Abstract
In the budding yeast Saccharomyces cerevisiae, the subunits of any given protein complex are either mostly essential or mostly nonessential, suggesting that essentiality is a property of molecular machines rather than individual components. There are exceptions to this rule, however, that is, nonessential genes in largely essential complexes and essential genes in largely nonessential complexes. Here, we provide explanations for these exceptions, showing that redundancy within complexes, as revealed by genetic interactions, can explain many of the former cases, whereas “moonlighting,” as revealed by membership of multiple complexes, can explain the latter. Surprisingly, we find that redundancy within complexes cannot usually be explained by gene duplication, suggesting alternate buffering mechanisms. In the distantly related Schizosaccharomyces pombe, we observe the same phenomenon of modular essentiality, suggesting that it may be a general feature of eukaryotes. Furthermore, we show that complexes flip essentiality in a cohesive fashion between the two species, that is, they tend to change from mostly essential to mostly nonessential, or vice versa, but not to mixed patterns. We show that these flips in essentiality can be explained by differing lifestyles of the two yeasts. Collectively, our results support a previously proposed model where proteins are essential because of their involvement in essential functional modules rather than because of specific topological features such as degree or centrality.
Keywords: essentiality, modularity, yeast, redundancy, genetic interactions, protein complexes
Introduction
Much of the work in cells is carried out by molecular machines known as protein complexes (Alberts 1998; Hartwell et al. 1999). These machines are composed of multiple proteins that are connected together in a single physical unit. In many ways, they are a canonical example of functional modules in biomolecular systems (Pereira-Leal et al. 2006): their protein components are densely connected in the protein interaction network, suggesting modularity in the network theory sense (Newman 2006); they are relatively independent, as many can be reconstituted in vitro in the absence of their surrounding network (Pereira-Leal et al. 2006); and they work together to carry out a common function.
The budding yeast Saccharomyces cerevisiae has proved to be an extraordinary resource for understanding the organization and behavior of these molecular machines. Two global experimental studies have sought to identify comprehensive maps of protein complexes in this organism (Gavin et al. 2006; Krogan et al. 2006), whereas others have curated the literature to identify hundreds of complexes with support from small-scale experiments (Pu et al. 2009). The availability of a genome wide set of gene deletion mutants in S. cerevisiae (Giaever et al. 2002) has also made this organism extremely useful in understanding how genotypes map to phenotypes. Perhaps, the easiest phenotype to measure in yeast is cell growth or viability, and most large-scale studies of the gene deletion collection have focused on it. Giaever et al. (2002) noted that approximately 81% of genes could be individually deleted in rich media without causing cell death—that is, they were nonessential. The remaining approximately 19% were essential for cell survival.
A disproportionate number of these essential genes were noted to belong to protein complexes (Hart et al. 2007; Michaut et al. 2011). Indeed, the relationship between a gene’s essentiality and the connectivity of its encoded protein has been the subject of much analysis, and a number of models have been proposed to explain the link between the two. Jeong et al. (2001) noted a strong correlation between essentiality and protein interaction degree (the “centrality-lethality” rule). Subsequent analyses have shown that although this correlation is weak for interaction networks that map binary or transient interactions (e.g., yeast two-hybrid) (Yu et al. 2008; Zotenko et al. 2008), it is robust for networks that map stable cocomplex interactions. However, although the result linking essentiality and interaction degree has been replicated a number of times, the interpretation of this link has been subject to much debate. It was initially suggested that proteins with a high degree are essential because of their role in maintaining the overall connectivity of the network (Jeong et al. 2001), and later that certain interactions are essential and high-degree proteins are simply more likely to disrupt an essential interaction (He and Zhang 2006). Both interpretations were convincingly rejected by Zotenko et al. (2008) who instead proposed that certain proteins were essential because of their involvement in “Essential Complex Biological Modules”—large groups of essential proteins, which are densely connected on the protein interaction network and are all involved in a common biological function. Similarly, others have shown that essential genes are concentrated into specific complexes and that protein complexes are either mostly essential or mostly nonessential, suggesting that essentiality is a property of molecular machines rather than individual components (fig. 1A) (Dezso et al. 2003; Hart et al. 2007; Semple et al. 2008; Wang et al. 2009). Wang et al. (2009) connected the two observations by noting that larger protein complexes are more likely to be essential, explaining why essential genes are more likely to have high cocomplex interaction degree.
