Abstract
Ascidians or sea squirts form a diverse group within chordates, which includes a few thousand members of marine sessile filter-feeding animals. Their mitochondrial genomes are characterized by particularly high evolutionary rates and rampant gene rearrangements. This extreme variability complicates standard polymerase chain reaction (PCR) based techniques for molecular characterization studies, and consequently only a few complete Ascidian mitochondrial genome sequences are available. Using the standard PCR and Sanger sequencing approach, we produced the mitochondrial genome of Ascidiella aspersa only after a great effort. In contrast, we produced five additional mitogenomes (Botrylloides aff. leachii, Halocynthia spinosa, Polycarpa mytiligera, Pyura gangelion, and Rhodosoma turcicum) with a novel strategy, consisting in sequencing the pooled total DNA samples of these five species using one Illumina HiSeq 2000 flow cell lane. Each mitogenome was efficiently assembled in a single contig using de novo transcriptome assembly, as de novo genome assembly generally performed poorly for this task. Each of the new six mitogenomes presents a different and novel gene order, showing that no syntenic block has been conserved at the ordinal level (in Stolidobranchia and in Phlebobranchia). Phylogenetic analyses support the paraphyly of both Ascidiacea and Phlebobranchia, with Thaliacea nested inside Phlebobranchia, although the deepest nodes of the Phlebobranchia–Thaliacea clade are not well resolved. The strategy described here thus provides a cost-effective approach to obtain complete mitogenomes characterized by a highly plastic gene order and a fast nucleotide/amino acid substitution rate.
Keywords: Tunicates, Ascidians, mitochondrial genome, mitogenomics, next-generation sequencing, Illumina, gene order, rearrangements, phylogeny, mixture models, genome assembly
Introduction
With thousands of described species, ascidians, or sea squirts (phylum: Chordata, subphylum: Tunicata, class: Ascidiacea) form a unique group of sessile marine non-vertebrate chordates (Shenkar and Swalla 2011; Shenkar et al. 2012). Because of their key systematic position as a vertebrate sister-clade (Delsuc et al. 2006; Singh et al. 2009), ascidians have a pivotal role in evolutionary developmental studies and have become important animal models in comparative genomics (Dahlberg et al. 2009). From the ecological point of view, their relatively short life cycle, their ability to thrive in eutrophic (nutrient-rich) environments and a lack of significant predators contribute to their success in newly introduced environments (Lambert 2001; Shenkar and Loya 2008). The corollary is that ascidians are among the worst marine invasive species and that their rate of introduction has increased during the past decade (Lambert 2009). It is thus essential to develop tools that enable us to distinguish nonindigenous from indigenous species and ascertain the source populations of the introduced species. Unfortunately, ascidian systematics is notoriously difficult, because species are mostly classified based on inner anatomical characters such as gonad or gut loop shape and positions, and branchial sac structures (Monniot et al. 1991). Consequently, misidentifications of ascidian species are frequent (Mastrototaro and Dappiano 2008; Lambert 2009). Molecular sequences provide a way to complement species identification, especially in situations where traditional morphology-based discrimination of taxa is inadequate (Geller et al. 2010). Molecular markers, and in particular mitochondrial (mt) DNA sequences, thus provide a powerful alternative to the morphological approach. As a case in point, mt DNA has been successfully used to unequivocally demonstrate the existence of two cryptic species in the cosmopolitan ascidian Ciona intestinalis (Iannelli, Pesole, et al. 2007). Notwithstanding, ascidians are fast-evolving species (Yokobori et al. 1999, 2005; Tsagkogeorga, Turon, et al. 2010), a feature that complicates the use of their molecular characters to infer their evolutionary history (Delsuc et al. 2006). More specifically, ascidian mt genomes are hypervariable in almost all genomic features, which include for example, extremely high rates of sequence divergence and rampant gene order rearrangements, even at low taxonomic levels such as in congeneric and cryptic species (Iannelli, Griggio, et al. 2007; Gissi et al. 2010). This extremely fast evolution of ascidian mt genomes makes their sequence amplification a challenging task, which in turn explains the paucity of these sequenced genomes. We thus aimed to develop a simple and efficient method by which complete ascidian mt genomes can be easily acquired.
