Abstract
DNA transfer from chloroplasts and mitochondria to the nucleus is ongoing in eukaryotes but the mechanisms involved are poorly understood. Mitochondrial DNA was observed to integrate into the nuclear genome through DNA double-strand break repair in Nicotiana tabacum. Here, 14 nuclear insertions of chloroplast DNA (nupts) that are unique to Oryza sativa subsp. indica were identified. Comparisons with the preinsertion nuclear loci identified in the related subspecies, O. sativa subsp. japonica, which lacked these nupts, indicated that chloroplast DNA had integrated by nonhomologous end joining. Analyzing public DNase-seq data revealed that nupts were significantly more frequent in open chromatin regions of the nucleus. This preference was tested further in the chimpanzee genome by comparing nuclear loci containing integrants of mitochondrial DNA (numts) with their corresponding numt-lacking preinsertion sites in the human genome. Mitochondrial DNAs also tended to insert more frequently into regions of open chromatin revealed by human DNase-seq and Formaldehyde-Assisted Isolation of Regulatory Elements-seq databases.
Keywords: endosymbiotic gene transfer, chloroplast, mitochondrion, double-strand break repair, open chromatin
Introduction
Endosymbiont DNAs have constantly bombarded the nucleus since the appearance of eukaryotes, and it is usual for nuclear genomes to contain multiple chromosomal integrants derived from cytoplasmic organellar genomes (Timmis et al. 2004). This process of DNA escape and integration has resulted in massive functional relocation to the nucleus of genes that once belonged to the prokaryotic ancestors of mitochondria and chloroplasts. Simple DNA transposition and functional gene relocation from the extant organellar genomes have both been demonstrated experimentally (Thorsness and Fox 1990; Huang et al. 2003; Stegemann et al. 2003) and found to occur at previously unexpectedly high frequencies. However, the mechanisms responsible for organellar DNA escape and incorporation into the nuclear genome have not been extensively investigated and relatively little is known about how relocated genes become functional.
DNA double-strand break (DSB) repair sometimes results in the integration of mitochondrial DNA into the nuclear genome in yeast (Ricchetti et al. 1999) and tobacco (Wang et al. 2012a), and chloroplast DNA incorporation is also implicated during repair of DSBs by nonhomologous end joining (NHEJ) (Lloyd and Timmis 2011). Bioinformatic analysis also suggests that NHEJ is involved in the formation of nuclear integrants of mitochondrial DNAs (numts) in primate genomes (Hazkani-Covo and Covo 2008).
Moreover, recent human numts were shown to insert preferentially into genes, especially into introns (Ricchetti et al. 2004) that are often flanked by regions of open chromatin (Tsuji et al. 2012). However, whether this is a general rule, and whether it applies to the plastid counterparts of numts (nuclear integrants of plastid DNAs [nupts]), has not been investigated. A recent study in tobacco showed that mild heat stress increases DNA transfer from chloroplast-to-nucleus (Wang et al. 2012a) suggesting the possibility that heterochromatin relaxation that is associated with heat stress (Pecinka et al. 2010) may be responsible. We have investigated the hypothesis that cytoplasmic organelle DNA tends to integrate preferentially into DNA in open chromatin regions by comparing nuclear organelle DNAs (norg) maps with chromatin status as revealed by DNase I hypersensitivity (DH) sites or Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) (Boyle et al. 2008; Song et al. 2011).
We identified recent nupts that are unique to Oryza sativa subsp. indica, which had inserted into the nuclear genome after its very recent divergence from O. sativa subsp. japonica. The mechanism involved in the plastid DNA integration into nuclear genome was analyzed. Furthermore, the nupt insertion sites were studied to determine whether they include, or are flanked by, DH sites in chromatin derived from seedling or callus tissues. A related study (Tsuji et al. 2012) found that numt loci in the human nucleus were often found in open regions of open chromatin. However, a problem with this approach is that it examines the chromatin status of numt junction sites after, rather than before insertion. Thus, there is no certainty that mitochondrial DNA inserted into pre-existing open chromatin, and it is possible that the numt insertion event may cause chromatin relaxation. To avoid this ambiguity, and to investigate the generality of norg insertion mechanisms, we identified chimpanzee-specific numts and characterized the equivalent loci in the human genome, allowing us to analyze the probable chromatin status before mitochondrial DNA insertion. Preinsertion loci were also analyzed for Oryza nupts, in the same manner as for chimpanzee-specific numts.
