Abstract
Sex chromosome divergence has been documented across phylogenetically diverse species, with amphibians typically having cytologically nondiverged (“homomorphic”) sex chromosomes. With an aim of further characterizing sex chromosome divergence of an amphibian, we used “RAD-tags” and Sanger sequencing to examine sex specificity and heterozygosity in the Western clawed frog Silurana tropicalis (also known as Xenopus tropicalis). Our findings based on approximately 20 million genotype calls and approximately 200 polymerase chain reaction-amplified regions across multiple male and female genomes failed to identify a substantially sized genomic region with genotypic hallmarks of sex chromosome divergence, including in regions known to be tightly linked to the sex-determining region. We also found that expression and molecular evolution of genes linked to the sex-determining region did not differ substantially from genes in other parts of the genome. This suggests that the pseudoautosomal region, where recombination occurs, comprises a large portion of the sex chromosomes of S. tropicalis. These results may in part explain why African clawed frogs have such a high incidence of polyploidization, shed light on why amphibians have a high rate of sex chromosome turnover, and raise questions about why homomorphic sex chromosomes are so prevalent in amphibians.
Keywords: sex chromosome, pseudoautosomal region, recombination, sex determination, African clawed frogs, Xenopus tropicalis
Introduction
Sex can be advantageous because it decouples beneficial from deleterious mutations via recombination, which increases the variance in fitness effects of linked mutations, and thus the efficiency with which natural selection operates. In species with genetic sex determination, developmental differences between the sexes are initiated by genetic differences between the sex chromosomes. In some lineages, the genes responsible for triggering sex determination vary, and the sex chromosomes (which carry the sex-determining region) are routinely reassigned from one to another ancestral pair of autosomal chromosomes (Fridolfsson et al. 1998; Ross et al. 2009; Evans et al. 2012; Pease and Hahn 2012). Ironically, suppression of recombination within a sex-specific region is often favored by natural selection, lest a sex-specific, sex-determining allele loses its sex specificity.
The origin of sex chromosomes could be initiated by sexual antagonism (van Doorn and Kirkpatrick 2007), and in many species, this is associated with cessation of recombination between a portion of the sex chromosomes that makes possible unisexual inheritance of a key genomic region that triggers sex determination. Cessation of recombination between the sex chromosomes can be achieved by reducing or eliminating homology (Charlesworth 1991), for example, through point mutations, inversion, deletion, or insertion of DNA. Strikingly, the extent of the nonrecombining region may increase overtime, although this is not necessarily the case (Charlesworth et al. 2005; Bergero and Charlesworth 2009). The expansion of nonrecombining regions may be influenced by the nature of evolution in nonrecombining genomic regions, which is influenced by Muller’s ratchet, background selection, Hill–Robertson effects, and genetic hitchhiking of deleterious alleles with beneficial mutations (Charlesworth B and Charlesworth D 2000). Suppressed recombination between sex chromosomes thus has important implications for genome evolution, speciation, and adaptation.
Sex chromosome “degeneration” can be associated with sex chromosome divergence resulting from suppressed recombination and involves the loss of coding regions, the accumulation of repetitive regions, and structural changes such as insertions, deletions, and inversions on the sex-specific chromosome (the Y or W). Thus, degenerate sex chromosomes have differences that extend beyond the fundamental difference in the presence or absence of a sex-determining allele. However, evolutionarily young sex chromosomes start out being similar to each other because they originated from an essentially identical pair of autosomal chromosomes. In the medaka fish, for example, the male-specific region of the Y chromosome contains a newly evolved sex-determining locus (dmrt1bY) and is only approximately 258,000-bp long (Kondo et al. 2006). Similarly, the sex chromosomes of the tiger pufferfish appear to be distinguished by only one nonsynonymous substitution (Kamiya et al. 2012). In theory, natural selection may drive the expansion of the sex-specific region of suppressed recombination (Charlesworth et al. 2005), and old sex chromosomes may evolve distinct suites of genes with unique, nonhomologous, and/or extensively diverged functions. In humans, for example, almost all the ancestral genes persist on the X chromosome but have been lost on the Y chromosome (Skaletsky et al. 2003). The limit of divergence is achieved if the sex-specific sex chromosome is lost altogether, as occurred in the Ryukyu spiny rat (Kuroiwa et al. 2010). In contrast, however, the sex chromosomes of ratite birds and boid snakes are old, but each pair is morphologically nondiverged (homomorphic), suggesting that the size of the region of suppressed recombination on the sex-specific sex chromosome does not necessarily expand over time (Matsubara et al. 2006; Tsuda et al. 2007).
Frog Sex Chromosomes
Most species of amphibians have homomorphic sex chromosomes (reviewed in Schmid et al. 2010). One possible explanation for the high incidence of homomorphic sex chromosomes in amphibians is that their sex chromosomes tend to be young because there has been frequent switching of the sex-determining locus during evolution. Consistent with this explanation is the inference that sex chromosomes have changed (“turned over”) approximately 32 times or more during evolution based on variation in male (XY) versus female (ZW) heterogamy (Evans et al. 2012; Schmid et al. 2010). Another explanation for widespread homomorphic sex chromosomes is that there is periodic recombination between the sex chromosomes over most of their length without changes in the sex-determining locus. In one group of hylid frogs, for instance, genomic regions that are tightly linked to the sex-determining locus are not substantially diverged between males and females, indicating that the sex chromosomes of these frogs recombine, at least occasionally, over most of their length (Stöck et al. 2011). This suggests that sex chromosomes of these frogs have large “pseudoautosomal” regions where inheritance of genetic information resembles autosomal genes and where recombination prevents divergence between the sex chromosomes. Thus, frequent turnover and recombination clearly both play a role in homomorphy of amphibian sex chromosomes, but which phenomenon plays the dominant role remains an open question.
The only known amphibian sex-determining gene is called DM-W and was discovered in the African clawed frog Xenopus laevis (Yoshimoto et al. 2008). DM-W is female specific and originated after divergence from the sister genus Silurana but before diversification of most or all extant species of Xenopus (Bewick et al. 2011). Because Silurana tropicalis (also known as X. tropicalis) lacks DM-W, sex determination in this species must be triggered by another as yet unidentified genetic locus. A high-quality draft genome sequence is available for S. tropicalis that was generated from a female (Hellsten et al. 2010), but the sex-specific region of this genome has not been characterized. Using amplified fragment length polymorphisms (AFLPs), Olmstead et al. (2010) identified 22 AFLPs linked to the sex-determining locus in the “golden” strain of S. tropicalis (table 1) and proposed that females are the heterogametic sex in this strain. Four of these 22 AFLPs placed to the distal tip of chromosome/linkage group 7 in a linkage map developed by Wells et al. (2011), also represented by scaffold 7 in version 7.1 of the S. tropicalis genome sequence. However, the linkage map contains a large (15 cM) gap between the most distal two markers where the sex-determining region is likely to reside (Wells et al. 2011). Many of the other sex-linked AFLPs identified by Olmstead et al. (2010) map to other major or small chromosome/linkage groups (table 1); this is presumably because some scaffolds are chimerical (e.g., a portion of scaffold 2 in version 7.1 is probably actually derived from S. tropicalis chromosome 7) and because linkage relationships between some small contigs (“orphan scaffolds”) and the larger scaffolds have not yet been established. Furthermore, additional experiments with other strains suggest that sex determination may occur through the action of multiple alleles at one locus or multiple tightly linked genes (Olmstead A, personal communication).
