Abstract
The transposon BuT5 caused two chromosomal inversions fixed in two Drosophila species of the repleta group, D. mojavensis and D. uniseta. BuT5 copies are approximately 1-kb long, lack any coding capacity, and do not resemble any other transposable element (TE). Because of its elusive features, BuT5 has remained unclassified to date. To fully characterize BuT5, we carried out bioinformatic similarity searches in available sequenced genomes, including 21 Drosophila species. Significant hits were only recovered for D. mojavensis genome, where 48 copies were retrieved, 22 of them approximately 1-kb long. Polymerase chain reaction (PCR) and dot blot analyses on 54 Drosophila species showed that BuT5 is homogeneous in size and has a widespread distribution within the repleta group. Thus, BuT5 can be considered as a miniature inverted-repeat TE. A detailed analysis of the BuT5 hits in D. mojavensis revealed three partial copies of a transposon with ends very similar to BuT5 and a P-element-like transposase-encoding region in between. A putatively autonomous copy of this P element was isolated by PCR from D. buzzatii. This copy is 3,386-bp long and possesses a seven-exon gene coding for an 822-aa transposase. Exon–intron boundaries were confirmed by reverse transcriptase-PCR experiments. A phylogenetic tree built with insect P superfamily transposases showed that the D. buzzatii P element belongs to an early diverging lineage within the P-element family. This divergent P element is likely the master transposon mobilizing BuT5. The BuT5/P element partnership probably dates back approximately 16 Ma and is the ultimate responsible for the generation of the two chromosomal inversions in the Drosophila repleta species group.
Keywords: transposon, MITE, inversions, Drosophila, transposase, expression
Introduction
Transposable elements (TEs) are DNA sequences able to proliferate and move to multiple sites in the genome. As a consequence of their mobility, TEs are a source of variation in gene and genome structure as well as size and organization of genomes (Kidwell and Lisch 2002). Therefore, the study of TEs can shed light on their ability to impact the genomes they inhabit (Kazazian 2004; Jurka et al. 2007; Fedoroff 2012). TEs that mobilize via an RNA intermediate are classified within class I and those which transpose directly, leaving the donor site, or via a DNA intermediate, within class II (Wicker et al. 2007; see also Kapitonov and Jurka 2008). Class II, or DNA transposons, is divided in two subclasses and subclass 1 comprises two orders, terminal inverted repeat (TIR) and Crypton. Canonical (autonomous) TIR transposons have TIRs and contain usually one (less often two) gene encoding the transposase, the protein that catalyzes their mobilization via a cut-and-paste mechanism. The numerous TIR transposon families have been grouped into 9–19 superfamilies based not only on phylogenetic relationships inferred from the transposase but also on TIR and target site duplication (TSD) features (Jurka et al. 2005, 2007; Feschotte and Pritham 2007; Wicker et al. 2007; Kapitonov and Jurka 2008; Bao et al. 2009; Yuan and Wessler 2011). The P superfamily comprises three transposons: P element (O’Hare and Rubin 1983), 1360 (also known as Hoppel or ProtoP) (Kapitonov and Jurka 2003; Reiss et al. 2003), and Galileo (Marzo et al. 2008).
The P element is a TIR transposon first discovered in Drosophila melanogaster (Bingham et al. 1982; Rubin et al. 1982) as the cause of the odd phenomenon of P-M hybrid dysgenesis (Kidwell and Novy 1979). The D. melanogaster P element is not only one of the first eukaryotic TEs to be discovered and molecularly characterized but also one of the most thoroughly studied (Rio 1991, 2002; Kidwell 1994; Engels 1996; Pinsker et al. 2001). The canonical P element of D. melanogaster is 2.9 kb in length and has 31-bp TIRs and a gene with four exons that encodes a 751 residues transposase. It also contains 11-bp sub-TIRs that act as transpositional enhancers and generates 8-bp TSD upon insertion (Rio 2002).
P-like elements are known to exist in a broad range of taxa, including protozoans such as Trichomona vaginalis (Kapitonov and Jurka 2009), several Dipterans (Perkins and Howells 1992; Lee et al. 1999; Sarkar et al. 2003), urochordata such as Ciona intestinalis (Kimbacher et al. 2009), and vertebrates (Hammer et al. 2005). In addition, P element has been repeatedly domesticated to generate cellular genes (Quesneville et al. 2005). For instance, the human genome contains 12 THAP-domain containing genes, and one of them (THAP9) has been recently shown to encode an active P-element transposase (Majumdar et al. 2013). In Drosophila, P element is widespread within the Sophophora subgenus (Daniels et al. 1990; Hagemann et al. 1992, 1994, 1996a, 1996b; Clark and Kidwell 1997) but seems much more scarce in the Drosophila subgenus (Loreto et al. 2001, 2012). An almost complete copy was isolated from D. mediopunctata in the tripunctata species group (Loreto et al. 2001), whereas relatively short fragments have been amplified by polymerase chain reaction (PCR) in other species of the tripunctata and cardini species groups (Loreto et al. 2012). The D. mediopunctata P element is 96.5% identical to that of D. melanogaster, and it has been suggested that it is the result of a horizontal transfer event (Loreto et al. 2001).
Miniature inverted-repeat TEs (MITEs) are small nonautonomous class II elements of a few dozen to a few hundred base pairs and flanked by TIRs. Their high copy numbers, homogeneous size, and high similarity within MITE families distinguish them from the typical defective nonautonomous transposons, which are usually unique copies (Feschotte et al. 2002; Guermonprez et al. 2008). Although MITEs were discovered in plants (Bureau and Wessler 1992, 1994), they have been found in a variety of organisms, including Drosophila (Smit and Riggs 1996; Tu 2000; Holyoake and Kidwell 2003; de Freitas Ortiz et al. 2010). Some MITEs have been found to be internal deletion derivatives of its autonomous partners and are likely to be mobilized by them (Feschotte and Mouchès 2000; Zhang et al. 2001). In other cases, however, MITEs share their terminal sequences with canonical elements, but their internal sequence does not have similarity to the master copy. The origin of these MITEs is obscure; they are the result of either profound changes in the original transposon sequence or the recruitment of unrelated sequences. As nonautonomous elements, MITEs depend on transposases encoded by canonical elements, but surprisingly MITEs can achieve higher copy numbers than their master transposons. The amplification success of MITEs has been attributed to different causes such as their promiscuity binding a range of related transposases, the gain of transposition enhancers, and loss of repressors when compared with autonomous TEs (Yang et al. 2009).
BuT5 was first described in the proximal breakpoint of a naturally segregating D. buzzatii inversion and tentatively classified as a class II TE (Cáceres et al. 2001). The reported copy was 1,039-bp long with 3-bp TIRs and imperfect 17-bp sub-TIRs and no coding capacity. Subsequently, similar BuT5 copies were observed at the breakpoints of two other polymorphic D. buzzatii inversions (Casals et al. 2003; Delprat et al. 2009). The three inversions were caused by ectopic recombination between copies of Galileo, a P-superfamily transposon (Marzo et al. 2008), and BuT5 was a secondary colonizer of the inversion breakpoints. The secondary colonization of breakpoints in recent polymorphic inversions (Casals et al. 2003; Delprat et al. 2009) and the relatively high abundance of BuT5 in different D. buzzatii strains (Casals et al. 2006) indicate current or recent transpositional activity of BuT5 in D. buzzatii. Furthermore, recent works in our group have revealed that BuT5 generated two recently fixed inversions in two repleta group species, 2s in D. mojavensis (Guillén and Ruiz 2012), and 2x3 in D. uniseta (Prada 2010). In both cases, each breakpoint harbors a copy of BuT5 and the exchanged TSDs between copies of the two breakpoints denote ectopic recombination as the generation mechanism (fig. 1). Therefore, BuT5 has had a significant role in the chromosomal evolution of the repleta group.
Fig. 1.—
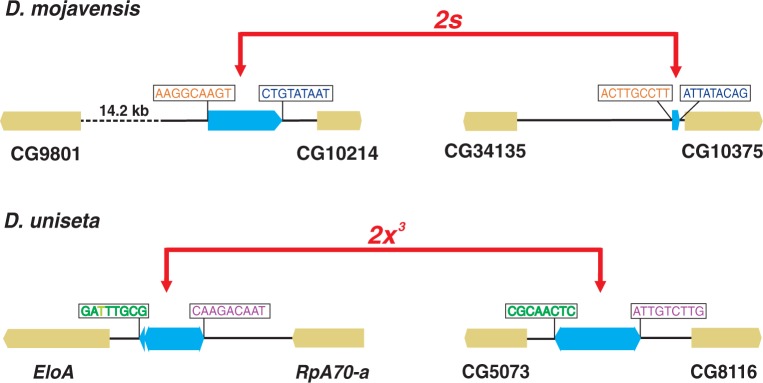
Molecular structure of breakpoint regions in two inversions generated by the transposon BuT5, 2s in Drosophila mojavensis (Guillén and Ruiz 2012), and 2x3 in D. uniseta (Prada 2010). BuT5 copies (blue rectangles) bounded by exchanged 8-bp or 9-bp TSD are found at the two breakpoints of each inversion, indicating ectopic recombination as the generating mechanism.