Materials and Methods
Identification of O. sativa Subsp. indica-Specific nupts
The nuclear, chloroplast, and mitochondrial genome sequences of O. sativa subsp. indica and O. sativa subsp. japonica were downloaded from NCBI (versions are described in the supplementary table S1, Supplementary Material online). The O. rufipogon nuclear genome was from Rice Haplotype Map Project Database (Huang et al. 2012). Nupts present in the subsp. indica genome were identified using BlastN (version 2.2.23) (Altschul et al. 1990). Local BlastN was carried out with the parameters previously described (Wang, Rousseau-Gueutin, et al. 2012). The same process was used to identify nupts in O. sativa subsp. japonica and O. rufipogon. Then, nupts that appeared only in O. sativa subsp. indica were selected and those that could not be located in O. sativa subsp. japonica and O. rufipogon genomes were eliminated from the study. A total of 14 nupts whose loci were clearly identified in O. sativa subsp. japonica and O. rufipogon were analyzed in detail.
NHEJ Analysis
The NHEJ analysis was as previously described (Hazkani-Covo and Covo 2008). In short, nupts were classified by known NHEJ patterns (microhomology and blunt end repair). Microhomology was identified only if the nucleotide(s) adjacent to the fusion point was shared among the nupt, the corresponding subsp. japonica and rufipogon nuclear sequences, and the plastomes of subsp. japonica or indica. If no microhomology was found, then the NHEJ was classified as a blunt-end repair. Any sequences of less than 10 nucleotides found at the junction sites, other than known nuclear, mitochondrial, or chloroplast DNA, was classified as a nontemplate insertion.
Open Chromatin Regions
To test whether insertion sites of norgs correlate with open chromatin regions, we downloaded the open chromatin data generated by DNase-seq for O. sativa subsp. japonica (Zhang et al. 2012) and human (Song et al. 2011) and the human database of FAIRE-seq (Song et al. 2011) from NCBI. Four cell lines were chosen for analysis, H1-ES has highest coverage of open chromatin by DNase-seq; and GM12878 has least coverage. HUVES has most coverage of open chromatin through FAIRE-seq, and HeLa-S3 has least coverage (Song et al. 2011). The position of individual norgs and their 1 kb flanking regions was superimposed on the coordinates of open chromatin to identify the norg chromatin status.
Results
Comparative Analysis of Nupt Integration Sites in Oryza Species Supports NHEJ-Mediated Chloroplast DNA Insertion
Using O. rufipogon, the wild progenitor of O. sativa (Khush 1997) as a control, we identified nupts that had inserted into the nuclear genome of O. sativa subsp. indica after its divergence from subsp. japonica (fig. 1). By comparing the same loci and their flanking genomic regions between the two subspecies, we were able to deduce the mechanism of DSB repair (Hazkani-Covo and Covo 2008). We reasoned that, if O. sativa subsp. indica contains a nupt that is absent at the equivalent loci in both O. sativa subsp. japonica and O. rufipogon, the latter two taxa will reveal the nupt preinsertion site. Therefore, the differences among the chromosomal nupt loci of these three Oryza taxa may be considered as record of the insertion process.
