MX, Shea S, Mayeux R. Hyperinsulinemia and risk of Alzheimer disease. Neurology. 2004;63:1187–1192. doi: 10.1212/01.wnl.0000140292.04932.87. [DOI] [PubMed] [Google Scholar]
- Mahoney TR, Liu Q, Itoh T, Luo S, Hadwiger G, Vincent R, Wang ZW, Fukuda M, Nonet ML. Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell. 2006;17:2617–2625. doi: 10.1091/mbc.E05-12-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Esiri MM. The pattern of acquisition of plaques and tangles in the brains of patients under 50 years of age with Down's syndrome. J Neurol Sci. 1989;89:169–179. doi: 10.1016/0022-510x(89)90019-1. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Bessinger K, Hellmann J, Murakami S. Aging-dependent and -independent modulation of associative learning behavior by insulin/insulin-like growth factor-1 signal in Caenorhabditis elegans. J Neurosci. 2005;25:10894–10904. doi: 10.1523/JNEUROSCI.3600-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315:418–425. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML. Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J Neurosci Methods. 1999;89:33–40. doi: 10.1016/s0165-0270(99)00031-x. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ott A, Stolk RP, van Harskamp F, Pols HA, Hofman A, Breteler MM. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology. 1999;53:1937–1942. doi: 10.1212/wnl.53.9.1937. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, Lockery SR. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature. 2001;410:694–698. doi: 10.1038/35070575. [DOI] [PubMed] [Google Scholar]
- Rankin CH, Abrams T, Barry RJ, Bhatnagar S, Clayton DF, Colombo J, Coppola G, Geyer MA, Glanzman DL, Marsland S, McSweeney FK, Wilson DA, Wu CF, Thompson RF. Habituation revisited: an updated and revised description of the behavioral characteristics of habituation. Neurobiol Learn Mem. 2009;92:135–138. doi: 10.1016/j.nlm.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. New York: Cold Spring Harbor Laboratory; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Schupf N, Kapell D, Nightingale B, Rodriguez A, Tycko B, Mayeux R. Earlier onset of Alzheimer's disease in men with Down syndrome. Neurology. 1998;50:991–995. doi: 10.1212/wnl.50.4.991. [DOI] [PubMed] [Google Scholar]
- Simón AM, Schiapparelli L, Salazar-Colocho P, Cuadrado-Tejedor M, Escribano L, López de Maturana R, Del Río J, Pérez-Mediavilla A, Frechilla D. Overexpression of wild-type human APP in mice causes cognitive deficits and pathological features unrelated to Abeta levels. Neurobiol Dis. 2009;33:369–378. doi: 10.1016/j.nbd.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, de la Monte SM. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer's disease–is this type 3 diabetes? J Alzheimers Dis. 2005;7:63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Wen JY, Kumar N, Morrison G, Rambaldini G, Runciman S, Rousseau J, van der Kooy D. Mutations that prevent associative learning in C. elegans. Behav Neurosci. 1997;111:354–368. doi: 10.1037//0735-7044.111.2.354. [DOI] [PubMed] [Google Scholar]
- Wiese M, Antebi A, Zheng H. Intracellular trafficking and synaptic function of APL-1 in Caenorhabditis elegans. PLoS One. 2010;5:e12790. doi: 10.1371/journal.pone.0012790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Sassa T, Kunoh K, Hosono R. A mutant exhibiting abnormal habituation behavior in Caenorhabditis elegans. J Neurogenet. 2002;16:29–44. doi: 10.1080/01677060213105. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hirotsu T, Matsuki M, Butcher RA, Tomioka M, Ishihara T, Clardy J, Kunitomo H, Iino Y. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science. 2010;329:1647–1650. doi: 10.1126/science.1192020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Mullane PC, Periz G, Wang J. TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling. Hum Mol Genet. 2011;20:1952–1965. doi: 10.1093/hmg/ddr076. [DOI] [PMC free article] [PubMed] [Google Scholar]