Abstract
The mammalian neocortex is parcellated into anatomically and functionally distinct areas. The establishment of area-specific neuronal diversity and circuit connectivity enables distinct neocortical regions to control diverse and specialized functional outputs, yet underlying molecular controls remain largely unknown. Here, we identify a central role for the transcriptional regulator Lim-only 4 (Lmo4) in establishing the diversity of neuronal subtypes within rostral mouse motor cortex, where projection neurons have particularly diverse and multi-projection connectivity compared with caudal motor cortex. In rostral motor cortex, we report that both subcerebral projection neurons (SCPN), which send projections away from the cerebrum, and callosal projection neurons (CPN), which send projections to contralateral cortex, express Lmo4, whereas more caudal SCPN and CPN do not. Lmo4-expressing SCPN and CPN populations are comprised of multiple hodologically distinct subtypes. SCPN in rostral layer Va project largely to brainstem, whereas SCPN in layer Vb project largely to spinal cord, and a subset of both rostral SCPN and CPN sends second ipsilateral caudal (backward) projections in addition to primary projections. Without Lmo4 function, the molecular identity of neurons in rostral motor cortex is disrupted and more homogenous, rostral layer Va SCPN aberrantly project to the spinal cord, and many dual-projection SCPN and CPN fail to send a second backward projection. These molecular and hodological disruptions result in greater overall homogeneity of motor cortex output. Together, these results identify Lmo4 as a central developmental control over the diversity of motor cortex projection neuron subpopulations, establishing their area-specific identity and specialized connectivity.
Introduction
Specialized neocortical areas that process motor output and sensory input rely on diverse projection neuron subtypes that are organized both radially into distinct cortical layers and tangentially in topographic patterns (Sur and Rubenstein, 2005; Rash and Grove, 2006). For example, subcerebral projection neurons (SCPN), which reside in layer V and send axons caudal from the cerebrum, and callosal projection neurons (CPN), which reside mostly in layers II/III and V and send axons to the contralateral hemisphere, acquire laminar and shared connectivity features via the influence of transcription factors broadly expressed within each class (Arlotta et al., 2005; Chen et al., 2005; Molyneaux et al., 2005, 2007, 2009; Alcamo et al., 2008; Britanova et al., 2008; Lai et al., 2008). In parallel, spatially restricted controls tangentially parcellate these broad classes into functionally specialized subpopulations through a process of arealization (Weimann et al., 1999; Joshi et al., 2008; Tomassy et al., 2010). This interplay ensures that projection neurons acquire unique connectivity appropriate for their area-specific functions (Stanfield et al., 1982; O'Leary and Stanfield, 1986, 1989; Arlotta et al., 2005; O'Leary and Sahara, 2008). Although molecular controls that specify broad subtype identity have recently begun to be characterized (Molyneaux et al., 2007; MacDonald et al., 2013), much less is known about area-specific controls over specialized subtype identity (Molnar and Cheung, 2006; Rash and Grove, 2006).
The motor cortex contains many anatomically specialized and regionally restricted neuronal subtypes. Motor cortex SCPN in rostromedial and superficial layer V (layer Va) are largely cortico-brainstem motor neurons (CBMN), which send axons to motor-related brainstem targets (Hevner et al., 2003; Brecht et al., 2004b), whereas those that occupy caudolateral or deep layer V (layer Vb) are largely corticospinal motor neurons (CSMN), which send axons to the spinal cord (Wise et al., 1979; Hevner et al., 2003; Arlotta et al., 2005; Molyneaux et al., 2007). Moreover, many rostral motor cortex CPN send a second backward projection (CPN/BPN), whereas their caudal counterparts primarily only send a single contralateral projection (Mitchell and Macklis, 2005). These intracortical backward projecting neurons (BPN) originate from the motor cortex, and target neurons in somatosensory and perirhinal cortices (Miyashita et al., 1994; Veinante and Deschênes, 2003; Mao et al., 2011). Here, for simplicity, we designate BPN specifically as neurons that send ipsilateral projections from motor to somatosensory cortex. Each of these subpopulations contributes to specific motor circuits and integrative motor and cognitive behaviors (Brecht et al., 2004a; Maggiolini et al., 2007, 2008; Mao et al., 2011), yet the underlying developmental controls are unknown.
In theory, candidate molecules might be expected to exhibit sharp regional boundaries of expression. For example, Bhlhb5 expression sharply delineates the caudal motor and somatosensory cortices, and regulates the acquisition of area-specific connectivity (Joshi et al., 2008). Similarly, the LIM-HD related transcriptional regulator Lmo4 sharply delineates frontal motor regions (Bulchand et al., 2003; Arlotta et al., 2005; Sun et al., 2005; Cholfin and Rubenstein, 2007, 2008), and exhibits neuronal subtype-specific expression (Arlotta et al., 2005; Azim et al., 2009).
Given its roles in regulating subtype identity (Lee et al., 2008; Azim et al., 2009; Joshi et al., 2009) and connectivity (Kashani et al., 2006; Huang et al., 2009), we hypothesized that Lmo4 might regulate aspects of rostral motor cortex projection neuron development. We find that Lmo4 is expressed by defined neuronal subpopulations within the motor cortex, in which it helps establish appropriate diversity and specificity of SCPN and CPN identities and connectivities. This work identifies Lmo4 as a critical developmental control over neocortical motor circuits.
Materials and Methods
Animals.
Immunocytochemical and anatomical characterizations of Lmo4 expression and projection neuron distributions were performed using C57BL/6 mice (Charles River Laboratories). Lmo4LacZ and Lmo4flox mice were generously provided by Dr. Lin Gan (University of Rochester, Rochester NY) (Deng et al., 2009). Whole-mount β-galactosidase detections of Lmo4 expression were performed using Lmo4LacZ/+ knock-in mice (Deng et al., 2009).
Mice with conditional forebrain deletion of Lmo4 were obtained by first crossing Lmo4flox/flox females with Emx1Cre/+ males (Jackson Laboratories). F1 generations were crossed with each other to obtain suitable breeding pairs. Emx1Cre/+Lmo4flox/+ or Emx1Cre/+Lmo4flox/flox males were crossed with Lmo4flox/+ or Lmo4flox/flox females to obtain Emx1Cre/+Lmo4flox/flox conditional knock-out mice (Lmo4-cKO). Emx1cre/+Lmo4+/+ and Emx1+/+Lmo4flox/flox mice were phenotypically indistinguishable from each other, and were used as controls (WT). Analyses of Lmo4-cKO and WT mice were performed on a mixed C57BL/6 and S129S6 background. Comparisons were performed on littermates of equal mixed strain background. All animal studies were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee, and performed in accordance with institutional guidelines.
β-Galactosidase whole-mount staining.
Brains were perfused with 4% paraformaldehyde (PFA) and postfixed for 2 h at 4°C. Brains were rinsed 3 times with PBS, then incubated at 37°C for 24–36 h with 0.1% X-Gal, 2 mm MgCl2, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, and 0.2% NP40 in PBS. Brains were postfixed overnight to stop the color reaction.
Immunocytochemistry and antibodies.
Immunocytochemistry was performed as previously described (Arlotta et al., 2005). Postnatal pups were transcardially perfused with PBS, then with 4% PFA, dissected, and postfixed in 4% PFA overnight. Brains were sectioned at 50 μm on a vibrating microtome (Leica). Floating sections were blocked with 4% goat or donkey serum, 0.3% BSA, 0.3% Triton X-100 (Sigma), and 0.025% sodium azide in PBS for 30 min. Primary antibodies were diluted in appropriate blocking solution, and incubated with sections overnight. The following day, sections were rinsed 3 times with PBS and incubated with appropriate secondary antibodies diluted in blocking solution for 3 h at room temperature. Sections were again rinsed 3 times with PBS and mounted using Fluoromount (Southern Biotechnology) for image acquisition.
Antibody dilutions were as follows: goat α-Lmo4 (1:200, Santa Cruz Biotechnology); goat α-Bhlhb5 (1:300, Santa Cruz Biotechnology); goat α-Lhx2 (1:200, Santa Cruz Biotechnology); rabbit α-Cux1 (1:200, Santa Cruz Biotechnology). Appropriate secondary antibodies from the Invitrogen Alexa Series were used (1:500, Invitrogen).
In situ hybridization.
Nonradioactive in situ hybridization was performed on 30-μm-thick sections. Brains were perfused and stored in 4% PFA for at least 3 d. Before sectioning, brains were equilibrated in 30% sucrose and 4% PFA in PBS. Brains were embedded in Optimum Cutting Temperature Formulation (Tissue-Tek), frozen, cut at 30 μm thickness on a CM3050 cryostat (Leica), and wet-mounted onto microscope slides. Sections were dried for 2 h at room temperature, fixed in 4% PFA for 15 min, and then rinsed 3 times in RNase-free PBS. The slides were hybridized overnight at 62°C to 70°C with DIG-labeled RNA probes in 50% Formamide (Sigma), 5× SSC (Promega), and 1% SDS (Sigma). The following day, slides were given three 45 min washes in 50% Formamide, 2× SSC, and 1% SDS at hybridization temperature. Slides were then rinsed 1 time in 2× SSC, and 1 time in 0.2× SSC for 20 min each, and incubated with an α-DIG-AP Fab fragment antibody (Roche) for 2–4 h at room temperature or overnight at 4°C. To develop the color, slides were first rinsed 4 times in 25 mm Tris HCl pH 7.5, 0.02% KCl, 0.8% NaCl, 0.5% Tween 20. The color reaction was performed at room temperature in 100 mm NaCl, 100 mm Tris HCl, pH 9.5, 50 mm MgCl2, 1% Tween 20, and NBT/BCIP (Roche). Reactions were closely monitored, and stopped with 4% formalin, and then 10 mm Tris-EDTA when background staining first became apparent. Primer sequences are as follows: Rorβ: F, 5′-GGAGATGCCACTCATCAGC-3′; R, 5′-CACGTCCTGGTTGGAGTACC-3′. Kitl: F, 5′-TGCATGGAAGAAAACGCAC-3′; R, 5′-GCTTACATTTAGGCTGCTC-3′. Lhx2: F, 5′-GATCTCGCCTGGAAACAGAG-3′; R, 5′-AAACAAGAAAGCGACCGTTG-3′. Igfbp4: F, 5′-GACGAAGCCATCCACTGC-3′; R, 5′-CTGGTGGCAGTCCAGCTC-3′. Igfbp5: F, 5′-GACCTAACCAGGACCTATTCTCC-3′; R, 5′-GACGAGACCTCTTTCCCTTCTAC-3′. Lmo3 and Clim1 probes were generated using primers, as previously described (Bach et al., 1997; Bulchand et al., 2003).