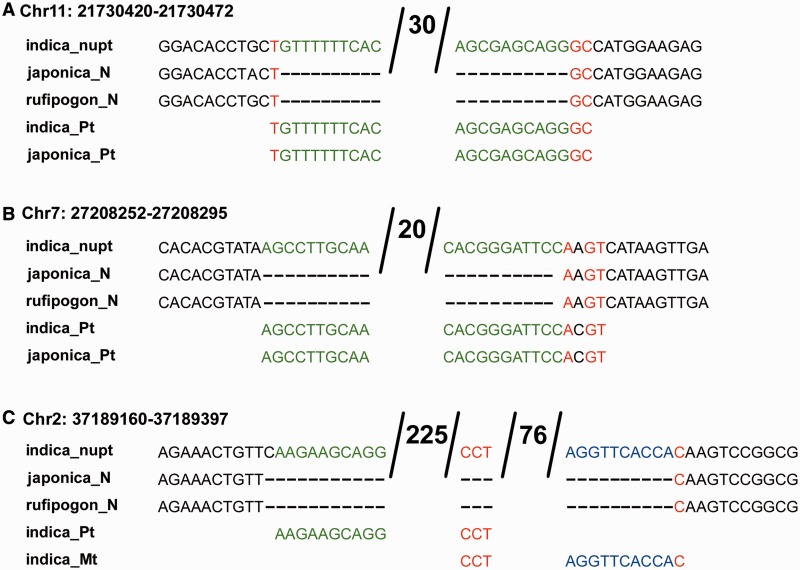
Fig. 1.—
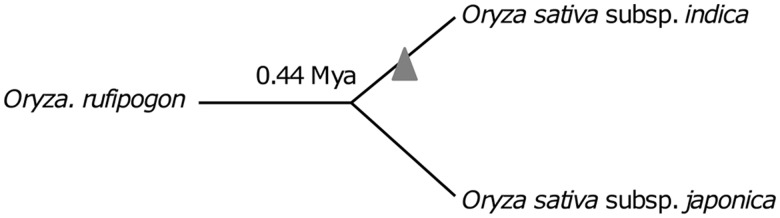
A phylogenetic tree of Oryza rufipogon, O. sativa subsp. Indica, and subsp. japonica showing the recent nupt insertions (gray triangle) used to investigate nupt insertion mechanisms. Subspecies indica and japonica diverged 0.44 Ma (Ma and Bennetzen 2004).
Among the 14 insertional events with their 14 × 2 molecular ligation points, eight involved perfect or slightly imperfect microhomology of more than 1 bp (fig. 2 and supplementary table S1, Supplementary Material online), with a single matching base seen at six other junctions (supplementary table S1, Supplementary Material online), implicating DSB repair by NHEJ. The remaining 14 junctions involved blunt-end ligation (supplementary table S1, Supplementary Material online). Consistent with the observations in primate numts (Hazkani-Covo and Covo 2008), only 2 of the 14 nupt insertions resulted in deletion of nucleotides, suggesting that DSB repair with cytoplasmic organelle DNA insertion reduces sequence loss when the break is healed. It is known that DSB repair of incompatible ends always involve deletion of a few nucleotides (Guirouilh-Barbat et al. 2004; Nick McElhinny et al. 2005; Lloyd et al. 2012).
Fig. 2.—
Examples of nupt-mediated DSB repair reflect NHEJ. Alignments show Oryza sativa subsp. indica loci containing a nupt, their corresponding nuclear sequence in subsp. japonica and O. rufipogon (TIGR database release 5), and the chloroplast DNA sequences (Pt) from subsp. indica and japonica. (A) A nupt insertion (shown in green) that involved short microhomology (1 or 2 bp shown in red) at both fusion points. The number 30 indicates nucleotides in the nupt that are identical to chloroplast DNA. (B) A nupt that involved imperfect microhomology at the right fusion point and blunt-end repair at the left fusion point. The number 20 indicates nucleotides in the nupt that are identical to chloroplast DNA. (C) A chimeric insertion containing a nupt of 225 bp (in green) and a numt of 76 bp (in blue). Complementary microhomology of AGG in the norg with CCT in chloroplast DNA and mitochondrial DNA is marked red. The subsp. indica mitochondrial DNA sequence (Mt) is shown in blue. The numbers 225 and 76 indicate nucleotides that are identical to chloroplast DNA and mitochondrial DNA, respectively.
Three chimeric insertions involving both mitochondrial- and chloroplast-derived sequences were observed, confirming their relative abundance as described previously (Lloyd and Timmis 2011; Wang et al. 2012a). The nupts examined show extensive variation, consisting of DNA fragments originating from different parts of the chloroplast circular genome, mixed chloroplast and mitochondrial DNA integrants (fig. 2), and mosaic DNA inserts containing nuclear, mitochondrial DNA, and chloroplast DNA (supplementary table S1, Supplementary Material online). Two of the loci reveal that mitochondrial and chloroplast DNA fragments also join together through microhomology (fig. 2C).