Despite BuT5 significance as a genome reshaping force and its recent transpositional activity, this element has not been classified to date. Consequently, its phylogenetic distribution, how it mobilizes or whether it is a MITE, a deletion derivative of a known DNA transposon, or a new type of TE is still unknown. To fill this gap, our objectives were to 1) study the interspecific distribution of BuT5 using both bioinformatic and experimental methods; 2) isolate a copy of the autonomous element that mobilizes BuT5; and 3) classify BuT5 and its master TE.
Materials and Methods
Bioinformatic Searches
BuT5 bioinformatic searches were carried out using BlastN (Altschul et al. 1997) against all National Center for Biotechnology Information (NCBI) available databases (December 2012). Searches were also made using CENSOR tool (Jurka et al. 1996) and TEs deposited in Repbase Update (Jurka et al. 2005). Default parameters were used in these searches. The first copy described BuT5_1 (Cáceres et al. 2001), from D. buzzatii, was used as a query. Basic Local Alignment Search Tool (Blast) results in other species were also used as queries in their genomes, performing species-specific searches. Significance thresholds used to retrieve sequences for analysis were an E value ≤ 10−10 for results from species different of the query and ≤10−25 for results from the same species of the query.
Experimental Searches
Primers
BuT5 and P-element primers BL, BR, P3, and P13 were designed based on sequences obtained by bioinformatic searches (from D. buzzatii and D. mojavensis). PriFi (Fredslund et al. 2005) was used to find the best regions to place the primers on multiple alignments. The rest of the primers were designed based on D. buzzatii P element (BU-73 strain). All primers were designed with Primer Designer v.1.01 (Scientific and Educational Software) and produced by Sigma-Aldrich, Inc. Sequences of primers used are provided in supplementary table S1, Supplementary Material online.
BuT5 Analysis in the Repleta Group
To detect BuT5, 85 Drosophila DNA samples of 41 species (supplementary table S2, Supplementary Material online) were screened by PCR with primers BL and BR (supplementary table S1, Supplementary Material online). PCRs were carried out in a volume of 26.5 µl including 50–100 ng of DNA, 0.83 U of DNA Taq polymerase (Roche), 0.04 mM of each dNTP, and 0.33 µM of both primers. Amplification conditions were 94 °C for 4 min, 30 cycles at 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min followed by a final extension step at 72 °C for 7 min. These PCR products were purified with the NucleoSpin Extract II (Macherey-Nagel) and were cloned using either the pGEM-T Easy Vector Kit (Promega) or the StrataClone Kit (Agilent Technologies). Approximately four clones per sample were selected and PCR amplified with primers SP6 and T7 (pGEM-T) or T3 and T7 (StrataClone). The PCR products presenting different electrophoresis mobility were purified with the Nucleospin Extract II Kit and sequenced by Macrogen Inc (Seoul, Korea) using universal primers. We also analyzed by dot blot 35 DNA samples from 29 species, specified in supplementary table S2, Supplementary Material online. Denatured DNA (200 ng) was transferred onto a nylon membrane (Roche) by using a Bio Dot apparatus (Bio-Rad) according to manufacturer’s specifications. The DNA was cross-linked by exposure to short-wavelength ultraviolet light. A D. mojavensis BuT5 clone (G035_2) was used as probe. It was labeled by PCR with digoxigenin-11-dUTP (PCR DIG Labeling Mix, Roche). Final reaction volume was 50 µl, including 2.5 U of Taq DNA polymerase (Roche) and its buffer, 0.2 mM of dNTP labeling mixture, 0.5 µM of primers BL and BR, and 50–100 ng of linearized DNA. Membrane prehybridization was done in DIG Easy Hyb (Roche) and 50 ng/ml of denatured DNA, MB grade from fish sperm (Roche) at 37 °C during 1 h. Denatured probe (10 µl) was added into 3.5 ml of fresh DIG Easy Hyb, and the hybridization was performed at 37 °C for 16 h. Then two washes were done with 2× SSC and 0.1% sodium dodecyl sulphate (SDS) at room temperature and two with 0.5× SSC and 0.1% SDS at 45 °C. DIG Wash and Block Buffer Set (Roche) was used for washing and blocking incubations according to manufacturer’s instructions, and detection was made with CDP-Star (Roche) also following the instructions. Membrane signals were quantified by Laboratori d'Anàlisi i Fotodocumentació, d'Electroforesis, Autoradiografies i Luminiscència of the Universitat Autònoma de Barcelona with ChemiDoc XRS (BioRad) and Quantity ONE 4.7 software (BioRad).
P-Element Sequence in D. buzzatii
P-element amplifications, with primers BL + P13, P3 + BR, and P1 + P15 were performed with Expand Long Template in 50 µl including 50–100 ng of DNA, 1 U of Enzyme mix, 0.02 mM of each dNTP, and 0.20 µM of both primers. Amplification conditions were established following manufacturer’s instructions. The BL + P13 2.8-kb band was identified and excised from a 1% agarose gel and cleaned up with the NucleoSpin Extract II Kit. The products of P3 + BR and P1 + P15 amplifications were directly cleaned up with the NucleoSpin Extract II Kit. PCR products were cloned with the StrataClone Kit (Agilent Technologies) following the manufacturer’s instructions. DNA of three clones per cloning reaction was retrieved using the GeneJET Plasmid Miniprep Kit (Thermo Scientific) and finally was sequenced by Macrogen Inc (Seoul, Korea) using primers T3 and T7. The 5′- and 3′-ends of the P element were isolated by inverse PCR (iPCR) from D. buzzatii strain BU-73. Digestion (HindIII) and ligation were performed following Berkeley Drosophila Genome Project iPCR protocol (available from http://www.fruitfly.org/about/methods, last accessed June 2, 2013). PCR was carried out with primers InvL and InvR (supplementary table S1, Supplementary Material online) under conditions similar to those described earlier for BL + P13, P3 + BR, and P1 + P15 amplifications. A single band was identified by electrophoresis in a 1% agarose gel. The DNA was cleaned up with the NucleoSpin Extract II Kit, and cloned with the StrataClone Kit (Agilent Technologies). Minipreps of 22 clones were performed with GeneJET Plasmid Miniprep Kit (Thermo Scientific). The plasmids were used as template for PCRs with primers BL and InvR, and the clones that yielded PCR products of different length were sequenced using primers T3 and T7 by Macrogen Inc (Seoul, Korea).
Transposase Gene Exon–Intron Boundaries
Total RNA was extracted from D. buzzatii adult females of strain BU-73 (Berna, Argentina). Forty-five female heads and 90 ovaries were extracted in physiological solution, and RNA was obtained for each part with the High Pure RNA tissue kit (Roche) according to the manufacturer’s instructions. Reverse transcriptase (RT)-PCRs were performed with the Transcriptor First-strand cDNA synthesis Kit (Roche) following the manufacturer’s instructions. To favor amplification of P element over other transcripts, P-element-specific primers, P15 or P4 (supplementary table S3, Supplementary Material online), were used in two separate retrotranscription reactions. After obtaining the cDNA, five experiments, each one with two nested PCR reactions, were done to increase the amount of specific product. The combination of primers used for these PCR reactions is detailed in supplementary table S3, Supplementary Material online. These amplifications were performed in a volume of 100 µl and using 10 µl of a 1:10 dilution of the previous reaction as template, 2.5 U of DNA Taq polymerase (Roche), 0.02 mM of each dNTP, and 0.20 µM of both primers. Products of the second PCRs were cloned with the StrataClone Kit (Agilent Technologies). Screening analyses were done with Miniprep or PCR (using primers T3 and T7) on 3–47 clones per cloning reaction. Plasmids containing fragments with different electrophoretic mobility were recovered using the GeneJET Plasmid Miniprep Kit (Thermo Scientific) and sequenced by Macrogen Inc (Seoul, Korea) with T3 and T7 primers.