Retrograde anatomical tracing.
Procedures for retrograde labeling of CSMN and SCPN (Arlotta et al., 2005), and CPN and BPN (Mitchell and Macklis, 2005) were previously described. Briefly, for retrograde labeling of CSMN, corticospinal tracts (CST) of deeply anesthetized postnatal day 2 (P2) pups were visualized using a Vevo 770 ultrasound backscatter microscopy system (Visual Sonics) at cervical vertebral level 1 (C1) to C2. Six 23 nl injections of cholera toxin subunit-B 647 (CTB647, 2 μg/μl) were deposited bilaterally into the CST using a Drummond Nanoject II. Mice were killed 5 or 7 d after injection. For retrograde labeling of CBMN, descending subcerebral projection tracts of deeply anesthetized P3 pups were visualized using a Vevo 770 ultrasound backscatter microscopy system (Visual Sonics) at the midbrain-hindbrain junction. Five 46 nl injections of CTB555 (2 μg/μl) were deposited bilaterally into the tracts. Mice were killed 4 or 6 d after injection (see Figs. 3 and 4 for schematics).
Figure 3.
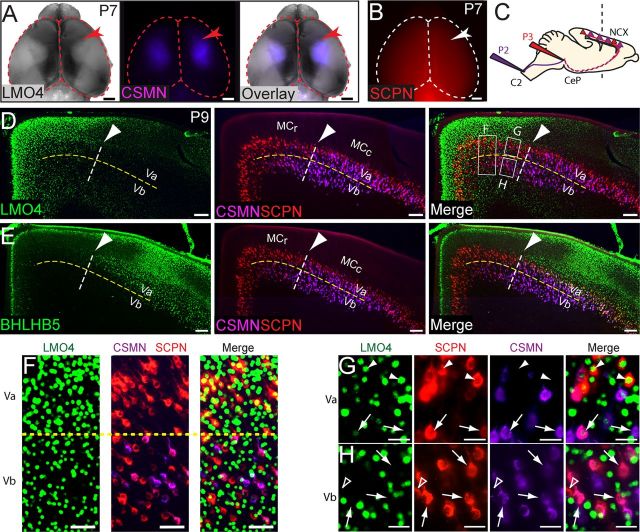
Lmo4 is preferentially expressed by subcerebral projection neurons that terminate in the brainstem rather than the spinal cord. A, Retrogradely labeled corticospinal motor neurons (CSMN) are sparser in the rostral motor cortex (MCr) than in the caudal motor cortex (MCc), marked by the expression of Lmo4 (red arrowhead marks MCc/MCr boundary). B, Retrogradely labeled subcerebral projection neurons (SCPN) are found throughout the entire neocortex (red), including in the MCr (white arrowhead marks MCc/MCr boundary). C, Schematic of labeling paradigm to label CSMN and cortico-brainstem motor neuron (CBMN) subpopulations. D, E, In the MCr, CSMN are excluded from layer Va, whereas in the MCc they are not (yellow line marks layer Va/Vb boundary). The boundary between distinct MCr and MCc SCPN populations precisely coincides with the Lmo4/Bhlhb5 boundary (white dashed line, arrowhead). F, In the MCr, layer Vb contains both CSMN (colabeled purple and red, Vb) and CBMN (red only, Vb), whereas layer Va contains mostly CBMN (red only, Va). Lmo4 is expressed in CBMN in layer Va (yellow in merge panel), but is expressed in few, if any, SCPN in layer Vb. G, High magnification reveals that in layer Va, CBMN (red only) express Lmo4 (green) at high levels (arrowheads), whereas CSMN (colabeled purple and red) express Lmo4 at low levels or not at all (arrows). H, In layer Vb, neither CSMN (colabeled purple and red, arrows) nor CBMN (red only, empty arrowhead) express Lmo4 (green). C2, Level of second cervical vertebra; P, postnatal day; NCX, neocortex; CeP, cerebral peduncle; Va, layer Va; Vb, layer Vb. Scale bars: A, B, 1 mm; D, E, 160 μm; F, 60 μm; G, H, 32 μm.
Figure 4.
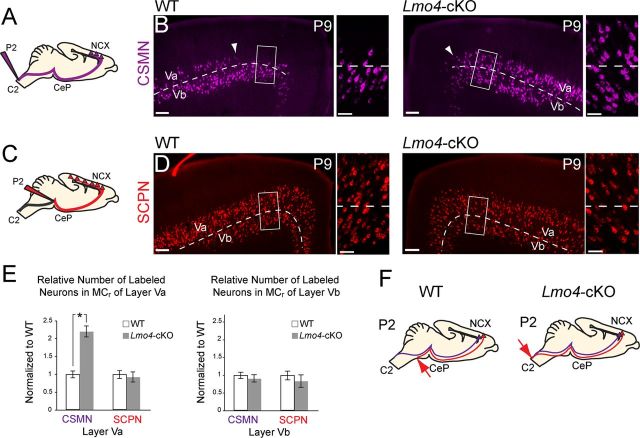
Lmo4 is required for cortico-brainstem motor neuron development in the rostral motor cortex. A, Schematic shows labeling paradigm to trace spinal cord projections from the rostral motor cortex (MCr). B, Corticospinal motor neurons (CSMN) are absent from medial layer Va in the MCr (WT, arrowhead shows medial extent). Neurons in medial layer Va aberrantly project to the spinal cord in Lmo4-cKO mice (Lmo4-cKO, arrowhead shows medial extent). Boxes denote regions of high magnification of layer Va and Vb MCr. C, Schematic shows labeling paradigm to trace all subcerebral projection neurons (SCPN) in the MCr. D, Equal densities of SCPN are labeled in WT and Lmo4-cKO animals. Boxes denote regions of high magnification of layer Va and Vb MCr. Boxed regions do not denote ROIs used for quantification. E, Quantification of CSMN and SCPN in layer Va and Vb. There is a 2.2-fold increase in the density of CSMN, but not SCPN, in layer Va of the Lmo4-cKO compared with WT (p < 0.005), whereas there is no change in layer Vb (n = 3 WT, n = 3 Lmo4-cKO). Measurements from the pia and midline were used to ensure that ROIs accurately and reproducibly delineate respective sublaminae within MCr. All quantifications are normalized to WT values. F, Summary of results. C2, level of second cervical vertebra; CeP, Cerebral peduncle; NCX, neocortex. Scale bars: B, D (low magnification), 160 μm; B, D (high magnification), 60 μm.
Dual retrograde labeling of CPN and BPN was performed with CTB555 and CTB647 (2 μg/μl), respectively. P7 pups were deeply anesthetized and secured in a neonatal-adapted Cunningham stereotaxic frame (Stoelting). For each injection site, five 4.6 nl injections were made at 250 μm depth, two 4.6 nl injections were made at 200 μm depth, one 4.6 nl injection was made at 150 μm depth, and two 4.6 nl injections were made at 100 μm depth. For CPN labeling, a 5 × 2 grid (∼3.5 mm × 0.5 mm) was defined along the midline. Five evenly spaced injections were made directly adjacent to the midline in the right hemisphere. The most caudal injection site was 1 mm caudal to bregma, and the most rostral injection-site was ∼1 mm caudal to the olfactory bulbs. A second column of injections was made 0.5 mm lateral to the first column. For BPN labeling, a 3 × 4 grid was defined above the presumptive left somatosensory and visual cortices. We injected broadly over the caudal cortex to ensure ample labeling of BPN that target somatosensory regions. To our knowledge, BPN are not known to connect motor to visual areas. Our labeling approach does not identify BPN that project from the motor to perirhinal cortex. Thus, we use BPN to specifically refer to neurons with an ipsilateral motor to somatosensory projection. The most caudomedial injection site was directly adjacent to lambda. The most rostrolateral injection site was 1.5 mm rostral to lambda and 2.5 mm lateral to the midline. All other injections were evenly spaced to complete the 3 × 4 grid. Mice were perfused 2 d after injection (see Figs. 5 and 6 for schematics).
Figure 5.