These results suggest that norg insertion precludes deletion at DSBs and involves both blunt-end repair and variable lengths of microhomology, whereas, if other filler DNAs are included in repairs, there is often some deletion at the preinsertion site (Lin and Waldman 2001a, 2001b). For example, capture of adeno-associated virus in I-SceI-induced breaks is associated with a high frequency of deletion (Miller et al. 2004; Hazkani-Covo and Covo 2008). Thus, our results suggest that organelle DNAs play a role in preserving genome integrity during potentially deleterious DSB repair (Hazkani-Covo and Covo 2008).
Norg Insertion Sites Prefer Open Chromatin Regions
Open chromatin regions often show nucleosome depletion that allows genomic DNA segments to be exposed to interacting molecules (Hogan et al. 2006; Kim et al. 2007; Song et al. 2011). The recent nupts we identified in subsp. indica provided an opportunity to examine whether open chromatin is more accessible to organellar DNA insertion. For each subsp. indica-specific nupt, we reasoned that the preinsertion sites located in the O. sativa subsp. japonica genome would represent the chromatin status before integration. Therefore, DNase-seq data for O. sativa subsp. japonica (Zhang et al. 2012) shed light on subsp. indica chromatin status before nupt insertion. Crosschecking subsp. japonica preinsertion sites together with 1 kb of flanking DNA revealed that 6 cases of the 14 examined were located in open chromatin regions in seedling chromatin, and 8 were seen in callus (table 1). The rice genome contains 420 Mb of DNA, of which only 5% and 7% is DNase I hypersensitive in seedling and callus chromatin, respectively (Zhang et al. 2012). Therefore, on average, one of these 14 recent insertions is expected in 30 Mb of the genome if the events are random. However, one of these recent nupts occurs in 16 kb (seedling) or 19 kb (callus) of DH DNA, indicating that nupt insertion strongly favors open chromatin (P < 0.0001, χ2 test).
Table 1.
Open Chromatin Data and Norg Descriptions Used in This Analysis
| Rice Nupt |
Primate Numt |
||||||
|---|---|---|---|---|---|---|---|
| Tissue | DH Sites in subsp. japonica (%) | Number of subsp. indica nupts. | Human Cell Line | Human DH Sites (%) | Number of Chimpanzee Numts Colocated | Human FAIRE Sites (%) | Number of Chimpanzee Numts Colocated |
| Seedling | 97,975 (5) | 6 | GM12878 | 103,075 (1.528) | 6 | 146,147 (0.728) | 10 |
| Callus | 155,025 (7) | 8 | HeLa-S3 | 142,403 (2.174) | 7 | 131,935 (0.694) | 2 |
| H1-ES | 138,025 (3.224) | 7 | 126,439 (0.695) | 6 | |||
| HUVEC | 133,091 (2.259) | 3 | 225,564 (1.723) | 5 | |||
Note.—Human cell line descriptions, GM12878, lymphoblast; Hela-S3, cervical carcinoma; H1-ES, human embryonic stem cells; HUVEC, human umbilical vein endothelial cells. Percentages (in parentheses) indicate the proportion of the rice or human genome identified as open chromatin regions. Example 1:6 of the 14 subsp. indica nupts (43%) examined were located in 5% of the genome identified as DH sites in subsp. japonica seedling tissues. Example 2:10 of the 52 (19%) chimpanzee numts examined were located in 0.728% of the human genome identified as FAIRE sites in human cell line GM12878.
To investigate whether norg insertion in eukaryotes favors open chromatin in mammalian systems, a similar analysis was carried out for 52 previously identified species-specific numts in the chimpanzee genome (version, panTro 2) (Hazkani-Covo and Covo 2008). The corresponding loci were then unequivocally located in the human genome (version, hg 18) (Karolchik et al. 2004), and 1 kb of flanking DNA was compared with open chromatin coordinates (table 1) defined by DNase-seq and FAIRE-seq in four different human cell lines (Song et al. 2011). In common with their nupt counterparts in Oryza, chimpanzee numt insertion sites strongly favored open chromatin in all the cell lines tested (P < 0.0001, χ2 test). Furthermore, different cell lines showed different degrees of preference for norg insertion, reflecting their known differences in chromatin status (Song et al. 2011). Consistent with these observations, more norgs correlate with open chromatin in parallel with the number of DH or FAIRE sites reported (table 1) in different human cell lines or in different plant tissues. We conclude that cytoplasmic organelle DNA preferentially inserts into open chromatin regions in diverse eukaryotes. Open chromatin regions are likely to be more accessible to the proteins involved in DNA breakage and repair, necessarily leading both to more cleavage and their more successful healing, sometimes with the incorporation of available mitochondrial or chloroplast DNA fragments.