Next-generation sequencing (NGS) technologies have revolutionized data acquisition in biology. Although sequencing protocols were originally developed for extracting a genome or transcriptome from a single organism, it is possible to mix several samples in a single flow cell (i.e., multiplex sequencing) as long as the sequences from the different samples can be subsequently separated. Standard multiplex methods allow pooling up to 96 different samples by introducing barcodes (or tags) during the DNA library preparation (Binladen et al. 2007). Following the sequencing step, reads are separated based on their barcode tags, such that assembly is performed for each sample separately. The advantage of this approach is the possibility to establish a trade-off between the total number of reads available from a single NGS run and the number of reads required to obtain a desired coverage for each individual sample. However, the disadvantage of such an approach is that it requires constructing separate genomic libraries for each sample, which can be costly. Several studies have suggested mixing several samples without barcoding them and separating the sequences only after the assembly step (Pollock et al. 2000; McComish et al. 2010; Timmermans et al. 2010; Dettai et al. 2012). We refer here only to nontheoretical studies. In Timmermans et al. (2010), the postassembly separation was based on bait sequences, which are short sequences (200–1,000 bp) obtained for each sample using Sanger sequencing. In McComish et al. (2010), the separation was performed by comparing the assembled contigs to a set of closely related reference mt genomes. Both Timmermans et al. (2010) and McComish et al. (2010) sequenced long polymerase chain reaction (PCR) amplified fragments covering the entire mitogenome. Unfortunately, the acquisition of long PCR fragments is extremely difficult in tunicates due to the pervasive gene order rearrangements. In addition, PCR artifacts can sometimes give rise to chimeric mt contigs (Timmermans et al. 2010).
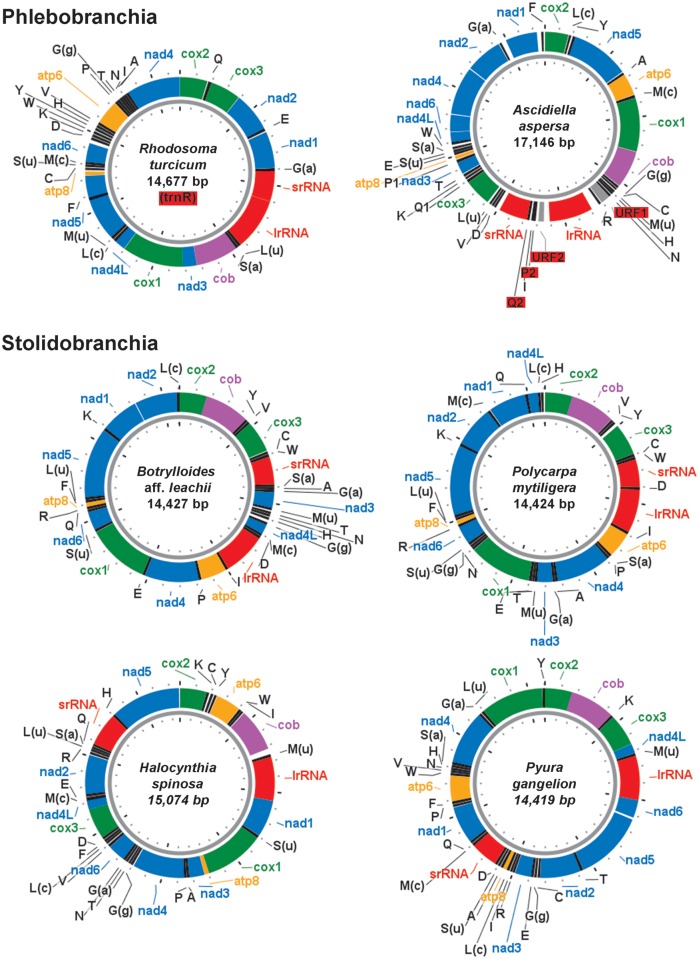
In this work, we chose to use the Illumina platform to sequence total genomic extracts of multiple species mixed together. Thus, both nuclear and mt DNA fragments of multiple species were sequenced together, and the mtDNA sequences were computationally retrieved through the assembly step. Our approach is similar to that used by Groenenberg et al. (2012), who obtained the complete mitogenome of a snail by Illumina sequencing and de novo assembly of the total DNA extracted from a single museum specimen. Following Timmermans et al. (2010), bait sequences were here used to identify the mt sequences of each sample rather than closely related sequences, as in McComish et al. (2010), since we sequenced, for example, the first representative of a family whose phylogenetic position is debated (e.g., Corellidae; Tsagkogeorga et al. 2009). The advantage of our “brute force” approach is that it neither depends on specific primers nor on enrichment protocols and it is blind to mt gene order. Using this approach, we successfully assembled five new complete mitogenomes: Rhodosoma turcicum (Phlebobranchia: Corellidae), Botrylloides aff. leachii and Polycarpa mytiligera (Stolidobranchia: Styelidae), Halocynthia spinosa, and Pyura gangelion (Stolidobranchia: Pyuridae) (fig. 1A and C–F, respectively). In addition, using the standard PCR and Sanger sequencing approaches, we obtained the mt genome of Ascidiella aspersa (Phlebobranchia: Ascidiidae) (fig. 1B). We describe and discuss our novel approach to mtDNA sequencing using NGS technology, together with the characteristics of these six new ascidian mt genomes in terms of genome organization and phylogenetic signal.