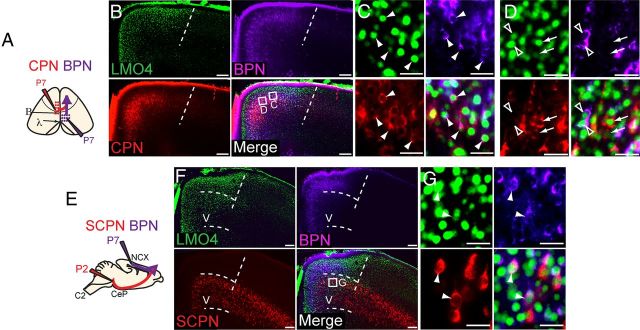
Lmo4 is expressed by multiple backward projecting neuronal subtypes in the rostral motor cortex. A, Labeling paradigm used to simultaneously trace callosal projection neurons (CPN) and backward projecting neurons (BPN). B, Many BPN and CPN are found in the area of high Lmo4 expression (medial to dashed line). C, Dual-projecting CPN/BPN (colabeled purple and red) express high levels of Lmo4 (arrowheads). D, Pure CPN (arrows, red only) and pure BPN (open arrowheads, purple only) express Lmo4. We cannot exclude the possibility that some pure CPN and pure BPN are dual-projecting neurons that did not get labeled by the second tracer dye injection. E, Schematic of paradigm used to simultaneously trace SCPN and BPN. F, Many BPN and SCPN are found in the area of high Lmo4 expression in layer V (medial to straight dashed line; curved dashed lines delineate layer V). G, Dual-projecting SCPN/BPN express Lmo4 (arrowheads). B, Bregma; λ, lambda; C2, second cervical vertebra; CeP, cerebral peduncle; NCX, neocortex; V, cortical layer V. Scale bars: B, F, 160 μm; C, D, G, 32 μm.
Figure 6.
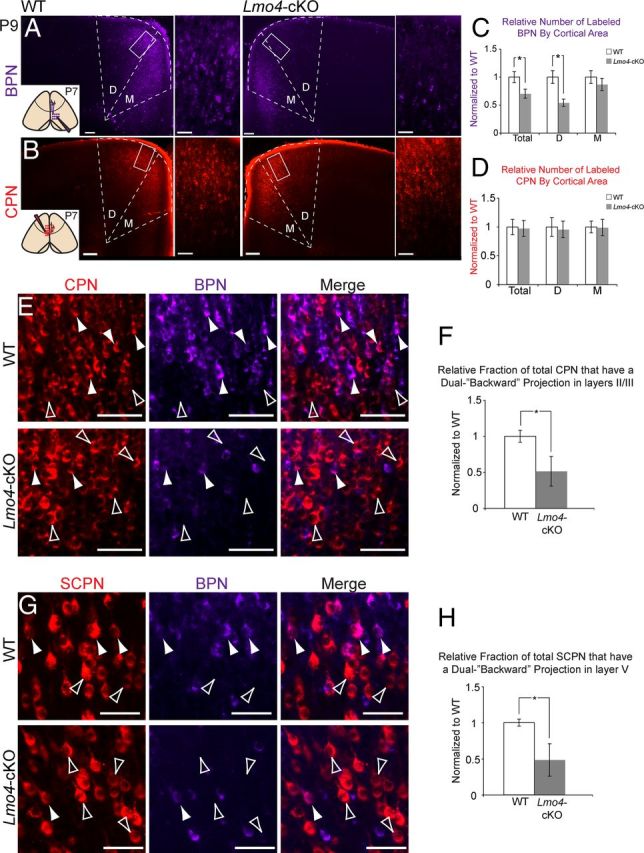
Lmo4 regulates the development of dual backward projecting callosal projection neurons and subcerebral projection neurons in the rostral motor cortex. A, Retrograde tracing from the ipsilateral caudal cortex (inset) at P7 labels backward projecting neurons (BPN). High densities of BPN are labeled in the dorsal (D) and medial (M) rostral motor cortex (MCr) of WT mice. In Lmo4-cKO mice, there is a striking reduction in BPN in the dorsal (D), but not medial (M), MCr. Boxes denote magnified regions of dorsal subdivisions. B, Callosal projection neurons (CPN) in the MCr are labeled by retrograde tracing from the homotopic region of the contralateral cortex (inset). In both WT and Lmo4-cKO mice, high densities of CPN are labeled throughout the entire MCr. C, Quantification of BPN density in WT and Lmo4-cKO MCr. There is a 30% reduction in BPN in the Lmo4-cKO mice (p < 0.05, Total). There is a 46% reduction in the density of BPN in the dorsal MCr (p < 0.01, D), whereas there is no significant reduction in the density of BPN in the medial MCr (M) (n = 5 WT, n = 5 Lmo4-cKO). D, Quantification of CPN density in WT and Lmo4-cKO MCr. There is no difference in the overall density of CPN between WT and Lmo4-cKO mice (n = 5 WT, n = 5 Lmo4-cKO). E, CPN/BPN (arrowheads) and pure CPN (open arrowheads) in layers II/III of the dorsal subdivision of WT and Lmo4-cKO MCr. F, Quantification of CPN/BPN density (fraction of CPN that send a dual backward projection) reveals a 49% reduction in Lmo4-cKO dorsal MCr (n = 5 WT, n = 4 Lmo4-cKO; p < 0.05). G, Dual projecting subcerebral projection neuron (SCPN)/BPN (arrowheads) and pure SCPN (open arrowheads) in dorsal subdivision of WT and Lmo4-cKO MCr. H, Quantification of SCPN/BPN density (fraction of SCPN that send a dual backward projection) reveals a 51% reduction in Lmo4-cKO dorsal MCr (n = 3 WT, n = 3 Lmo4-cKO; p < 0.05). All quantifications are normalized to WT values. Scale bars: A, B (low magnification), 160 μm; A, B (high magnification), E, G, 60 μm.
To quantify retrograde labeling experiments, we took advantage of the highly stereotyped size of cortical regions and layers across mice, using Adobe Photoshop to generate grids that define regions of interest (ROIs) for each tracing paradigm. Because cortical and laminar thickness is unaffected in Lmo4-cKO animals (data not shown), the same grids were used between genotypes. The corpus callosum and anterior commissure were used as landmarks to accurately match the rostrocaudal level of analyzed sections.
ROIs were defined as follows. CSMN and SCPN ROI (see Fig. 4E): to restrict quantification to within the MCr, the lateral boundary of the ROI was placed 800 μm lateral to the midline, significantly more medial than the MCc/MCr boundary. The division between layer Va and layer Vb was designated as 640 μm below the pia, and was anatomically defined as the dorsal-ventral boundary in layer V that separates CSMN-dense and CSMN-sparse sublaminae. ROIs in layer Va and layer Vb were 160 μm × 320 μm. Total CPN and BPN ROI (see Fig. 6A–D): ROIs outlined nearly the entire dorsal and medial subdivisions of the MCr. The ends of the long axes of grids were placed at the apex of the corpus callosum and the apex of the pia (see Fig. 6A,B). CPN/BPN ROI (see Fig. 6E,F): ROI was a 180 μm × 180 μm box placed 60 μm below the pia, in the dorsal subdivision of MCr, within layers II/III. SCPN/BPN ROI (see Fig. 6G,H): ROI was a 224 μm × 224 μm box placed at the apex of layer V.
For quantifications depicted in figure histograms, WT and Lmo4-cKO values are represented as fractions of WT values, such that WT is 1. For CSMN, SCPN (see Fig. 4E), total BPN (see Fig. 6C), and total CPN (see Fig. 6D), values were obtained by counting the total number of labeled neurons within each ROI. For CPN/BPN (see Fig. 6F) and SCPN/BPN (see Fig. 6H), values were obtained by dividing the number of double-labeled neurons by total CPN or total SCPN, respectively, within each ROI.
Results
Lmo4 expression delineates the rostral motor cortex
We used heterozygous mice that express β-galactosidase (βgal) under the control of the Lmo4 promoter (Lmo4LacZ/+) to characterize Lmo4 area-specific expression during cortical development (Deng et al., 2009). Low levels of Lmo4 expression have been detected in the developing forebrain as early as embryonic day 11.5 (E11.5) (Asprer et al., 2011). We began our whole-mount expression analysis at E12.5, when Lmo4 is expressed in the neocortical preplate, and the earliest wave of postmitotic neurons is being generated (Huang et al., 2009). At this age, Lmo4 expression is weak and localized in the presumptive sensory-motor area (Fig. 1A). By E15.5, during mid corticogenesis, Lmo4 is expressed along the cortical midline, with graded expression toward lateral regions (Fig. 1B). By E17.5, as the last wave of cortical neurons is generated, Lmo4 expression transitioned from a graded pattern to sharply defined expression in a subregion of the presumptive motor area in the rostral neocortex (Fig. 1C, arrowhead). It also sharply delineates the more caudal presumptive visual area, albeit with lower level expression (Fig. 1C, arrow). This expression pattern is maintained through P0, with expression intensity increasing slightly in the motor region (Fig. 1D, arrowhead) (Cholfin and Rubenstein, 2007, 2008). By P4, the areal specificity of Lmo4 expression becomes more sharply defined; it is excluded from the presumptive sensorimotor cortex (Fig. 1E, S1), and sharply delineates medial subsections of the presumptive motor and visual areas (Fig. 1E, M1 and V1). By P7, Lmo4 acquires uniform and strong expression throughout the entire rostromedial portion of the motor cortex (Fig. 1F, M1), intermediate level expression in the primary visual area (Fig. 1F, V1), high expression medial to the primary visual area, and remains absent from the primary somatosensory area (Fig. 1F, S1).
Figure 1.