Fig. 1.—
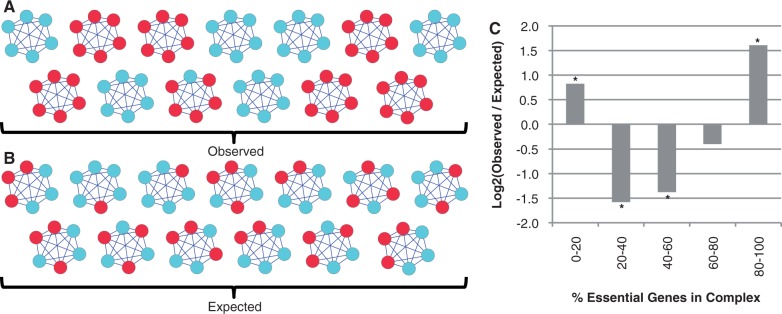
Comparing the observed distribution of essential genes in Saccharomyces cerevisiae to that expected at random. (A) Distribution of essential genes among complexes in S. cerevisiae. For clarity, only nonoverlapping complexes with six subunits are shown. Essential genes are shown in red, nonessential in blue. (B) An example of randomly distributed essential genes among the same set of complexes shown in (A). (C) Comparing the observed distribution of essential genes in complexes to that seen in 1,000 randomizations (see Materials and Methods). The log 2 ratio of observed to expected is shown here; positive values indicate observed frequency above random, and negative values indicate frequency below random. Stars indicate values that are more extreme than those seen in any of the randomizations.
More generally, the observation that entire complexes appear essential, henceforth referred to as modular essentiality, indicates that phenotypes can be associated with whole complexes and are not restricted to individual genes. Related work in yeast has shown that members of the same protein complex respond in a coherent fashion to the same drugs (Hillenmeyer et al. 2010), whereas other analyses in humans have suggested that genes whose protein products belong to the same complex are more likely to result in the same disease phenotype (Fraser and Plotkin 2007; Lage et al. 2007; Oti and Brunner 2007).
In this work, we seek to address two outstanding questions related to modular essentiality:
First—if modularity as an organizing principle explains essentiality in protein complexes, why do exceptions arise? that is, why are there nonessential subunits in largely essential complexes and why are there essential subunits in largely nonessential complexes? We show that redundancy within complexes, as revealed by genetic interactions, can explain many of the former cases, whereas “moonlighting,” as revealed by membership of multiple complexes, can explain the latter.
Second—is the phenomenon of modular essentiality conserved across species? There is evidence in a number of model organisms that members of the same complex are more likely to share the same phenotype when perturbed (Lage et al. 2007; Lee et al. 2008); however, the lack of a comprehensive set of gene deletions in any organism other than S. cerevisiae has thus far prevented a global analysis. Using comprehensive essentiality data from the distantly related fission yeast Schizosaccharomyces pombe (Kim et al. 2010), we find strong evidence of modular essentiality, suggesting that it may be a general feature of eukaryotes. Furthermore, we show that complexes flip essentiality between the two species in a cohesive fashion, that is, they change from mostly essential to mostly nonessential or vice versa and that these flips can be associated with larger scale biological differences between the two yeasts.
Materials and Methods
Essentiality Data
Saccharomyces cerevisiae data were obtained from the OGEE database (Chen et al. 2012), filtered to only include the results from Giaever et al. (2002). Schizosaccharomyces pombe essentiality data, and the orthology mapping between Sch. pombe and S. cerevisiae, were obtained from Kim et al. (2010). Mouse essentiality data were obtained from the OGEE database (Chen et al. 2012).
Gene Duplications
A list of duplicate gene pairs resulting from whole-genome duplication was obtained from Byrne and Wolfe (2005). This list was extended to include a list of small-scale duplications identified using the method of Rost (1999). An initial list of duplicates was identified by performing an all against all Basic Local Alignment Search Tool comparison (E = 10). These were subsequently filtered, such that the length of the aligned region (L) was ≥80% of the length of the longer protein, and the identity of the aligned region was greater than or equal to a minimum identity threshold. For pairs with an aligned region greater than 150 a.a., the threshold was 30%, otherwise it was equal to n + 480 L−0.32(1 + exp(−L/1,000)), where n = 6, as per Gu et al. (2002).
Genetic Interactions
The quantitative genetic interactions used for our analysis were obtained from a recently collated interactome containing the majority of quantitatively measured S. cerevisiae interactions (Ryan et al. 2012). All interactions with a score less than −2.3 were considered to be a strong negative genetic interaction. To better assess the relationship between homology and within-complex redundancy, a more complete list of genetic interactions (including those from low-throughput and nonquantitative screens) was obtained from the BioGRID (Stark et al. 2011) version 3.1.8.8. Interactions annotated “Negative Genetic,” “Synthetic Growth Defect,” or “Synthetic Lethality” were included in this list if both genes were identified as nonessential in our data.
Protein Domain Data
Protein domain annotations were obtained from the precalculated InterProScan results (domain.tab) hosted at the Saccharomyces Genome Database (Cherry et al. 2012). Annotations matching “HMMPfam” were extracted, resulting in 31,924 protein pairs sharing a Pfam domain.
Drug Sensitivity Data
Drug sensitivity data were obtained from the heterozygous fitness screen of Hillenmeyer et al. (2008). Only drug–gene interactions with P < 0.01 were considered.
Gene Ontology Data
Gene Ontology annotations were obtained from the Saccharomyces Genome Database (Cherry et al. 2012), and only unique annotations from the “Biological Process” ontology were counted.