Table 1.
Genomic Regions of Silurana tropicalis That Are Putatively Sex Linked Based on Linkage Study of Olmstead et al. (2010) and Sequencing of Chromosome Arm 7p by Seifertova et al. (submitted)
| AFLP | Recombination | v4 | v7.1 | 7p? | Portion Sex Linked |
|---|---|---|---|---|---|
| E33.M72.143 | 0 | 605:241571-241691 | 7:4966286-4966166 | Yes | 4435335-5175370 |
| E33.M81.275 | 0 | 494:27646-27898 | No hits | — | NA |
| E33.M90.327 | 0 | No hits | 211:76840-76535 | Yes | ALL |
| E38.M93.218 | 0 | 953:138210-138402 | 278:109297-109490 | No | ALL |
| No name | 0 | 494:31633-32115 | No hits | — | NA |
| No name | 0 | 494:27541-27902 | No hits | — | NA |
| No name | 0 | 379:817889-818000 | 2:149826489-149826600 | Yes | 149787496-150105127 |
| No name | 0 | 736:292586-293067 | 78:248343-247864 | Yes | ALL |
| No name | 0.3 | 605:245039-245621 | 7:4963243-4962661 | Yes | 4435335-5175370 |
| No name | 0 | 605:116800-117215 | 22:1040592-1040177 | Yes | ALL |
| E40.M52.572 | 0.4 | 859:57522-58049 | 7:3195527-3196069 | Yes | 76939-3370464 |
| E33.M61.177 | 0.5 | 1778:6156-6312; 1778:9971-9815 | 144:136541-136385 | Yes | ALL |
| E33.M61.177 | 0.5 | 1778:6156-6312; 1778:9971-9815 | 662:22621-22465 | No | NONE because 114 is on 7p |
| No name | 0.5 | 810:261995-262744 | 94:264236-264985 | Yes | ALL |
| No name | 0.5 | 810:261995-262744 | 94:263563-263460 | Yes | ALL |
| E32.M94.406 | 1.9 | Multiple hits | 22:58856-59241 | Yes | ALL |
| E37.M52.423 | 1.9 | 810:325559-325959 | 144:95531-95931 | Yes | ALL |
| E37.M52.423 | 1.9 | 1151:130316-130719 | 144:95931-95531 | Yes | ALL |
| E41.M83.506 | 1.9 | 810:276803-277288 | 94:279867-279382 | Yes | ALL |
| E32.M35.552 | 2.6 | Multiple hits | No hits | — | NA |
| E37.M60.232 | 2.6 | 6092: 2392-2601 | 7931:611-828 | Yes | ALL |
| E32.M59.335 | 2.9 | 735:292141-292443 | 7:7903155-7902853 | Yes | 6687308-9940823 |
Note.—NA, not applicable. AFLP refers to the name of the AFLP from Olmstead et al. (2010) if provided. Recombination refers to the recombination rate with the sex-determining locus from that study. Scaffold and position of AFLPs are provided for genome assembly version 4.0 (v4) and 7.1 (v7.1). Sex-linked portions that were included in categories in tables 2 and 3 based on the level of recombination (Portion sex linked, with NA meaning not applicable) either refer to base pair positions of a contig within a larger scaffold that is not interrupted by unknown sequence or the entire scaffold was assumed to be sex linked (ALL).
The goal of this study is to further characterize the sex chromosomes of S. tropicalis in terms of the size and level of divergence of the sex-specific region, and to compare molecular evolution and expression of sex-linked and nonsex-linked genes. To this end, we used restriction-site associated DNA (RAD) tags (Baird et al. 2008), a reduced representation next-generation sequencing approach, to genotype millions of homologous nucleotide positions in male and female individuals including positions that are monomorphic in both sexes, polymorphic in one or both sexes, and positions in which a genotype inference (i.e., homozygous or heterozygous) was only possible in one sex due either to sex specificity of the genotyped position or differences in coverage of that position between the sexes. The RAD tag approach produces sequences of thousands of small regions that are adjacent to a rare cutting restriction enzyme site. Because the sequenced portions of the genome are associated with restriction enzyme sites, many homologous sequences are obtained from multiple individuals. Missing data among individuals can arise in unusual cases where mutation generates polymorphism in the presence or absence of the restriction enzyme sites or because of variation among individuals in the depth of sequencing coverage for a particular region. Our analysis incorporated information on sex-linked regions from Olmstead et al. (2010), information from a laser-dissected chromosome arm 7p from a male individual (Seifertova et al., submitted), which is linked to the sex-linked region identified by Olmstead et al. (2010), and the most recent genome assembly (version 7.1, reference accession PRJNA12348). This study thus provides, for the first time, a comprehensive perspective on the extent of sex chromosome divergence in this species by evaluating the distribution of homozygous and heterozygous genotypes, molecular evolution, and gene expression of sex chromosomes in the context of the rest of the genome.
Materials and Methods
Four female and four male S. tropicalis individuals were obtained from Xenopus Express (Brooksville, FL). Sex was confirmed by dissection and species assignment achieved by comparing between 809 and 812 bp of mitochondrial DNA sequence from a portion of the 16S gene from each sample to homologous sequence data from all other known species of African clawed frog (Evans et al. 2011). We performed a phylogenetic analysis on these 8 sequences, 27 sequences from individuals used in the polymerase chain reaction (PCR) screen detailed earlier, all Silurana sequences from Evans et al. (2004), 6 S. tropicalis samples from Ghana (obtained from tissue archive at the Burke Museum, University of Washington, accession numbers UWBM5957–8, UWBM5961–63, and UWBM5969), and sequences from 6 individuals from the “golden” strain used by Olmstead et al. (2010) that were provided by Richard Harland. We used an X. laevis sequence from South Africa as an outgroup in this analysis, and the total alignment length was 817 bp. Model selection for phylogenetic analysis was accomplished using MrModeltest2 (Nylander 2004). Phylogenetic analysis was performed with MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001) using the best-fit model based on the Akaike Information Criterion, with two independent Markov chain Monte Carlo (MCMC) runs, each for 2,000,000 generations. Convergence of the MCMC runs on the posterior distribution was assessed by inspecting parameter trends and effective sample sizes using Tracer version 1.5 (Rambaut and Drummond 2007). Based on these analyses, a burn-in of 500,000 generations was discarded before constructing a consensus tree with MrBayes.