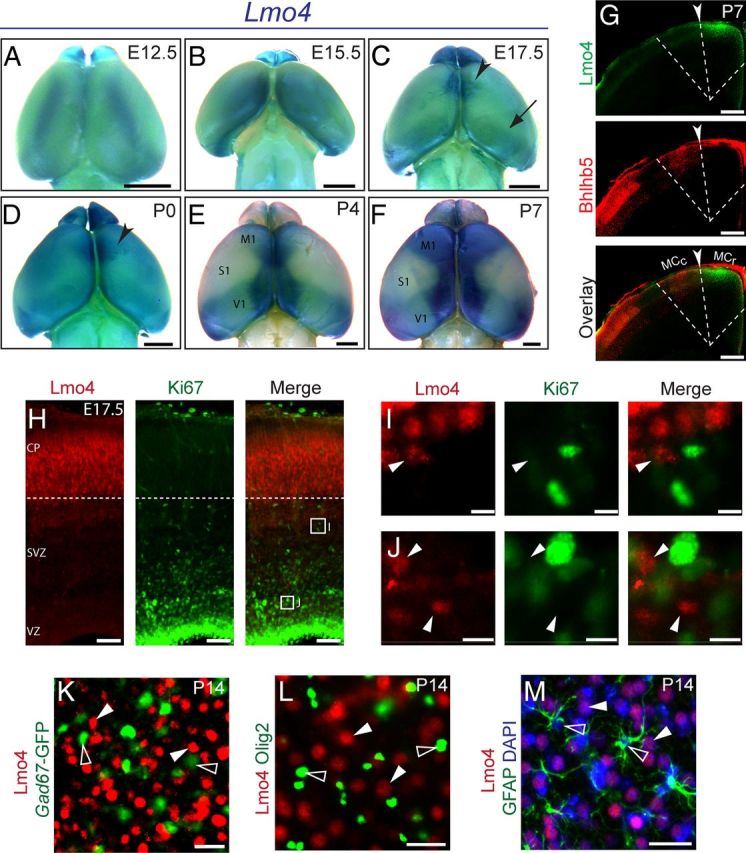
Lmo4 is expressed by postmitotic neurons in the rostral motor cortex. A–F, βgal (visualized by whole-mount X-gal reactivity) expressed from the Lmo4 locus reveals the temporal course of Lmo4 expression. Lmo4 progressively acquires sharp areal boundaries (arrowheads) along the midline and in motor cortex, while it is excluded from somatosensory cortex (S1). G, Overlay of Lmo4 and Bhlhb5 immunofluorescence on serial sections at P7 reveals Lmo4 expression (green) sharply delineates the rostral motor cortex (MCr), and is excluded from the Bhlhb5+ caudal motor cortex (MCc) (G, arrowhead and dashed line). H, Lmo4 (red) is expressed at high levels by postmitotic neurons in the cortical plate. I, J, Cells expressing low levels of Lmo4 in the SVZ (I, arrowhead) and ventricular zone (VZ) (J, arrowheads) do not express the mitotic marker Ki67. K, GAD67-GFP+ cortical interneurons (green, open arrowheads) do not express Lmo4 (red, closed arrowheads). L, Oligo2+ cortical oligodendrocytes (green, open arrowheads) do not express Lmo4 (red, closed arrowheads). M, GFAP+ cortical astrocytes (green, empty arrowheads) do not express Lmo4 (red, closed arrowheads). CP, Cortical plate; M1, Primary motor cortex; S1, primary somatosensory cortex; SVZ, subventricular zone; V1, primary visual cortex; VZ, ventricular zone. Scale bars: A–F, 1 mm; G, 320 μm; H, 60 μm; I, J, 8 μm; K–M, 16 μm.
Notably, as Lmo4 expression is refined postnatally in the motor cortex, a strikingly complimentary Bhlhb5 expression pattern emerges; Bhlhb5 is excluded from the rostral motor cortex (MCr) but highly expressed in the caudal motor cortex (MCc), where it controls area-specific neuronal subtype development (Joshi et al., 2008). We performed immunocytochemistry on coronal sections to more precisely localize Lmo4 and Bhlhb5 cortical expression. It is possible to visualize both the MCc and the MCr in the same coronal plane because the boundary between them is obliquely oriented in a caudomedial to rostrolateral direction (Joshi et al., 2008). Because antibodies to Bhlhb5 and Lmo4 were raised in the same animal, we overlaid serial sections. This analysis indicates that medial Lmo4 expression sharply abuts lateral Bhlhb5 expression in the motor cortex (Fig. 1G, arrowhead and dashed line). Thus, Lmo4 expression marks the MCr, which contains regions controlling facial, neck, and forelimb movement, and Lmo4 is excluded from the MCc, which contains regions controlling trunk and hindlimb movement.
We next characterized Lmo4 cell type-specific expression during cortical development using immunocytochemistry. Lmo4 has been reported to be present in Neurogenin 2 (Ngn2)-expressing mitotic progenitors at E11.5 and E14.5 (Asprer et al., 2011). During late corticogenesis at E17.5, our analysis indicates that Lmo4 is expressed in the developing cortical plate (CP) (Fig. 1H) (Azim et al., 2009; Huang et al., 2009). Though Lmo4 mRNA (Huang et al., 2009; Pinto et al., 2009) and low levels of Lmo4 protein can be detected in the subventricular zone (SVZ) (Fig. 1H–J), Lmo4+ cells do not express the mitotic protein Ki67+ in the SVZ or ventricular zone (VZ) at E17.5 (Fig. 1I,J, arrowheads), agreeing with previous results (Azim et al., 2009). Therefore, at least during later stages of cortical development, Lmo4 expression is restricted to postmitotic populations. Lmo4 expression is not detected in Gad67+ inhibitory interneurons (Fig. 1K, open arrowheads), Olig2+ oligodendrocytes (Fig. 1L, open arrowheads), or GFAP+ astrocytes (Fig. 1M, open arrowheads), indicating that Lmo4 expression is restricted to pallium-derived excitatory cortical projection neurons.
The establishment of the rostral motor cortex boundary does not require Lmo4 function
Given the restricted areal and subtype-specific expression of Lmo4, we hypothesized that: (1) Lmo4 might establish the position of the boundary between the MCr and MCc, and/or (2) Lmo4 might regulate aspects of neuronal subtype identity within the MCr. To test these hypotheses, we used a conditional loss-of-function approach. Because Lmo4LacZ/LacZ mutant mice die during mid-embryonic development from failed neural tube closure (Hahm et al., 2004; Tse et al., 2004; Lee et al., 2005), we bred Emx1Cre/+ Lmo4flox/flox mice with Lmo4flox/flox and Lmo4flox/+ females (Deng et al., 2009) to generate progeny with conditional loss of Lmo4 function exclusively in the dorsal pallium (Emx1Cre/+ Lmo4flox/flox; Lmo4-cKO). We used Emx1Cre/+ Lmo4+/+ and Emx1+/+ Lmo4flox/flox mice as WT controls because these mice were phenotypically indistinguishable from each other.
To assess whether Lmo4 function is necessary to establish the boundary between the MCr and MCc, we performed immunocytochemistry and in situ hybridization for genes that are specifically expressed in the MCc and somatosensory areas, but excluded from the MCr. We performed this analysis at P7, when areal boundaries are well established. If Lmo4 regulates the position of this boundary, then the expression of these genes would be predicted to change.
We first examined Bhlhb5 in the Lmo4-cKO cortex. Bhlhb5 expression is sharply excluded from the MCr (Figs. 1G, 2A) (Joshi et al., 2008). In Lmo4-cKO brains, the medial boundary of Bhlhb5 expression is maintained (Fig. 2A, arrowheads), strongly suggesting that Lmo4 is not required to maintain the molecular boundary between the MCr and MCc. We next examined genes known to be expressed in the somatosensory cortex, but not in motor cortex. Cux1 expression is high in the somatosensory cortex, but is sharply excluded from frontal cortical regions, where Lmo4 expression is high (Ferrere et al., 2006). Cux1 maintains this sharp boundary in layers IV and II/III in Lmo4-cKO cortices (Fig. 2B, arrowheads). Lmo3, a Lim-only transcriptional regulator expressed in somatosensory cortical regions (Cholfin and Rubenstein, 2007, 2008), maintains normal expression in Lmo4-cKO mice (Fig. 2C, arrowheads). Similarly, expression of the orphan nuclear receptor Rorβ, which is normally expressed in layer IV of the somatosensory area (Nakagawa and O'Leary, 2003), is unaffected in the Lmo4-cKO cortex (Fig. 2D, arrowheads). Finally, superficial layer Kitl expression is excluded from the medial cortex (Cholfin and Rubenstein, 2007, 2008), and this sharp boundary is maintained with loss of Lmo4 function (Fig. 2E, arrowheads). Together, these analyses indicate that cardinal features of gene expression of the MCc and somatosensory regions are unchanged in Lmo4-cKO mice. Thus, Lmo4 is not required to establish or maintain the molecular MCr/MCc boundary.
Figure 2.
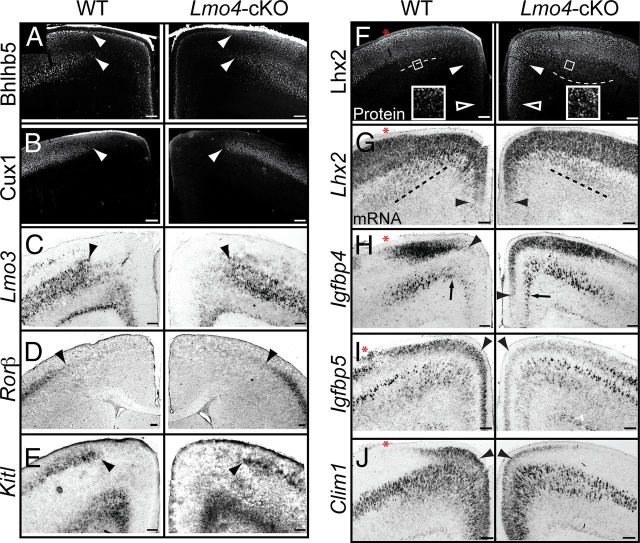
Lmo4 regulates molecular identities within the rostral motor cortex. A, Expression of Bhlhb5 (visualized by immunofluorescence) at the border of the caudal motor cortex (MCc) is unchanged in Lmo4-cKO cortex (arrowheads). B–E, Expression of Cux1 (visualized by immunofluorescence), Lmo3, Rorβ, and Kitl (visualized by in situ hybridization) at the border between somatosensory and motor cortex is unchanged in Lmo4-cKO mice (arrowheads). F, Lhx2 protein is ectopically expressed in the medial and ventral subdivisions of the rostral motor cortex (MCr) (solid and open arrowheads), and in layer V (dashed lines and insets) in Lmo4-cKO cortex. G, Lhx2 mRNA expression is unaffected in the dorsal (dashed lines) and medial (arrowheads) subdivisions of the MCr in Lmo4-cKO mice. Dashed lines in F and G represent the ventral boundary of Lhx2 expression in layer V. H, Igfbp4 expression, normally confined to the dorsal subdivision of the MCr, is ectopically expressed in the medial subdivision in layers II/III (arrowheads) and V (arrows) in Lmo4-cKO cortex. I, J, The expression of Igfbp5 and Clim1 is significantly reduced in layers II/III of the MCr in Lmo4-cKO cortex (arrowheads). Clim1 expression is preferentially affected in the dorsal MCr. Red asterisks denote the MCr/MCc border. In I, a more rostral section, in which the MCr extends further laterally, is shown to more clearly illustrate the reduction of Igfbp5 expression in layers II/III of the MCr. More caudally, as the MCc becomes more prominent, Igfbp5 expression is weaker in layers II/III of WT MCr. n ≥ 3 for all conditions. Scale bars, 160 μm.
