Abstract
Expression of the mammalian pyruvate kinase M (PKM) gene provides an important example of mutually exclusive splicing. We showed previously that the hnRNP proteins A1, A2 and PTB play a critical role in this process. Here we provide evidence that concentration-dependent interactions involving a network of these proteins are sufficient to determine the outcome of PKM splicing. At high concentrations, such as found in most cancer cells, hnRNP A1 binding to two sites in the upstream regulated exon (exon 9) orchestrates cooperative interactions leading to exon 9 exclusion. At lower concentrations, binding shifts to downstream intronic sites such that exon 9 is included and exon 10 largely excluded, with any mRNA including both exons degraded by nonsense-mediated decay. Together our results provide a mechanism by which a small number of general factors control alternative splicing of a widely expressed transcript.
Introduction
Alternative splicing is a common mechanism for regulating gene expression and generating proteomic diversity1,2. Whether an alternative exon is included or excluded during splicing is often determined by cis-regulatory elements recognized by trans-acting regulatory proteins3. Two groups of classical AS regulators are the serine/arginine-rich (SR) proteins, which usually function as splicing activators, and hnRNP proteins, which often act as splicing repressors. SR proteins frequently function by recognizing exonic splicing enhancers (ESEs), while hnRNP proteins bind to exonic or intronic splicing silencers (E/ISSs) to inhibit splicing.
Disruption of alternative splicing is a frequent cause of disease4–8. This can occur in several ways, including mutation of RNA sequence elements or deregulation of RNA binding proteins4,5. Mutations can destroy or weaken essential splicing signals or regulatory elements, while changes in expression of hnRNP, or other splicing factors, can have significant effects on AS patterns. Indeed, a variety of diseases have been shown to involve changes in the expression levels of splicing factors4,5,9.
Regulation of PKM mRNA splicing provides an important example of an AS event critical for disease, and which reflects changes in the levels of hnRNP splicing regulators10–12. The PKM gene encodes a primary transcript that contains two mutually exclusive exons (MEEs), exon 9 and exon 10, and inclusion of one or the other leads to two different isoforms, PKM1 and PKM2, respectively13. PKM2 is expressed in embryonic cells while PKM1 is expressed in most adult tissues14,15. Reversion from PKM1 to PKM2 is observed in most cancers, partially explaining the Warburg effect observed in tumor cells16 and ensuring maximal turmorigenicity15,17. We recently described a pathway that regulates PKM splicing10. Expression of three hnRNP proteins, hnRNP A1 (A1), hnRNP A2 (A2) and PTB, was found to be upregulated in cancer cells by the oncogene c-Myc, which promotes formation of PKM2. These three hnRNP proteins, all previously shown to function as splicing repressors and to be overexpressed in a variety of cancers5,10,18–20, exclude exon 9 by binding to sequences flanking exon 9. A subsequent study also provided evidence that A1/A2/ PTB are important for the PKM splicing switch12. But a full understanding of the molecular mechanism underlying PKM mutually exclusive alternative splicing (MEAS) has been lacking.
In this study, we provide additional insights into how PKM AS is regulated. First, using a minigene construct that accurately recapitulates PKM splicing in HeLa cells, we identified additional PTB and A1/A2 (A1 and A2 are highly similar, so we refer to them as “A1/A2”) ISSs in intron 9 necessary for full exclusion of exon 9. More importantly, we found two A1 binding sites in exon 9 that function cooperatively to facilitate A1 binding to a previously described ISS in intron 9 (ref. 10), and showed that they play a critical role in exon 9 exclusion when A1/A2/PTB (the three proteins are frequently coregulated, so we name them as “A1/A2/PTB”) levels are high. When the levels of these proteins were reduced by RNAi, exon 9 was now as expected included, but exon 10 was excluded in a manner dependent on additional A1/A2/PTB binding sites in introns 9 and 10 that were effectively occupied despite the decreased concentration of these proteins. This concentration-dependent mechanism, coupled with nonsense mediated decay, functions to prevent the appearance of PKM mRNA containing both exon 9 and exon 10.
Results
Intronic hnRNP binding sites inhibit exon 9 inclusion
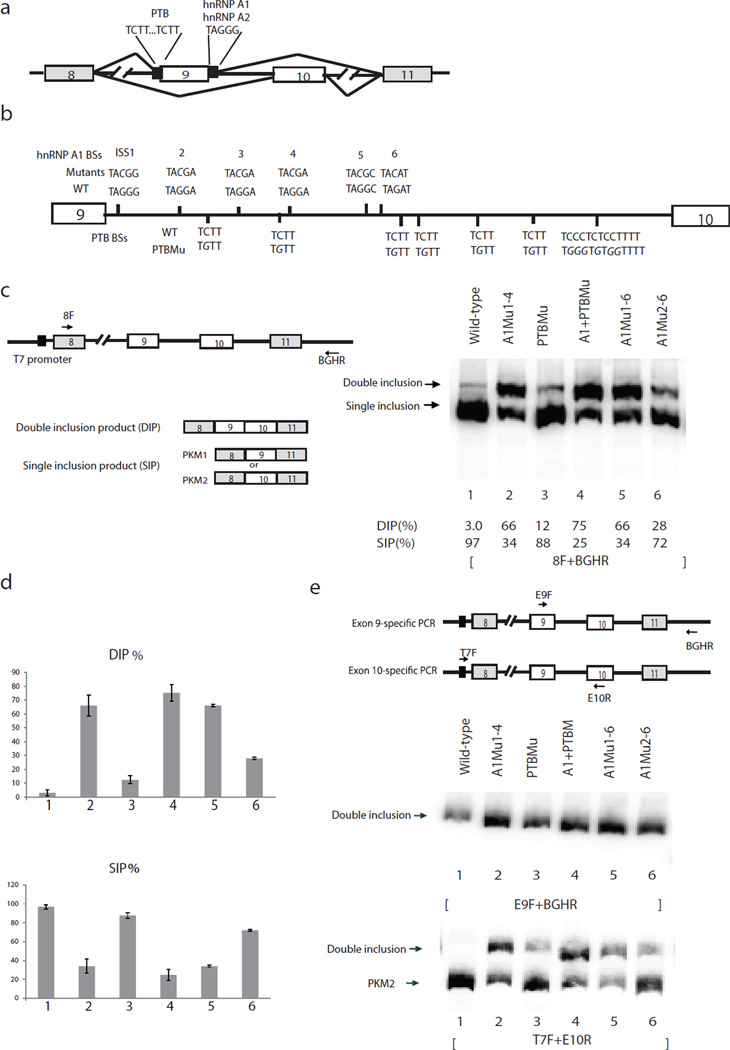
We previously showed that A1/A2 and PTB inhibit PKM exon 9 inclusion by binding to intronic sequences flanking exon 9. PTB recognizes two UCUU elements upstream of the 3’ splice site (ss) of exon 9 and A1/A2 bind to UAGGGC (ISS1), which is immediately downstream of the exon 9 5’ ss10 (Fig. 1a). In order to investigate whether additional intronic sequences are involved in regulating PKM splicing, we constructed a minigene splicing construct containing PKM sequences from exon 8 to exon 11 with 200–400 nucleotide (nt) intronic sequences flanking each exon and with an intact 401 nt intron 9 (Fig. 1a). This construct accurately recapitulates PKM alternative splicing in HeLa cells (see below).
Figure 1.
Mutations of intron 9 sequences derepress exon 9 inclusion. (a) Schematic diagram of PKM splicing construct containing exon 8 to exon 11. “//” indicates deletions of intron sequences. Mutually exclusive AS of exon 9 and exon 10 is indicated. Solid black boxes flanking exon 9 indicate binding sites for hnRNP A1/A2 and PTB, described previously10. (b) Schematic diagram of PKM intron 9. Vertical lines indicate putative A1/A2 (above the line indicating intron 9) and PTB (below the line) binding sites (BSs). Mutations of BSs are indicated above or below wild-type BSs in italic. (c) Schematic diagram of splicing construct and possible products are indicated on the left panel. Black arrows indicate primers used to amplify PKM AS products. RT−PCR assays of RNA isolated from transient transfections of wild-type and mutated splicing constructs. The positions of splicing products are indicated on the left. The percentages of DIP (DIP(%)) and SIP (SIP(%)) in total products (DIP(%))are indicated under the lane numbers. (d) Bar graphs show percentages of DIP (left) and SIP (right) using wild-type and mutated splicing constructs with standard deviation, n=3. Lane numbers correspond to lane numbers in panel. DIP, double inclusion product. SIP, single inclusion product. c, and the same lane numbers represent the same constructs. (e) Left panel, scheme indicates positions of exon 9- and exon 10-specific primers. E9F, which anneals to exon 9, and vector-specific primer BGHR were used to amplify exon 9-containing products. Vector-specific primer T7F and E10R were used to amplify exon 10-containing products. Right, RT-PCR assays with primers that amplify only exon 9-containing products to analyze splicing products from intron 9-mutated splicing constructs. Splicing constructs are indicated above, and splicing products are indicated on the left. Lane numbers correspond to those in panel c, and the same lane numbers represent the same constructs.