Genomic DNA was extracted from liver using QIAGEN DNeasy kit, purified using QIAGEN’s spin purification protocol, and RAD tag library preparation performed by Floragenex, Inc (Eugene, OR). For each individual, two libraries were generated—one used the restriction enzyme SbfI and another used NotI. The RAD tag libraries were multiplexed on three Illumina flow cells using individual barcodes, and Illumina sequencing was performed at the University of Oregon. These data have been deposited in GenBank (accession number SRP022004).
Illumina sequence reads were sorted by barcode with RADtools v1.2.4 using the “fuzzy_MID” option, which assigns reads with barcode errors to the nearest barcode (Baxter et al. 2011). Data from each individual were independently aligned to the S. tropicalis version 7.1 genome using bwa-0.6.2 (Li and Durbin 2009) and samtools.0.1.18 (Li et al. 2009). The “MarkDuplicates” function in picard (http://picard.sourceforge.net) was used to mark putative PCR-amplified duplicates, which were then excluded from the genotyping analysis with an aim of minimizing genotyping error. The Genome Analysis Toolkit (GATK) version 2.2-15 was then used to realign indels using the “RealignerTargetCreator” and “IndelRealigner” functions (McKenna et al. 2010; DePristo et al. 2011). The “FixMateInformation” function of picard was then used to adjust mate pair alignments.
Following “Best Practices” guidelines on the GATK website and forum (http://gatkforums.broadinstitute.org/) for analysis of genomes that lack known single-nucleotide polymorphisms (SNPs), the “UnifiedGenotyper,” “BaseRecalibrator,” and “PrintReads” functions of GATK were used to iteratively genotype, recalibrate base quality scores, and generate new input (bam) files, using the genotype files generated from “UnifiedGenotyper” as known polymorphic positions to be ignored for base recalibration in each iteration. Convergence was reached by the fifth iteration, in that variable positions recovered from this analysis were 99.8% identical to those from the fourth iteration. The “VariantFiltration” and “SelectVariants” functions of GATK were then used to identify and exclude genotyped positions that 1) were within 10 bp of an insertion/deletion, 2) had a Phred genotype quality score (Ewing and Green 1998) of less than 30, which means that we removed positions that had a probability of error of greater than 0.001, or 3) had more than one-tenth of the reads mapping equally well to another position and where there were at least four of these reads.
The S. tropicalis genome assembly 7.1 consists of 7,730 scaffolds aggregated from 55,234 contigs connected by “N”s within each scaffold. The total number of bases is 1,437,594,934, of which 5% (n = 71,599,926) are “N”s. This assembly includes 14 large “super scaffolds” that were assembled using meiotic map, synteny, and cytological data, corresponding to the 10 haploid chromosomes, with some chromosomes being represented by multiple scaffolds (3a and 3b; 5a and 5b; and 8a, 8b, and 8c). The rest of the scaffolds are “orphan scaffolds” whose chromosomal locations are not yet known. We divided the genomic regions into five mutually exclusive groups based on 1) the inferred level of recombination with the sex-determining region by Olmstead et al. (2010), 2) the linkage groups in the genome assembly 7.1 (table 1), and 3) the results of the Illumina sequencing of the dissected petite arm of chromosome 7 (Seifertova et al., submitted). The first of the five groups (“completely sex linked”) included contigs from assembly 7.1 that contain regions that had no recombination (0%) with the sex-determining region in Olmstead et al. (2010). This means that recombination between an AFLP polymorphism and the sex-determining region was not observed in any of 300 individuals assayed by Olmstead et al. (2010). The second group (“partially sex-linked”) included contigs from assembly 7.1 that contain regions that had a recombination rate of more than 0% and less than 3.0% in Olmstead et al. (2010). The third group (“chromosome 7p”) contained sections of scaffolds in assembly 7.1 that are located on chromosome 7p according to Seifertova et al. (submitted), and not in the “completely sex linked” or “partially linked” categories. The fourth group (“non-7p chromosomes”) contained the remaining sections on the chromosome-scale scaffolds in Assembly 7.1, including the portion of scaffold 7 that did not map to chromosome 7p. The fifth group (“other orphans”) contained orphan scaffolds in assembly 7.1 that 1) have not been linked to a chromosome, 2) have no evidence of sex linkage according to Olmstead et al. (2010), and 3) did not map to chromosome arm 7p according to Seifertova et al. (submitted). More specific information on the scaffold or scaffold portions in each of these groups is provided in table 1.
Genome-Wide Distribution of Genotypes in Female and Male S. tropicalis
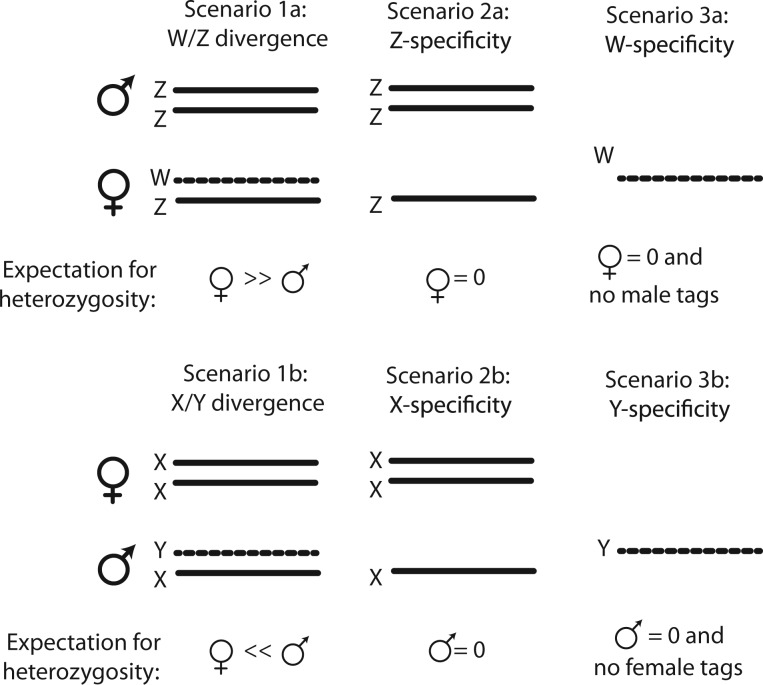
For genomic regions in each of the six categories described earlier, we tabulated genotype patterns for three scenarios (fig. 1) in 500,000-bp windows across the S. tropicalis genome; smaller windows were examined at the ends of scaffolds or when a scaffold was smaller than 500,000 bp. Genotype patterns in each sex (i.e., the distribution of homozygous or heterozygous positions) are relevant to sex chromosome evolution in the following ways. First, divergence between the sex chromosomes due to suppressed recombination generates positions that are either heterozygous in all females and no males (for a ZW sex-determining system) or heterozygous in all males and no females (for an XY sex-determining system). We call these patterns “Scenario 1a” and “Scenario 1b,” respectively (fig. 1). We note that in “Scenario 1a” regions, some positions can also be heterozygous in males due to polymorphism on the Z chromosome, and in “Scenario 1b,” some positions can also be heterozygous in females due to polymorphism on the X chromosome. In any case, in genomic regions consistent with Scenario 1, heterozygosity observed in all samples from one sex is expected to exceed heterozygosity observed in all samples from the other sex. We, therefore, searched for regions with heterozygosity present in all females or in all males. For both of these statistics, we ignored positions that are heterozygous in all genotyped individuals. To account for variation in coverage in males and females, for each window, we divided these counts by the total number of positions in each window for which genotype calls were made in at least one female and at least one male.