Sequence Analysis
Sequence analysis was performed with Geneious v5.1.3 (Biomatters Ltd.), and alignments were done with MUSCLE (Edgar 2004) through Geneious. Search for open reading frames (ORFs) with a minimum size of 100 bp was made with Geneious software. Predicted ORFs were subsequently used in BlastX searches against NCBI nonredundant protein sequences database. Gblocks (Castresana 2000) was used to select the conserved bocks of the alignment of BuT5 sequences over 800 bp, keeping 87% of the original alignment length. To use less stringent condition, parameters were set as follows: minimum number of sequence for a flank position: 44, maximum number of contiguous nonconserved positions: 8, minimum length of a block: 5, and allowed gap position: “with half.” MEGA 5 software (Tamura et al. 2011) was used to reconstruct BuT5 phylogeny using maximum likelihood method and the best fit model according to jModelTest (Posada 2008), general time reversible model with a discrete gamma distribution (four discrete categories). Bootstrap test was performed with 1,000 replicates. The phylogeny of the P superfamily transposases was based on 31 putatively complete protein sequences from insects and the human THAP9 (NM_024672) and aligned with MUSCLE. P-like, Galileo, and 1360 sequences were taken from Repbase (Jurka et al. 2005) and Marzo et al. (2008). The alignment, with 1,192 positions, was used to conduct phylogenetic analyses with neighbor joining and maximum likelihood methods on MEGA 5. Bootstrap test was performed with 1,000 replicates.
P-element transposase gene introns were manually predicted using BlastX and NCBI Conserved Domains search. BlastX alignment of D. buzzatii P-element complete copy with D. bifasciata O-type P-element transposase (AAB31526, E value = 8e-91) revealed discontinuities coincident with stop codons and frameshift mutations. NCBI Conserved Domains search tool (Marchler-Bauer et al. 2011) provided information regarding which virtually translated residues were part of transposase domains. BlastN searches were also used to refine the first predictions by comparing the transposase generated with other P-element transposases.
Results
BuT5 Bioinformatic Searches
We carried out BlastN (Altschul et al. 1997) searches using as query BuT5-1 (Cáceres et al. 2001) against all NCBI nucleotide databases (including 2,428 bacterial, 122 archaeal, and 426 eukaryotic genomes). We retrieved 36 previously published D. buzzatii BuT5 sequences (supplementary table S4, Supplementary Material online) plus 48 new BuT5 sequences from the genome of D. mojavensis (supplementary table S5, Supplementary Material online), a relative of D. buzzatii that belongs to the repleta group (Drosophila subgenus). No hits were significant in any of the other Drosophila genomes or the other genomes searched. In addition, no results were recovered from searches in Repbase Update (Jurka et al. 2005).
Only two other sequences from D. buzzatii had a size similar to that of BuT5-1 (1,039 bp), the rest being fragments less than 800-bp long likely resulting from deletions. The three longest copies have 3-bp TIRs, imperfect (two mismatches) 17-bp sub-TIRs (fig. 2), and TSD 8-bp or 9-bp long (Cáceres et al. 2001; Casals et al. 2003; Delprat et al. 2009). Twenty-two out of the 49 BuT5 copies retrieved from the D. mojavensis genome were over 800-bp long (mean ± standard deviation [SD] = 1,017.4 ± 23.2) and had a pairwise identity of 93.2%. Fifteen of them had both 3-bp TIRs, and 14 had 16-bp imperfect (two mismatches) sub-TIRs (fig. 2). Seventeen D. mojavensis BuT5 copies were flanked by TSDs: 4 8-bp long and 13 9-bp long (one has two mismatches). The BuT5 consensus sequence of D. mojavensis, built with the 22 longer copies, has 67.3% pairwise identity to the BuT5 consensus sequence of D. buzzatii, built with the three longer copies previously isolated. However, the identity between BuT5 consensuses of both species is higher at the terminal regions (fig. 2), where the first 65 bp shows 90.8% identity and the last 32 bp, 90.6%. This suggests that the size and the terminal features of BuT5 are particularly conserved between D. mojavensis and D. buzzatii copies.
Fig. 2.—
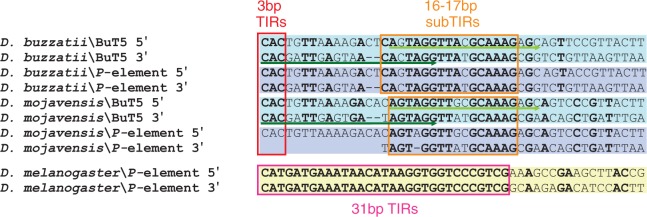
Alignment of the 5′- and 3′-terminal regions of BuT5 and P element from Drosophila buzzatti and D. mojavensis. For comparison, the D. melanogaster P-element terminal sequences are included but not aligned. The red box indicates BuT5 and P-element TIRs, the orange box BuT5 and P-element sub-TIRs, and the pink box the D. melanogaster P-element TIRs. Green arrows indicate the primers BL (dark green) and BR (light green).
BuT5 Experimental Searches
A pair of degenerated primers, BR and BL, was designed to match BuT5 ends, which are conserved between D. buzzatii and D. mojavensis, to increase the chances of successful interspecific amplification. PCR screening was done with 85 DNA samples of 41 species from the Drosophila repleta species group (supplementary table S2, Supplementary Material online). PCR products were cloned and sequenced and 86 clones from 26 species were confirmed as BuT5 copies. However, as the primers were inside the element, some features such as TIRs or TSDs could not be retrieved from these copies. Sequences over 800 bp (61 from 19 species) had a mean size (±SD) of 959.2 bp (±46.8) that amounts to 1,014.2 bp if the unsequenced element ends are taken into account. To complement the PCR search, a dot blot analysis was carried out with 20 PCR-negative repleta group samples (15 species) plus samples from D. nannoptera and D. wassermani, two species in the cactophilic nannoptera species group (Pitnick and Heed 1994), and samples from D. buzzatii and 12 species with available genome sequences as controls (supplementary table S2, Supplementary Material online). Dot blot confirmed as negative three species of the repleta group (D. hydei, D. nigrospiracula, and D. pegasa) but yielded positive for the other 12 PCR-negative species. Results were also negative for the two species of the nannoptera group and for all species with sequenced genome except D. mojavensis.
In summary, BuT5 was detected, either by PCR or dot blot, in 38 of the initial 41 species of repleta group, belonging to four of the six described subgroups (samples were not available for subgroups fasciola and inca) (fig. 3). BuT5 is present in most lineages, including the most basal branch of the repleta group (D. eremophila and D. mettleri), estimated to have shared their last common ancestor 16 Ma (Oliveira et al. 2012).
Fig. 3.—
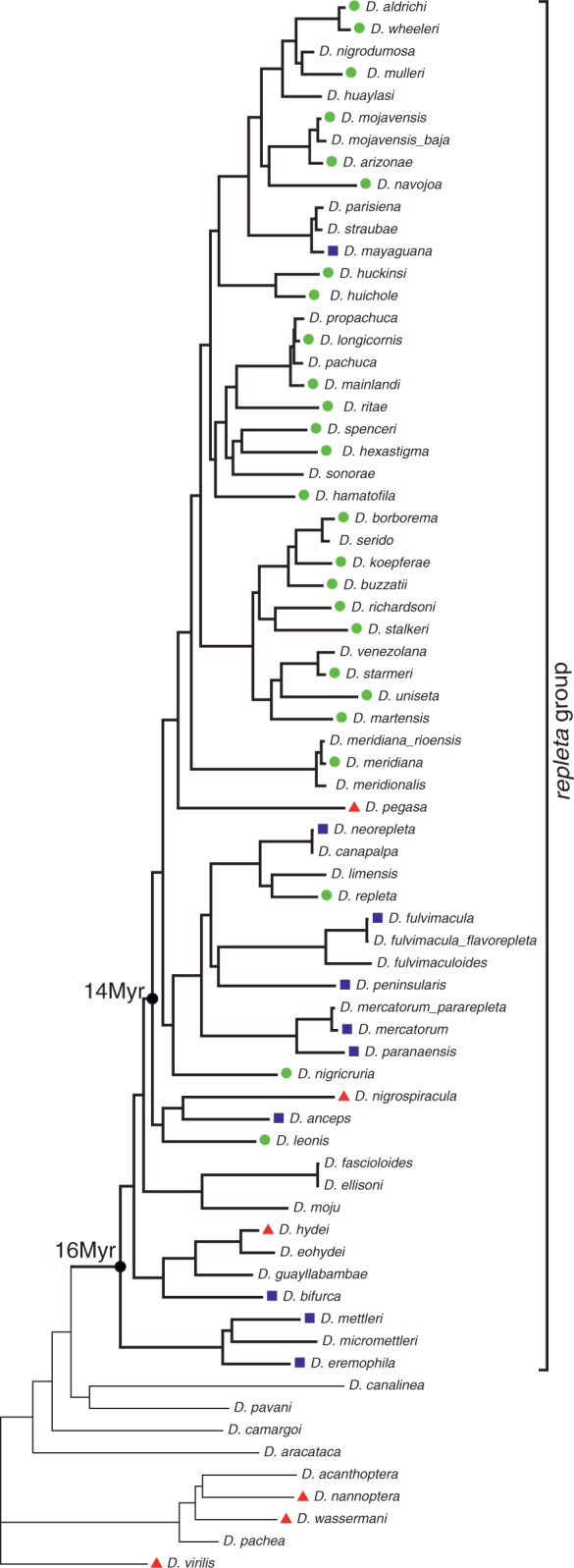
Distribution of the transposon BuT5 plotted onto the repleta group phylogeny (taken from Oliveira et al. 2012). Green dots denote species with BuT5 sequences recovered by PCR; blue squares and red triangles indicate positive and negative results for dot blot, respectively.