Fig. 1.—
Ascidian species sequenced in this work. (A) Rhodosoma turcicum (Corellidae), (B) Ascidiella aspersa (Ascidiidae), (C) Botrylloides aff. leachii (Styelidae), (D) Polycarpa mytiligera (Styelidae), (E) Halocynthia spinosa (Pyuridae), and (F) Pyura gangelion (Pyuridae).
Materials and Methods
Tissue Samples Origin
The origin of the tissue samples is indicated in supplementary table S1, Supplementary Material online. Samples were deposited at the Steinhardt National Collection of Natural History, Zoological Museum at Tel Aviv University (Israel) except for the A. aspersa sample. None of the field studies in Italy or Israel involved endangered or protected species. The A. aspersa sample was collected in a free area of the Venice Lagoon, which is neither privately owned nor protected in any way. The sampling in Israel was approved by the Israel Nature and Parks Authority (permit 2005/21942 and 2005/23512).
Sequencing of the Ascidiella mt Genome
Total DNA of A. aspersa was isolated from the muscle of a single individual using the Puregene Tissue kit (Gentra Systems) according to the manufacturer’s protocol. The complete mt genome was amplified in four long overlapping fragments ranging from 3.8 to 5.3 kbp, and one short fragment of 1.3 kbp. All amplifications were performed with the Expand High Fidelity PCR System (Roche Applied Science) in 25 µl reaction mixture according to the manufacturer’s instructions. Initial PCR reactions were carried out using several combinations of heterologous primers designed on the most conserved regions of the mt protein-coding genes. Only reactions that gave a bright single band during the electrophoretic analysis were further processed: the sequences of these amplicons were used to design the species-specific primers necessary to amplify the remaining portions of the mtDNA. Supplementary table S2, Supplementary Material online, provides the list of Ascidiella amplicons covering the entire mtDNA and the primer sequences. Amplicons were directly sequenced using a primer walking strategy. In addition, two small fragments of 0.5 and 1.3 kbp were also cloned using the TOPO-TA Cloning kit (Invitrogen) and their sequences were obtained as the consensus of three different clones each. This strategy enabled confirmation of the low-quality sequence surrounding two homopolymeric stretches more than 8 bp. Sanger sequencing was performed by the Eurofins MWG operon company (Ebersberg, Germany).
Illumina Sequencing
For Illumina sequencing, genomic DNA was isolated from gonads following the protocol detailed in Fulton et al. (1995). All samples, except R. turcicum, yielded high quality DNA, as observed on an agarose gel. The total DNA extracts of the five species were then pooled. Specifically, 1.5 µg from each DNA extract was used in the mix, except for R. turcicum, for which double the amount was used due to significantly lower DNA quality. The mixed DNA sample was then sent for a single library construction and sequencing to the Genome Sequencing & Analysis Core Resource of Duke University (Durham, NC). Paired-end sequencing of 100 bp reads derived from fragments of average length of 195 bp was performed on a HiSeq 2000 platform.
Amplification of the COI Baits
Amplifications of the COI baits were performed in two steps using the total DNA extract of each species as template. Specifically, for each sample, a first amplification round was performed with external primers, followed by a re-amplification round of the initial PCR product using nested primers. Different primer pairs were used for most species. The primer pairs, the primer sequences, and the length of the fragment amplified are detailed in supplementary table S3, Supplementary Material online.
For P. gangelion, we failed to amplify a clean COI sequence with the degenerate primers used for the other species. Specific primers were thus designed based on the SOAPdenovo-Trans contig (the Pyura contig was assigned in preliminary analysis using as bait the COI sequence of Pyura dura FJ528619 available in public databases). The COI sequence of P. gangelion was then amplified in a single PCR reaction using the genomic DNA as a template. The sequence obtained was identical to the one obtained from the Illumina read assemblies.
Bioinformatics Analysis of the Raw Data
A total of 201,057,100 paired-end reads, 100-bp long, were produced by the HiSeq 2000 sequencing platform. We then discarded all reads that were marked to have failed chastity and purity quality filtering by the Consensus Assessment of Sequence and Variation (CASAVA) pipeline used by the Illumina sequencing platform. As a result, 10,052,019 pairs of reads were removed (∼5%). We additionally used the FASTX-Toolkit (http://cancan.cshl.edu/labmembers/gordon/fastq_illumina_filter/, last accessed June 14, 2013) to remove reads for which more than 10% of the bases had a phred quality score below 20. This resulted in the removal of an additional 17,783,809 pairs of reads, leaving 173,220,272 read pairs for all subsequent analyses (∼86% of the initial read count).