Apart from the elements identified previously10, sequence examination (Fig. 1b) and UV crosslinking assays (Supplementary Fig. 1) revealed a number of additional A1/A2 and PTB binding sites in intron 9. To examine whether these sites are important for PKM splicing, we mutagenized several of them and examined effects on PKM splicing by transfecting wild-type and mutated splicing constructs into HeLa cells, isolated total cellular RNA and performed RT-PCR assays. Two major bands were detected (indicated in Fig. 1c, right panel). One, which we dubbed SIP (single inclusion product), contained either exon 9 or 10 (see below), while the other, DIP (double inclusion product), contained both of the MEEs.
We determined the efficiency of exon inclusion with wild-type and mutant constructs by dividing the amount of each product by the sum of both products, and are graphed in Figure 1d. With the wild-type construct, the majority (~97%) of the pre-mRNA produced was spliced into an SIP, while only 3.0% was DIP (Fig. 1c, lane 1). Additionally, similar to endogenous PKM splicing in HeLa cells, all SIP contained exon 10 (see Fig. 1e below). Notably, TAG to TAC mutations in putative A1/A2 binding sites8,10,21,22 drastically increased DIP, from 3.0% to 66% (Fig. 1c and 1d, lanes 2 and 5). In addition, C to G mutations in putative PTB binding sites increased the amount of DIP, from 3.0% to 12%, indicating that these sequences contribute to exon 9 exclusion (Fig. 1c and d, lanes 1 and 3). Finally, mutations including both A1/A2 and PTB binding sites displayed the greatest effect, increasing DIP from 3.0% in wild-type to 75% (Fig. 1, lane 4). These findings suggest that efficient exon 9 exclusion involves multiple A1/A2 and PTB binding sites in intron 9. To determine the amounts of exon 9 inclusion (PKM1) in the SIP, we designed exon 9-;specific and E10-specific primers to amplify exon 9 or exon 10-specific SIP and DIP, respectively (Fig. 1e, left panel). Notably, in all cases all the exon 9-included product was DIP (Fig. 1e, middle panel), suggesting that the release of repression of exon 9 inclusion by mutation of intron 9 ISSs led to increased DIP but not exon 9-containing SI.
Exonic sequences are involved in PKM splicing regulation
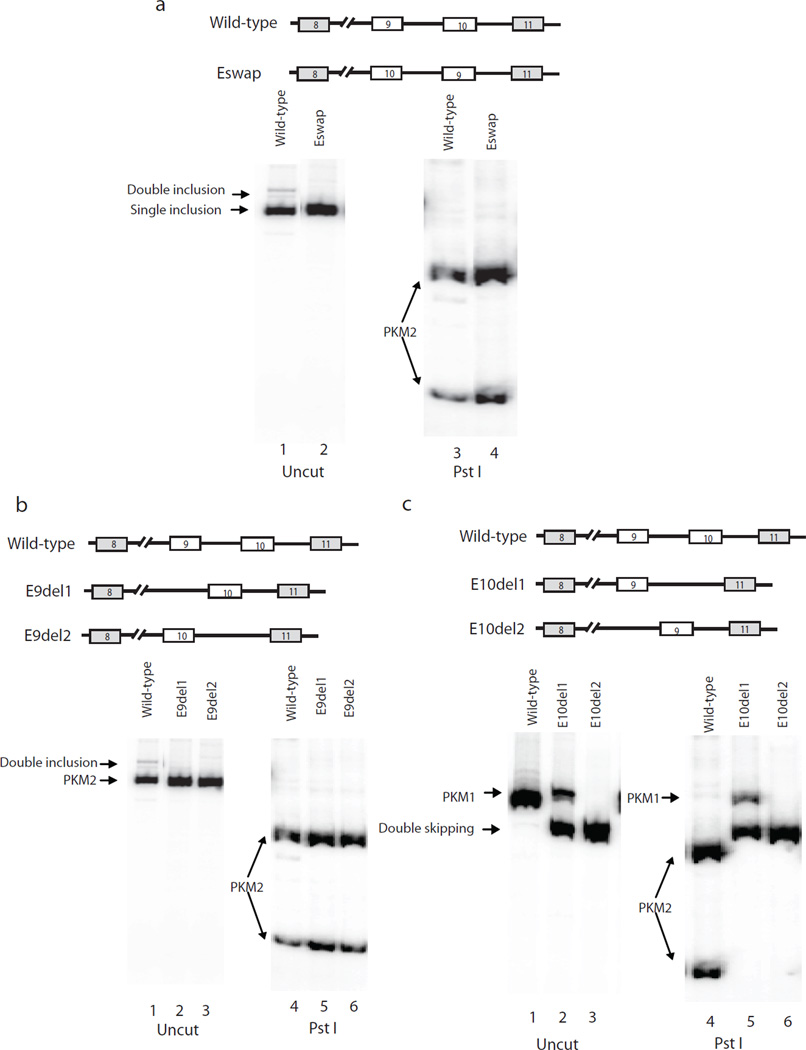
We next set out to investigate whether cis-elements in exons function in PKM splicing regulation. We first precisely swapped exon 9 and exon 10 in the wild-type splicing construct to determine if changing the intronic sequences flanking the exons would affect splicing (Eswap; Fig. 2a upper panel). Surprisingly, exon swapping did not alter the outcome of PKM splicing (Fig. 2a, lanes 2 and 4). We conclude that despite the importance of intronic sequences in intron 9 (and 8), these ISSs are not sufficient to exclude any adjacent exon. We next examined exon 9 and exon 10 splicing independent of each other. First, we precisely deleted exon 9 or exon 10 from wild-type and Eswap constructs (Fig. 2b and 2c, top panels), and found that exon 10 was included efficiently in both positions (Fig. 2b) while exon 9 was largely excluded in both cases (Fig. 2c). Thus inclusion or exclusion of either exon was independent of its position, but rather dependent on exonic sequences.
Figure 2.
Exonic sequences are involved in PKM alternative splicing regulation. (a) Top panel, diagram of wild-type and Eswap constructs, in which the positions of exon 9 and exon 10 are switched. Lower panel, RT-PCR assays of RNAs extracted from transfected HeLa cells, using primers 8F and BGHR (lower left). Bands representing PKM2 are indicated with arrows. PCR products were digested with Pst I (lower right). (b) Top panel, diagrams indicate splicing constructs with exon 9 deleted from wild-type (E9del1) and from Eswap (E9del2). RT-PCR assays of RNA isolated from HeLa cells tranfected with indicated plasmids, with primers 8F and BGHR (lower left). PCR products were digested with Pst I (lower right). PKM splicing products are indicated on the left. (c) Top panel, diagrams indicate constructs with exon 10 deleted from wild-type (E10del1) and from Eswap (E10del2). RT-PCR assays of RNA isolated from HeLa cells tranfected with indicated splicing constructs with primers 8F and BGHR (lower left). PCR products were digested with Pst I (lower right). PKM splicing products are indicated on the left. Double skipping, products that contains only exon 8 and exon 11.