Fig. 1.—
Genotypic scenarios for sex-linked regions. Expectations for heterozygosity depend on which sex is heterogametic and the region of the sex chromosome (pseudoautosomal region vs. sex-determining region). Female heterogamy is potentially associated with female-biased heterozygosity (Scenario 1a), male-only heterozygosity (Scenario 2a), or female-only homozygosity with no male genotypes (Scenario 3c). Corresponding scenarios (Scenarios 1b, 2b, and 3b) apply to the opposite sex for male heterogamy.
Another genotypic scenario for sex chromosomes is that a genomic region may be present only on the Z chromosome (with female heterogamy) or only on the X chromosome (with male heterogamy) (Scenarios 2a and 2b; fig. 1). No counterpart exists on the W chromosome (or Y chromosome) due to deletion, insertion, or divergence. To detect such a genomic region, we searched for regions with heterozygous positions present in one sex but not the other. For such positions, we required a genotype call in at least one individual of each sex but heterozygous calls to be present in only one sex. To account for variation in coverage in males and females, for each window we divided these counts by the total number of positions in each window for which genotype calls were made in at least one female and at least one male.
A third genotypic scenario for sex chromosomes is that a genomic region may be present only on the W chromosome or only on the Y chromosome (Scenarios 3a and 3b, fig 1). Thus, we searched for positions that had genotype calls only in females (or only in males) and that are all homozygous. To account for variation in coverage, we standardize the counts in each window by the sum of the number of positions in each window for which genotype data are available for 1) at least one female and at least one male, 2) at least one female but no males, and 3) at least one male but no females. Thus, by evaluating these three genotype scenarios in genomic windows across the S. tropicalis genome assembly, we attempted to identify genomic windows that either had significantly more heterozygous positions in one sex (Scenario 1), that had heterozygous positions only in one sex (Scenario 2), or that had homozygous positions in only one sex and no homologous genotypes in the other (Scenario 3). Higher values for each ratio are suggestive of genotype patterns characteristic of diverged sex chromosomes.
Expression and Molecular Evolution
As described in Chain et al. (2011), we estimated gene expression levels based on sequences across 26 expressed sequence tag (EST) libraries from the following tissues or developmental stages: egg, gastrula, neurula, embryo, tailbud, tadpole, metamorphosis, adipose tissue, bone, brain, head, heart, intestine, kidney, limb, liver, lung, ovary, oviduct, skeletal muscle, skin, spleen, stomach, tail, testis, and thymus. We summarized patterns of gene expression across EST libraries using the nonindependent “total,” “intensity,” and “evenness” statistics described in Chain et al. (2011). The “total” expression of a gene (T) is the proportion of times that a gene was sequenced in each EST library (Li) summed across all libraries (T = ∑Li). The “intensity” of expression (I) is the mean expression level from the perspective of a gene and is calculated following this equation: I = ∑Li2/∑Li. “Evenness” of expression (E) can be thought of as the “effective number” of tissues in which a gene is expressed and is calculated following this equation: E = T/I.
For a subset of the sex-linked and nonsex-linked genes, we also calculated the rate ratio of nonsynonymous to synonymous substitutions per site (dN/dS) along the S. tropicalis lineage using PAML version 4.5 (Yang 1997). This ratio was calculated using a maximum likelihood model that individually estimates dN/dS for each branch in a phylogeny, following Chain et al. (2011). Our phylogeny was estimated from sequences from S. tropicalis, X. laevis, and using sequences from another pipid frog (Pipa carvalhoi or Hymenochirus curtipes) as an outgroup. To avoid undefined values, we added 0.02 to all dS values before calculating dN/dS, following Chain et al. (2011). We made this adjustment a priori by looking only at dS values, to make better use of the data. Because extreme values for dN and dS were occasionally estimated, we excluded from the analysis genes with an estimated dN or dS value above 2, and any genes whose available data comprised less than 100 synonymous positions.
We used a one-sided permutations to test whether the expression and molecular evolutionary statistics differed between genes that either (a) were or (b) were not on the same chromosome as the sex-determining locus. The permutations randomly divided the set of (a + b) values into two groups of size a and b and then calculated the difference between the averages of each group. We repeated this 1,000 times to generate a distribution for the null hypothesis that the values were drawn from the same underlying distribution, and then compared this with the observed differences, which is the test statistic of each test. A significant difference was inferred if the observed difference was greater than 95% of the differences from the permutations. Because these tests are one sided, the operands of the test statistic (i.e., the minuend and subtrahend of each difference) were defined according to specific expectations for sex chromosome degeneration discussed later.