Protein Complex Definitions
Saccharomyces cerevisiae complex definitions were obtained from a manually curated list of 408 complexes supported by small-scale experiments from the literature (Pu et al. 2009). This set covers 56% of the essential genes in budding yeast (588/1,088).
Mouse complex definitions were obtained from the CORUM database (Ruepp et al. 2010).
Schizosaccharomyces pombe complexes were identified from the Gene Ontology annotations maintained at PomBase (Wood et al. 2012). Descendants of the term “macromolecular complex” were included if the term name contained the word “complex” or “subunit.” This list was manually expanded to include a number of complexes that followed different naming conventions—for example, “signalosome” and “U1 SnRNP.” Only associations annotated with “IDA” or “IPI” were included—indicating that the complex definition is supported by direct experimental evidence in Sch. pombe and not just inferred by sequence similarity. Finally, this list was pruned to remove complexes with identical subunit composition but different names, for example, “RNA-directed RNA polymerase complex” and “nuclear RNA-directed RNA polymerase complex.” The full list of 104 complexes is available in supplementary table S1, Supplementary Material online.
Assessing the Modular Distribution of Essential/Flipped Genes
Using a methodology similar to Hart et al. (2007), we assessed the extent to which essentiality was concentrated in specific complexes. The percentage of essential genes in each complex was calculated, sorted into five equal-sized bins, and compared with an expected background generated by randomly assigning essential genes to the same set of complexes. The same methodology was used to assess the distribution of genes that “flipped” essentiality.
Results
We first confirmed the observation of modular essentiality in S. cerevisiae using an up to date manually curated set of “gold-standard" protein complexes (Pu et al. 2009) and the method of Hart et al. (fig. 1, see Materials and Methods). We found, as in previous studies, that complexes are mostly essential or mostly nonessential, but that it is not an all or nothing phenomenon. Indeed, only approximately 26% (105/401) of complexes consist of solely essential or solely nonessential subunits. Despite the success of the concept of modular essentiality in explaining the behavior of protein complex subunits, the extensive presence of exceptions is troubling. If protein complexes truly behave as a single unit, with their activity dependant on the coordinated function of all subunits, then why would some subunits behave differently to others?
Redundancy Explains Many of the Nonessential Members of Essential Complexes
One possible explanation for the presence of nonessential genes in largely essential complexes is redundancy—the ability of one gene to functionally compensate for the loss of another (fig. 2A). Such relationships are ubiquitous in biological systems (Nowak et al. 1997) and can be identified in high throughput in yeast by comparing the growth of mutants with two gene deletions to the growth of mutants with each gene deleted individually (Baryshnikova et al. 2010; Collins et al. 2010). Negative genetic interactions, also known as synthetic sick or synthetic lethal, are identified when the double mutant grows significantly worse than expected and indicate a degree of functional redundancy between the two deleted genes.
Fig. 2.—
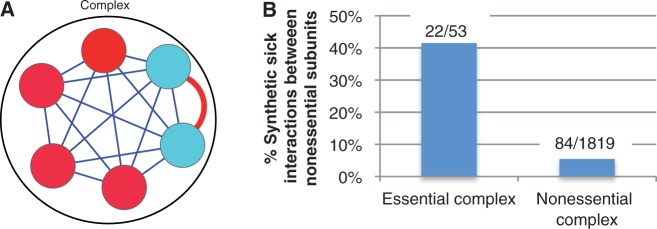
Redundancy explains nonessential genes in largely essential complexes. (A) Cartoon example of redundancy within a protein complex. Red circles represent essential genes, blue circles represent nonessential genes, and the red edge indicates a redundant relationship. (B) Interactions between the nonessential members of essential complexes are more likely to be synthetic sick than interactions between nonessential members of nonessential complexes (P = 1.9 × 10−15, two sided Fisher’s exact test). Complexes with ≥60% essential genes are considered essential and complexes with <40% essential genes are considered nonessential. Counts above bars indicate the number of synthetic sick interactions over the total number of measured interactions.
If redundancy explains the presence of nonessential genes within essential protein complexes, then essential complexes should be enriched for negative genetic interactions. Using a set of quantitatively measured genetic interactions, we found that this is indeed the case with 42% (22/53) of the measured interactions in essential complexes (≥60% of subunits essential) being negative compared with 5% (84/1,819) of those within nonessential complexes (fig. 2B). Note that we would not expect 100% of the measured interactions between nonessential genes in essential protein complexes to be negative, as each gene need only be redundant with one other complex subunit. Indeed, approximately 65% of the genes covered by our data set feature a negative genetic interaction with at least one other member of the same complex.