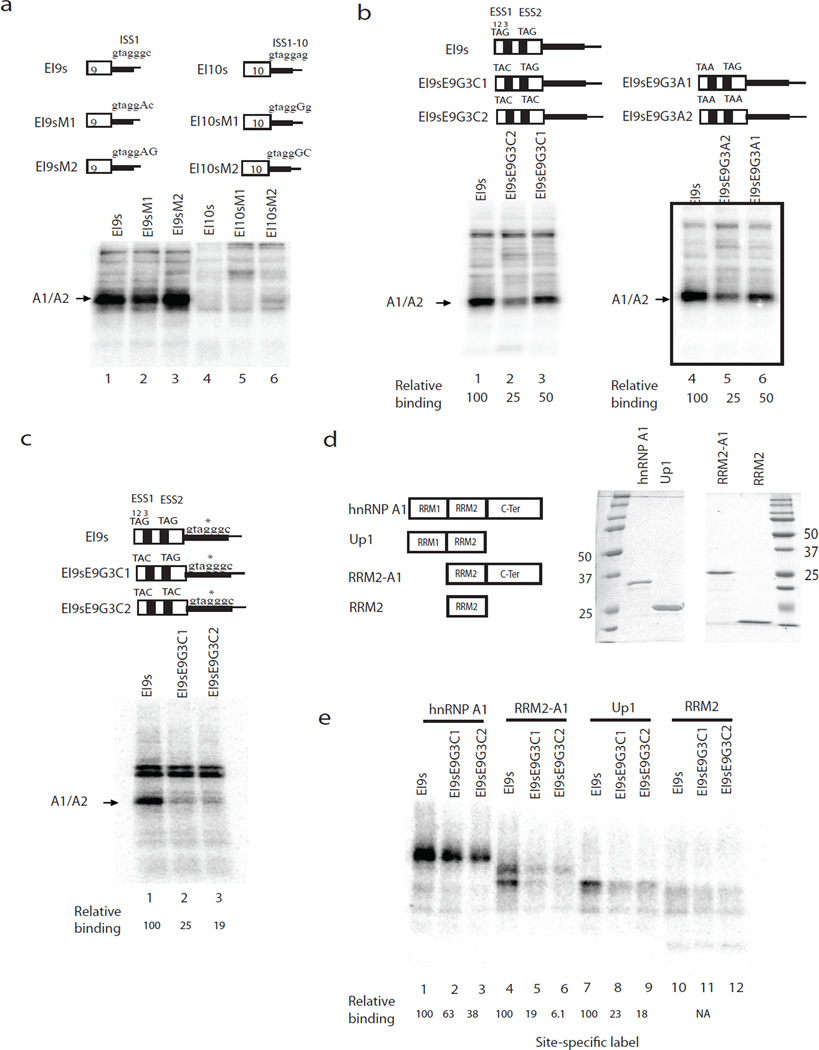
We next set out to examine the possible role of exonic sequences in protein binding to PKM RNA regulatory sequences, such as ISS1. Our previous studies showed that an RNA consisting of the 3’ part of exon 9 and 5’ part of intron 9, called EI9, was bound by A1/A2, while the corresponding EI10 RNA was not10. However, the apparent A1/A2 binding site, ISS1, is very similar to the corresponding sequence in intron 10, ISS1-10 (Fig. 3a, top panel). We hypothesized that exonic sequences contribute to the binding profiles of EI9 and EI10. To test this idea, we carried out UV crosslinking assays with HeLa nuclear extract22, initially using uniformly labeled exon 9- or exon 10-containing RNAs. We prepared two RNAs for crosslinking, EI9s and EI10s, which contain the exonic region of EI9 or EI10 and a very short intronic extension that contains only ISS1, ISS1-10, or mutated derivatives (Fig. 3a, top panel). Similar to EI9s, both EI9sM1 and EI9sM2 were bound strongly by A1/A2 (Fig. 3a, lanes 1–3). Moreover, mutations “improving” ISS1-10 did not promote binding of A1/A2 to the EI10s substrates (lanes 4–6). Our results suggest that sequences in exon 9 cooperate with ISS1 to enable A1/A2 binding to EI9s but not EI10s.
Figure 3.
hnRNP A1/A2 bind cooperatively to exonic and intronic elements. (a) UV crosslinking of EI9s, EI10s and variants in HeLa nuclear extract. Upper panel, diagrams of UV-crosslinking substrates EI9s, EI10s and variants. ISS1 and the corresponding sequence in intron 10 are indicated above each RNA. Mutations are indicated with capital letters. Lower panel, RNA constructs shown in the upper panel were incubated with NE, crosslinked, and analyzed by SDS-PAGE. The position of A1 is indicated on the left. (b) Upper panel, Schematic diagrams of EI9s and variants. Putative exonic A1 binding sites are indicated as black box and sequence indicated above. TAG was mutated into TAC or TAA as indicated. Lower panel, UV crosslinking assays with RNAs shown in the upper panel. A1/A2 positions are indicated on the left. Quantification of A1/A2 binding is indicated below. (c) Site-specific label of EI9s and variants with exonic TAG mutations. Upper panel, diagrams of EI9s and its variants. Asterisks indicate the nucleotide labeled with 32P. Site-specifically labeled EI9s and mutants were incubated in NE, crosslinked, and analyzed by SDS-PAGE. A1/A2 is indicated on the left. Asterisk, background, non-specific band. Quantification of the binding of A1/A2 is indicated below. (d) Left panel, schematic diagrams indicate A1 and truncated derivatives. A1, full-length hnRNP A1. UP1, contains the N-terminal RRM1 and RRM2. RRM2-A1, hnRNP A1 with RRM1 deleted. RRM2 contains only the second RRM domain. Right panel, recombinant A1 and truncations are purified from E. coli, analyzed on SDS-PAGE, and commassie stained. (e) UV-crosslinking assays with SSL EI9s and variants. RNAs were incubated with 0.3 µM A1 (lanes 1–3), RRM2-A1 (lanes 4–6), Up1 (lanes 7–9) and RRM2 (lanes 10–12), UV crosslinked and analyzed by SDS-PAGE. Quantification of binding is indicated at the bottom.
We next examined exonic sequences and found two evolutionarily conserved TAG motifs (which we refer to as ESS1 and ESS2) in exon 9 that are absent in the homologous exon 10 (66% sequence identity). hnRNP A1 has been suggested to bind cooperatively to RNAs with multiple binding sites23–26, so we propose that the binding of A1/A2 to EI9s requires all three elements, ESS1, ESS2 and ISS1. To investigate this idea, we mutated the TAG to TAC or TAA in ESS1 or in both ESSs (Fig. 3b, top), and first analyzed uniformly labeled RNAs by UV crosslinking. Consistent with the effects of a similar mutation (G3C) in ISS110, both mutations in ESS1 decreased A1/A2 binding to EI9s RNA by half (Fig. 3b, lanes 3 and 6), while the double mutant reduced the amount of A1 crosslinked to ~25% (Fig. 3b, lanes 2 and 5). Thus, although both ISS1 and ISS1-10 contain putative A1/A2 binding sites, exon 9 harbors two elements that enable A1/A2 to interact with EI9s RNA. To extend these results, we next used site-specifically labeled EI9s and the ESS mutants, in which the G at position 3 in ISS1 was 5’ labeled with 32P. Importantly, the results (Fig. 3c) show that while EI9s was bound strongly by A1, binding was reduced to ~25% when one ESS was mutated and to ~19% when both were altered. These results indicate that the two ESSs in exon 9 facilitate A1/A2 binding to ISS1.
The above experiments suggest that A1/A2 bind the exon 9 region cooperatively. To investigate this further, we purified A1 and several truncations from E. coli (Fig. 3d) and performed UV-crosslinking assays with site-specifically labeled wild-type and mutant EI9s RNAs (Fig. 3e). These results indicate first that hnRNP A1 alone is sufficient for binding to ISS1, and second that the C-terminal domain is not essential for cooperative binding; RRM1 can be sufficient. While the C-terminal domain has been previously shown to be involved in protein-protein interactions23,27 and it contributes to optimal binding to EI9s, our results indicate that cooperativity can be achieved in its absence.