Lmo4 regulates molecular identities within the rostral motor cortex
To test the alternative hypothesis that Lmo4 regulates aspects of subtype identity within the MCr, we used in situ hybridization and immunocytochemistry to analyze the expression of genes that are expressed within the MCr. We examined genes with either uniform expression throughout the MCr or with differential expression between its dorsal and medial subdivisions.
Lhx2 is a LIM-HD transcription factor that is critical for the organization of the dorsal telencephalon and cerebral cortex (Bulchand et al., 2001; Monuki et al., 2001; Mangale et al., 2008; Chou et al., 2009). At P7, Lhx2 protein exhibits a lateral-high to medial-low expression pattern in layers II/III. It is expressed at high levels in WT dorsal MCr (Fig. 2F, dashed line), at lower levels in WT medial MCr (Fig. 2F, solid arrowhead), and is absent from the most ventral part of WT medial MCr (Fig. 2F, open arrowhead). Lhx2 expression is also high more laterally in the MCc. In contrast to WT cortex, in Lmo4-cKO cortex, Lhx2 protein is expressed highly throughout the medial and ventral MCr (Fig. 2F, open and solid arrowheads). In the dorsal subdivision, Lhx2 protein is normally confined to layers II/III, with only minor expression in the most superficial portion of layer V. In Lmo4-cKO cortex, many cells in layer V ectopically express Lhx2 (Fig. 2F, dashed lines and insets). Thus, in the absence of Lmo4 function, Lhx2 is highly expressed throughout the entire MCr, and it fails to acquire its normal graded expression pattern in layers II/III. Lhx2 expression is not perturbed laterally, in the MCc, of Lmo4-cKO mice. Interestingly, Lhx2 mRNA expression is equivalent in the dorsal and medial subdivisions of WT and Lmo4-cKO MCr (Fig. 2G, arrowheads and dashed line). This distinction between Lhx2 mRNA and Lhx2 protein in WT but not in Lmo4-cKO motor cortex suggests that Lmo4 imparts posttranscriptional regulation on the translation or stability of Lhx2 protein.
Igfbp4 regulates both Wnt and IGF signaling pathways (Ning et al., 2008; Zhu et al., 2008), and is expressed in the dorsal, but not medial, subdivision of the MCr in layers V and II/III (Tomassy et al., 2010). Igfbp4 is expressed by both CSMN and CPN (Arlotta et al., 2005). In the Lmo4-cKO cortex, Igfbp4 is ectopically expressed in the medial subdivision of both layers V (Fig. 2H, arrows) and II/III (Fig. 2H, arrowheads). Moreover, because Igfbp4 expression invades medial territories but not the more lateral MCc, these data further indicate that Lmo4 is not required to establish or maintain the MCr/MCc boundary.
We next examined the expression of Igfbp5 and Clim1, which are normally present throughout the entire extent of layers II/III of the MCr (Fig. 2I,J). In the Lmo4-cKO cortex, expression of Igfbp5, a gene that modulates IGF signaling (Pera et al., 2001; Salih et al., 2004), is essentially absent from layers II/III in the MCr (Fig. 2I, arrowheads). As expected, Igfbp5 expression is unchanged in the more lateral MCc, where Lmo4 is not normally expressed. Clim1, a transcriptional cofactor of LIM-domain containing proteins that can form a protein complex with Lmo4 (Kashani et al., 2006), shows significantly reduced expression in layers II/III of the dorsal, but not medial, subdivision of the MCr (Fig. 2J, arrowheads).
These data reveal both ectopic and reduced molecular expression in the MCr in the absence of Lmo4 function. Together, our findings indicate that Lmo4 regulates the acquisition of appropriate molecular identities specifically within the MCr.
Lmo4 establishes the diversity of subcerebral projection neuron subtypes in layer V of the rostral motor cortex
Abnormal molecular expression in the MCr in the absence of Lmo4 function suggests that projection neuron identity and connectivity in MCr might be disrupted. Therefore, we investigated what types of projection neurons in the MCr express Lmo4. We first focused on SCPN subtypes; SCPN in the MCr include CSMN and CBMN, all of which project caudal from the cerebrum. Overlay of whole-mount Lmo4 expression and retrogradely labeled CSMN, via injection of cholera toxin subunit-B (CTB) into the cervical spinal cord, reveals that most CSMN are excluded from the MCr (Fig. 3A, arrowheads; left panel reproduced from Fig. 1). In contrast, retrograde labeling of all SCPN by injection into the pyramidal tract at the level of cerebral peduncles, reveals that SCPN are evenly distributed across the entire cortex (Fig. 3B), and thus, many SCPN that project to the brainstem, but not to the spinal cord, are located in medial cortical areas coincident with high levels of Lmo4 expression.
Because CSMN are sparse in the most medial portion of the motor cortex (Fig. 3A), we investigated whether the Lmo4/Bhlhb5 molecular boundary between MCr and MCc delineates the boundary between CSMN-abundant and CSMN-sparse regions of motor cortex. We first developed a labeling approach to distinguish CSMN from CBMN. We define CBMN as any cortico-brainstem projecting neuron residing in the motor cortex (Molyneaux et al., 2007). By injecting CTB647 into the cervical spinal cord at P2, we labeled CSMN at early stages of spinal cord innervation (Fig. 3C, purple). We then injected CTB555 at P3 at the level of the cerebral peduncles, near the midbrain/hindbrain junction, to label the broader class of SCPN (Fig. 3C, red), including CSMN and CBMN. Using ultrasound guidance, we preferentially injected the pyramidal tract, although dye was also likely deposited into neighboring brainstem regions. This approach labels CSMN with both CTB647 and CTB555, whereas brainstem-projecting neurons are labeled only by CTB555.
These analyses indicate that the anatomical composition of SCPN populations changes precisely at the Lmo4/Bhlhb5 boundary (Fig. 3D,E, white dashed line). Medial to this boundary, in the MCr, CSMN (purple) are rarely found in layer Va (Fig. 3D–F, Va, above yellow dashed line), while they are common in layer Vb (Fig. 3D–F, Vb, below yellow dashed line). Lateral to the boundary, in the MCc, CSMN populate both layer Va and Vb (Fig. 3D,E, right of white dashed line). The anatomical distinction between layer Va and layer Vb likely underlies electrophysiological and functional distinctions between neurons in these sublaminae (Hevner et al., 2003; Anderson et al., 2010; Mao et al., 2011).
Previous reports have found that Lmo4 is progressively excluded from CSMN as they develop (Arlotta et al., 2005; Azim et al., 2009). Similarly, we find that CSMN in MCr do not express Lmo4, or that Lmo4 is expressed at very low levels by the few CSMN in layer Va (Fig. 3F, cells colabeled purple and red, G, arrows) or the abundant CSMN in layer Vb (Fig. 3F, cells colabeled purple and red, H, arrows). However, Lmo4 is highly expressed by CBMN in layer Va of MCr (Fig. 3F, yellow cells in merged panel, G, arrowheads). Interestingly, Lmo4 expression is largely excluded from CBMN in layer Vb of MCr (Fig. 3H, open arrowhead, red only).
Given that Lmo4 is selectively expressed by layer Va CBMN, but not by CSMN, and that Lmo4 acquires its sharp expression boundary in the medial motor cortex between E15.5 and P0 (Fig. 1B–D), at the same time that axons of SCPN in this area are beginning to reach their brainstem or spinal cord targets, we hypothesized that Lmo4 might control the developmental bifurcation between CBMN and CSMN in the MCr. Retrograde labeling of CSMN via CTB injection into the cervical spinal cord of Lmo4-cKO mice (Fig. 4A) reveals that, in the absence of Lmo4 function, many neurons in layer Va of MCr, which normally lacks CSMN, inappropriately project to the spinal cord (Fig. 4B, arrowheads). Retrograde labeling of the broad SCPN class, including CBMN (Lmo4+) and CSMN (Lmo4−), via injection of CTB into the pyramidal tracts at the level of the cerebral peduncles (Fig. 4C), reveals that the overall number of SCPN in layers Va and Vb of the MCr is unchanged in the absence of Lmo4 function (Fig. 4D); however, strikingly more of these neurons project to the spinal cord.