One plausible explanation for redundancy is gene duplication (Guan et al. 2007; Ihmels et al. 2007; Musso et al. 2007; Dean et al. 2008; Szklarczyk et al. 2008; Li et al. 2010; VanderSluis et al. 2010). To assess the extent to which gene duplication can explain this within-complex redundancy, we assembled a list of paralogs from small-scale and whole-genome duplications (see Materials and Methods). Surprisingly, we found that none of the 22 negative genetic interactions within essential protein complexes can be explained by gene duplication. Indeed, examination of all previously reported negative genetic interactions (Stark et al. 2011) (including those from low-throughput and nonquantitative screens) suggests that redundancy between cocomplexed pairs can only rarely be explained by homology. Of 315 negative genetic interactions reported to occur between members of the same protein complex, only 67 can be explained by gene duplication. Of these 67 interactions, 50 occur between members of the large or small cytosolic ribosomal complexes, suggesting that, with the notable exception of the ribosome, homology between redundant cocomplexed pairs is the exception rather than the rule.
Expanding our analysis to include all proteins that share a Pfam (Punta et al. 2012) domain (see Materials and Methods), rather than strict paralogs, we find that only 89 of the 315 negative genetic interactions occurring between members of the same complex share a protein domain. Again, the majority (51) of these domain-sharing pairs are ribosomal proteins, suggesting that shared domains cannot explain the apparent redundancy within complexes.
Moonlighting Proteins Explain Many of the Essential Genes in Nonessential Complexes
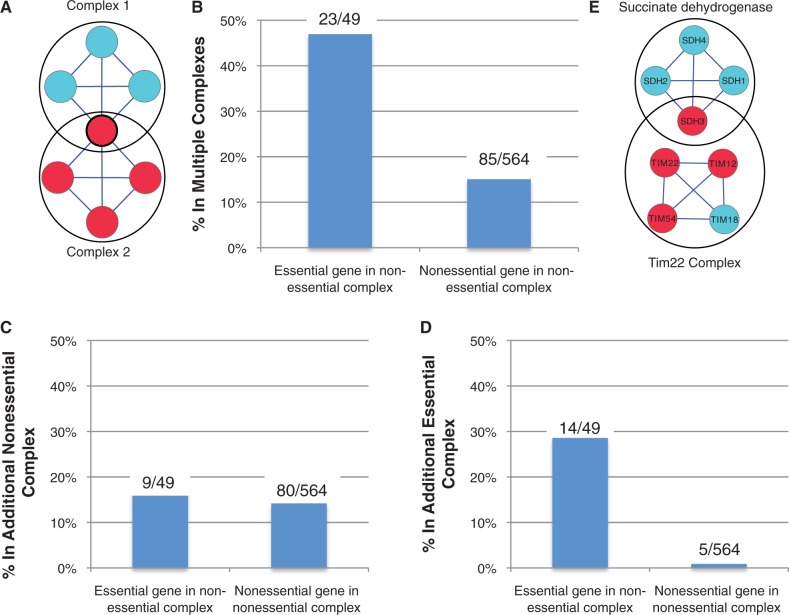
A possible explanation for the existence of essential genes in otherwise nonessential complexes is that these essential genes have additional roles outside the complex—that is, they are “moonlighting” (Jeffery 2003). If this hypothesis is correct, we reasoned that the protein products of these moonlighting genes would be more likely to be members of multiple protein complexes (fig. 3A). We tested this hypothesis by comparing the essential genes in nonessential complexes (<40% subunits essential) to the nonessential genes in the same complexes and discovered that the essential genes were significantly more likely to be members of multiple complexes (fig. 3B). These moonlighting proteins may be essential for two reasons—their participation in an essential complex in addition to their role in the nonessential complex or their participation in two distinct nonessential complexes. To distinguish between the two cases, we further stratified the groups and asked whether essential genes in nonessential complexes are more likely to be a member of an additional nonessential complex than nonessential genes in the same complexes. We found a slight but not statistically significant difference between the two groups (fig. 3C, P = 0.40, Fisher’s exact test), suggesting that it is not simply membership of multiple complexes that renders a protein essential. When restricting the analysis to membership of an additional essential complex, we find a very significant difference between the two groups—with essential genes being approximately 30 times more likely to belong to an additional essential complex than nonessential genes (fig. 3D, P = 5.7 × 10−13).
Fig. 3.—
Moonlighting explains essential proteins in nonessential complexes. (A) Cartoon example of a moonlighting protein present in both Complex 1 and Complex 2. Colors as in figure 2. (B) Essential genes in nonessential complexes are more likely to belong to multiple complexes than nonessential genes in nonessential complexes (P = 6.3 × 10−7 two-sided Fisher’s exact test). Complexes with <40% essential genes are considered nonessential, and the cytosolic ribosomal subunits genes are excluded from this analysis, as their behavior is better explained by gene duplication. Labels above bars indicate actual gene counts. (C) Essential genes in nonessential complexes are no more likely to belong to an additional nonessential complex than nonessential genes in nonessential complexes (P = 0.40 two-sided Fisher’s exact test). Details as in (B). (D) Essential genes in nonessential complexes are also more likely to belong to an essential complex than nonessential genes in nonessential complexes (P = 5.67 × 10−13 two-sided Fisher’s exact test). Details as in (B). (E) SDH3, the only essential member of the succinate dehydrogenase complex, has recently been identified as a having an additional role as part of the essential Tim22 complex.