The exon 9 A1/A2 sites are critical for exon exclusion
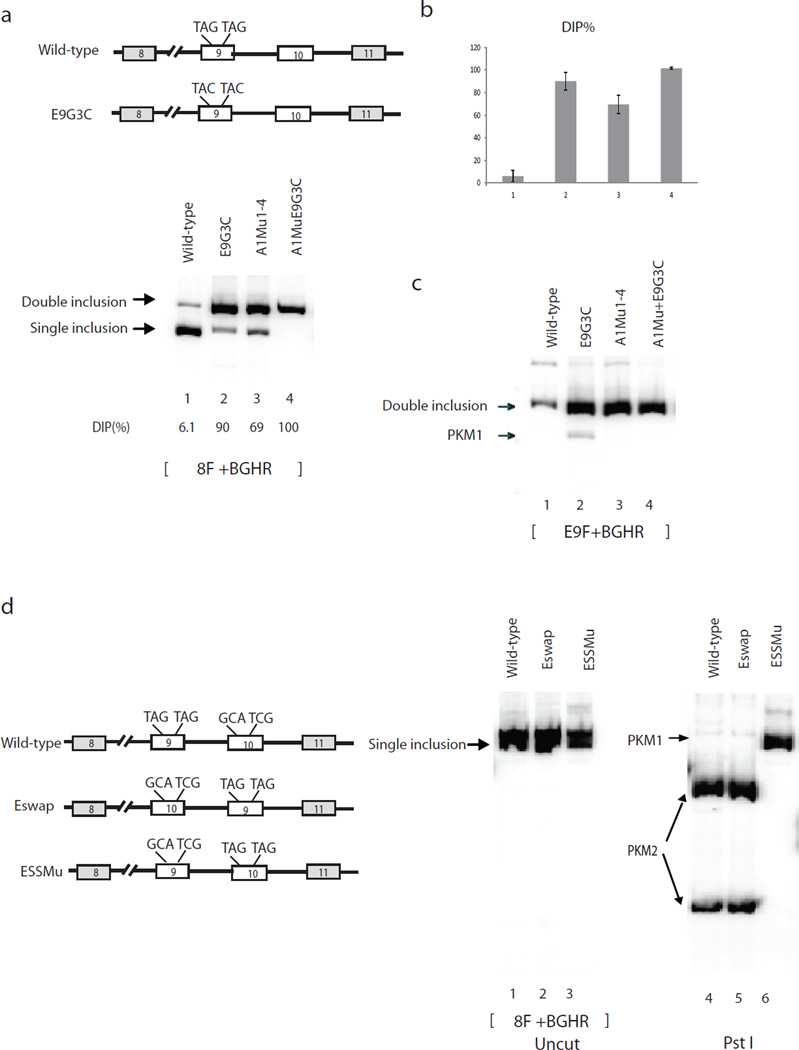
We next wanted to test whether the two exon 9 A1 binding sites defined above are important in regulating PKM splicing. To this end, we mutated both TAGs to TACs in the wild-type and A1Mu1-4 constructs (Fig. 4a). Strikingly, the ESS mutations in the wild-type construct sharply increased DIP, from 6.1% to 90% (Fig. 4a and 4b, lanes 1 and 2), while the same mutations in A1Mu1-4 also increased DIP, from 69% in A1Mu1-4 to 100% (Fig. 4a and 4b, lanes 3 and 4). In addition, we found that 8.3% of the exon 9-containing products were in the form of PKM1 with the ESS mutant, compared to 0% in wild-type (Fig. 4c, lanes 1 and 2). Together, these results indicate that the ESSs in exon 9 play a significant role in exon 9 exclusion.
Figure 4.
Exonic hnRNP A1/A2 binding sites are critical for exon exclusion. (a) Left top, schematic indicates E9G3C mutations in exon 9. Right, RT-PCR assays of RNAs isolated from plasmids transfected HeLa cells. PCR products are indicated on the left. Splicing constructs used for transfection are indicated on the top. The percentages of DIP in total products are indicated under lane numbers. (b) Bar calphalongraphs show percentages of DIP and SIP with standard deviation calculated from three independent experiments. Lane numbers correspond to lane numbers in Figure 3a, and the same lane numbers represent the same constructs. (c) RT-PCR with splicing constructs with ESS mutations using exon 9-specific primers. (d) Left panel, diagrams depict splicing constructs. ESSs of exon 9 and corresponding sequences of exon 10 are indicated on top of the exons. Right panel, RT-PCR assays of RNAs extracted from HeLa cells transfected with wild-type (lanes 1, 4, 7), Eswap (lanes 2, 5, 8) and ESSMu (lanes 3, 6, 9). PCR products were digested with Pst I (lanes 4–6) or Tth111 I (lanes 7–9). Splicing products are indicated with arrows.
We next wanted to investigate whether other exonic sequences besides the ESSs in exon 9 are important for PKM splicing regulation. One way to address this is to create TAG elements in exon 10 while simultaneously mutating the TAGs in exon 9 into the corresponding sequences in exon 10, and determining whether this is sufficient to switch PKM splicing from PKM2 to PKM1 (ESSMu, Fig. 4b, left panel, sequences indicated on top). Strikingly, we found that the mutations in ESSMu completely switched splicing from PKM2 (exon 10 inclusion) to PKM1 (exon 9 inclusion) (Fig. 4d, lanes 3 and 6). These results indicate that the two TAG elements are the only exonic sequences critical for determining whether exon 9 or exon 10 is included in proliferating cells.
Intron 9 and 10 sequences prevent double inclusion
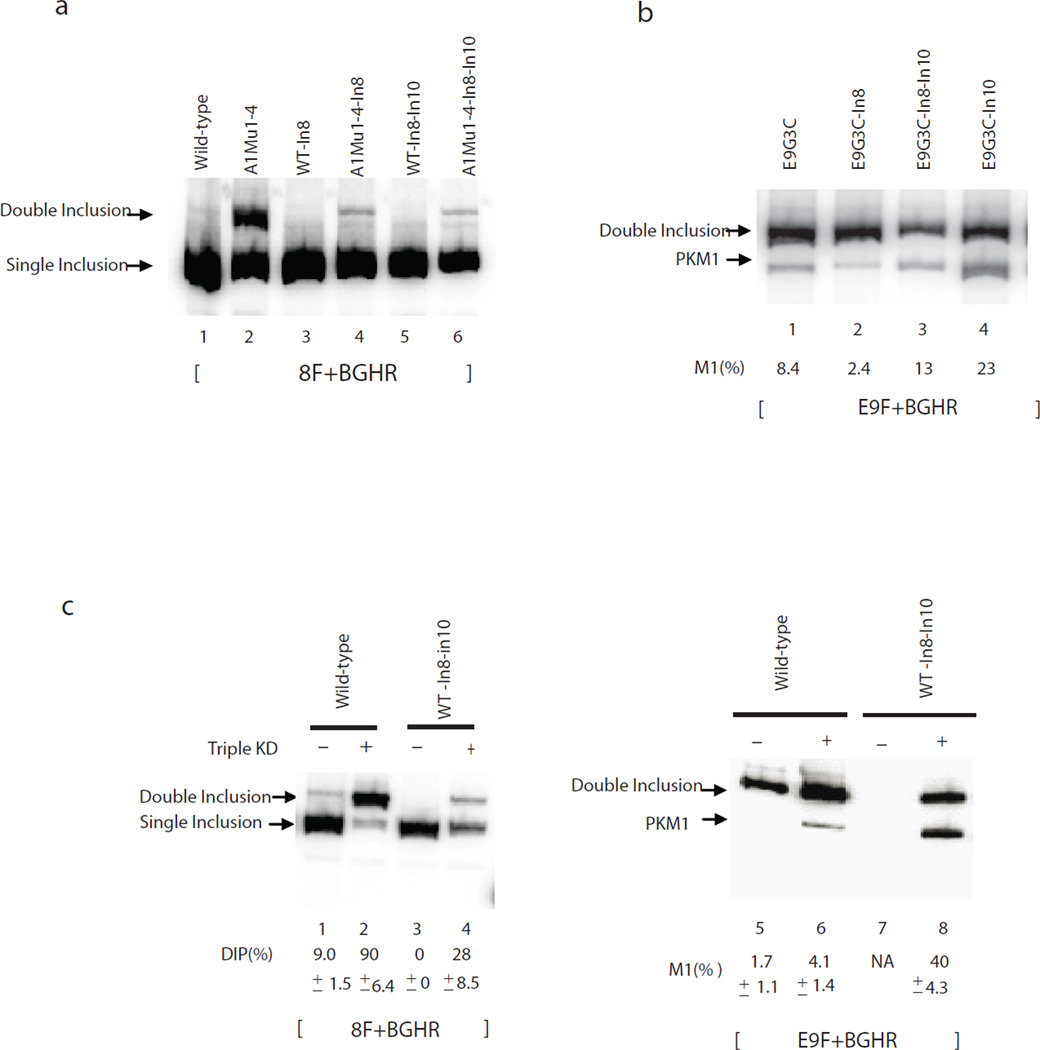
The above experiments have provided a detailed picture of how exon 9 is excluded from PKM mRNA to generate PKM2 mRNA. However, when exon 9 inclusion was increased by mutating ISSs or ESSs, most of the product detected was in the form of DIP, rather than SIP (PKM1) RNA. One explanation for this is that sequences in intron 8 and/or 10 missing in the minigene construct contribute to exon 10 exclusion. To investigate this, we first made a construct containing full-length intron 8 (WT-In8) and several derivatives containing mutations that increase exon 9 inclusion. However, none of these displayed an increase in the amount of PKM1 mRNA, although the amount of DIP was somewhat decreased (Fig. 5a, lanes 3 and 4 and Supplementary Fig. 2), perhaps due to the presence of additional repressive PTB sites in the added sequences28. We next inserted additional intron 10 sequences into two intron 8-containing constructs, to create WT-In8-In10 and E9G3C-In8-In10. Although addition of In10 did not by itself markedly change PKM splicing relative to that obtained with WT-In8 (Fig. 5a, lanes 5 and 6), we observed an increase in PKM1 levels (i.e., M1(%)), from 2.4% to 13%, with E9G3C-In8-In10 compared with E9G3C-In8 (Fig. 5b, lanes 2 and 3).