Analysis of BuT5 Sequences
As a result of the bioinformatic and experimental searches, we retrieved 86 BuT5 sequences over 800-bp long from 19 species. These were used to build a phylogenetic tree using maximum likelihood methods (fig. 4). The BuT5 sequence recovered from D. nigricruria was used to root the tree because this species is the most distant one and does not belong to the mulleri, longicornis, or buzzatii complexes. The BuT5 phylogenetic tree (fig. 4) is broadly concordant with that of the host species (fig. 3) and mirrors the relationship between the mulleri, longicornis, and buzzatii complexes (Wasserman 1992; Ruiz and Wasserman 1993; Oliveira et al. 2005), yet sequences of the longicornis complex do not form a monophyletic cluster. Consequently, BuT5 has been vertically transmitted, and no clear-cut evidence for horizontal transfer was found.
Fig. 4.—
Maximum likelihood phylogenetic tree built with 86 BuT5 sequences longer than 800 bp, recovered by PCR (sample code, clone number, and species name) or bioinformatic searches (boldface; copy number, species name and TSD length) from 19 species of the repleta group. Bootstrap values over 80 are shown at nodes.
We estimated the age of the BuT5 copies in D. mojavensis with the formula t = K/r (Kapitonov and Jurka 1996), where K is the average divergence of the copies from their consensus sequence and r the neutral substitution rate (0.0111 substitutions per bp per Myr; Tamura 2004). For the 22 most complete BuT5 copies isolated from the D. mojavensis genome, K = 0.0267 and t = 2.4 Myr. However, there is evidence for more recent transposition events. For a subset of five closely related copies, K = 0.004 and t = 0.36 Myr.
We searched putative ORFs in all BuT5 copies by several methods. ORF longer than 100 bp showed no similarity to previously described proteins, corroborating that BuT5 has no coding capacity (Cáceres et al. 2001). Furthermore, most BuT5 copies had a size similar to that of the original BuT5-1 copy (∼1 kb) or were smaller (partial copies). The TIR, lack of coding capacity, abundance, and homogeneous size of BuT5 allow us to consider it tentatively as a MITE.
On the other hand, similarity searches with BuT5 in the D. mojavensis genome revealed two nearby significant hits in scaffold_6541 spaced by approximately 3 kb. When the intervening sequence was explored using BlastX against protein databases, we found a significant similarity to the transposase of P element (O-type) from D. bifasciata (AAB31526.1, amino acid identity 47%, E value: 3e-91). The total sequence, including the terminal segments similar to BuT5, was 3,221-bp long. This sequence was used to search against the D. mojavensis genome with BlastN, finding two other sequences with similarity to the P element (supplementary table S6, Supplementary Material online). The consensus of the three copies has a size of 3,254 bp and shows similarity to the BuT5 ends only (fig. 5). We hypothesized that this P element could represent the autonomous transposon family mobilizing BuT5.
Fig. 5.—
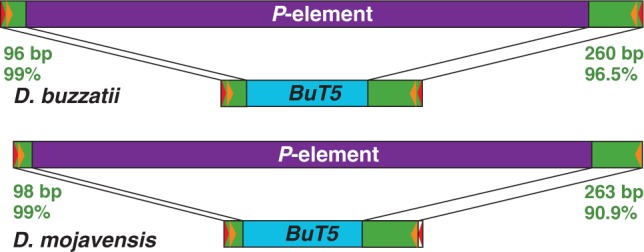
A comparison of P element and BuT5 structures in Drosophila buzzatii and D. mojavensis. In both cases, the two elements show high similarity at both ends (green) but no similarity in the interior segment (purple or blue). Red and orange triangles denote the TIRs and sub-TIRs, respectively.
Isolation and Characterization of P Element in D. buzzatii
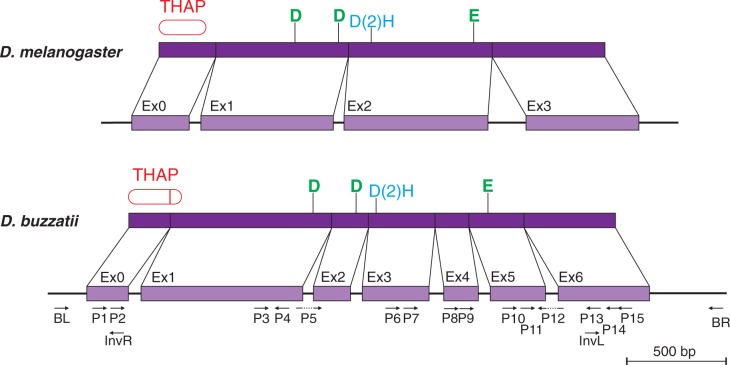
To test the hypothesis that the P element detected in D. mojavensis represents the master copy mobilizing BuT5, we searched for similar elements in 42 strains of D. buzzatii from different geographical origins, 7 strains of D. mojavensis, and 1 strain of D. uniseta. Two overlapping PCR reactions were carried out with primers BL + P13 and P3 + BR (fig. 6), because primers BL + BR placed at the ends of BuT5 retrieve only copies of this MITE (see earlier). Only one D. buzzatii strain (BU-73 from Berna, Argentina) produced positive results for both reactions. The two sequences were 2,694-bp and 2,389-bp long and overlapped 1,755 bp. An additional PCR with primers P1 + P15 (fig. 6) generated a 2,583-bp product that covers the central part of the element. Finally, both ends of the P element were isolated by iPCR with primers InvL + InvR (fig. 6). Only one band, approximately 6.3-kb long, was observed. The retrieved sequences overlap 362 bp at the 5′-end and 637 bp at the 3′-end with the other P-element sequences and are 100% identical to them in the overlapping segments. The complete P element in D. buzzatii is 3,386-bp long and is flanked by 9-bp TSDs (CTAGTAGGT). This P element and D. buzzatii BuT5 consensus sequence show 99% identity over the first 96 bp at the 5′-end and 96.5% over the last 260 bp at the 3′-end (fig. 5).
Fig. 6.—
Comparison of the transposase gene in Drosophila buzzatii P element (bottom) with that of the D. melanogaster canonical P element (top). The exon–intron structure of the D. buzzatii gene was corroborated by RT-PCR experiments using the primers shown below the gene. The structure of the protein is also shown with the DNA-binding THAP domain (Roussigne et al. 2003; Clouaire et al. 2005) and the catalytic motives DDE and D(2)H (Yuan and Wessler 2011) highlighted in each case.
On the other hand, only one strain of D. mojavensis (G091) was positive for one of the reactions (BL + P13). We built a consensus sequence of 3,260 bp for D. mojavensis P element using the three copies from the sequenced genome (supplementary table S6, Supplementary Material online) and the 2,686-bp sequence recovered by PCR. This consensus sequence has 99% identity to BuT5 D. mojavensis consensus over 98 bp at the 5′-end and 90.9% over 263 bp at the 3′-end (fig. 5). The highly congruent observations in D. buzzatii and D. mojavensis support our hypothesis.
A BlastX search with the D. buzzatii P element against the NCBI protein database corroborated the similarity with D. bifasciata O-type P-element transposase (AAB31526, amino acid identity 34%, E value = 8e-91), but the BlastX alignment showed clear-cut discontinuities. A tentative manual annotation using BlastX hits and the results of a search against NCBI Conserved Domain Database predicted a transposase-coding gene comprising eight exons and seven introns that encoded a protein of 761 residues. The subsequent expression experiments (see later) corroborated six of the seven introns (but with modifications) and rejected one of them (that keeps the reading frame and does not contain STOP codons). Therefore, the transposase gene of the D. buzzatii P element comprises seven exons and encodes an 822-aa protein.
Expression Analysis
RT-PCR experiments using diverse primers (fig. 6) were carried out to assess the expression of the P element and test the manual annotation of the transposase gene. Ovaries and female heads were used to extract total RNA. Then single-stranded cDNA was generated from the two RNA samples using one of two P-element-specific primers (P4 or P15). Finally, five amplification reactions were performed to amplify several TE fragments enclosing the predicted exon–intron junctions (supplementary table S3, Supplementary Material online). These PCRs were carried out in two consecutive rounds with the primers of the 2nd round designed within the product of the 1st round. PCR products were cloned, and a screening of the clones was performed through PCR or Miniprep. Those clones with a different electrophoretic mobility were sequenced (supplementary table S3, Supplementary Material online).