Results
Mitochondrial DNA Variation within S. tropicalis, Including the “Golden” Strain
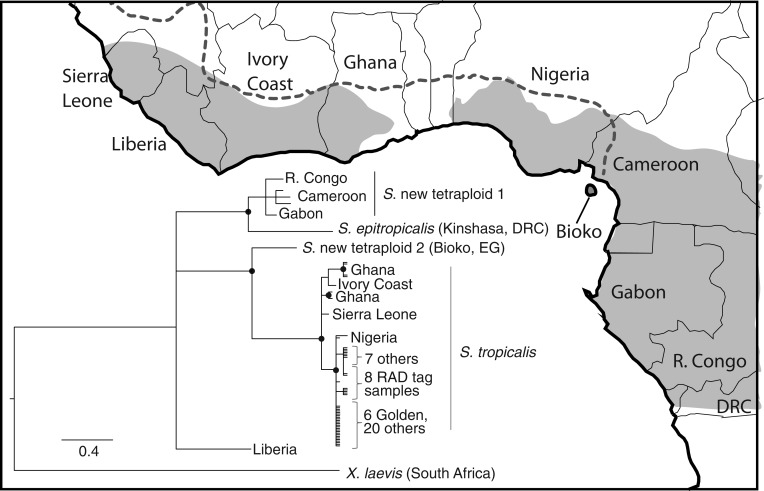
We analyzed phylogenetic relationships among approximately 810-bp region of mitochondrial DNA from the commercially obtained S. tropicalis individuals that we used for RAD tags and PCR screens, six individuals from the golden strain used by Olmstead et al. (2010), and several other wild-caught S. tropicalis individuals and individuals from other Silurana species. An identical mitochondrial DNA sequence was obtained from the six golden strain individuals, one of the samples we used for RAD tag sequencing (a female), 20 of the samples we used for PCR screens (9 females and 11 males), and one individual sampled from Nigeria. Mitochondrial sequences from five samples used in the RAD tag sequencing (2 females and 3 males) were identical to each other and differed from the golden strain sequence by one nucleotide substitution. Mitochondrial sequences from two other samples used in RAD tag sequencing (1 female and 1 male) and seven samples used in PCR screens (4 females and 3 males) were identical to each other and differed from the golden strain mitochondrial sequence by a different single-nucleotide substitution than the previously mentioned sequence present in five of the RAD tag samples. Mitochondrial sequences from another sample from Nigeria differed from the golden strain mitochondrial sequence by two nucleotide substitutions. Phylogenetic analysis of these and other sequences indicates that the commercially obtained S. tropicalis samples used in this study form a well-supported clade that includes two sequences from Nigeria and the six sequences from the golden strain of S. tropicalis (fig. 2). This clade is possibly common in individuals east of the Dahomey Gap, a savannah corridor that interrupts the West African rain forest (Salzmann and Hoelzmann 2005).
Fig. 2.—
Phylogenetic analysis of mitochondrial DNA sequences suggest that the “golden” strain used by Olmstead et al. (2010) and samples used in this study (8 for RAD tag analysis and 27 others for PCR assays) originate from Nigeria. Nodes with ≥95% posterior probability are indicated with a black circle. Species names, including those undescribed, follow Evans et al. (2004). Abbreviated country names include the Republic of the Congo (R. Congo), the Democratic Republic of the Congo (DRC), and Equatorial Guinea (EG). The scale bar refers to the number of substitutions per site, gray areas on the map indicate the distribution of tropical forest in West Africa, and the dotted line indicates the approximate distribution of Silurana tropicalis inferred by Tinsley et al. (1996).
Reduced Representation Genome-Wide Genotyping from RAD Tags
We used a reduced representation genome sequencing approach called “RAD tags” to sequence many small but homologous portions of the S. tropicalis genome in four female and four male individuals. An average of 9,696,525 Illumina reads were mapped in each individual, with the average number of reads mapped per female or per male being 9,445,507 and 9,947,543 reads, respectively. After excluding positions in the reference sequence with no data, within an individual the average depth of coverage was 18.4 reads per position. Genotypes were called for a total of 19,624,824 positions, and 193,199 SNPs (0.98%) were detected. For each position at which at least one genotype was called, an average of 7.14 out of 8 individuals were genotyped.
If S. tropicalis has a large female-specific genomic region on the W chromosome, we expected a higher proportion of the Illumina reads from females to map to the genome assembly because this assembly was generated from a female individual. Contrary to this expectation, a slightly higher proportion of reads from males (average per male individual 89.2%, range: 87.8–90.8%) than from females (average per female individual 87.9%; range: 85.0–90.8%) mapped to this genome assembly, arguing against there being a large female-specific region in the S. tropicalis genome. Another indication of a large female-specific genomic region on the W chromosome would be a substantially higher number of positions genotyped in females than in males. Out of a total of 19,624,824 positions that were genotyped with high confidence in at least one individual, slightly more genotypes were recovered in females than in males: 912,738 (4.7%) positions were genotyped only in one or more females, and 462,792 (2.4%) positions genotyped only in one or more males. However, in the 10 largest scaffolds, the number of genotype calls in at least one female was consistently 1.1–3.3% higher than the number of genotype calls in at least one male, with scaffold 7 having 2.5% more genotype calls in females than males. This suggests that the higher number of unique genotype calls in females is primarily a technical artifact related to differences in coverage among individuals in the RAD tag libraries. The RAD tag data did not provide high-quality genotypes from any positions on 5,721 scaffolds, which together comprise 36,133,437 bp (∼2.1% of the genome).
Genome-Wide Genotype Patterns and Nucleotide Diversity Similar in Males and Females
We searched 500,000-bp windows for various genotypic patterns consistent with sex chromosome divergence expected under female or male heterogamy (fig. 1). In general, this effort failed to identify any regions with a pronounced genotypic signature of sex chromosome divergence expected by female heterogamy (table 2). One exception was a significant excess of windows with female-only homozygous genotypes (Scenario 3a) in orphan scaffolds, but we suspect this was an artifact related to the broader coverage in females. Most notably, portions of linkage groups 2 and 7 that were categorized as “partially sex-linked” and “completely sex-linked” to the sex-determining region based on Olmstead et al. (2010) did not exhibit a genotypic pattern consistent with degenerate sex chromosomes based on the RAD tag genotypes.
Table 2.
Putatively Sex-Linked Regions Do Not Exhibit Genotypic Hallmarks Expected for Diverged Sex Chromosomes
| Region | Number of Genotype Calls | Scenario 1a: All Females Heterozygous | Scenario 1b: All Males Heterozygous | Scenario 2a: All Females Homozygous | Scenario 2b: All Males Homozygous | Scenario 3a: All Females Homozygous; Males No Genotype | Scenario 3b: All Males Homozygous; Females No Genotype |
|---|---|---|---|---|---|---|---|
| “Non-7p chromosomes” | 17,026,460 | 0.00013 (0.00000–0.00087) | 0.00018 (0.00000–0.00123) | 0.00124 (0.00000–0.00399) | 0.00179 (0.00000–0.00504) | 0.04980 (0.00390–0.15189) | 0.02670 (0.00050–0.09960) |
| “Chromosome 7p” | 830,811 | 0.00019 | 0.00019 | 0.00072 | 0.00456 | 0.10066 | 0.03760 |
| “Other orphans” | 1,618,864 | 0.00034 | 0.00040 | 0.00305 | 0.00524* | 0.33587* | 0.18522* |
| “Partially sex-linked” | 132,177 | 0.00024 | 0.00014 | 0.00182 | 0.00263 | 0.06098 | 0.03920 |
| “Completely sex linked” | 16,512 | 0.00026 | 0.00003 | 0.00228 | 0.00308 | 0.02848 | 0.01176 |
Note.—Analysis of 500,000-bp genomic windows indicates that genotype patterns of genomic regions linked to sex do not resemble scenarios expected for sex chromosomes substantially more than other parts of the genome. Numbers indicate average values for three genotypic scenarios depicted in figure 1 and described in Materials and Methods section. For the “Non-7p chromosomes,” 95% confidence intervals are in parentheses. Asterisks indicate values that are higher than the 95% confidence intervals from the “non-7p chromosomes.”