Figure 5.
Full-length intron 10 is required for exon 10 exclusion. (a) RT-PCR assays with T7 and 11R primers. Splicing products are indicated on the left. In8, the intron 8 sequences that were not included in wild-type. In10, the remainder of intron 10 sequences that is not included in wild-type. (b) RT-PCR assays with exon-specific primers. Splicing products are indicated on the left. M1(%) represents the percentage of M1 in all exon 9 included. (c) Lanes 1–4, PCR with 8F and BGHR primers to amplify PKM1, PKM2 and DIP products and DI/total% is indicated below. Lanes 5–8, exon 9-specific primers, the percentages of M1 in all exon 9 included products are indicated below.
We next wished to investigate whether intronic sequences contribute to exon 10 exclusion when A1/A2/ PTB levels are reduced. To this end, we transfected WT-In8-In10 and wild-type constructs into PTB/A1/A2 siRNA- or control siRNA-treated HeLa cells (knockdown efficiency was estimated by western blotting; Supplementary Fig. 3). PTB/ A1/A2 knockdown greatly increased the amount of DIP in wild-type, from 9% to 90% (Fig. 5c, lanes 1 and 2), with the de-repressed exon 9 found mainly in DIP (96%) rather than PKM1 (4.1%) (Fig. 5c, lane 6). WT-In8-In10 showed less DIP in both control and knockdown experiments than wild-type, due to the repressive function of full-length intron 8 (Fig. 5c, compare DIP in lanes 3 and 4 to lanes 1 and 2). Interestingly, however, following PTB/A1/A2 depletion, the amount of PKM1 produced from WT-In8-In10 increased substantially, from 4.1% to 40% (lanes 6 and 8). The enhanced PKM1 splicing appeared to be due solely to the expanded exon 10 sequences, as splicing of WT-In8 in PTB/A1/A2-depleted cells did not result in increased PKM1 production (Supplementary Fig. 2). To confirm this, we constructed and analyzed WT-In10 and E9G3C-In10, which contain intact intron 10 but not intron 8. WT-In10 showed a similar effect in reducing DIP and increasing SIP (38% PKM1) when PTB/A1/A2 were depleted (Fig. 5d, lanes 6 to 8) as did WT-In8-In10 (lane 7) (35% of PKM1). Likewise, E9G3C-In10 showed increased PKM1 compared to E9G3C, from 8.4% to 23% (Fig. 5b, lanes 1 and 4). Together, these results indicate that full-length intron 10 enhances PKM1 production by reducing exon 10 inclusion when A1/A2/PTB levels are reduced. We were unable to identify a specific region of intron 10, as constructs containing various intron 10 truncations failed to produce PKM1 (Supplementary Fig. 4)
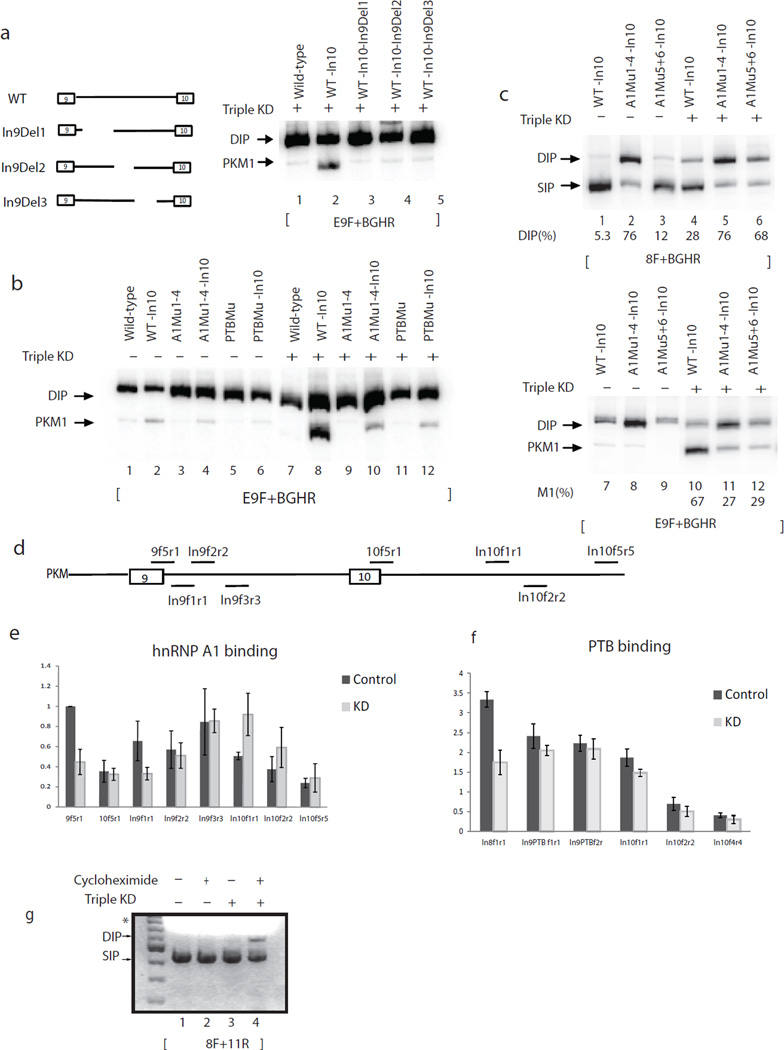
We next asked whether intron 9 sequences play a similar role in preventing double inclusion. First, several internal deletions of intron 9 in WT-In10 resulted in large reductions in PKM1 RNA, with a concomitant increase in DIP when PTB/A1/A2 levels were reduced (Fig. 6a), indicating that PKM1 production enhanced by full-length intron 10 also depends on full-length intron 9. We next cloned full-length intron 10 into the various intron 9 mutants analyzed in Figure 1 and examined if the A1/A2 and PTB binding sites in intron 9 play a role in exon 10 exclusion when PTB/A1/A2 levels are reduced. Consistent with the intron deletion results, these mutations greatly reduced PKM1 levels and increased DIP when PTB/A1/A2 levels were reduced (Fig. 6b, compare lane 8 and lanes 10 and 12 and Supplementary Fig. 5). Interestingly, we found that mutation of the two most distal A1/A2 binding sites in intron 9 did not affect exon 9 exclusion when PTB/A1/A2 levels were high (Fig. 6c, lane 3; see also Fig. 1e), but was sufficient to reduce exon 10 exclusion when PTB/A1/A2 levels were low (Fig. 6c, lanes 10 to 12).
Figure 6.
Two mechanisms prevent double inclusion PKM mRNA when hnRNP A1/A2 and PTB levels are low. (a–f) A1 and PTB repress exon 10 inclusion by binding to introns 9 and 10 when their levels are reduced. (a) Left panel, scheme indicates the positions of deletions in intron 9. Right panel, RT-PCR assays with exon 9-specific primers. Splicing products are indicated on the left. (b) RT-PCR assays with exon 9-specific primers and total RNA extracted from HeLa cells transfected with wild-type, WT-In10, A1Mu1-4, A1Mu1-4-In10, PTBMu and PTBMu-In10 in either control or A1/A2 and PTB depleted conditions. Splicing products are indicated on the left. (c) RT-PCR assays with primers 8F and BGHR (top panel) and exon 9-specific primers (lower panel) and total RNA extracted from HeLa cells transfected with WT-In10, A1Mu1-4-In10 and A1Mu5+6-In10 in either control or A1/A2 and PTB depleted conditions. Splicing products are indicated on the left. (d) Scheme indicates the positions of amplicons by primers used in CLIP assays. (e) CLIP of A1 in HeLa cells treated with control or A1/A2/PTB siRNAs. Primers used are indicated under each column. IPs were performed using antibody against A1 (4B10). The precipitated RNAs were subjected to RT-PCR. Data show the mean ± S.D. of triplicates from three independent experiments. (f) CLIP of PTB in HeLa cells treated with control or A1/A2/PTB siRNAs. Primers used are indicated under each column. IPs were performed using antibody against PTB (BB7). The precipitated RNAs were subjected to RT-PCR. Data show the mean ± S.D. of triplicates from three independent experiments. (g) PKM double inclusion mRNA is subject to NMD. RT-PCR with RNA extracted from HeLa cells treated with cycloheximide (as indicated) after PTB and A1/A2 were depleted by siRNAs (as indicated), using primers 8F and 11R to amplify endogenous PKM. PCR products were resolved in 1.5% (w/v) agarose gel and stained with ethidium bromide. Products are indicated on the left. *, the bright smear is from the loading dye.