Velvet assembler was used with a k-mer length of 61, and all remaining parameters were left at their default values. ABySS assembler was used with a k-mer length of 64, a minimum number of 10 pairs required for building contigs, and all other parameters remained with their default values. SOAPdenovo assembler was used with a k-mer length of 63, the mode that uses reads for both contig and scaffold assembly (asm_flags = 3), and all other parameters remained with their default values. SOAPdenovo-Trans was similarly used, leaving a k-mer length of 31. Trinity assembler was used with the inchworm k-mer method, and all the server resources (stack size, CPU time, file size, data size, core dump size, memory usage, and virtual memory usage) were set to unlimited, following author recommendations, to facilitate completion of the assembly task.
Coverage Analysis
Coverage statistics of the mt genomes were computed with the software Geneious Pro version 5.4 (http://www.geneious.com, last accessed June 14, 2013), by mapping single-mate reads (i.e., the reads were considered individually and not as pairs) on the mitochondrial contigs obtained with SOAPdenovo-Trans. The following mapping parameters were used: a minimum of 24 consecutive read bases that must perfectly match the reference sequence, a maximum 10% of single mismatches over the read length, a minimum of 80% of base identity in the overlapping region, and a maximum of 10% of gaps with a maximum gap size of 3 nucleotides.
Mitochondrial Genome Annotation
Mitochondrial genes were annotated by similarity to orthologous genes of metazoans, taking advantage of the BlastN/BlastP service of the MitoZoa database (D'Onorio de Meo et al. 2012). The start codon of a protein-coding gene was defined as the first ATG or the first nonstandard initiation codon (Wolstenholme 1992) that does not cause overlap with the upstream gene and maximizes the similarity to orthologous ascidian proteins. According to the punctuation model of mt transcript maturation (Ojala et al. 1981), incomplete T or TA stop codons were hypothesized only if immediately adjacent to a downstream tRNA gene and are assumed to be completed by transcript polyadenylation (Gissi and Pesole 2003). Transfer RNA genes were identified by their potential cloverleaf secondary structure using the programs tRNAscan-SE (Lowe and Eddy 1997) and ARWEN (Laslett and Canback 2008). Moreover, tRNAs with unusual structure, such as those lacking an arm, were searched using specific patterns designed with the PatSearch program (Pesole et al. 2000). All the above-predicted tRNA sequences were manually checked through multiple sequence alignment to orthologous tRNAs of other ascidians and deuterostome representatives. Thus, the final tRNA boundaries were defined based on sequence similarity and on the presence of a conserved cloverleaf secondary structure. The boundaries of the two rRNA genes were inferred as abutted to the flanking genes. As an exception, in A. aspersa we hypothesized the presence of small noncoding (NC) regions upstream rrnL and downstream both rrnL and rrnS. Indeed, these sequences lack similarity to other ascidian rRNA genes. The new mtDNA sequences were deposited in the EMBL-EBI European Nucleotide Archive under accession numbers HF54855–HF548561.
Comparative Analyses
Gene order rearrangements were analyzed on two data sets, with and without tRNA genes. For each pair of mt genomes, the difference in gene order was quantified by dividing the breakpoint distance (BD, i.e., the number of gene adjacencies present in one mt genome and absent in the other, Blanchette et al. [1997]) by the number of genes shared by that pair. The obtained normalized breakpoint distance (BDn) ranges from zero (no rearrangements) to one (almost random permutations) and can be compared among different gene data sets or taxonomic groups because it is independent of the gene content. In cases in which duplicated genes were found in the same mt genome, the copy showing the highest similarities and/or the same position compared with a homolog in a closely related species was considered as the ortholog, and subsequently retained in the calculation of the pairwise BDn. The web-based CREx (Bernt et al. 2007) was used to calculate all pairwise breakpoint distances. Gene blocks conserved between different ascidian mt genomes were detected using the GeneSyn program (Pavesi et al. 2004).
Direct repeats longer than 10 bp were identified using the REPFIND program (Betley et al. 2002) and web-server (http://cagt.bu.edu/page/REPFIND_submit, last accessed June 14, 2013). Transfer RNA genes of all 19 available tunicate species (including the two sequences for C. lepadiformis) were manually aligned based on their secondary structure. To identify cases of tRNA gene recruitment, two different alignments were analyzed: one including the entire tRNA sequence and the other excluding the tRNA loop regions, as these often produce unreliable alignments. Following Lavrov and Lang (2005), neighbor-joining phylogenetic trees were then reconstructed based on uncorrected pairwise distances (p distances) using PAUP* 4.0b10 (Swofford 2000). Bootstrap branch support values were computed using 100 replicates. The placement of a tRNA in a clade including only tRNAs of a different category with a bootstrap value more than 50% was considered as a statistically significant signal of tRNA gene recruitment.