This phenomenon is widespread and is not explained by a small number of essential moonlighting proteins present in many nonessential complexes: membership of multiple complexes accounts for approximately 50% of the essential genes in nonessential complexes (excluding the cytosolic ribosome). Nevertheless, it is reasonable to ask what accounts for the rest. It is possible that these genes are actually members of other complexes, but these associations are not yet experimentally determined or documented in databases. An example of this is SDH3, which in our set of complex definitions is only annotated as a member of the succinate dehydrogenase complex (Oyedotun and Lemire 2004). SDH3 is the only essential member of this complex, suggesting that it has an additional role outside the complex, and indeed, it has recently been identified as a member of the essential TIM22 mitochondrial inner membrane protein insertion complex (Gebert et al. 2011) (fig. 3E).
If undocumented membership of another complex was a common explanation for essential genes annotated as belonging a single mostly nonessential complex, then we would expect these genes to display more phenotypes when perturbed than nonessential genes in the same complex due to their additional functions. We tested this using data from a large-scale haploinsufficiency screen (Hillenmeyer et al. 2008), finding that they display more drug sensitivities than nonessential genes in nonessential complexes (median of 33 drug interactions vs. 27). Furthermore, we find that essential genes in nonessential complexes have a greater number of biological process annotations in the Gene Ontology (median of 9 vs. 6) (Ashburner et al. 2000). Both of these observations support the hypothesis that these essential genes have additional functions outside the nonessential complex.
Schizosaccharomyces pombe Complexes Display Modular Essentiality
Having confirmed the phenomenon of modular essentiality in S. cerevisiae, we next sought to test whether this observation is conserved across species. The fission yeast S. pombe is separated from S. cerevisiae by approximately 400 Myr of evolution (Sipiczki 2000) and is the only other eukaryote for which comprehensive essentiality data are available (Kim et al. 2010). These two species differ greatly in their basic biology, with Sch. pombe appearing more similar to metazoan cells in many respects—including aspects of mRNA splicing (due to the extensive presence of introns in Sch. pombe), gene expression controlled in part by the RNA interference machinery (absent in S. cerevisiae), chromosome structure and epigenetic mechanisms, and cell-cycle regulation by the G2/M transition control (Wood 2006; Rhind et al. 2011). Furthermore, since their divergence, S. cerevisiae has undergone a whole-genome duplication (Wolfe and Shields 1997). Consequently, features conserved in both species are likely to be broadly conserved across eukaryotes.
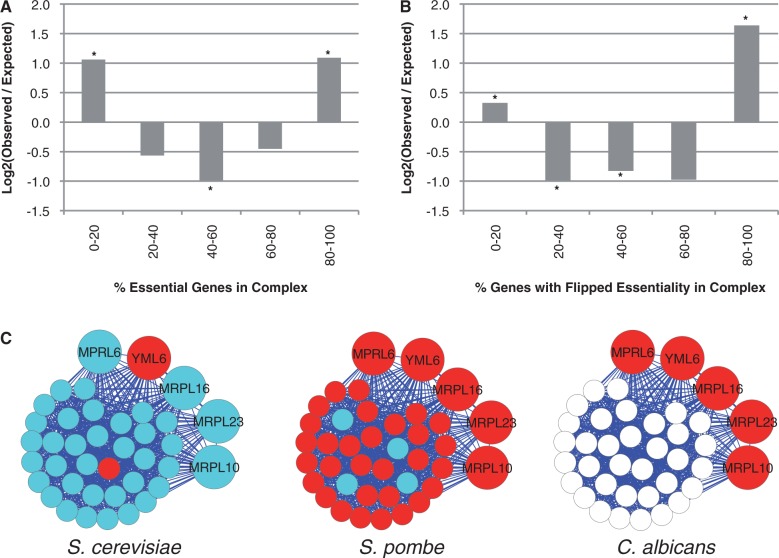
We assembled a list of 104 complexes with experimental support in Sch. pombe (see Materials and Methods) including nine complexes that are absent in S. cerevisiae. By performing the same analysis described in figure 1, we found that Sch. pombe complexes are disproportionately mostly essential or mostly nonessential (fig. 4A), confirming that modular essentiality is conserved across species. Interestingly, eight of the nine complexes that are absent in S. cerevisiae are completely nonessential.
Fig. 4.—
Schizosaccharomyces pombe complexes exhibit modular essentiality, and complexes flip essentiality between the two yeast species. (A) Experimentally characterized S. pombe complexes exhibit modular essentiality. The observed distribution of essential genes is compared with that expected at random, as in figure 1C (See Materials and Methods). (B) Protein complexes flip essentiality between S. pombe and S. cerevisiae. The observed distribution of “flipped” genes is compared with that expected at random, as in figure 1C. Only genes present in a single copy in both species are used for this analysis. The complex definitions are the same as those used for S. cerevisiae, as they have greater coverage than S. pombe. (C) The mitochondrial ribosomal large subunit. This complex is essential in S. pombe and Candida albicans but nonessential in S. cerevisiae. White circles correspond to genes whose essentiality status has yet to be experimentally determined. Genes whose orthologs have unknown essentiality status in C. albicans are unlabeled and shown as smaller circles.