Considerable caution is needed in the interpretation of the average genotype frequencies in genomic windows for the “other orphans” category because in many cases the scaffold is smaller than the window size (500,000 bp), and the resulting truncated genomic windows are therefore expected to have an increased variance in the frequency of various genotypic patterns. Additionally, average genotype frequencies in these genomic windows could fail to detect small scaffolds that have genotypic patterns consistent with sex chromosome divergence. For example, “other orphans” had higher than expected values for Scenarios 2b and 3b, which are consistent with male heterogamy, and this is probably related to the small size of these scaffolds and consequent increase in the sex-specific genotypes in truncated windows for these scaffolds.
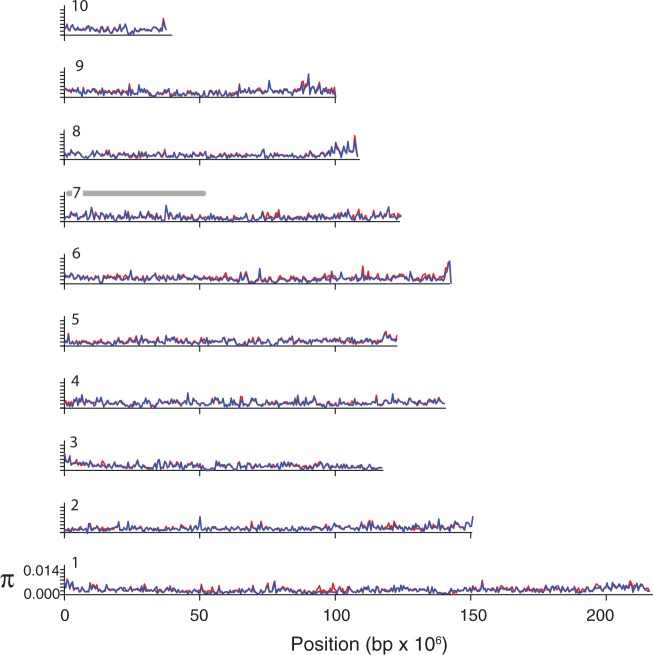
Additional insights are gained by examining nucleotide diversity in each sex within 500,000-bp windows. If a portion of the sex chromosomes is substantially diverged, we expected much higher average nucleotide diversity per site in one sex (females for female heterogamy) in genomic windows spanning this diverged region. However, average nucleotide diversity per site is essentially identical in males and females throughout these scaffolds, including chromosome arm 7p, which is linked to the sex-determining region (fig. 3). To explore the possibility that there could be variation within the RAD tag samples in sex chromosome divergence that corresponds with the three mitochondrial DNA haplotype groups detailed earlier, we explored nucleotide diversity in male and female individuals from each group. This analysis also did not identify a pronounced signature of sex chromosome divergence (supplementary figs. S1 and S2, Supplementary Material online).
Fig. 3.—
Average nucleotide diversity per site (π) in 500,000-bp windows is similar in males (blue) and females (red) throughout much of the Silurana tropicalis genome. Plots are labeled with numbers referring to scaffolds 1–10, which collectively comprise approximately 75% of the genome. A gray bar on scaffold/chromosome 7 indicates the petite arm based on the linkage map of Wells et al. (2011), which carries the sex-determining region in the S. tropicalis golden strain (Olmstead et al. 2010).
Genotype Patterns Based on Sanger Sequencing
We amplified 65 portions (amplicons) of genomic regions identified by Olmstead et al. (2010) to be linked to the sex-determining region, including 18 and 46 amplicons from “completely sex-linked” and “partially sex-linked” regions, respectively (table 1, supplementary table S1 and fig. S3, Supplementary Material online). None had female-specific amplifications, allowing us to dismiss Scenario 3 (fig. 1) for all these regions. We sequenced 45 of these amplifications in multiple male and female individuals. SNPs or insertion/deletion polymorphisms were shared between males and females in at least one amplicon for essentially all scaffolds (no polymorphism was observed in amplicons from scaffold 144). This suggests that Scenario 1 is unlikely for these regions, with the caveat being that a heterozygous position could arise in both sexes in a region consistent with Scenario 1 through convergent evolution on the W and Z.
We also used PCR to examine an additional 173 regions that exhibited signs of sex linkage based on our analyses of the RAD tag data, including regions of chromosome 7p and elsewhere as detailed in supplementary figure S3 and table S1, Supplementary Material online. None had sex-specific amplifications, allowing us to dismiss Scenario 3 for all these regions. We sequenced 94 of these amplifications from male and female individuals. Thirty of these were not polymorphic in any of the individuals we sequenced. Forty-eight had polymorphisms shared between males and females, allowing us to conclude that Scenario 1 is unlikely for these regions. Five had polymorphisms in both sexes with none being shared across sexes. Ten had polymorphisms only in females and two had polymorphisms only in males.
Two amplifications were of particular interest. An amplification on scaffold 7 that spanned positions 10,128,301–10,129,920 was highly polymorphic in females but not males, although no polymorphism was fixed in females (out of seven females and three males sequenced; supplementary table S1, Supplementary Material online). This region failed to amplify in four females and three males. Another amplification, which targeted a region on scaffold 163 had 31 polymorphisms in three females but only one polymorphism in three males, but essentially all the female polymorphisms were present in only one individual.
Gene Expression and Molecular Evolution
Expression was detected in a total of 37,790 transcripts in at least one of the 26 EST libraries we surveyed (table 3). On the basis of studies of recently diverged neosex chromosomes in fruit flies (Drosophila) (reviewed in Bachtrog 2013), we expected expression of genes situated near the sex-determining locus to be expressed 1) at a lower total level, 2) higher intensity, 3) lower evenness, and to have 4) higher dN/dS compared with genes in other parts of the genome (i.e., the “non 7p chromosomes”). For the most part, these expectations were not met for genes that were demonstrably very close to the sex-determining region, with the one exception that the intensity of “completely sex-linked” genes were individually significantly higher than the “non 7p chromosomes” (P < 0.05, table 3). Evenness of “other orphans” was also significantly lower than “non 7p chromosomes” as was total expression of “other orphans.” dN/dS was significantly higher only in “chromosome 7p” compared with “non-7p chromosomes” but the magnitude of this difference was small. No expression data were recovered from genes on Scaffold 22, which is tightly linked to the sex-determining region (Olmstead et al. 2010), even though it was 1,156,260-bp long. We examined this scaffold using Xenbase (Bowes et al. 2009) and found that it contained a cluster of olfactory receptors, which (not surprisingly) were not highly expressed in any of the EST libraries we examined.
Table 3.