Quantitative analyses of CSMN and SCPN densities in layer Va support these observations. In layer Va of Lmo4-cKO mice, there is a 2.2-fold increase in the number of neurons sending a projection to the spinal cord (p < 0.005), although the overall density of SCPN in layer Va is unchanged (Fig. 4E). Because the number of layer Va SCPN is unchanged in Lmo4-cKO mice, it is unlikely that the aberrant spinal cord projecting neurons result from overproduction of CSMN in layer Va or from fate-conversion of any non-SCPN subpopulation. The numbers of CSMN and SCPN in layer Vb are both unchanged (Fig. 4E). Together, these data reveal that, in the absence of Lmo4 function, neurons in layer Va of the MCr that would have normally projected to the brainstem aberrantly project to the spinal cord, similar to SCPN near them in layer Vb (Fig. 4F). Thus, in addition to the disrupted molecular identity of SCPN, the connectivity of SCPN in MCr becomes more homogenous in the absence of Lmo4 function.
Lmo4 establishes diversity of dual-projecting neuron subtypes in rostral motor cortex
Because Lmo4 is also expressed outside of layer V in the MCr, we investigated whether Lmo4 functions similarly in establishing neuronal diversity of intracortical projection neuron classes. Dual retrograde labeling of CPN and BPN, via injection of CTB555 into the medial contralateral cortex and CTB647 into the ipsilateral somatosensory cortex labels dual-projecting CPN/BPN (Fig. 5A). Although there is developmental exuberance of dual-projecting neurons, many CPN/BPN maintain simultaneous contralateral and ipsilateral projections into adulthood in both rodent and primate cortex (Schwartz and Goldman-Rakic, 1982; Mitchell and Macklis, 2005).
The overwhelming majority of labeled BPN reside within the Lmo4+ area of motor cortex, but not in more lateral regions (Fig. 5B, left of dashed line), confirming previous findings of BPN predominance in medial frontal cortex (Mitchell and Macklis, 2005; Mao et al., 2011). As previously described (Mitchell and Macklis, 2005), this dual CPN/BPN labeling indicates that many CPN in this area also send a dual backward projection (Fig. 5C, arrowheads). These analyses also identify many “pure” CPN (Fig. 5D, arrows) and “pure” BPN (Fig. 5D, empty arrowheads), though the possibility exists that some of these are dual-projecting neurons that were not labeled by the second injection. In the MCr, essentially all labeled pure CPN, pure BPN, and CPN/BPN express Lmo4 (Fig. 5C,D).
In addition, a small population of dual-projecting neurons that send both subcerebral projections and ipsilateral backward projections from motor to sensory cortex has been reported in rat (Veinante and Deschênes, 2003). To first determine whether similar neurons exist in mice, we performed dual retrograde labeling from the cerebral peduncles at P2 with CTB555, and from the ipsilateral presumptive somatosensory areas with CTB647 at P7 (Fig. 5E). This approach labels both SCPN and BPN in layer V (Fig. 5F,V), and a small subpopulation of Lmo4+ SCPN that send dual backward projections (Fig. 5G, arrowheads). These data indicate that Lmo4 is expressed by multiple dual backward projecting neuronal subtypes that reside in the MCr and that send projections terminating in somatosensory areas.
Because CPN/BPN and SCPN/BPN are largely restricted to the MCr, the dual backward projections appear to be an area-specific feature. To determine whether Lmo4 regulates the development of such BPN features in general, we performed retrograde labeling of BPN in Lmo4-cKO mice (Fig. 6A, inset). In striking contrast to MCr in WT mice (Fig. 6A, WT), there is a dramatic decrease in the overall number of neurons sending a backward projection to the somatosensory area in both deep and superficial layers of the MCr in Lmo4-cKO mice (Fig. 6A, Lmo4-cKO). There is no apparent difference in the overall number of labeled CPN (Fig. 6B) or SCPN (Fig. 4D,E) between WT and Lmo4-cKO mice. Analyzing the MCr by subdivision, the reduction in BPN is most apparent in the dorsal subdivision (Fig. 6A, Area D), roughly corresponding to the facial or vibrissal regions of the motor cortex (Brecht et al., 2004b), but not in the medial subdivision (Fig. 6A, Area M), roughly corresponding to the cingulate cortex. There is no apparent difference in the overall number of labeled CPN in either the medial or dorsal subdivision of the MCr (Fig. 6B, “D” and “M”).
To more deeply investigate these findings, we performed quantitative analysis of CPN and BPN densities. This reveals a 30% decrease in the density of labeled BPN in the Lmo4-cKO MCr (Fig. 6C, Total) (p < 0.05). Subdividing the MCr, there is a 46% reduction in the density of BPN in the dorsal subdivision (Fig. 6C, “D”) (p < 0.01), but no significant reduction in the medial subdivision (Fig. 6C, “M”). There are no significant differences in the density of labeled CPN in either MCr subdivision, or throughout the entire MCr (Fig. 6D).
Because the numbers of BPN are selectively reduced in MCr (Fig. 6A,C), although the overall numbers of CPN (Fig. 6B, D) and SCPN (Fig. 4) are unaffected, our data suggest that many CPN and SCPN fail to acquire appropriate MCr-specific dual-projection features in the absence of Lmo4 function. Consequently, it might be expected that the density of dual-projecting CPN/BPN and SCPN/BPN might be reduced in the absence of Lmo4 function. To test this hypothesis, we quantified the fraction of CPN and SCPN that send a dual backward projection from the dorsal subdivision of WT and Lmo4-cKO MCr. Compared with WT mice, there is a 49% reduction in the density of CPN that send a dual-projection (Fig. 6E,F, solid arrowheads) (p < 0.05). Similarly, there is a 51% reduction in the density of SCPN that send a dual-projection (Fig. 6G,H, solid arrowheads) (p < 0.05). Our CPN/BPN quantification was performed in layers II/III, whereas our SCPN/BPN quantification was performed in layer V, thus confirming our observations that BPN development is perturbed in both superficial and deep layers. Our labeling approach reveals that backward projections do not appropriately target the somatosensory cortex. Therefore, it is possible that Lmo4-cKO BPN misproject to non-somatosensory regions or fail to project altogether. Overall, the dramatic ∼50% reduction of backward projections across multiple subtypes indicates that Lmo4 is required for the appropriate development of MCr-specific CPN and SCPN dual connectivity.
Together, these data reveal that Lmo4 is a critical postmitotic control over the development of projection neuron diversity in MCr. In the absence of Lmo4 function, the molecular identity of MCr neurons is substantially disrupted, the diversity of their projection targets is reduced, and the complex associative connectivity of the neocortex is dramatically diminished.
Discussion
Here, we show that Lmo4 is required to establish projection neuron subtype diversity within the motor cortex. Lmo4 expression sharply delineates the MCr, which is populated by multiple related, but anatomically distinct, subpopulations. In the absence of Lmo4 function, the molecular identity of neurons in the MCr is disrupted. Moreover, the anatomical diversity of both SCPN and CPN populations is dramatically perturbed. Fewer SCPN exclusively target the brainstem, although more target the spinal cord, and many SCPN and CPN fail to send a dual backward projection (∼50% of each subtype). Therefore, Lmo4-deficient MCr projection neuron populations send axons to fewer target areas, resulting in substantially less associative connectivity and hodological diversity, with much more homogenous motor cortex output.
Lmo4 controls area-specific neuronal subtype diversity
During development, Lmo4 expression is progressively sharpened along areal boundaries. Lmo4 has been reported to demarcate the dorsomedial subdivision of the frontal cortex, functioning in patterning programs downstream of transcription factors, such as Emx2, and morphogens including Fgf8 and Fgf17 (Cholfin and Rubenstein, 2007; 2008). Loss of Lmo4 function perturbs the expression of regional cortical markers (Huang et al., 2009).
Extending these findings, we identify important functions for Lmo4 in the area-specific molecular and anatomical development of projection neuron subtypes within the rostral neocortex. Two expression patterns highlight molecular perturbations in the Lmo4-cKO MCr. First, Lhx2 and Igfbp4 do not respect the boundary separating dorsal and medial subdivisions. Second, Igfbp5 and Clim1a fail to be expressed with the appropriate patterns.
Anatomical perturbations in the Lmo4-cKO MCr are broadly characterized by reduced projection target diversity. Lmo4-deficient SCPN largely target the spinal cord, instead of bifurcating into distinct populations that either exclusively target the brainstem or continue into the spinal cord. Most Lmo4-deficient SCPN and CPN send only a primary projection to subcerebral or contralateral targets, respectively, and thus fail to connect with ipsilateral sensory cortical areas through a second backward projection. This results in substantially reduced associative connectivity, thought to underlie integrative motor, sensory, planning, and cognitive functions.
It is interesting that the resulting anatomical phenotypes of projection neurons in Lmo4-cKO MCr resemble WT MCc projection neurons. Though the observed phenotypes probably represent a misspecification of boundaries within the MCr, rather than an ectopic expansion of MCc identity into MCr, it is possible, however, that MCc neuron identity represents a default pathway upon which Lmo4 acts to generate functionally specialized MCr subtypes. Alternatively, the MCr and MCc might contain complementary differentiation programs. In support of this idea, Bhlhb5-deficient CSMN do not reach the spinal cord, and instead stop in the brainstem (Joshi et al., 2008), a phenotype complementary to Lmo4-deficient CBMN, which abnormally traverse through the brainstem into the spinal cord.
Notably, in contrast to morphogens and transcription factors that specify cortical area size and shape at the progenitor stage (O'Leary et al., 2007), we find that Lmo4 is excluded from mitotic cortical progenitors during mid to late stages of corticogenesis, and, thus, its function is likely restricted to postmitotic neurons. However, it was recently shown that Lmo4 is a coactivator of neurogenin 2 (Ngn2) in developing cortical progenitors at earlier stages (Asprer et al., 2011). Conditional removal of Lmo4 from postmitotic neurons, while leaving its progenitor functions intact, might clarify which aspects of cortical development are controlled by Lmo4 at progenitor and postmitotic stages.