It is possible that modular essentiality is a feature conserved in single-celled fungi but absent in metazoans due to the increased complexity requirements of multiple cell types. However, analysis of the essentiality of an incomplete set of knock-out mice suggests that the distribution of essential genes within complexes also follows a similar, albeit weaker, pattern in this species (supplementary fig. S1, Supplementary Material online). As the coverage of knockout collections in metazoans increases, we will be able to assess the conservation of this trend better.
Protein Complexes “Flip" Essentiality between the Two Species
Eighty-three percent of the one-to-one orthologs between Sch. pombe and S. cerevisiae have conserved essentiality, that is, they are nonessential in both species or essential in both species. The remaining 17% of genes are nonessential in one species and essential in the other. Interestingly, Kim et al. (2010) found that genes that change essentiality are enriched in certain functional categories. This enrichment would still be statistically significant if only 50% of the subunits of a large complex changed essentiality, but such behavior would not be consistent with the idea of modular essentiality. Instead, if essentiality is truly a modular behavior, we should see that complexes “flip" essentiality as a single unit. By analyzing the distribution of flipped genes within complexes and comparing it to that expected at random, we found that complexes do behave in a modular fashion, with either most or few of their subunits changing essentiality (fig. 4B).
We find that for 14 of the complexes, 80% or more of their subunits flip essentiality in a coherent fashion between the two species. A complete list of these complexes is given in table 1. These complexes range in size from the large mitochondrial ribosomal subunit, which features 36 genes that flip, to smaller complexes such as the SBF transcription complex, whose two subunits flip. Furthermore, we note that most complexes (64%) show no change at all in essentiality between species—for example, the DNA-directed RNA polymerase III complex, whose 17 subunits are essential in both species, and the Elongator complex, whose six subunits are nonessential in both species.
Table 1.
Protein Complexes That Flip Essentiality between Schizosaccharomyces pombe and Saccharomyces cerevisiae
| Complex Name | Fraction Flipped | No. Essential in S. pombe | No. Essential in S. cerevisiae | Complex Size | Associated Lifestyle Difference |
|---|---|---|---|---|---|
| Mitochondrial ribosomal small subunit | 0.83 | 23 | 3 | 24 | Mitochondrial translation |
| Mitochondrial ribosomal large subunit | 0.85 | 36 | 2 | 40 | Mitochondrial translation |
| Clathrin | 1.00 | 2 | 0 | 2 | Golgi associated |
| GARP complex | 1.00 | 4 | 0 | 4 | Golgi associated |
| H+-transporting ATPase, Golgi | 1.00 | 11 | 0 | 11 | Golgi associated |
| Mdm12p/Mmm1p/Mdm10p complex | 1.00 | 3 | 0 | 3 | Mitochondrial translation (absence results in loss of mtDNA) |
| DASH complex | 1.00 | 0 | 9 | 9 | Spindle/Kinetochore |
| Gcd10p/Gcd14p complex | 1.00 | 0 | 2 | 2 | |
| Mtr2p/Mex67p complex | 1.00 | 0 | 2 | 2 | |
| TORC 2 complex | 1.00 | 0 | 3 | 3 | |
| Uba2p/Aos1p complex | 1.00 | 0 | 2 | 2 | |
| Anthranilate synthase complex | 1.00 | 2 | 0 | 2 | |
| SBF complex | 1.00 | 2 | 0 | 2 | |
| Ula1p/Uba3p complex | 1.00 | 2 | 0 | 2 |
Complexes that function in processes or organelles that have well-documented differences between the two yeasts account for half of the complexes that flip essentiality (table 1). For example, the most significant difference was seen in the mitochondrial translation machinery. Saccharomyces cerevisiae cells can survive without mtDNA as “petite" mutants, in contrast to Sch. pombe (“petite negative") (Jiang et al. 2011). If this difference in lifestyle causes the mitochondrial translation machinery to flip essentiality between species, then one would expect this machinery to be essential in other petite negative species. Candida albicans is one such species—Phylogenetically it is closer to S. cerevisiae than Sch. pombe, but it displays a petite negative phenotype. Comprehensive essentiality data are not available for C. albicans; however, a number of pilot studies (Becker et al. 2010; Noble et al. 2010) have established the status of a few hundred genes with S. cerevisiae orthologs, allowing us to test this hypothesis. Consistent with the idea that changes in lifestyle are associated with flips in essentiality, we find that all the members of the large mitochondrial ribosomal subunit in C. albicans that have been experimentally tested are essential (fig. 4C).