Phylogenetic Reconstructions
The sequences of the 13 mitochondrial protein-coding genes for the 20 available tunicate species, including the six new species obtained in this study, were recovered from the whole mitogenomic sequences. Following the taxonomic sampling of Singh et al. (2009), the sequences of 17 non-tunicate deuterostomes were added to the data set to reconstruct phylogenetic relationships. The tree was rooted using a protostome (i.e., the mollusk Haliotis rubra) as an outgroup.
Given the wide taxonomic scale of our sampling and the high evolutionary rate of the ascidian mt genomes, phylogenetic inference at the nucleotide level would be inadequate because of saturation and erosion of the evolutionary signal due to multiple substitutions (see saturation analysis in supplementary file S1, Supplementary Material online). Therefore, we performed analyses at the amino-acid level to attenuate the saturation problem. The sequences of each independent gene were aligned and translated using MACSE version 0.9_beta1 (Ranwez et al. 2011), which allows the use of different genetic codes while respecting the open reading frames. Subsequently, ambiguous regions of the protein sequence alignments were filtered using TrimAl v1.4rev7 (Capella-Gutiérrez et al. 2009) under the parameters set in the automated1 option. This yielded a total of 3,038 unequivocally aligned amino acid sites, which were used as input for the Bayesian phylogenetic inference. Sequence alignments are available in the Dryad repository: doi:10.5061/dryad.ph920.
Bayesian phylogenetic analyses were performed with Phylobayes 3.3b (Lartillot et al. 2009) under the CAT + GTR + Γ model. The site-heterogeneous CAT mixture model (Lartillot and Philippe 2004) accounts for site-specific amino acid replacement preferences, making it well suited for phylogenomic studies. Four Markov chains Monte Carlo (MCMC) were run and sampled every 10 cycles. Convergence of the chains was monitored through the evolution of the likelihood and model parameters across generations using GNUPLOT (http://www.gnuplot.info/, last accessed June 14, 2013) and confirmed with the bpcomp utility included in Phylobayes. Specifically, each chain was stopped after sampling more than 9,000 trees, that is, when the maximum difference in posterior probability for a given node, as estimated by the 4 independent MCMCs, reached less than 0.1, which is the advised value for a correct convergence. The first 1,000 trees of each MCMC were treated as the burn-in step and thus excluded, and the majority-rule consensus tree was computed from the remaining 4 × 8,000 = 32,000 combined trees. We also verified that for each run the parameters “rel_diff” were less than 0.1 and “effsize” were higher than 100.
Results
Assembling Mitogenomes
The complete mt genome of A. aspersa was amplified in four long overlapping fragments and sequenced using the Sanger method. For most metazoans this is a fast and simple strategy, but due to highly diverged mt gene order in ascidian species, a large set of forward and reverse primers needed to be tested to find the optimal ones for A. aspersa. Additionally, due to the fast ascidian substitution rate “ascidian-specific” primers are often more than 36-fold degenerate primers (Gissi et al. 2010). Consequently, the PCR product quantities are often too low for direct sequencing or cloning, thus requiring fragment re-amplification by nested-PCRs with new sets of primers. We thus considered a novel approach to sequence new ascidian mt genomes: Illumina sequencing of mixed total genomic extracts, followed by a single assembly run from which complete mt genomes are identified among all assembled contigs. The Illumina sequencing of the five pooled DNA samples and subsequent read-quality filtering yielded 173,220,272 paired-end 100 bp reads (see Materials and Methods). The analysis of the coverage of the COI bait sequences indicated average values higher than 50× for all species (table 1). Coverage was found to be nonuniform among species (e.g., Pyura and Rhodosoma have the highest and lowest coverages of 2,000× and 50×, respectively), which is not surprising as mtDNAs of the different species in the pooled DNA probably had different molar quantities (see Materials and Methods). Despite the fact that the same quantity of DNA (∼1.5 µg) was mixed for most species, the relative amount of mtDNA compared to nuclear DNA can vary among species, tissues, and even egg maturity stages for gonads. Even assuming that similar mt:nuclear DNA ratios were purified from all our specimens, nuclear-genome size differences among species could account for differences in the molar quantity of mtDNA present in the processed mixed DNA sample. In addition, DNA molecules that are at different degradation states may give rise to variable coverages. Specifically, our DNA library consisted of fragments of average length of 195 bp (among which 100 bp were sequenced on both sides). This implies that any DNA sample, which is highly degraded and contains fragments shorter than 195 bp, will be poorly covered. This observation could explain the lowest coverage of Rhodosoma, whose starting DNA had low quality (see Materials and Methods).