Positive expression results were produced in heads in four of the five amplification reactions. Two different products, with and without intron 1, were recovered from the first PCR with primers P2 + P4 (fig. 6). In the second PCR with primers P5 + P12, both located in exon–exon junctions, a product with introns 2, 3, 4, 5, and 6 spliced was produced (fig. 6). The third PCR with primers P7 + P12 gave rise to three differently spliced products, one keeping introns 4 and 5, one without introns 4 and 5, and one without intron 4 and a 3′ alternatively spliced intron 5. The fourth PCR with primers P9 + P12 was negative in heads (but positive in ovaries, see later). Finally, the fifth PCR with primers P11 + P14 gave a single product retaining intron 6.
In ovaries, positive expression results were produced in three of the five amplification reactions. A single product was obtained in the first PCR (P2 + P4) that kept intron 1. For the second PCR (P5 + P12), no products were obtained from ovarian samples. The third PCR (P7 + P12) did not produce any products. In the fourth PCR (P9 + P12), only one product with intron 5 present and intron 6 spliced was obtained. The fourth reaction (P11 + P14), in ovaries, gave rise to two different forms, one retaining exon 6 and the other without this last intron.
In other words, we have detected splicing of all six introns in heads (somatic tissue) but not in ovaries (somatic and germline cells), where only the splicing of the last intron was detected.
Phylogenetic Analysis
To place our D. buzzatii P element in the context of other P elements from Drosophila and other Dipterans, an alignment was made with the nucleotide sequences of the P-element fragments generated by Clark and Kidwell (1997) and Loreto et al. (2012). The nucleotide identity between our P element and those previously isolated from Drosophila, Scaptomyza, Lordiphosa, and Lucilia genera is very low, and the alignment was not very reliable. When a phylogenetic tree (not shown) was generated with these sequences, the D. buzzatii P-element branch diverged early and was well separated from all other P-element sequences. Given the high nucleotide divergence observed, we turned to the protein phylogeny to compare our P element with all members of the P superfamily.
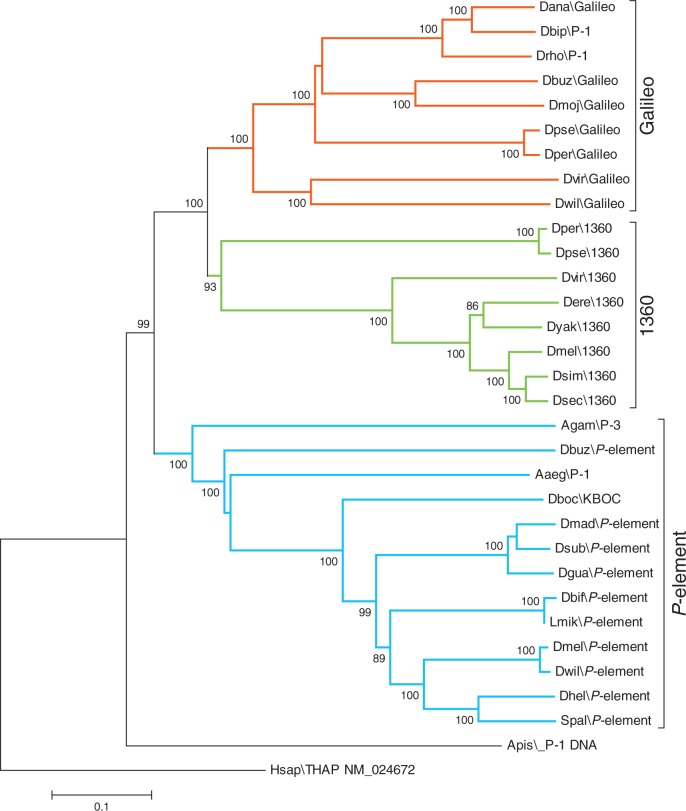
The complete 822-aa predicted transposase from the P element of D. buzzatii was used in a phylogenetic analysis that included 30 other P-like transposase sequences from insects (Drosophila, Aedes, Anopheles, Acyrthosiphon, and Scaptomyza genera), and the human THAP9 protein (Majumdar et al. 2013) was used as outgroup. The alignment of the 32 transposases is provided in supplementary figure S1, Supplementary Material online. Phylogenetic trees with the same topology were generated both with the neighbor joining method (fig. 7) and the maximum likelihood method (not shown). The tree presents three major monophyletic clades, grouping each of the families described in the P superfamily, namely Galileo, 1360, and P element. The predicted transposase for D. buzzatii falls into the well-supported P family clade, unambiguously placing our TE within this family.
Fig. 7.—
Neighbor joining phylogeny built with an alignment of 31 P-element transposases from insects and the human THAP9 protein as outgroup. Galileo family is highlighted in red, 1360 in green, and P element in blue. Bootstrap values over 80 are shown.
Discussion
A Divergent P Element in the D. repleta Species Group
We have identified and fully characterized a P element in D. buzzatii, a member of the repleta species group of the subgenus Drosophila. This element has also been found (but only partially characterized) in D. mojavensis, another member of the same species group. These two species diverged approximately 12 Ma (Oliveira et al. 2012) suggesting that this transposon is widespread within the repleta group (see later). The P element that we have characterized is very divergent from all the previously detected P elements in Drosophila, but phylogenetic analyses (fig. 7) indicate that it should be placed in this family rather than in the other two closely related families (1360 and Galileo) of the P superfamily. This finding suggests that P elements may be more widespread within the Drosophila genus than previously thought and have gone undetected in previous bioinformatic or experimental searches due to their divergence from canonical D. melanogaster P element (Loreto et al. 2012).
The D. buzzatii P element is divergent from other Drosophila P elements in three regards: 1) nucleotide and protein sequence; 2) transposase-encoding gene structure; and 3) expression pattern.
The nucleotide sequence of D. buzzatii P element is quite dissimilar from that of D. melanogaster P element and all the previously described P elements in Drosophila. In DNA transposons, terminal regions bear important features for their mobilization and are accordingly evolutionarily conserved. Drosophila melanogaster P element has TIRs of 31-bp and sub-TIRs of 11-bp (that overlap the THAP-domain-binding sites). The TIRs of P elements in D. bifasciata (M and O types) and Scaptomyza pallida are 31- or 32-bp long, respectively (Hagemann et al. 1994). Similarly, the TIRs of Anopheles gambiae autonomous P element range from 27 to 31 bp (Quesneville et al. 2006). In contrast, the D. buzzatii and D. mojavensis P elements have 3-bp TIRs and 16–17 bp sub-TIRs (fig. 2). The distance between the beginning of the TIR and the end of the sub-TIR in repleta group P elements is 32 bp at the 5′-end and 30 bp at the 3′-end. Therefore, it seems that this segment is equivalent to the 31 bp of D. melanogaster P-element TIRs. We did not find an equivalent for the D. melanogaster P-element 11-bp sub-TIRs in the repleta group P element.
The D. buzzatii P element encodes a transposase with 822 residues. This transposase shows a 34.8% identity with that of the D. melanogaster P element and comparable identity values with other Drosophila P-element transposases. The highest identity seems to be that with the Aedes aegypti P-1 element transposase (36.9%). These data emphasize the high divergence between the repleta P element and all other described P elements. However, the repleta P-element transposase contains the typical protein domains of P-transposases, N-terminal DNA-binding domain, and C-terminal catalytic domain (fig. 6). The D. melanogaster P-element transposase contains a zinc-dependent DNA-binding domain evolutionarily conserved in an array of different cellular proteins and named THAP domain (Roussigne et al. 2003; Clouaire et al. 2005). It includes a metal-coordinating C2CH signature plus four other residues (P, W, F, and P) that are very conserved as well as eight residues that make contact with the double-helix major groove (M, Y, L, H, N, and Q) and the minor groove (R and R) (Sabogal et al. 2010). We searched in the D. buzzatii P-element transposase for domains using conserved domains search (Marchler-Bauer et al. 2011) and found a significant match (3.4E-21) with the THAP domain (PF05485) in the N-terminus (positions 1–91). The D. buzzatii P-element THAP domain contains the eight conserved residues in positions C3, C8, P23, W32, C51, H54, F55, and P82. However, only three of the eight residues that make contact with the double-helix seem to be present (M1, N47, and R72). This is not unexpected because there is variability in the residue composition of the THAP domain (Sabogal et al. 2010). The D. buzzatii P-element transposase contains also a putative catalytic domain in the C-terminus with the DDE triad and D(2)H motif (Yuan and Wessler 2011) conserved in positions D313/D386/E610 and D419(2)H422, respectively (fig. 6).