Discussion
In this work, we have shown that modular essentiality is an organizing principle conserved across two very distantly related species, suggesting that it may be a general feature of eukaryotic systems. Furthermore, we have provided explanations for exceptions to this principle. Finally, we have shown that protein complexes flip essentiality as a coherent unit between two species. Overall, these results reinforce the concept that protein complexes are a fundamental functional unit of cell biology and that phenotypes can be associated with whole protein complexes rather than individual genes.
From Essentiality to Other Phenotypes
Modular essentiality is consistent with the tendency of members of protein complexes to display coherent “monochromatic" genetic interactions (Segrè et al. 2005; Bandyopadhyay et al. 2008; Michaut et al. 2011), to display sensitivity to the same drugs (Fraser and Plotkin 2007; Hillenmeyer et al. 2010; Kapitzky et al. 2010), and to be associated with the same disease (Fraser and Plotkin 2007; Lage et al. 2007; Oti and Brunner 2007). Our explanations for exceptions to modular essentiality may be applied in these contexts as well—providing explanations for why perturbation of one member of a complex may cause a different disease or exhibit different drug sensitivity to other members of the same complex
Understanding Exceptions in Greater Detail
We found that essential genes in otherwise nonessential complexes tend to be part of a second complex. This sharing of subunits could come about for two reasons—either the protein in question could have a unique structural feature (e.g., a catalytic activity or a structural recognition domain) that is required by both complexes (“moonlighting") or it could have distinct structural features required in each complex. Distinguishing between the two cases will require the integration of structural models (Kim et al. 2006) or through genetic perturbation of individual interacting domains (Dreze et al. 2009; Ear and Michnick 2009; Zhong et al. 2009).
Furthermore, we found that nonessential genes in otherwise essential complexes frequently exhibit negative genetic interactions with another member of the same complex. Previous work found that complexes that are enriched in negative genetic interactions tend to contain at least one essential gene (Bandyopadhyay et al. 2008; Wilmes et al. 2008; Baryshnikova et al. 2010); however, these analyses included genetic interactions between hypomorphic alleles of essential genes, making the interpretation of these results difficult. Here, by asking the inverse question (“do nonessential genes in essential complexes display negative genetic interactions?”) and only analyzing genetic interactions between nonessential genes, we find support for the idea that these nonessential subunits are redundant.
We observed that only approximately 5% of the genetic interactions in nonessential protein complexes are negative. However, many complexes are only essential in the presence of some drug or stress, so many negative genetic interactions may only be observed under specific conditions. Indeed, it has been noted that certain genetic interactions (termed conditional genetic interactions) are only observed in specific conditions (St Onge et al. 2007; Bandyopadhyay et al. 2010; Guénolé et al. 2013). Consequently, we note that this is likely a lower bound on the estimate of the frequency of redundancy within nonessential complexes.
Homology between Redundant Proteins in the Same Complex as the Exception Rather than the Rule
Several groups have analyzed the retention of duplicate genes in protein complexes and the roles these duplicates play (Musso et al. 2007; Pereira-Leal et al. 2007; Szklarczyk et al. 2008). Others have analyzed the extent to which duplicate gene pairs display functional redundancy (Guan et al. 2007; Ihmels et al. 2007; Dean et al. 2008; Li et al. 2010; van Wageningen et al. 2010; VanderSluis et al. 2010) and noted that duplicates coding for proteins in the same complex are frequently redundant (Li et al. 2010). Here, we find that duplicate genes are responsible for only a small fraction of the redundancy within complexes. This suggests that although duplicate genes within the same complex may be frequently redundant, the majority of redundancy within complexes cannot be explained by duplication. The mechanism by which nonhomologous genes perform their redundant roles is unclear; however, we have recently shown that these redundant relationships within complexes are highly conserved across species (Ryan et al. 2012) suggesting that they merit further investigation.
Function, Not Topology, as the Primary Determinant of Essentiality
There is strong evidence that protein–protein interactions, especially those between members of the same complex, are highly conserved across species (Shevchenko et al. 2008; van Dam and Snel 2008; Leducq et al. 2012). That a protein’s interaction partners may remain relatively constant while the requirement of that protein for survival may change suggests that topological features are not the primary determining factor of essentiality. Indeed, the observation that so many changes in essentiality between species fall into specific functional categories (mitochondrial translation, kinetochore, Golgi associated) lends strong support to the proposal that it is involvement in specific biological processes that renders a protein essential, rather than network connectivity (Zotenko et al. 2008).
Using Protein Complexes to Understand Differences between Species
We were interested in how the essentiality of protein complexes differs between species. Previous work has highlighted the utility of protein complexes as an abstraction to understand the evolution of cell-cycle regulation (Jensen et al. 2006), phosphorylation (Beltrao et al. 2009), genetic interactions (Roguev et al. 2008; Frost et al. 2012; Koch et al. 2012; Ryan et al. 2012), drug–gene interactions (Kapitzky et al. 2010), and transcription factor binding (Tan et al. 2007). In this work, we found that protein complexes “flip" essentiality as a coherent unit between species. As gene deletion collections become available in additional species, especially those more closely related than Sch. pombe and S. cerevisiae, we may be able to compare the relative contributions of lifestyle changes and other factors. It may also be possible to define a core set of complexes that are essential in all eukaryotes under all conditions, for example, those involved in core processes such as transcription, and a set of “conditionally essential" complexes that may be associated with specific environments or morphology.