Table 1.
Results of the Blast Searches Using the COI Baits as Queries against the Different Assemblies
| ABySS |
SOAPdenovo |
Trinity 1/75 |
SOAPdenovo-Trans |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | COI Bait Length (bp) | No. of COI Reads | COI Coverage | No. of COI- Contigs | Length (bp) | No. of Reads (% of Total Filtered Reads) | No. of COI- Contigs | Length (bp) | No. of Reads (% of Total Filtered Reads) | No. of COI- Contigs | Length (bp) | No. of Reads (% of Total Filtered Reads) | No. of COI- Contigs | Length (bp) | No. of Reads (% of Total Filtered Reads) |
| Botrylloides aff. leachii | 1,156 | 9,009 | 740 ± 83 | 1 | 14,429 | 106,776 ± 11,976 | 1 | 14,429 | 106,776 ± 11,976 | 3 | 6,185 | 45,768 ± 5,134 | 1 | 14,429 | 106,776 ± 11,976 |
| (0.03 ± 0.003) | (0.03 ± 0.003) | (0.01 ± 0.001) | (0.03 ± 0.003) | ||||||||||||
| Polycarpa mytiligera | 600 | 1,082 | 167 ± 32 | 0 | 0 | — | 1 | 14,487 | 24,194 ± 4,636 | 2* | 785 | 1,310 ± 250 | 1 | 14,454 | 24,138 ± 4,626 |
| (0.006 ± 0.001) | (0.0003 ± 7 × 10−5) | (0.007 ± 0.001) | |||||||||||||
| Pyura gangelion | 1,214 | 26,786 | 2,102 ± 267 | 14 | 2,767 | 58,162 ± 7,388 | 4 | 7,653 | 160,866 ± 20,434 | 1 | 5,412 | 113,760 ± 14,450 | 1 | 14,443 | 303,592 ± 38,562 |
| (0.02 ± 0.002) | (0.05 ± 0.006) | (0.03 ± 0.004) | (0.09 ± 0.01) | ||||||||||||
| Rhodosoma turcicum | 643 | 365 | 52 ± 8 | 1 | 14,734 | 7,662 ± 1,178 | 1 | 14,734 | 7,662 ± 1,178 | 1 | 868 | 452 ± 70 | 1 | 14,708 | 7,648 ± 1,176 |
| (0.002 ± 0.0003) | (0.002 ± 0.0003) | (0.0001 ± 2 × 10−5) | (0.002 ± 0.0003) | ||||||||||||
| Halocynthia spinosa | 1,149 | 7,192 | 594 ± 53 | 2 | 5,547 | 32,950 ± 2,940 | 1 | 14,271 | 84,770 ± 7,564 | 3 | 4,761 | 28,280 ± 2,524 | 1 | 15,121 | 89,818 ± 8,014 |
| (0.01 ± 0.0008) | (0.02 ± 0.002) | (0.008 ± 0.0007) | (0.03 ± 0.002) | ||||||||||||
Note.—COI reads: number of single-mate reads (i.e., the reads were considered individually and not as pairs) mapped to the COI bait (with at least 90% identity over 100 bp); COI coverage: mean coverage of the COI baits and standard deviations; COI-contigs: number of contigs with at least 95% identity over 50 bp with the COI bait sequences; Length: length of the single COI-contig or of the super-contig obtained from the assembly of multiple COI-contigs plus the COI bait sequence (e.g., in the case of the Pyura gangelion ABySS assembly, the 14 COI-contigs and the COI bait sequence can be assembled into a super-contig of 2,767 bp); Reads: number of single-mate reads (and percentage of the total 2 × 173,220,272 filtered single-mate reads used for each assembly) estimated to comprise the contig based on COI coverage and length. Cases for which the complete mt genome has not been identified with the COI baits are indicated in bold.
*Contigs do not overlap.
Since ascidian mt genomes are short (usually less than 15,000 bp) and were sequenced to high coverage, we expected the assembly programs to efficiently assemble the complete mt genome of each species into a single separate contig. We thus anticipated that BlastN searches (Altschul et al. 1990), using the COI bait sequences obtained beforehand as queries, would allow us to easily assign these single mt contigs to a given species based on the sequence identity criterion.