Average Expression and Molecular Evolution Statistics for Silurana tropicalis Genes in Five Genomic Categories
| Region | Number of Genes (Expression) | Total | Intensity | Evenness | Number of Genes (dN/dS) | dN/dS |
|---|---|---|---|---|---|---|
| “Non-7p chromosomes” | 35,135 | 0.00065 | 0.00015 | 3.19655 | 9183 | 0.2702 |
| “Chromosome 7p” | 2,114 | 0.00079 | 0.00019 | 3.17054 | 546 | 0.2850* |
| “Other orphans” | 260 | 0.00040* | 0.00012 | 2.59443* | 55 | 0.2670 |
| “Partially sex-linked” | 246 | 0.00090 | 0.00018 | 3.37658 | 71 | 0.2751 |
| “Completely sex-linked” | 35 | 0.00101 | 0.00049* | 2.56020 | 8 | 0.2615 |
Note.—See Materials and Methods for description of statistics.
*Values that are individually significantly different from the “Non-7p chromosomes” (P < 0.05, one-sided permutation tests).
Discussion
To explore sex chromosome divergence in an amphibian, we used genotype calls from approximately 20 million positions, information about sex linkage, EST databases, and molecular evolutionary analyses to further characterize the sex chromosomes of the Western tropical frog S. tropicalis. Phylogenetic analysis of mitochondrial DNA sequences suggests our samples originated in Nigeria, which is also the source of the female individual from which the genome sequence was generated (Hellsten et al. 2010). Additionally, our analysis also suggests that the golden strain analyzed by Olmstead et al. (2010) is from Nigeria.
Known sequences in the S. tropicalis genome sequence assembly version 7.1 comprise approximately 80.4% of the approximately 1.7 Gbp genome, and scaffolds, including “N”s, comprise approximately 84.5% of the genome. Thus, the RAD tag data could not be compared with 15–20% of the genome because of gaps in the genome sequence. Because of variation in coverage, high confidence genotype calls were not made on scaffolds that together comprise an additional 2.1% of the genome. Thus, in this study, we lack information from a nontrivial portion of this genome.
Mindful of these substantial gaps in genome sequence and the uncertainty in linkage relationships among many unassembled (orphan) scaffolds, we leveraged information from a targeted sequencing effort of chromosome arm 7p and also the linkage analysis by Olmstead et al. (2010) to guide our analysis. The dearth of genotypic patterns consistent with divergent sex chromosomes, and particularly patterns that are consistent with female heterogamy (table 2), and the similar level of pairwise nucleotide diversity in males and females throughout the petite arm of chromosome 7 (fig. 3) argues strongly against there being a large sex-specific region of the S. tropicalis chromosomes. This inference is consistent with the findings of Uno et al. (2008) who detected no sex differences in C-banded heterochromatin in S. tropicalis.
On the basis of studies of fruit flies (reviewed in Bachtrog 2013), we expected genes linked to the sex-determining locus to potentially exhibit lower total expression and higher specificity (i.e., higher intensity and lower evenness as defined in Materials and Methods section). We also expected molecular evolution of these genes to be consistent with relaxed purifying selection. However, on the basis of a small sample size, we only observed a significant increased expression intensity of “completely linked” genes compared with the rest of the genome, with none of these expectations met in “partially sex-linked” genes (table 3). Some of these expectations were also met in orphan scaffolds, which have undetermined linkage relationships with respect to the sex-determining locus, and regions of chromosome arm 7p. It is not clear that these latter observations are related in any way to linkage to the sex-determining region.
Caveats exist in our interpretation of these data. First, nonrecombining portions of the genome tend to accumulate repetitive sequences that can be difficult to sequence and map. For this reason, the sex-specific portion of the S. tropicalis genome may be under-represented in the current genome assembly and/or our mapped Illumina reads. Second, it is conceivable that there is polymorphism in the sex-determining mechanism (Olmstead A, personal communication). Polymorphism in genetic sex determination could occur at a single locus wherein multiple, differently functioned sex-determining alleles are segregating at a single locus, which have distinct and not necessarily transitive dominance relationships. Polymorphism in genetic sex determination could also occur at multiple loci distributed on the same or different chromosomes. Sex determination in zebrafish, for example, appears to be orchestrated by genes on different chromosomes (Anderson et al. 2012). Genotypic patterns expected with these types of polymorphisms are unclear and could include a dearth or absence of pronounced sex chromosome divergence. A third caveat to our conclusions is that polymorphism among females could also potentially exist in the extent of divergence between the W and Z chromosomes. Under this scenario, it is conceivable that there could be variation among populations in the extent of recombination along the sex chromosomes and consequently the extent and magnitude of divergence between the sex chromosomes. Further exploration of these possibilities will be assisted by the identification of the sex-determining locus in S. tropicalis, the completion of high-quality sequencing and assembly of sex-linked regions, and the exploration of variation within and among populations in sex determination and sex chromosome evolution.
Polyploidization, Dosage Compensation, and Sex Chromosome Turnover
Within a species, the propensity to undergo genome duplication and sex chromosome evolution is potentially interrelated. For example, polyploidization might be less common in species with divergent sex chromosomes where one has degenerated because, after duplication, the degenerate ancestral sex chromosome would segregate as a new autosomal chromosome, and the resulting homozygous null genotypes could be detrimental (Evans et al. 2012). Sex chromosome degeneration also creates imbalances in allelic copy number between the sexes, which can lead to the evolution of dosage compensation—a factor that is also potentially relevant to polyploid speciation (Orr 1990). Dosage compensation is a process that equalizes expression levels in each sex of a gene that has a different number of alleles in each sex. This could evolve in a species with female heterogamy, for example, through inactivation of one of the Z alleles in males or through upregulation of the Z allele in females. Orr (1990) proposed that dosage compensation in species with a degenerate sex chromosome could act as a barrier to polyploid speciation because dosage compensation would be disrupted when a newly formed triploid individual backcrosses with a diploid parental individual. Our analyses suggest that the sex-specific region of S. tropicalis is small, that sex chromosome divergence is minimal, and therefore that dosage compensation associated with degeneration of the sex-specific sex chromosome would have evolved in very few genes or none at all. Together these features of the sex chromosomes may have facilitated (or at least not impeded) polyploidization in Silurana, which occurred at least once (reviewed in Evans 2008). Interestingly, the sister genus Xenopus has a newly evolved sex-determining gene called DM-W (Yoshimoto et al. 2008; Bewick et al. 2011). Species in this group also probably have minimally diverged sex chromosomes and have undergone polyploid speciation multiple times (Evans 2008). Clearly, however, this is not the only consideration in the ability of species to tolerate polyploidization because many amphibian groups that have homomorphic sex chromosomes lack polyploid species.