Supplementary Material
Supplementary table S1 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank A. Roguev, D. Greene, D.C. Shields, and P. Beltrao for helpful discussion. This work was supported by the Irish Research Council for Science and Engineering Technology to C.J.R., by the NIH (GM084448, GM084279, GM081879, and GM098101) to N.J.K., and by Science Foundation Ireland (08/SRC/I1407) to P.C. and G.C.
Literature Cited
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JM, et al. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A. 2010;107:22044–22049. doi: 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-HH, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z, Oltvai ZN, Barabási A-L. Bioinformatics analysis of experimentally determined protein complexes in the yeast Saccharomyces cerevisiae. Genome Res. 2003;13:2450–2454. doi: 10.1101/gr.1073603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M, et al. “Edgetic” perturbation of a C. elegans BCL2 ortholog. Nat Methods. 2009;6:843–849. doi: 10.1038/nmeth.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ear PH, Michnick SW. A general life-death selection strategy for dissecting protein functions. Nat Methods. 2009;6:813–816. doi: 10.1038/nmeth.1389. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Plotkin JB. Using protein complexes to predict phenotypic effects of gene mutation. Genome Biol. 2007;8:R252. doi: 10.1186/gb-2007-8-11-r252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A-C, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gebert N, et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional Analysis of Gene Duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénolé A, et al. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol Cell. 2013;49:346–358. doi: 10.1016/j.molcel.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TT, Lee I, Marcotte ER. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2:e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, et al. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010;11:R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Collins S, Schuldiner M, Krogan N, Weissman J. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol. 2007;3:86. doi: 10.1038/msb4100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- Jensen LJJ, Jensen TSS, De Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jiang H, et al. Identification and characterization of the mitochondrial RNA polymerase and transcription factor in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:5119–5130. doi: 10.1093/nar/gkr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitzky L, et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol. 2010;6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-U, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- Koch EN, et al. Conserved rules govern genetic interaction degree across species. Genome Biol. 2012;13:R57. doi: 10.1186/gb-2012-13-7-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Leducq J-B, et al. Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genet. 2012;8:e1003161. doi: 10.1371/journal.pgen.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- Li J, Yuan Z, Zhang Z. The cellular robustness by genetic redundancy in budding yeast. PLoS Genet. 2010;6:e1001187. doi: 10.1371/journal.pgen.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaut M, et al. Protein complexes are central in the yeast genetic landscape. PLoS Comput Biol. 2011;7:e1001092. doi: 10.1371/journal.pcbi.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Zhang Z, Emili A. Retention of protein complex membership by ancient duplicated gene products in budding yeast. Trends Genet. 2007;23:266–269. doi: 10.1016/j.tig.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Newman M. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Oti M, Brunner HG. The modular nature of genetic diseases. Clin Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004;279:9424–9431. doi: 10.1074/jbc.M311876200. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy E, Kamp C, Teichmann S. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy ED, Teichmann SA. The origins and evolution of functional modules: lessons from protein complexes. Philos Trans R Soc Lond B Biol Sci. 2006;361:507–517. doi: 10.1098/rstb.2005.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Ruepp A, et al. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrè D, DeLuna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Semple JI, Vavouri T, Lehner B. A simple principle concerning the robustness of protein complex activity to changes in gene expression. BMC Syst Biol. 2008;2:1. doi: 10.1186/1752-0509-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, et al. Chromatin central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1:reviews1011–reviews1011.4. doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA, Snel B. Complex fate of paralogs. BMC Evol Biol. 2008;8:337. doi: 10.1186/1471-2148-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Shlomi T, Feizi H, Ideker T, Sharan R. Transcriptional regulation of protein complexes within and across species. Proc Natl Acad Sci U S A. 2007;104:1283–1288. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam TJP, Snel B. Protein complex evolution does not involve extensive network rewiring. PLoS Comput Biol. 2008;4:e1000132. doi: 10.1371/journal.pcbi.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderSluis B, et al. Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol. 2010;6:429. doi: 10.1038/msb.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wageningen S, et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143:991–1004. doi: 10.1016/j.cell.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol Cell Proteomics. 2009;8:1361–1381. doi: 10.1074/mcp.M800490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wood V. Schizosaccharomyces pombe comparative genomics; from sequence to systems. In: Sunnerhagen P, Piskur J, editors. Comparative genomics: using fungi as models. Vol. 15. Berlin (Germany): Springer; 2006. pp. 233–285. [Google Scholar]
- Wood V, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotenko E, Mestre J, O’Leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.