We assembled the raw data using three de novo genome assemblers, Velvet (Zerbino and Birney 2008; Zerbino et al. 2009), ABySS (Simpson et al. 2009), and SOAPdenovo (Li et al. 2010). Although McComish et al. (2010) successfully used Velvet, in our case it failed to run to completion as the 128 GB of RAM available on our assembly dedicated server were insufficient. The two other assemblers yielded different numbers of contigs and contig lengths, with SOAPdenovo providing fewer and longer contigs than ABySS (supplementary table S4, Supplementary Material online), in agreement with previous observations (Lin et al. 2011; Henson et al. 2012). Surprisingly, neither SOAPdenovo nor ABySS successfully reconstructed all five mt genomes. In table 1, we present the number and lengths of the contigs matching the COI baits, hereafter termed “COI-contigs.” ABySS successfully assembled in a single COI-contig only two mt genomes: those of Botrylloides and Rhodosoma. In addition to Botrylloides and Rhodosoma, SOAPdenovo assembled in a single COI-contig the mt genomes of Polycarpa and Halocynthia. However, the long COI-contig of Halocynthia contained numerous gaps in the sequence (indicated by Ns). As for Pyura, several COI-contigs were detected: 4 in the SOAPdenovo assembly and 14 in the ABySS assembly. However, these contigs did not assemble into a complete mt genome, suggesting that the genomes of these species are spread over several contigs. Surprisingly, no COI-bait contig was detected for Polycarpa in the ABySS assembly even though the coverage of the COI bait estimated from read mapping was higher for this species than for Rhodosoma (table 1).
A DNA library that contains different molecules in extremely variable copy numbers resembles a transcriptome library where variable gene expression levels give rise to transcripts with variable copy numbers. Our pooled DNA library presents this situation, as nuclear and mitochondrial reads are present in different numbers within and among species, indicated by the different COI-bait coverages observed among species (table 1). Unlike de novo genome assemblers, de novo transcriptome assemblers do not assume uniform read coverage among different molecules. Thus, due to the structure of our DNA libraries, we reasoned that de novo transcriptome assemblers might perform better than de novo genome assemblers and correctly assemble the mt genomes into single contigs. We thus used two de novo transcriptome assemblers: SOAPdenovo-Trans (k-mer = 31) and Trinity (minimum contig length = 100, k-mer method = inchworm). Confirming our intuition, SOAPdenovo-Trans was able to efficiently assemble all five mt genomes, each in a single contig (table 2). On the other hand, Trinity failed to run to completion due to insufficient RAM. Only at a read dilution of 1/75 (i.e., randomly choosing 1 out of every 75 reads) was Trinity able to complete the assembly task, but these diluted data were insufficient for proper assembly of all five mt genomes (table 2). In summary, these results suggest that de novo transcriptome assemblers are more appropriate for assembling small mt genomes present in variable copy numbers in total DNA mixed-samples.
Table 2.
Results of Blast Searches Using the Complete Mitochondrial Genomes as Queries against the Different Assemblies
| ABySS |
SOAPdenovo |
Trinity 1/75 |
SOAPdenovo-Trans |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | mtDNA Length (bp) | No. of Reads | Coverage | % Reads | No. of Contigs | Average Contig Length (min, max) | No. of Contigs | Average Contig Length (min-max) | No. of Contigs | Average Contig Length (min-max) | No. of Contigs | Contig Length |
| Botrylloides aff. leachii | 14,427 | 92,063 | 633 ± 145 | 0.027 | 1 | 14,429 | 1 | 14,429 | 8 | 1,814 (371–4,159) | 1 | 14,429 |
| Polycarpa mytiligera | 14,425 | 26,467 | 182 ± 38 | 0.008 | 0 | 0 | 1 | 14,487 | 34 | 284 (104–829) | 1 | 14,454 |
| Pyura gangelion | 14,419 | 304,982 | 2,095 ± 346 | 0.088 | 73 | 472 (101–3412) | 70 | 297 (101–7600) | 4 | 3,632 (5,412–2,183) | 1 | 14,443 |
| Rhodosoma turcicum | 14,677 | 7,759 | 52 ± 12 | 0.002 | 1 | 14,734 | 1 | 14,734 | 15 | 163 (106–280) | 1 | 14,708 |
| Halocynthia spinosa | 15,074 | 88,779 | 583 ± 111 | 0.026 | 16 | 1,921 (116–8,114) | 24 | 724 (107–14,721) | 14 | 1,331 (170–3,212) | 1 | 15,121 |
Note.—Reads: number of single-mate reads (i.e., the reads were considered individually and not as pairs) mapped to the complete mt sequences (at least 90% identity over 100 bp); Coverage: mean coverage of the complete mt genomes and standard deviations. % Reads: percentage of mitochondrial single-mate reads of the total 2 × 173,220,272 high quality filtered reads used in the assembly. Number of contigs: number of contigs with at least 95% identity over 100 bp with the mt genome sequences; Average contig length (min-max): average, minimum and maximum bp length of the contigs matching complete mt sequences. Cases for which contigs did not cover the corresponding complete mt genome are depicted in bold.