Reduced levels of hnRNP A1 and PTB exclude exon 10
The results above suggest the intriguing possibility that A1/A2 and PTB have dual roles in PKM splicing regulation. When PTB/A1/A2 levels are high, the proteins bind strongly to sites in and around exon 9 to repress its inclusion, but when PTB/A1/A2 levels are reduced, PTB/A1/A2 binding to the exon 9-proximal sites is more dramatically reduced than is binding to the intronic sites flanking exon 10, and this functions to prevent exon 10 inclusion. To test this hypothesis, we performed crosslinking-immunoprecipitation (CLIP)29 using an hnRNP A1 antibody to detect A1 binding in cells at different positions on the PKM pre-mRNA (Fig. 6d) in both control and PTB/A1/A2 siRNA-treated HeLa cells. A1 binding was quantified by dividing RT-PCR values from IP signals by input RNA signals, and binding at the exon 9 5’ ss in control siRNA-treated cells was set to 1.0. As expected, A1 binding to the exon 9 5’ss was higher than to the exon 10 5’ss, consistent with its role in repressing exon 9 inclusion, and binding of A1 to the 5’ss of exon 9 was reduced by about 50% after A1/A2 levels were reduced (Supplementary Fig. 6). Interestingly, however, A1 binding to a region including A1 binding sites 5 and 6 (amplified by In9f3r3) did not decrease after A1/A2 depletion, while A1 binding to three regions in intron 10 containing consensus A1 binding sites actually increased (Fig. 6e and Supplementary Fig. 6). In vitro UV crosslinking experiments with purified A1 established that the differences in A1 crosslinking in vivo were not due to intrinsic differences in affinity (Supplementary Fig. 7).
We also examined binding of PTB to introns 8 and 9. As expected, binding of PTB to the site immediately upstream of exon 9 (In8f1r1) was reduced after PTB/A1/A2 knockdown. However, binding to sites in intron 9 (In9PTBf1r1 and In9PTBf2r2) was not reduced (Fig. 6f and Supplementary Fig. 8). These results suggest that, as with A1/A2, exon 9-proximal PTB binding was more sensitive to reduction in PTB levels than PTB binding deep within introns. The fact that the hnRNP proteins remained bound to the intron 9/10 sites after their partial depletion, along with the functional importance of the corresponding PTB/A1/A2 consensus sites in preventing double inclusion, point to a role for these proteins when present at reduced levels in prevention of DIP by inhibiting exon 10 inclusion (see Discussion).
Nonsense-mediated decay helps prevent PKM DI mRNA
While the above data shows that sequences in the introns flanking exon 10 function to reduce double inclusion, a significant amount of DIP mRNA was nonetheless still detected. In contrast, double included product is never detected from the endogenous. Exon 9 and10 are both 167 nt long, so a double inclusion mRNA results in a frameshift and generation of a premature stop codon. This suggests that PKM DIP would be subject to nonsense mediated decay (NMD). NMD has been shown to influence alternative splicing outcome in several studies30–33. To test this hypothesis, we treated HeLa cells with cycloheximide, a translation inhibitor known to block NMD, when PTB/A1/A2 were depleted using siRNAs and measured PKM mRNA by RT-PCR (Fig. 6g). A PKM DI product indeed appeared in the presence but not the absence of cycloheximide (lanes 3 and 4). No DI mRNA was detected when cells were treated with cycloheximide but not siRNAs (lane 2). This result was confirmed by depleting Upf1, a factor required for NMD34, together with PTB and hnRNP A1/A2 depletion (data not shown). We conclude that NMD helps to prevent accumulation of DI PKM transcript in cells in which exon 9 exclusion is de-repressed.
Discussion
In this study, we have provided a detailed analysis of the mechanism involved in regulation of an important MEAS event, the switch that determines whether PKM1 or PKM2 mRNA is made. Our data suggest that this mechanism is on the one hand simple, requiring only the activity of several well-characterized RNA binding splicing repressors, while on the other hand complex, involving multiple cis elements in and around the two alternative exons that respond differently to varying levels of these proteins. Importantly, our experiments indicate that the splicing repressors A1/A2 and PTB are sufficient to control exon 9 exclusion and exon 10 inclusion in proliferating cells. Perhaps most interestingly, we show that PTB/A1/A2 play a second role in PKM splicing. When their levels are reduced, they are displaced from binding sites flanking exon 9, but remain bound to intronic regions flanking exon 10, where they act to prevent exon 10 inclusion. Exon 10 exclusion however is not complete, and NMD functions to remove DIP (Fig. 7). Below we discuss these mechanisms in more detail, as well as the implications of our results with respect to alternative splicing control more generally.
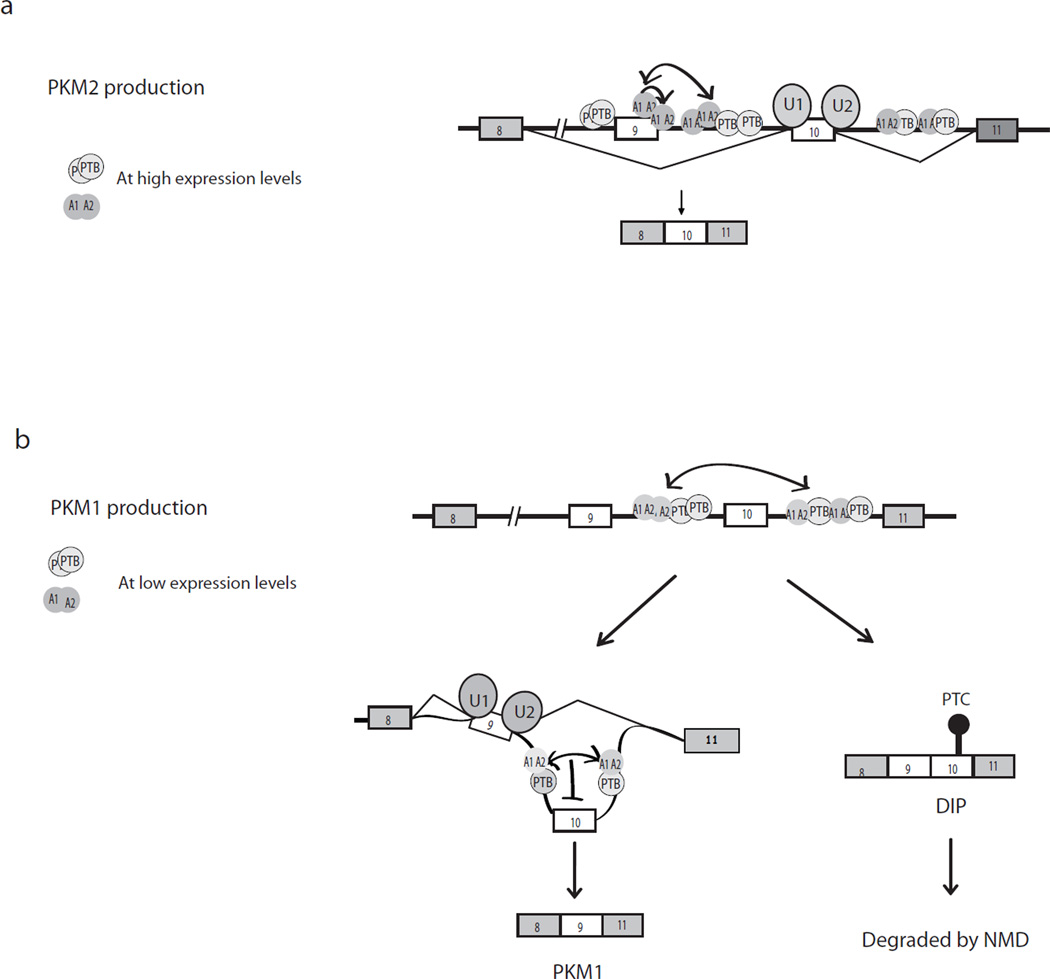
Figure 7.