Moreover, in addition to its restricted expression in rostral motor cortex, we show that Lmo4 is also expressed in more caudal neocortical areas. Given the roles we describe here, as well as other Lmo4-dependent aspects of cortical subtype differentiation (Kashani et al., 2006; Huang et al., 2009), it will be interesting to investigate potential developmental functions for Lmo4 in more caudal visual areas.
LIM-HD signaling during neuronal development
Combinatorial interactions between LIM-HD related proteins are important for neuronal specification and differentiation (Shirasaki and Pfaff, 2002; Gill, 2003). Lmo and Clim proteins function by directly nucleating or disrupting transcriptional complexes that often contain Lhx transcription factors (Milan and Cohen, 1999; de la Calle-Mustienes et al., 2003; Lee et al., 2008; Joshi et al., 2009; Song et al., 2009). LIM-HD related proteins are expressed with area (Bulchand et al., 2003) and subtype specificity (Arlotta et al., 2005; Molyneaux et al., 2007, 2009; Azim et al., 2009) in the developing neocortex, suggesting that combinatorial LIM interactions might be important for cortical arealization and neuronal diversification.
Previous analysis has found that Lhx2 is expressed by CPN (Arlotta et al., 2005; Molyneaux et al., 2009) and that Clim1a and Lmo4 are transiently coexpressed in developing layer V, gradually segregating with postmitotic neuronal development into distinct projection neuron subtypes (Azim et al., 2009). Because Lmo4 is expressed by both SCPN and CPN in the MCr, it is ideally situated to interact with both Lhx2 and Clim1a during neuronal subtype differentiation.
We found that the transcription factor Lhx2 acquires a lateral-high to medial-low pattern of expression, complementary to that of Lmo4 at P7. In the absence of Lmo4 function, Lhx2 protein is ectopically expressed in the medial MCr and cingulate cortex. Because Lhx2 mRNA transcript is expressed normally in these medial areas, it is likely that Lmo4 exerts posttranscriptional repression over Lhx2 protein translation or stability. Drosophila Lmo regulates Apterous (Lhx2 homolog) by displacing it from a Clim protein complex, thus making it accessible to proteosome degradation (Weihe et al., 2001). Though the role of Lhx2 in postmitotic neocortical development remains unknown, it is possible that suppression of Lhx2 expression in the medial MCr is important for projection neuron development, perhaps analogous to the required downregulation of Lhx transcription factors during spinal motor neuron differentiation (Kania et al., 2000; Sharma et al., 2000). The exact mechanisms by which Lmo4 might interact with Lhx2 and Clim1a proteins remain to be identified.
Evolutionary specialization of rostral motor cortical circuits
Mammalian cortical areas differ greatly in relative size, cellular composition, and connectivity based on species-specific demands and constraints (Krubitzer and Kaas, 2005). The evolution of rodent vibrissae appears to have produced specialized circuitry in the MCr that enables complex rhythmic movement of whiskers and, in turn, precise sensory abilities (Diamond et al., 2008). CBMN, CPN, and BPN underlie critical activities of the vibrissal system (Brecht et al., 2004a; Maggiolini et al., 2007, 2008; Mao et al., 2011), implicating Lmo4 in its development.
Cortico-brainstem circuits
Vibrissa motor output is directly controlled by CBMN that project from the vibrissa motor cortex (VMC) and monosynaptically connect with brainstem motor neurons (Brecht et al., 2004a; Grinevich et al., 2005). The VMC is located in the dorsal subdivision of the MCr (Brecht et al., 2004b), where Lmo4 regulates CBMN development (Fig. 4, Fig. 6). Though Lmo4 is expressed in layer Va but not layer Vb CBMN, intracortical micro-stimulation (ICMS) at all dorsoventral positions in layer V of the VMC evokes whisker movements (Brecht et al., 2004b).
Our anatomical characterization of CBMN in the dorsal MCr suggests that they might use different mechanisms to achieve target specificity than do other SCPN. Most SCPN achieve target specificity by selectively pruning exuberant branches (Stanfield et al., 1982; O'Leary and Stanfield, 1985, 1986; O'Leary, 1987). Our results reveal that axons of layer Va CBMN in the MCr do not innervate the spinal cord at P2, when layer Vb CSMN in the MCr have just begun to enter the spinal cord (Fig. 3). Therefore, it is possible that these CBMN directly and specifically recognize their brainstem targets, rather than pruning exuberant spinal cord projections. Such a direct recognition system encodes the establishment of monosynaptic sensory-motor connections in the spinal reflex circuit (Pecho-Vrieseling et al., 2009). It is interesting to speculate that the mechanisms underlying target selection might be different for cortical projection neurons that directly synapse onto brainstem motor neurons (cortico-vibrissa motor neurons; potentially also the small subset directly synapsing onto spinal motor neurons) versus those that target interneuron circuits.
Callosal circuits
CPN have also been shown to functionally integrate into sensory-motor circuits by sharpening functional boundaries between vibrissal and forelimb regions within the MCr (Maggiolini et al., 2007, 2008). Moreover, it has been suggested that CPN, and in particular those with a dual backward projection, might coordinate the characteristic bilateral rhythmic whisking patterns of rodents (Veinante and Deschênes, 2003). In the absence of Lmo4 function, our results reveal that MCr CPN remain, but no longer project to the same diversity of targets. Appropriate development of CPN diversity, like SCPN diversity, likely critically underlies the establishment of a broad motor repertoire.
Backward projection circuits
The integration of motor behavior and sensory input is particularly relevant to the rodent vibrissal system, because this system provides major sensory input, at the same time as it exhibits complex motor output. The MCr contains specialized intracortical circuits that underlie sensory-motor integration. The coordination of the dorsal premotor and parietal areas through long-range ipsilateral connectivity enables primates to integrate sensory and motor information, allowing them to execute coordinated and planned motor behaviors (Pesaran et al., 2006, 2008). Recently, it was shown that BPN in rodents are functionally integrated in the sensory-motor loop between the VMC and somatosensory cortex (Mao et al., 2011). We found that Lmo4 is expressed by multiple dual backward projecting neuronal subtypes that reside in the MCr, and that send projections terminating in somatosensory areas. Although the precise functions of these dual-projecting neurons has not been fully elucidated, they are likely involved in sensory-motor integration (Veinante and Deschênes, 2003). Accurate decoding of sensory inputs by the somatosensory cortex requires real-time knowledge of the position and movement of sensory organs, such as whiskers and skin (Diamond et al., 2008; Curtis and Kleinfeld, 2009). Lmo4+ backward projecting neurons, and in particular, the dual-projecting CPN/BPN and SCPN/BPN subpopulations, might be capable of controlling bilateral motor output, and simultaneously relaying information about position and movement to somatosensory cortex (Veinante and Deschênes, 2003; Ahrens and Kleinfeld, 2004; Mao et al., 2011). We find that, in the absence of Lmo4 function, there is a substantial reduction in the number of BPN in the MCr, strongly suggesting that this arm of the circuit is disrupted in Lmo4-cKO mice. Consistent with this interpretation, previous studies have shown that Lmo4-deficient mice exhibit sensory-motor behavioral deficits (Huang et al., 2009). Thus, Lmo4 appears to be a central control over this evolutionarily emergent sensory-motor integration system.
Together, our data reveal that Lmo4 establishes molecular and anatomical diversity of motor control projection neurons within the MCr. This diversity of neuronal connectivity centrally includes the development of dual projection neurons that integrate sensory and motor information within the same hemisphere and across hemispheres. Generation of the full repertoire of neuronal subtypes within the MCr is necessary for complex movements; thus, Lmo4 is critical for the development of cortical circuits that integrate and execute motor behavior.
Footnotes
This work was supported by grants from the National Institutes of Health (NS049553 and NS041590; additional infrastructure provided by NS045523 and NS075672), the Spastic Paraplegia Foundation, the ALS Association, the Harvard Stem Cell Institute, the Jane and Lee Seidman Fund for CNS Research, and the Emily and Robert Pearlstein Fund for Nervous System Repair (to J.D.M.). E.A. was partially supported by a National Institutes of Health predoctoral NRSA (F31 NS060421). S.J.S. was partially supported by a National Institutes of Health predoctoral NRSA (F31 NS063516). H.P. was partially supported by an International Brain Research Organization Fellowship, and a McKnight Brain Research Institute/Regeneration Project Fellowship. We thank Dr. Lin Gan for generously providing mouse lines, members of the Macklis Laboratory for thoughtful discussion, and Aaron Wheeler, Patrick Davis, and Jessica Kim for technical assistance.
The authors declare no competing financial interests.