The THAP domain of D. melanogaster P-element transposase can bind to four experimentally verified binding sites within the P-element sequence, two in the 5′-end, and two in the 3′-end (table 1). Seemingly, the transposase THAP domain is able to recognize one binding site on each side, whereas the repressor THAP domain can recognize the four THAP-domain-binding sites (Kaufman et al. 1989; Lee et al. 1996). Because the D. buzzatii transposon belongs to the P-element family, and its transposase encodes a THAP domain, we searched for THAP-domain-binding sites in the P-element terminal regions similar to the consensus for THAP-domain-binding sites (Sabogal et al. 2010). We found in the D. buzzatii P element two sequences (one on each end) identical to two of the D. melanogaster P-element-binding sites, although they are placed in reverse orientation (table 1). We did not find any other putative THAP-domain-binding sites, and therefore, if there are other binding sites in the D. buzzatii P element, they must be divergent from the D. melanogaster consensus.
Table 1.
Transposase-Binding Sites in P Element
| Start | THAP Domain Binding Site | End | TE | Region |
|---|---|---|---|---|
| D. melanogaster | ||||
| 61 | TAAGTGTA | 54 | P element | 5′-end |
| 2,859 | TAAGTGGA | 2,866 | P element | 3′-end |
| 136 | TAAGGGTT | 129 | P element | 5′-end |
| 2,763 | TAAGGGTT | 2,777 | P element | 3′-end |
| D. buzzatii | ||||
| 90 | TAAGTGTA | 97 | P element | 5′-end |
| 3,185 | TAAGGGTT | 3,178 | P element | 3′-end |
| 90 | TAAGTGTg | 97 | BuT5 | 5′-end |
| 841 | TAAGGGTT | 834 | BuT5 | 3′-end |
Note.—Four experimentally verified naturally occurring binding sites for the P-element transposase in Drosophila melanogaster (Kaufman et al. 1989; Lee et al. 1996; Sabogal et al. 2010). Putative binding sites observed in D. buzzatii P element and BuT5. Each of the D. buzzatii sequences is identical to one of the D. melanogaster-binding sites except for a mismatch in the last nucleotide of the BuT5 5′-end binding site (lowercase). Coordinates are given in 5′→3′ orientation for D. melanogaster P element (O'Hare and Rubin 1983) and for D. buzzatii P element (this work) and BuT5 (Cáceres et al. 2001).
The structure of the transposase encoding gene in the repleta P element is quite unusual, with seven exons (0–6) and six introns (1–6). The transposase-encoding gene in D. melanogaster P element has four exons (0–3) and three introns (IVS1-3). This structure seems conserved in other Drosophila P elements (Haring et al. 1998). Anopheles gambiae P elements seem to have three or four exons different from those in D. melanogaster (Quesneville et al. 2006). The locations of the repleta P-element introns along the transposase sequence are different from those of the D. melanogaster P-element introns (i.e., none of them coincide; see fig. 6) and also different from those of A. gambiae. Thus, they seem evolutionarily independent. Because Galileo and 1360, the other two members of the P superfamily (fig. 7), do not contain introns (Marzo et al. 2008), it seems reasonable to assume that the ancestor of the P superfamily transposons was an intronless element that has acquired different introns along the several branches of the P-element phylogeny (fig. 7). Introns can be acquired via several different mechanisms although “intron duplication” has long been favored as the most likely source of new spliceosomal intron positions (Rodríguez-Trelles et al. 2006).
P-element expression has been extensively studied, and several regulatory mechanisms have been described (for reviews see Rio 1991; Castro and Carareto 2004). In D. melanogaster, transposase is only produced in the germline. In the somatic tissue, the P-element transcripts are not completely spliced, retaining the third intron (IVS3), which possess a termination codon in the first 9 bp. The resultant truncated protein of 66 kDa acts as a repressor of the transposase excision activity. This form was also found in other species (Haring et al. 1998) and proposed to occur irrespective of the P-element type and host species. However, this regulation mechanism, or a similar one based on the retention of specific introns, is not applicable to D. buzzatii P element. Drosophila buzzatii P elements not only have a different exonic structure but we have also detected splicing of all introns in the somatic tissue (head). Thus, seven-exon P element shows differences to the other Drosophila P elements also regarding regulation.
BuT5 Is a MITE Associated with the P Element
MITEs are short nonautonomous elements with TIRs, no coding capacity, and capable of reaching high copy numbers in plant genomes (Feschotte et al. 2002). Similar to nonautonomous elements, MITEs transpose using transposases encoded by autonomous elements. A distinctive feature of MITEs is their homogeneity in size and sequence, which differentiates them from typical nonautonomous elements, which are usually unique defective copies (Guermonprez et al. 2008). Moreover, some MITEs are not just deletion derivatives of complete transposons and unlike other nonautonomous TEs have internal sequences unrelated to their master copies (Feschotte et al. 2003). BuT5 is a relatively short repetitive sequence without coding capacity; it has very short TIRs and is flanked by TSDs. Given this and the remarkably high identity between BuT5 and the repleta P element at the 5′- and 3′-ends and the lack of similarity of the internal sequences (fig. 5), it is reasonable to consider BuT5 as a MITE, probably mobilized by the P-element transposase.
The size of BuT5 copies seems fairly homogeneous. In the genome of D. mojavensis, we retrieved 22 BuT5 copies with a mean size of approximately 1 kb and a 93.2% pairwise identity. The rest of copies were smaller and likely to bear partial deletions. The size of BuT5 in D. buzzatii is similar, as it is that of copies isolated from other species of the repleta group (see earlier). BuT5 is also quite abundant. In D. buzzatii, BuT5 was found (using in situ hybridization to polytene chromosomes) to be the most abundant of a set of seven transposons, with a basal density of 10−2 copies per genome and chromosomal band (Casals et al. 2006). BuT5 is particularly abundant as secondary colonizer of the inversion breakpoints, indicating that it is (or has been until very recently) transpositionally active (Delprat et al. 2009). It is true that these copy numbers are not close to the thousands of MITEs detected in plant genomes (Feschotte et al. 2002). However, in Drosophila, MITEs are not as abundant as in plants (Holyoake and Kidwell 2003; Dias and Carareto 2011; Depra et al. 2012), possibly because the genome size is considerably smaller and autonomous TEs are not as abundant either (Bartolomé et al. 2002; Tenaillon et al. 2010).
The terminal sequences of BuT5 and P element show high nucleotide identity (>90%), whereas their internal sequences do not have any similarity (figs. 2 and 5). Similarity between terminal sequences of a transposon and a MITE has been previously found and seemingly indicates that the autonomous element is responsible for the MITE origin and amplification because these sequences are known to be important for transposition (Turcotte et al. 2001; Feschotte et al. 2002, 2003). Thus, we can hypothesize that the P element is the autonomous transposon responsible for BuT5 mobilization. If BuT5 were mobilized by the P-element transposase, we would expect that it contains THAP-domain-binding sites. We searched for such motifs in the three D. buzzatii BuT5 longest sequences (copies 1, 6, and 9 in supplementary table S4, Supplementary Material online). All three contain in the 3′-end an identical sequence and in the 5′-end a nearly identical one (one mismatch) to one of the D. melanogaster THAP-domain-binding sites (table 1). Significantly, in the D. buzzatii P element, the putative 5′-THAP-domain-binding site is located in the limit of the segment conserved between BuT5 and P element, whereas the 3′-binding site is embedded within the conserved segment (fig. 5). That is, the similarity of BuT5 and P element in the 5′-end is lost precisely after the putative THAP-domain-binding site. These observations provide strong support for our hypothesis.
The size of the TSDs is a function of the transposase that catalyzes the element mobilization (Craig 2002) and is one of the key features that characterize each superfamily, yet several superfamilies have variable TSD size (Wicker et al. 2007). The D. melanogaster P element generates 8-bp TSDs (O'Hare and Rubin 1983) and the other two families of the P superfamily, 1360 (Kapitonov and Jurka 2003; Reiss et al. 2003), and Galileo (Cáceres et al. 1999), generate 7-bp TSDs. In D. melanogaster, the length of P-element TSDs is highly conserved; only 2% of TSDs from natural insertions were not 8-bp long (Liao et al. 2000; Linheiro and Bergman 2008, 2012). In contrast, a great number of P-element insertions have been studied, and the conservation of the TSDs and target site motives sequence (TSMs), although significant, is low when compared with consensuses for TSDs and TSMs of other TE families.
Two different lengths of BuT5 TSDs (8 and 9 bp) were previously recovered from D. buzzatii (Cáceres et al. 2001; Delprat et al. 2009) and now have been detected flanking highly similar copies in the D. mojavensis genome (4 are 8 bp and 13 are 9 bp). The only P-element copy isolated from D. buzzatii is flanked by 9-bp TSDs (CTAGTAGGT). Although some superfamilies have variable TSD size, each element family usually has a single TSD size. However, there are exceptions. For instance, both the prokaryotic insertion sequence ISRm3 and the maize element Popin show TSDs of 8 or 9-bp (Wheatcroft and Laberge 1991; Rhee et al. 2009). We consider the two sizes of TSDs are more likely the product of transposase flexibility in the staggered double-strand break (DSB) rather than the result of cross-mobilization events (Yang et al. 2009) that would imply the maintenance of BuT5 and two transposases over at least 12 Myr. Because 9-bp TSDs are shared between BuT5 and D. buzzatii P element, we consider these results as consistent with the notion that P element mobilizes BuT5.