The extent of sex chromosome degeneration is also relevant to the chances a species experiences future sex chromosome turnover—a change in which pair of chromosomes carries the trigger for sex determination (Charlesworth and Mank 2010). If sex chromosome turnover occurs in a species with a diverged and degenerate sex chromosome, the ancestral degenerate chromosome could segregate autosomally, and some individuals could inherit two copies and be homozygous for degenerate alleles (Charlesworth and Mank 2010). Thus, sex chromosome turnover may be more likely in species that have sex chromosomes that are not substantially degenerated.
If sex chromosome turnover were common, this could maintain homomorphy of sex chromosomes. Recent work on sex chromosomes in African clawed frogs has established nonhomology between the sex chromosomes of X. laevis and S. tropicalis (Uno et al., submitted) and the recent appearance of a novel sex-determining locus in X. laevis (Yoshimoto et al. 2008; Bewick et al. 2011). Thus, it appears that the origin of a new sex-determining gene in X. laevis was associated with a reassignment of sex chromosomes without necessarily involving a change in heterogamy, and that recent sex chromosome turnover can account for sex chromosome homomorphy in this species (Tymowska 1991). In other species, including S. tropicalis, it is also possible that the sex-determining mechanism of nondiverged sex chromosomes could be old, but that divergence is prevented by periodic recombination, which possibly could be facilitated by breeding individuals that are phenotypically sex reversed (the “fountain of youth” hypothesis; Perrin 2009). Because we do not yet know the sex-determining gene(s) of S. tropicalis or other frogs that might have inherited this sex determination system from a recent common ancestor (e.g., genera Hymenochirus, Pseudhymenochirus, or Pipa), we cannot determine at this time whether turnover or recombination best accounts for the apparent homomorphy of the sex chromosomes of this species. Additional identification of sex-determining genes in amphibians, and analysis of their evolutionary histories and genomic context, is thus an exciting direction for future research.
Supplementary Material
Supplementary figures S1–S3 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Richard Harland, Mustafa Khokha, James Evans, and Maura Lane for assistance with samples and information on the “golden” strain of S. tropicalis. They thank three anonymous reviewers for helpful comments on an earlier draft of this manuscript. They additionally thank Brian Golding and Sharcnet (www.sharcnet.ca) for access to computational facilities. This work was supported by grants from the National Science and Engineering Research Council of Canada, an Ontario Internal Prestige Scholarship, an Early Researcher Award from the Ontario Ministry of Economic Development and Innovation, and McMaster University. Support for sequencing of the laser dissected of chromosome arm 7p was provided by UK Medical Research Council U117560482 and U117597137, the Grant Agency of Charles University in Prague (407311), the Grant Agency of Czech Republic (P502/11/P522), the Ministry of Education, Youth and Sports of Czech Republic (MSM0021620858), UNCE (204013), SVV (265211), and the Ministry of Agriculture of the Czech Republic (MZE 0002716202).
Literature Cited
- Anderson JL, et al. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One. 2012;7:e40701. doi: 10.1371/journal.pone.0040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NS, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SW, et al. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One. 2011;6:e19315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus) Evolution. 2011;65:698–712. doi: 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- Bowes JB, et al. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2009;36:D761–D772. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain FJJ, Dushoff J, Evans BJ. The odds of duplicate gene persistence after polyploidization. BMC Genomics. 2011;12:599. doi: 10.1186/1471-2164-12-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Mariais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Mank JE. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics. 2010;186:9–31. doi: 10.1534/genetics.110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ. Genome evolution and speciation genetics of allopolyploid clawed frogs (Xenopus and Silurana) Front Biosci. 2008;13:4687–4706. doi: 10.2741/3033. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of clawed frogs: phylogeography on sub-Saharan Africa and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Evans BJ, et al. Description of a new octoploid frog species (Anura: Pipidae: Xenopus) from the Democratic Republic of the Congo, with a discussion of the biogeography of African clawed frogs in the Albertine Rift. J Zool. 2011;283:276–290. doi: 10.1111/j.1469-7998.2010.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Pyron RA, Wiens JJ. Polyploidization and sex chromosome evolution in amphibians. In: Soltis PS, Soltis DE, editors. Polyploidy and genome evolution. Springer Verlag; 2012. pp. 385–410. [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Fridolfsson A, et al. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci U S A. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, et al. The genome of the western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kamiya T, et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubipes (Fugu) PLoS Genet. 2012;8:e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, et al. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006;16:815–826. doi: 10.1101/gr.5016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa A, Ishiguchi Y, Yamada F, Shintaro A, Matsuda Y. The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma. 2010;119:519–526. doi: 10.1007/s00412-010-0275-8. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, et al. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. Uppsala University: Evolutionary Biology Centre; 2004. MrModeltest v2. [Google Scholar]
- Olmstead AW, Lindberg-Livingston A, Degitz SJ. Genotyping sex in the amphibian, Xenopus (Silurana) tropicalis, for endocrine disruptor bioassays. Aquat Toxicol. 2010;98:60–66. doi: 10.1016/j.aquatox.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Orr HA. “Why polyploidy is rarer in animals than in plants” revisited. Am Nat. 1990;136:759–770. [Google Scholar]
- Pease JB, Hahn MW. Sex chromosomes evolved from independent ancestral linkage groups in winged insects. Mol Biol Evol. 2012;29:1645–1653. doi: 10.1093/molbev/mss010. [DOI] [PubMed] [Google Scholar]
- Perrin N. Sex reversal: a fountain of youth for sex chromosomes? Evolution. 2009;63:3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.5. 2007. Available from: http://beast.bio.ed.ac.uk/Tracer.
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann U, Hoelzmann P. The Dahomey Gap: an abrupt climatically induced rain forest fragmentation in West Africa during the late Holocene. Holocene. 2005;15:190–199. [Google Scholar]
- Schmid M, et al. The chromosomes of Terraranan frogs. Cytogenet Genome Res. 2010;130–131:1–568. doi: 10.1159/000301339. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Stöck M, et al. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 2011;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley RC, Loumont C, Kobel HR. Geographical distribution and ecology. In: Tinsley RC, Kobel HR, editors. The biology of Xenopus. Oxford: Clarendon Press; 1996. pp. 35–59. [Google Scholar]
- Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y. Comparison of the Z and W sex chromosomal architectures in elegant chrested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma. 2007;116:159–173. doi: 10.1007/s00412-006-0088-y. [DOI] [PubMed] [Google Scholar]
- Tymowska J. Polyploidy and cytogenetic variation in frogs of the genus Xenopus. In: Green DS, Sessions SK, editors. Amphibian cytogenetics and evolution. San Diego (CA): Academic Press; 1991. pp. 259–297. [Google Scholar]
- Uno Y, et al. Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Ranidae and Xenopodinae. Chromosome Res. 2008;16:999–1011. doi: 10.1007/s10577-008-1257-z. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- Wells DE, et al. A genetic map of Xenopus tropicalis. Dev Biol. 2011;354:1–8. doi: 10.1016/j.ydbio.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.