References
- Ahrens KF, Kleinfeld D. Current flow in vibrissa motor cortex can phase-lock with exploratory rhythmic whisking in rat. J Neurophysiol. 2004;92:1700–1707. doi: 10.1152/jn.00020.2004. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Sheets PL, Kiritani T, Shepherd GM. Sublayer-specific microcircuits of corticospinal and corticostriatal neurons in motor cortex. Nat Neurosci. 2010;13:739–744. doi: 10.1038/nn.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Asprer JS, Lee B, Wu CS, Vadakkan T, Dickinson ME, Lu HC, Lee SK. LMO4 functions as a coactivator of neurogenin 2 in the developing cortex. Development. 2011;138:2823–2832. doi: 10.1242/dev.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim E, Shnider SJ, Cederquist GY, Sohur US, Macklis JD. Lmo4 and Clim1 progressively delineate cortical projection neuron subtypes during development. Cereb Cortex. 2009;19:i62–i69. doi: 10.1093/cercor/bhp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I, Carrière C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Brecht M, Schneider M, Sakmann B, Margrie TW. Whisker movements evoked by stimulation of single pyramidal cells in rat motor cortex. Nature. 2004a;427:704–710. doi: 10.1038/nature02266. [DOI] [PubMed] [Google Scholar]
- Brecht M, Krauss A, Muhammad S, Sinai-Esfahani L, Bellanca S, Margrie TW. Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J Comp Neurol. 2004b;479:360–373. doi: 10.1002/cne.20306. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, Molnár Z, Tarabykin V. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–175. doi: 10.1016/S0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci U S A. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J Comp Neurol. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O'Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis JC, Kleinfeld D. Phase-to-rate transformations encode touch in cortical neurons of a scanning sensorimotor system. Nat Neurosci. 2009;12:492–501. doi: 10.1038/nn.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Lu Z, Cortés M, Andersen B, Modolell J, Gómez-Skarmeta JL. Xenopus Xlmo4 is a GATA cofactor during ventral mesoderm formation and regulates Ldb1 availability at the dorsal mesoderm and the neural plate. Dev Biol. 2003;264:564–581. doi: 10.1016/j.ydbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Deng M, Pan L, Xie X, Gan L. Requirement for Lmo4 in the vestibular morphogenesis of mouse inner ear. Dev Biol. 2010;338:38–49. doi: 10.1016/j.ydbio.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond ME, von Heimendahl M, Knutsen PM, Kleinfeld D, Ahissar E. ‘where’ and ‘what’ in the whisker sensorimotor system. Nat Rev Neurosci. 2008;9:601–612. doi: 10.1038/nrn2411. [DOI] [PubMed] [Google Scholar]
- Ferrere A, Vitalis T, Gingras H, Gaspar P, Cases O. Expression of cux-1 and cux-2 in the developing somatosensory cortex of normal and barrel-defective mice. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:158–165. doi: 10.1002/ar.a.20284. [DOI] [PubMed] [Google Scholar]
- Gill GN. Decoding the LIM development code. Trans Am Clin Climatol Assoc. 2003;114:179–189. [PMC free article] [PubMed] [Google Scholar]
- Grinevich V, Brecht M, Osten P. Monosynaptic pathway from rat vibrissa motor cortex to facial motor neurons revealed by lentivirus-based axonal tracing. J Neurosci. 2005;25:8250–8258. doi: 10.1523/JNEUROSCI.2235-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner deaf-1. Mol Cell Biol. 2004;24:2074–2082. doi: 10.1128/MCB.24.5.2074-2082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kawase-Koga Y, Zhang S, Visvader J, Toth M, Walsh CA, Sun T. Transcription factor Lmo4 defines the shape of functional areas in developing cortices and regulates sensorimotor control. Dev Biol. 2009;327:132–142. doi: 10.1016/j.ydbio.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61:839–851. doi: 10.1016/j.neuron.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/S0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, Goebbels S, Nave KA, Ghosh A. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krubitzer L, Kaas J. The evolution of the neocortex in mammals: how is phenotypic diversity generated? Curr Opin Neurobiol. 2005;15:444–453. doi: 10.1016/j.conb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Fame RM, Azim E, Shnider SJ, Molyneaux BJ, Arlotta P, Macklis JD. Specification of cortical projection neurons: transcriptional mechanisms. In: Rubenstein JLR, Rakic P, editors. Developmental Neuroscience: A Comprehensive Reference. Oxford: Elsevier; 2013. In press. [Google Scholar]
- Maggiolini E, Veronesi C, Franchi G. Plastic changes in the vibrissa motor cortex in adult rats after output suppression in the homotopic cortex. Eur J Neurosci. 2007;25:3678–3690. doi: 10.1111/j.1460-9568.2007.05622.x. [DOI] [PubMed] [Google Scholar]
- Maggiolini E, Viaro R, Franchi G. Suppression of activity in the forelimb motor cortex temporarily enlarges forelimb representation in the homotopic cortex in adult rats. Eur J Neurosci. 2008;27:2733–2746. doi: 10.1111/j.1460-9568.2008.06248.x. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao T, Kusefoglu D, Hooks BM, Huber D, Petreanu L, Svoboda K. Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron. 2011;72:111–123. doi: 10.1016/j.neuron.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999;4:267–273. doi: 10.1016/S1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Macklis JD. Large-scale maintenance of dual projections by callosal and frontal cortical projection neurons in adult mice. J Comp Neurol. 2005;482:17–32. doi: 10.1002/cne.20428. [DOI] [PubMed] [Google Scholar]
- Miyashita E, Keller A, Asanuma H. Input-output organization of the rat vibrissal motor cortex. Exp Brain Res. 1994;99:223–232. doi: 10.1007/BF00239589. [DOI] [PubMed] [Google Scholar]
- Molnár Z, Cheung AF. Towards the classification of subpopulations of layer V pyramidal projection neurons. Neurosci Res. 2006;55:105–115. doi: 10.1016/j.neures.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/S0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Ning Y, Schuller AG, Conover CA, Pintar JE. Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol Endocrinol. 2008;22:1213–1225. doi: 10.1210/me.2007-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD. Remodelling of early axonal projections through the selective elimination of neurons and long axon collaterals. Ciba Found Symp. 1987;126:113–142. doi: 10.1002/9780470513422.ch8. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Stanfield BB. Occipital cortical neurons with transient pyramidal tract axons extend and maintain collaterals to subcortical but not intracortical targets. Brain Res. 1985;336:326–333. doi: 10.1016/0006-8993(85)90661-4. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Stanfield BB. A transient pyramidal tract projection from the visual cortex in the hamster and its removal by selective collateral elimination. Brain Res. 1986;392:87–99. doi: 10.1016/0165-3806(86)90235-x. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Stanfield BB. Selective elimination of axons extended by developing cortical neurons is dependent on regional locale: experiments utilizing fetal cortical transplants. J Neurosci. 1989;9:2230–2246. doi: 10.1523/JNEUROSCI.09-07-02230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Pecho-Vrieseling E, Sigrist M, Yoshida Y, Jessell TM, Arber S. Specificity of sensory-motor connections encoded by Sema3e-Plxnd1 recognition. Nature. 2009;459:842–846. doi: 10.1038/nature08000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Wessely O, Li SY, De Robertis EM. Neural and head induction by insulin-like growth factor signals. Dev Cell. 2001;1:655–665. doi: 10.1016/S1534-5807(01)00069-7. [DOI] [PubMed] [Google Scholar]
- Pesaran B, Nelson MJ, Andersen RA. Dorsal premotor neurons encode the relative position of the hand, eye, and goal during reach planning. Neuron. 2006;51:125–134. doi: 10.1016/j.neuron.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaran B, Nelson MJ, Andersen RA. Free choice activates a decision circuit between frontal and parietal cortex. Nature. 2008;453:406–409. doi: 10.1038/nature06849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L, Drechsel D, Schmid MT, Ninkovic J, Irmler M, Brill MS, Restani L, Gianfranceschi L, Cerri C, Weber SN, Tarabykin V, Baer K, Guillemot F, Beckers J, Zecevic N, Dehay C, Caleo M, Schorle H, Götz M. AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci. 2009;12:1229–1237. doi: 10.1038/nn.2399. [DOI] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr Opin Neurobiol. 2006;16:25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Salih DA, Tripathi G, Holding C, Szestak TA, Gonzalez MI, Carter EJ, Cobb LJ, Eisemann JE, Pell JM. Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc Natl Acad Sci U S A. 2004;101:4314–4319. doi: 10.1073/pnas.0400230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, Goldman-Rakic PS. Single cortical neurones have axon collaterals to ipsilateral and contralateral cortex in fetal and adult primates. Nature. 1982;299:154–155. doi: 10.1038/299154a0. [DOI] [PubMed] [Google Scholar]
- Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Song MR, Sun Y, Bryson A, Gill GN, Evans SM, Pfaff SL. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136:2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield BB, O'Leary DD, Fricks C. Selective collateral elimination in early postnatal development restricts cortical distribution of rat pyramidal tract neurones. Nature. 1982;298:371–373. doi: 10.1038/298371a0. [DOI] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Tomassy GS, De Leonibus E, Jabaudon D, Lodato S, Alfano C, Mele A, Macklis JD, Studer M. Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proc Natl Acad Sci U S A. 2010;107:3576–3581. doi: 10.1073/pnas.0911792107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, Boehm T, Rabbitts TH. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veinante P, Deschênes M. Single-cell study of motor cortex projections to the barrel field in rats. J Comp Neurol. 2003;464:98–103. doi: 10.1002/cne.10769. [DOI] [PubMed] [Google Scholar]
- Weihe U, Milán M, Cohen SM. Regulation of apterous activity in drosophila wing development. Development. 2001;128:4615–4622. doi: 10.1242/dev.128.22.4615. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Zhang YA, Levin ME, Devine WP, Brûlet P, McConnell SK. Cortical neurons require Otx1 for the refinement of exuberant axonal projections to subcortical targets. Neuron. 1999;24:819–831. doi: 10.1016/S0896-6273(00)81030-2. [DOI] [PubMed] [Google Scholar]
- Wise SP, Murray EA, Coulter JD. Somatotopic organization of corticospinal and corticotrigeminal neurons in the rat. Neuroscience. 1979;4:65–78. doi: 10.1016/0306-4522(79)90218-5. [DOI] [PubMed] [Google Scholar]
- Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, Naito AT, Nishi J, Ueno H, Umezawa A, Minamino T, Nagai T, Kikuchi A, Asashima M, Komuro I. IGFBP-4 is an inhibitor of canonical wnt signalling required for cardiogenesis. Nature. 2008;454:345–349. doi: 10.1038/nature07027. [DOI] [PubMed] [Google Scholar]