Evidence has been provided supporting BuT5 recent mobilization and the implication of P-element transposase in this process. The finding of a putatively complete P-element copy in D. buzzatii that is transcribed and spliced suggests P-element activity. However, an autonomous transposon will need to transpose in the germline, and we found splicing of all six introns in female heads but not in ovaries. This observation does not necessarily imply that P element is not active in D. buzzatii as transposition could be restricted to the male germline or to other developmental stages different from those we studied here. A more thorough expression analysis is required to solve this question.
BuT5 and the P Element, 16 Myr Partnership
We have found that the MITE BuT5 is widespread in the Drosophila repleta species group (fig. 3) and has most likely been vertically transmitted (fig. 4). A recent and comprehensive analysis proposed that diversification of the main repleta group lineages occurred approximately 16 Ma (Oliveira et al. 2012). Therefore, BuT5 would be at least 16 Myr old. In previous works, BuT5 was detected as a secondary colonizer in very recent chromosomal inversion breakpoints (Delprat et al. 2009). Additionally, in this work, we have found BuT5 copies from the D. mojavensis sequenced genome with noteworthy similarity (99.3%). Both findings reveal that BuT5 has been recently active in two species that diverged approximately 12 Ma (Oliveira et al. 2012). Because BuT5 is a nonautonomous element that cannot move by itself but requires the transposase of the P element, we can infer that the P element has been recently active in these two species, D. buzzatii and D. mojavensis. Similarly, the presence and conservation of BuT5 ends in many species of the repleta group indicates a widespread distribution of the P element within this species group. Most likely, the partnership of BuT5 and P-element traces back to at least 16 Ma.
BuT5 has been found to be the transposon directly responsible for inversions 2s in D. mojavensis (Guillén and Ruiz 2012) and 2x3 in D. uniseta (Prada 2010) that were generated by ectopic recombination between copies inserted in opposite orientation at two chromosomal sites (fig. 1). However, BuT5 is a nonautonomous element that does not encode for a transposase and thus requires the P-element transposase for its mobilization. Thus, if BuT5 is the main actor, P element must be considered as a necessary accomplice. P element is not only necessary for BuT5 mobilization, that is, for the insertion of the two BuT5 copies in their chromosomal sites, but it is also likely to have taken part in the ectopic recombination event. Ectopic recombination begins with the generation of a DSB followed by the DNA ends searching for homologous sequences for DSB repair. P-element transpose by a cut-and-paste mechanism that involves the binding of the transposase to the element TIRs and the excision of the element generating a DSB at the donor site followed by the integration of the element into a different chromosomal site (Beall and Rio 1997; Tang et al. 2007). Hence, DSBs produced during normal or aberrant transposition events may provide the required initial step for ectopic recombination events.
The P element is well known for its potential to induce chromosomal rearrangements in the laboratory D. melanogaster (Berg et al. 1980; Engels and Preston 1981, 1984; Gray 2000). In contrast, no direct evidence has been found so far for the generation of natural chromosomal inversions by P elements. None of the eight inversions that are polymorphic in natural populations of D. melanogaster were seemingly generated by TEs (Corbett-Detig et al. 2012). In D. willistoni, a species with a rich inversion polymorphism, P-element hybridization sites in the polytene chromosomes often coincide with inversion breakpoints (Regner et al. 1996), but this provides only circumstantial evidence as TEs can be secondary colonizers of inversions breakpoints (Cáceres et al. 2001; Delprat et al. 2009). We have shown that BuT5 and its master transposon P element generated two inversions recently fixed in D. mojavensis and D. uniseta, two species of the repleta group (fig. 1). Therefore, this is the first unequivocal demonstration of the role of P elements in Drosophila chromosomal evolution.
Supplementary Material
Supplementary tables S1–S6 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Cédric Feschotte, Josefa González, Yolanda Guillén, Mar Marzo, Marta Puig, and Rosemary Thwaite for critical reading of previous versions of the manuscript. They also thank UC San Diego Drosophila Species Stock Center, Deodoro Oliveira, and William Etges for sharing with them samples of Drosophila species. This work was supported by grants BFU2008-04988 and BFU2011-30476 from Ministerio de Ciencia e Innovación (Spain) to A.R. and by a PIF-UAB fellowship to N.R.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 2009;26:983–993. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 2002;19:926–937. doi: 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- Beall EL, Rio DC. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 1997;11:2137–2151. doi: 10.1101/gad.11.16.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg R, Engels W, Kreber R. Site-specific X-chromosome rearrangements from hybrid dysgenesis in Drosophila melanogaster. Science. 1980;210:427–429. doi: 10.1126/science.6776625. [DOI] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Puig M, Ruiz A. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 2001;11:1353–1364. doi: 10.1101/gr.174001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Casals F, Cáceres M, Ruiz A. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol Biol Evol. 2003;20:674–685. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- Casals F, González J, Ruiz A. Abundance and chromosomal distribution of six Drosophila buzzatii transposons: BuT1, BuT2, BuT3, BuT4, BuT5, and BuT6. Chromosoma. 2006;115:403–412. doi: 10.1007/s00412-006-0071-7. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Castro JP, Carareto CMA. Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica. 2004;121:107–118. doi: 10.1023/b:gene.0000040382.48039.a2. [DOI] [PubMed] [Google Scholar]
- Clark JB, Kidwell MG. A phylogenetic perspective on P transposable element evolution in Drosophila. Proc Natl Acad Sci U S A. 1997;94:11428–11433. doi: 10.1073/pnas.94.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, et al. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Cardeno C, Langley CH. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics. 2012;192:131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL. Mobile DNA: an introduction. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington: American Society for Microbiology Press; 2002. pp. 3–11. [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990;124:339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Freitas Ortiz M, Lorenzatto KR, Corrêa BRS, Loreto ELS. hAT transposable elements and their derivatives: an analysis in the 12 Drosophila genomes. Genetica. 2010;138:649–655. doi: 10.1007/s10709-010-9439-y. [DOI] [PubMed] [Google Scholar]
- Delprat A, Negre B, Puig M, Ruiz A. The transposon Galileo generates natural chromosomal inversions in Drosophila by ectopic recombination. PloS One. 2009;4:7883. doi: 10.1371/journal.pone.0007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depra M, Ludwig A, Valente V, Loreto E. Mar, a MITE family of hAT transposons in Drosophila. Mob DNA. 2012;3:13. doi: 10.1186/1759-8753-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias ES, Carareto CMA. msechBari, a new MITE-like element in Drosophila sechellia related to the Bari transposon. Genome Res. 2011;93:381–385. doi: 10.1017/S0016672311000371. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. P elements in Drosophila. In: Saedler H, Gierl A, editors. Transposable elements. Berlin (Germany): Springer; 1996. pp. 103–123. [Google Scholar]
- Engels WR, Preston CR. Identifying P factors in Drosophila by means of chromosome breakage hotspots. Cell. 1981;26:421–428. doi: 10.1016/0092-8674(81)90211-7. [DOI] [PubMed] [Google Scholar]
- Engels WR, Preston CR. Formation of chromosome rearrangements by P factors in Drosophila. Genetics. 1984;107:657–678. doi: 10.1093/genetics/107.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Mouchès C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler SR. Miniature inverted-repeat transposable elements and their relantionship to established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington: American Society for Microbiology Press; 2002. pp. 1147–1158. [Google Scholar]
- Fredslund J, Schauser L, Madsen LH, Sandal N, Stougaard J. PriFi: using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 2005;33:516–520. doi: 10.1093/nar/gki425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YHM. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Guermonprez H, Loot C, Casacuberta JM. Different strategies to persist: the pogo-like Lemi1 transposon produces miniature inverted-repeat transposable elements or typical defective elements in different plant genomes. Genetics. 2008;180:83–92. doi: 10.1534/genetics.108.089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén Y, Ruiz A. Gene alterations at Drosophila inversion breakpoints provide prima facie evidence for natural selection as an explanation for rapid chromosomal evolution. BMC Genomics. 2012;13:53. doi: 10.1186/1471-2164-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann S, Haring E, Pinsker W. A new P element subfamily from Drosophila tristis, D. ambigua, and D. obscura. Genome. 1996a;39:978–985. doi: 10.1139/g96-122. [DOI] [PubMed] [Google Scholar]