A model for PKM mutually exclusive splicing. (a) PKM2 mRNA production. In the presence of elevated levels of A1/A2 and PTB, as in proliferating or cancer cells, A1/A2 bind to two ESS elements in exon 9 and ISS1 in intron 9 through cooperative binding (one-directional arrow), excluding exon 9 (wild-type). PTB and additional A1/A2 molecules bind to both intron 8 (PTB only) and intron 9. This further inhibits exon 9 inclusion, through formation of an inhibitory network of hnRNPs via protein-protein interactions (two-directional arrow). (b) PKM1 mRNA production. When A1/A2 and PTB are expressed at low levels, as in differentiated cells, binding in and around exon 9 is unable to compete with the splicing machinery, exon definition occurs and exon 9 is included. However binding to sites in the distal part of intron 9 and in intron 10 persists or is even increased at these low levels of A1/A2/PTB, and interactions between these molecules now excludes exon 10, likely via RNA looping. Unlike with exon 9 in the presence of high hnRNP levles, exon 10 exclusion is not complete. Therefore, double inclusion mRNA containing both exon 9 and exon 10 is produced, but is degraded by NMD.
While the details of the mechanism of exon 9 exclusion by cooperative binding of A1/A2 differ from other well-studied examples, overall it is consistent with a number of previous observations, and appears to hinge on the ability of hnRNP A1 to dimerize or multimerize. For example when A1 binds to two intronic binding sites flanking HNRNPA1 exon 7b, it “loops out” the intervening RNA to repress exon inclusion35,36. However, PKM exon 9 exclusion is unlikely to involve an RNA loop as the critical A1 binding sites, the two ESSs and the site overlapping the 5’ ss, are in close juxtaposition. It is likely that hnRNP A1, through cooperative interactions, forms a multimer that spans the 5’ ss and sterically blocks U1 snRNP binding. Although it has been shown that ISSs situated near a 5’ ss can interfere with U1 snRNP function but not binding37, in our case the overlapping nature of the sites strongly suggests that binding is inhibited.
Another repression model, known as the propagation model, is also based on A1 cooperative binding, and suggests that A1 molecules spread along RNAs following binding to a high affinity site26,27. While this is unlikely to apply to PKM splicing, as the principal mechanism of exon 9 exclusion involves cooperative binding to closely spaced sites, our finding that additional A1 sites situated downstream in intron 9 are important for full repression suggests that a more extensive array of bound A1 molecules contributes to exclusion. Although the cooperative binding of A1 just described plays a critical role, crosstalk between A1/A2 and PTB proteins also contributes. For example, our data, here and previously10, indicates that interactions involving a number of PTB sites in intron 8 contribute to repression by interfering with U2AF binding to the exon 9 3’ ss10. Our findings together indicate that the repressive context formed by the network of hnRNP binding sites is more extensive than we previously envisioned. Our data also show that the precise location of the exon within the inhibitory network is secondary to the presence or absence of ESS sequences within the exon, indicating a high degree of flexibility in the evolution of splicing regulation.
An additional significant aspect of our work is our demonstration that A1/A2 and PTB play dual roles in PKM splicing, depending on their expression levels. How might the switch from repression of one exon to another occur? We suggest the following model (Fig. 7): When A1/A2 levels are high, the proteins bind to multiple sites in intron 9 and intron 10, and also bind strongly to exon 9 and the exon 9 5’ss, resulting in exon 9 exclusion. However, at lower levels, they are less able to compete with the splicing machinery for binding to the exon 9 5’ss, exon definition occurs and the hnRNP proteins are displaced. In contrast, A1/A2 bound to sites deep in intron 9 are not subject to competition with the splicing machinery, and occupancy is therefore not decreased, even when A1/A2 levels are sharply reduced. PTB behaves in a similar manner, with exon 9-proximal binding more sensitive to low PTB levels, while intronic binding is maintained. Importantly, A1 binding to intron 10 sites is actually increased when A1/A2 levels are reduced. Given the high degree of cooperativity in A1 RNA binding, shown here and elsewhere, we hypothesize that this increased binding results from cooperative interactions with A1 molecules bound to intron 9. When A1/A2 levels are high, A1/A2 molecules bound in and around exon 9 interact with A1/A2 bound to intron 9, contributing to exon 9 exclusion and precluding their interaction with molecules bound to the intron 10 sites. This may also be favored simply by the fact that the upstream sites are transcribed first and therefore available for binding before the downstream sites. After displacement of A1/A2 from the exon 9 5’ ss, intron 9-bound A1/A2 molecules interact with and promote A1/A2 binding to the intron 10 sites, explaining the increased binding of A1/A2 to intron 10 observed at lower PTB/A1/A2 levels. Such cross-exon interactions would likely form an RNA loop, resulting in exon 10 exclusion. An important implication of these results is the likelihood that exonic hnRNP binding is more sensitive to changes in cellular hnRNP concentration than binding to intronic sites, due to competition with the splicing machinery. This concept is likely to have broad implications for regulation of alternative splicing in general.
Accumulation of double inclusion PKM mRNA is also limited by NMD38. Up to one third of alternative splicing events create a PTC38. While our data has established a role for this process in PKM splicing regulation, an interesting question is whether NMD is commonplace in MEAS regulation. To address this, we searched the MEAS events identified by Wang et al.1, and analyzed the 34 genes in which two alternative spliced exons were annotated in the UCSC Genome browser. Among the 34 MEAS events, 25 transcripts would contain a PTC that triggers NMD when both exons are included. Of the nine transcripts that would not create a PTC, six of them contain a very short intron between the two ME exons (from 1 nt to 64 nts). Given that a minimum size of ~66 nt is required for splicing in mammalian cells39, this small size is by itself sufficient to prevent DIP. This analysis suggests that NMD and short intron size provide two mechanisms generally used to prevent DI during MEAS. As we found with PKM, this implies that regulation of inclusion/exclusion of only one of the two ME exons must be stringently regulated during MEAS.
In conclusion, we have provided a detailed picture of how MEAS can be regulated. We have shown that hnRNP A1/A2 and PTB are sufficient to regulate PKM splicing, and that this occurs through a network of binding sites that respond differently to varying levels of the proteins. It will be of interest in the future to learn how widely this simple mechanism, or related ones, is used to regulate MEAS.
Methods
Plasmid constructs
Ultraviolet (UV) crosslinking substrates EI9s and EI10s were prepared by inserting an HpaI site downstream of ISS1 and ISS1-10 in EI9 and EI10 constructs10 by PCR-based site-directed mutagenesis with Pfu Turbo (Stratagene). The primers were:
EI9HpaIF, ggagtctggcaggtagggcccgttaactaagggcaggtaacactg;
EI9HpaIR, cagtgttacctgcccttagttaacgggccctacctgccagactcc;
EI10HpaIF, caagtctggcagtaggaggcgttaacggcagcggctccctggaatg;
EI10HpaIR, cattccagggagccgctgccgttaacgcctcctacctgccagacttg.
EI9s and EI10s mutants were prepared with site-directed mutagenesis. The constructs for synthesizing the 5’ RNA oligos for PCR-based site-specific labeling were prepared by inserting a BstZ17I site at the 4th position intron 9 in EI9 construct. The primers used were:
EI9BSTZF: gacggagtctggcaggtatacgggccctaagggcaggt;
EI9BSTZR: acctgcccttagggcccgtatacctgccagactccgtc.
The wild-type splicing construct was prepared as described10. Intronic and exonic mutants were prepared by PCR-based site-directed mutagenesis with primers containing desired mutations in the center and complementary sequences on both sides of the point of mutations. E9del1/2, E10del1/2 and Eswap were prepared by site-directed mutagenesis.
Transfection and splicing assays
We transiently transfected PKM-derived plasmids into HeLa cells with Lipofectamine 2000 (Invitrogen) as described7. After 24 hours, we isolated total RNA and reverse-transcribed 5 µg RNA, using 0.5 µg of oligo (dT)15 primer and Maxima reverse transcriptase (Fermentas). Resultant cDNAs were PCR amplified with PKM-specific forward primer 8F and a plasmid-specific reverse primer BGHR. Primer sequences are as follows: 8F, ctgaaggcagtgatgtggcc; BGHR, tagaaggcacagtcgaggct; E9F, cgcggatccttcttcttataagtgtttagcagcagct; T7F, gactcactatagggagaccc; E10R, cgggatccctgccagacttggtgaggacg; 11R, acccggaggtccacgtcctc.
For semi-quantitative PCR assays, we amplified 1 µl of cDNAs in 50 µl of standard reaction mixtures containing 0.6 µCi [α-32P]dCTP (3,000 Ci mmol−1; Perkin Elmer) for 26 cycles. PCR products were resolved on gels buffered with 4% polyacrylamide and 90 mM Tris, 89 mM boric acid and 2 mM EDTA, pH 8.3, visualized by autoradiography and quantitated by using a PhosphorImager (Molecular Dynamics). Due to the slight difference in the number of Cs in the PCR products, the percent DI calculated was slightly overestimated. However the effect was small (~10%) and did not affect our conclusions.