Discussion
NHEJ has been suggested to associate with chloroplast DNA insertion into nuclear genome (Lloyd and Timmis 2011), and this is supported by the current comparative analysis using Oryza genome sequence data. Moreover, NHEJ-mediated DSB repair that includes chloroplast DNA insertion may protect genome integrity by precluding deletions, though insertional mutagenesis may be a by-product. However, mutation may be alleviated as approximately half of de novo nupts are unstable and may be very quickly deleted from the genome (Sheppard and Timmis 2009), though the precision or otherwise of excision remain to be established. The frequency of DNA transfer from chloroplast to nucleus is different in different tissues (Huang et al. 2003; Stegemann et al. 2003; Sheppard et al. 2008), and it is known that the frequency of organellar DNA transposition is positively correlated with the amount of available organelle DNA (Wang et al. 2012a, 2012b). Here, we describe that DNA transfer from organelle to nucleus also tends to occur in regions of open chromatin. Analyses of human chromatin data (Tsuji et al. 2012) suggested that human numt insertion sites are often colocalized with open chromatin regions. This analysis cannot rule out the possibility that open chromatin status results from, rather than being responsible for, numt insertion. Therefore, we studied the preference of chimpanzee-specific numt insertions by analyzing the preinsertion status revealed in human chromatin. This provided direct evidence that norgs insert into open chromatin (this article), and it is probable that the loci remain transcriptionally active after insertion (Tsuji et al. 2012). Thus, chromatin state appears to be a significant contributor to the successful relocation of cytoplasmic organellar genes to the nucleus. Moreover, as norgs are also present that are not in nonopen chromatin, it may be that the accessibility of chromatin is modified by stress (Pecinka et al. 2010), accounting for the significant increase in stable integration of plastid DNA after mild heat treatment (Wang et al. 2012a).
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Zhipeng Qu for helpful discussion of this topic. This work was supported by the Australian Research Council’s Discovery Projects funding scheme (grant number DP0986973) and by China Scholarship Council to D.W.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- Kim A, Song SH, Brand M, Dean A. Nucleosome and transcription activator antagonism at human beta-globin locus control region DNase I hypersensitive sites. Nucleic Acids Res. 2007;35:5831–5838. doi: 10.1093/nar/gkm620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001a;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001b;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AH, Timmis JN. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol. 2011;28:2019–2028. doi: 10.1093/molbev/msr021. [DOI] [PubMed] [Google Scholar]
- Lloyd AH, Wang D, Timmis JN. Single molecule PCR reveals similar patterns of non-homologous DSB repair in tobacco and Arabidopsis. PLoS One. 2012;7:e32255. doi: 10.1371/journal.pone.0032255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36:767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Pecinka A, et al. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, Timmis JN. Instability of plastid DNA in the nuclear genome. PLoS Genet. 2009;5:e1000323. doi: 10.1371/journal.pgen.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, et al. Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol. 2008;148:328–336. doi: 10.1104/pp.108.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci U S A. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness PE, Fox TD. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990;346:376–379. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Tsuji J, Frith MC, Tomii K, Horton P. Mammalian NUMT insertion is non-random. Nucleic Acids Res. 2012;40:9073–9088. doi: 10.1093/nar/gks424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Lloyd AH, Timmis JN. Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc Natl Acad Sci U S A. 2012a;109:2444–2448. doi: 10.1073/pnas.1117890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Lloyd AH, Timmis JN. Nuclear genome diversity in somatic cells is accelerated by environmental stress. Plant Signal Behav. 2012b;7:595–597. doi: 10.4161/psb.19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Rousseau-Gueutin M, Timmis JN. Plastid sequences contribute to some plant mitochondrial genes. Mol Biol Evol. 2012;29:1707–1711. doi: 10.1093/molbev/mss016. [DOI] [PubMed] [Google Scholar]
- Zhang W, et al. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012;22:151–162. doi: 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.