- Hagemann S, Haring E, Pinsker W. Repeated horizontal transfer of P transposons between Scaptomyza pallida and Drosophila bifasciata. Genetica. 1996b;98:43–51. doi: 10.1007/BF00120217. [DOI] [PubMed] [Google Scholar]
- Hagemann S, Miller WJ, Pinsker W. Identification of a complete P-element in the genome of Drosophila bifasciata. Nucleic Acids Res. 1992;20:409–413. doi: 10.1093/nar/20.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann S, Miller W, Pinsker W. Two distinct P element subfamilies in the genome of Drosophila bifasciata. Mol Gen Genet. 1994;244:168–175. doi: 10.1007/BF00283519. [DOI] [PubMed] [Google Scholar]
- Hammer SE, Strehl S, Hagemann S. Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol Biol Evol. 2005;22:833–844. doi: 10.1093/molbev/msi068. [DOI] [PubMed] [Google Scholar]
- Haring E, Hagemann S, Pinsker W. Transcription and splicing patterns of M- and O-type P elements in Drosophila bifasciata, D. helvetica, and Scaptomyza pallida. J Mol Evol. 1998;46:542–551. doi: 10.1007/pl00006335. [DOI] [PubMed] [Google Scholar]
- Holyoake AJ, Kidwell MG. Vege and Mar: two novel hAT MITE families from Drosophila willistoni. Mol Biol Evol. 2003;20:163–167. doi: 10.1093/molbev/msg023. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P. Censor—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. First examples of protozoan P DNA transposons. Repbase Rep. 2009;9:2162. [Google Scholar]
- Kaufman PD, Doll RF, Rio DC. Drosophila P element transposase recognizes internal P element DNA sequences. Cell. 1989;59:359–371. doi: 10.1016/0092-8674(89)90297-3. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. The evolutionary history of the P family of transposable elements. J Hered. 1994;85:339–346. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Transposable elements as sources of genomic variation. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington: American Society for Microbiology Press; 2002. pp. 59–90. [Google Scholar]
- Kidwell MG, Novy JB. Hybrid dysgenesis in Drosophila melanogaster: sterility resulting from gonadal dysgenesis in the P-M system. Genetics. 1979;92:1127–1140. doi: 10.1093/genetics/92.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbacher S, Gerstl I, Velimirov B, Hagemann S. Drosophila P transposons of the urochordata Ciona intestinalis. Mol Genet Genomics. 2009;282:165–172. doi: 10.1007/s00438-009-0453-7. [DOI] [PubMed] [Google Scholar]
- Lee CC, Mul YM, Rio DC. The Drosophila P-element KP repressor protein dimerizes and interacts with multiple sites on P-element DNA. Mol Cell Biol. 1996;16:5616–5622. doi: 10.1128/mcb.16.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Clark JB, Kidwell MG. A P element-homologous sequence in the house fly, Musca domestica. Insect Mol Biol. 1999;8:491–500. doi: 10.1046/j.1365-2583.1999.00147.x. [DOI] [PubMed] [Google Scholar]
- Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro RS, Bergman CM. Testing the palindromic target site model for DNA transposon insertion using the Drosophila melanogaster P-element. Nucleic Acids Res. 2008;36:6199–6208. doi: 10.1093/nar/gkn563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro RS, Bergman CM. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PloS One. 2012;7:e30008. doi: 10.1371/journal.pone.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto ELS, Zambra FMB, Ortiz MF, Robe LJ. New Drosophila P-like elements and reclassification of Drosophila P-elements subfamilies. Mol Genet Genomics. 2012;287:531–540. doi: 10.1007/s00438-012-0691-y. [DOI] [PubMed] [Google Scholar]
- Loreto EL, Valente VL, Zaha A, Silva JC, Kidwell MG. Drosophila mediopunctata P elements: a new example of horizontal transfer. J Hered. 2001;92:375–381. doi: 10.1093/jhered/92.5.375. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Singh A, Rio DC. The human THAP9 gene encodes an active P-element DNA transposase. Science. 2013;339:446–448. doi: 10.1126/science.1231789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo M, Puig M, Ruiz A. The Foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus. Proc Natl Acad Sci U S A. 2008;105:2957–2962. doi: 10.1073/pnas.0712110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K, Rubin GM. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Oliveira DCSG, et al. Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of the Drosophila repleta species group. Mol Phylogenet Evol. 2012;64:533–544. doi: 10.1016/j.ympev.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Oliveira DCSG, O'Grady PM, Etges WJ, Heed WB, DeSalle R. Molecular systematics and geographical distribution of the Drosophila longicornis species complex (Diptera: Drosophilidae) Zootaxa. 2005;1069:1–32. [Google Scholar]
- Perkins HD, Howells AJ. Genomic sequences with homology to the P element of Drosophila melanogaster occur in the blowfly Lucilia cuprina. Proc Natl Acad Sci U S A. 1992;89:10753–10757. doi: 10.1073/pnas.89.22.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsker W, Haring E, Hagemann S, Miller W. The evolutionary life history of P transposons: from horizontal invaders to domesticated neogenes. Chromosoma. 2001;110:148–158. doi: 10.1007/s004120100144. [DOI] [PubMed] [Google Scholar]
- Pitnick S, Heed WB. New species of cactus-breeding Drosophila (Diptera: Drosophilidae) in the nannoptera species group. Ann Entomol Soc Am. 1994;87:307–310. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Prada CF. 2010. Evolución cromosómica del cluster Drosophila martensis: origen de las inversiones y reutilización de los puntos de rotura. [PhD thesis]. [Barcelona (Spain)]: Universitat Autònoma de Barcelona. [Google Scholar]
- Quesneville H, Nouaud D, Anxolabehere D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol. 2005;22:741–746. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- Quesneville H, Nouaud D, Anxolabehere D. P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC Genomics. 2006;7:214. doi: 10.1186/1471-2164-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner LP, Pereira MSO, Alonso CEV, Abdelhay E, Valente VLS. Genomic distribution of P elements in Drosophila willistoni and a search for their relationship with chromosomal inversions. J Hered. 1996;87:191–198. doi: 10.1093/oxfordjournals.jhered.a022984. [DOI] [PubMed] [Google Scholar]
- Reiss D, Quesneville H, Nouaud D, Andrieu O, Anxolabéhère D. Hoppel, a P-like element without introns: a P-element ancestral structure or a retrotranscription derivative? Mol Biol Evol. 2003;20:869–879. doi: 10.1093/molbev/msg090. [DOI] [PubMed] [Google Scholar]
- Rhee Y, Lin H, Buell R, Childs K, Kaeppler S. A c2 allele of maize identified in regenerant-derived progenyfrom tissue culture results from insertion of a novel transposon. Maydica. 2009;54:429–437. [Google Scholar]
- Rio DC. Regulation of Drosophila P element transposition. Trends Genet. 1991;7:282–287. doi: 10.1016/0168-9525(91)90309-E. [DOI] [PubMed] [Google Scholar]
- Rio DC. P transposable element in Drosophila melanogaster. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington: American Society for Microbiology Press; 2002. pp. 484–518. [Google Scholar]
- Rodríguez-Trelles F, Tarrío R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- Roussigne M, et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28:66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Wasserman M. Evolutionary cytogenetics of the Drosophila buzzatii species complex. Heredity. 1993;70:582–596. doi: 10.1038/hdy.1993.85. [DOI] [PubMed] [Google Scholar]
- Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat Struct Mol Biol. 2010;17:117–123. doi: 10.1038/nsmb.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, et al. P elements are found in the genomes of nematoceran insects of the genus Anopheles. Insect Biochem Mol Biol. 2003;33:381–387. doi: 10.1016/s0965-1748(03)00004-3. [DOI] [PubMed] [Google Scholar]
- Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tang M, Cecconi C, Bustamante C, Rio DC. Analysis of P element transposase protein-DNA interactions during the early stages of transposition. J Biol Chem. 2007;282:29002–29012. doi: 10.1074/jbc.M704106200. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Tu Z. Molecular and evolutionary analysis of two divergent subfamilies of a novel miniature inverted repeat transposable element in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol. 2000;17:1313–1325. doi: 10.1093/oxfordjournals.molbev.a026415. [DOI] [PubMed] [Google Scholar]
- Turcotte K, Srinivasan S, Bureau T. Survey of transposable elements from rice genomic sequences. Plant J. 2001;25:169–179. doi: 10.1046/j.1365-313x.2001.00945.x. [DOI] [PubMed] [Google Scholar]
- Wasserman M. Cytological evolution of the Drosophila repleta species group. In: Krimbas CB, Powell JR, editors. Drosophila inversion polymorphism. Boca Raton (FL): CRC press; 1992. pp. 455–552. [Google Scholar]
- Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yang G, Holligan-Nagel D, Feschotte C, Hancock C, Wessler S. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- Yuan Y-W, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A. 2011;108:7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci U S A. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.