RNA interference
We carried out RNA interference as described10 with minor modifications. Briefly, we reverse transfected with 25 pmol of hnRNPA1 duplex RNA and 12.5 pmol of the other duplex RNAs into HeLa cells, cultured in DMEM supplemented with 10% FBs (Hyclone), at 1–2 × 104 cells per well in 24-well plates with lipofectamine RNAi Max reagent (Invitrogen). The next day, siRNA transfection was repeated and cells were split into 12-well plates. siGENOME non-targeting siRNA (Dharmacon) was used as a control to ensure that parallel experiments had equal amounts of RNA. To express splicing constructs in PTB and hnRNP A1/A2 depleted cells, we transfected plasmids after 48 hours of the initial siRNA knockdown. Seventy-two hours after the initial siRNA transfection, we collected cells for total RNA isolation and immunoblotting. We used the following siRNAs (Dharmacon; the sense strand sequences are given): hnRNPA1, cagcugaggaagcucuuca; hnRNPA2, ggaacaguuccguaagcuc; PTB, gccucaacgucaaguacaa.
UV crosslinking assays and immunoprecipitation
We carried out ultraviolet (UV) crosslinking assays as described in7, with some modifications. EI9s and EI10s were synthesized in vitro by T7 RNA polymerase (Promega) with [32P]-UTP from HpaI-linearized plasmids. We incubated ~10 fmol (1×105 c.p.m.) of RNA with 2 µl of HeLa nuclear extract in 10 µl reaction mixtures. Reaction mixtures were incubated at 30°C for 15 min, and samples were then irradiated with UV light using a Stratalinker (Stratagene), treated with RNaseA (USB), and proteins resolved by 10% (w/v) SDS/PAGE. We carried out site-specific labeling as described in ref. 40. The 3’ RNA oligo was purchased from Dharmacon and the 5’ RNA oligos was transcribed in vitro with T7 RNA polymerase from BstZ17I-linearized plamids. The 3' oligo fragment was 5’-end-labeled with [γ-32P]-ATP and T4 polynucleotide kinase (NEB). The two RNAs were annealed to a DNA bridging oligonucleotide complementary to the 3’ end of the 5’' RNA fragment and the 5' end of the 3' RNA fragment and ligated using T4 DNA ligase for 4 hours at room temperature. The ligation product was purified following electrophoresis on a 10% (w/v) denaturing polyacrylamide gel. The sequences of the 3’ oligo and bridging DNA are as follows: 3’ oligo, gggcccuaag; bridging DNA, cccttagggccctacctgccagactccgtcagaactatcaaagc. UV crosslinking-IP was described in ref. 10.
Recombinant proteins
hnRNP A1 cDNA and truncations were cloned intothe pGEX6p-1 vector. GST-tagged hnRNP A1, UP1, RRM2-A1 and RRM2 were purified from E. coli using glutathione–Sepharose 4B (GE healthcare), and the GST tag was cleaved off with Prescission protease (GE healthcare). Purity and concentration of proteins were determined by Coomassie blue staining of SDS gels.
Crosslinking-immunoprecipitation
HeLa cells were cross-linked in vivo, and the RNA-binding targets for hnRNP A1 and PTB were obtained as described41,42, with the following modifications. In brief, 15 cm dishes were cross-linked at 400 mJ cm−2, cells were collected and total cell extracts were prepared in lysis buffer (1×PBS, 0.5% (v/v) Triton-X100. After DNase treatment and a mild digestion with RNase, the extracts were immunoprecipitated for 4 h at 4 °C with the monoclonal antibody 4B10 (Immuquest) coupled to protein A–Sepharose beads (GE) and BB7 (ATCC) bound to protein G-Sepharose beads (GE). After extensive washing with wash buffer (1×PBS, 0.5% (v/v) Triton-X100, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate for 4B10 and PK buffer (50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10mM EDTA), the cross-linked RNAs were eluted with 4mg ml−1 Proteinase K (USB) in PK buffer and then PK buffer containing 7M Urea. RNAs were then precipitated, treated with DNase RQ1 (Promega) and purified again before RT-PCR analysis as described in “Transfection and splicing assays”, except that random hexamer was used in the RT step. The primers used in PCR were as follows: 9f5, gcagcagctttgatagttctgacgg; 9r1; ggctggttatcctaaca; 10f5, tggggccataatcgtcctcacca; 10r1, ccactgagcagggcatt; In8f1, tgttgtgtctcgtttttttcctcctcc; In8r1, ctcacgagctatctgtaaggtttagg; I9PTBf1, cacttggtgaaggactggtttctgtgg; I9PTBr1, gaggagcccaatcactggagattctg; I9PTBf2, gttcctcaccagctgtccggtga; I9PTBr2, ggacagagctttgtcagagctttgtc; In9f1, ggccctaagggcaggtaacac; In9r1, ggttgggcctggctgtcttc; In9f2, ccagcctcttgctccacctg; In9r2, gacctccctggcggtgttc; In9f3, gaccaaagggtcctgtggctc; In9r3, cagtccttcaccaagtgcctgtg; In10f1, ggcccaggacattgagttaatg; In10r1, ccctgtggacagggacaagg; In10f 2, cccactcttggctgaacagc; In10r2, gcaagaactggaccttttaagaaaggt; In10f4, ccagttcttgctgtttggctattttaag; In10r4, cctgcacttcacgcttggcc; In10f5, gggcctttatagcccagttgct; In10r5,ctgggaatcaacatcaggcttctct.
Supplementary Material
Acknowledgements
We thank T. Kashima for hnRNP A1/A2 siRNA, E. Rosonina for help with semi-quantitative PCR assays, D. Campigli Di Giammartino for discussion regarding the RNA immunoprecipitation experiments, P. Richard for help with 5’ labeling, and members of the Manley laboratory for discussions. This work was supported by a grant from the NIH R01 GM048259 (J. L. M.).
Footnotes
Author Contributions
M.C., C. J. D. and J. L. M. conceived the project. M. C. and J. L. M. designed the experiments. C. J. D. discussed the experiments and results and performed the experiment in Fig. 6g and M. C. performed the rest of the experiments. M. C. and J. L. M. wrote the paper.
References
- 1.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010 doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 7.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 8.Kashima T, Rao N, Manley JL. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci U S A. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 10.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, David CJ, Manley JL. Tumor metabolism: hnRNP proteins get in on the act. Cell Cycle. 2010;9 doi: 10.4161/cc.9.10.11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takenaka M, et al. Isolation and characterization of the human pyruvate kinase M gene. Eur J Biochem. 1991;198:101–106. doi: 10.1111/j.1432-1033.1991.tb15991.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Zhang J, Manley JL. Turning on a Fuel Switch of Cancer: hnRNP Proteins Regulate Alternative Splicing of Pyruvate Kinase mRNA. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2010 doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 17.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 18.Zerbe LK, et al. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: implications for pre-mRNA processing. Mol Carcinog. 2004;41:187–196. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]
- 19.Pino I, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–143. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 20.Moran-Jones K, Grindlay J, Jones M, Smith R, Norman JC. hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res. 2009;69:9219–9227. doi: 10.1158/0008-5472.CAN-09-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 23.Cartegni L, et al. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 24.Cobianchi F, Karpel RL, Williams KR, Notario V, Wilson SH. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1. Large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988;263:1063–1071. [PubMed] [Google Scholar]
- 25.Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA. 2002;8:1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 27.Okunola HL, Krainer AR. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol. 2009;29:5620–5631. doi: 10.1128/MCB.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 30.McGlincy NJ, et al. Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics. 2010;11:565. doi: 10.1186/1471-2164-11-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones RB, et al. The nonsense-mediated decay pathway and mutually exclusive expression of alternatively spliced FGFR2IIIb and -IIIc mRNAs. J Biol Chem. 2001;276:4158–4167. doi: 10.1074/jbc.M006151200. [DOI] [PubMed] [Google Scholar]
- 34.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 35.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchison S, LeBel C, Blanchette M, Chabot B. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5' splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277:29745–29752. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, et al. Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Fu XY, Colgan JD, Manley JL. Multiple cis-acting sequence elements are required for efficient splicing of simian virus 40 small-t antigen pre-mRNA. Mol Cell Biol. 1988;8:3582–3590. doi: 10.1128/mcb.8.9.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Method References
- 40.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: the 2'-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 41.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.