Finally, we verified that assembly errors in the form of contigs consisting of incorrectly ordered genes do not exist in any of our mt genome assemblies. This was achieved by conducting several Blast searches to verify that none of the assembly programs (i.e., SOAPdenovo, ABySS, or Trinity) produced mt-contigs with a different gene order arrangements than the one found by SOAPdenovo-Trans (supplementary file S2, Supplementary Material online).
Overall Features of the mt Genomes
The sizes of the six ascidian mt genomes assembled here range from the 14,419 bp of P. gangelion to the 17,146 bp of A. aspersa (supplementary table S5, Supplementary Material online). A detailed analysis of the NC regions and of the overlap between adjacent genes is reported in supplementary file S3, Supplementary Material online. Among the overlapping gene pairs, we carefully investigated the presence of the cox2-cob overlap. Indeed, previous studies suggested that this is a tunicate-specific feature related to transcriptional constraints, because in C. intestinalis spA and in Halocynthia roretzi, the cox2-cob overlapping genes are transcribed in a single mature bicistronic mRNA (Gissi and Pesole 2003; Gissi et al. 2010). Remarkably, the presence of a cox2-cob gene adjacency is always coupled with the presence of a cox2-cob overlap (supplementary table S6, Supplementary Material online). Moreover, if present, the cox2-cob overlap is the longest gene overlap, except in two cases (the Pyuridae H. roretzi and Microcosmus sulcatus; supplementary table S6, Supplementary Material online). The newly sequenced mt genomes add new insights regarding the evolutionary conservation of both the cox2-cob overlap and adjacency in ascidians. Our new sequences reveal that the cox2-cob gene block is less frequent in Phlebobranchia and Stolidobranchia than previously suggested (Gissi et al. 2010). In particular, cox2-cob has been found only in five out of the seven available Phlebobranchia, and only in five of the eight available Stolidobranchia. Moreover, in these taxa this gene block appears to have been lost or acquired a number of times, independently, in three different families (i.e., Ascidiidae, Pyuridae, and Styelidae) and even within the genus Halocynthia (supplementary table S6, Supplementary Material online).
The assembled mt genomes of the six ascidians were found to encode the small and large subunit rRNAs (rrnS and rrnL) and the 13 protein-coding genes of the mt respiratory apparatus, including the small atp8 gene, which was initially considered absent in tunicates (Yokobori et al. 1999). Surprisingly, the mt genome of A. aspersa was found to contain two extra unassigned ORFs (URFs 1 and 2), which show no sequence similarity to any known proteins (marked in red in fig. 2). URF-1, located between trnN and trnR, is 267 bp long, whereas URF-2, located between rrnL and trnP2, is 162-bp long. Moreover, the 3′-end of URF-1 is approximately 97% identical to the 3′-end of URF-2, giving rise to two copies of a 121-bp long direct repeats located approximately 2 kbp apart. The lack of similarity to other known mt and non-mt sequences, and the presence of a repeated region, suggest that these URFs are not protein coding sequences. In this case, these sequences should form part of the two longest NC regions of the mt genome of A. aspersa (403- and 297-bp long, containing URF-2 and URF-1, respectively). No repeats of similar size were found in the other mt genomes. The only repeats detected in the other assembled mt genomes are low-complexity repeats with a maximum length of 25 bp (data not shown).
Fig. 2.—
Organization of the assembled tunicate mt genomes. Red background highlights extra and lost (in brackets) genes. tRNA genes are marked in black by their one-letter code, except for G(a), Gly(AGR); G(g), Gly(GGN); L(u), Leu(UUR); L(c), Leu(CUN); M(c), Met(CAU); M(u), Met(UAU); S(a), Ser(AGY); and S(u), Ser(UCN). P2 and Q2 indicate the extra trnP-2 and trnQ-2 genes of A. aspersa. rRNAs are marked in red. ATP synthase genes are marked in orange. NADH dehydrogenase genes (Complex I) are marked in blue. The cytochrome b (complex III) is marked in purple. Cytochrome c oxidase genes (Complex VI) are marked in green. Noncoding regions are marked in white. Ticks are set every 400 bp. All genes are transcribed clockwise.
Concerning tRNAs, the canonical tunicate complement consists of 24 tRNA genes, in accordance with the usage of a modified mt genetic code (Durrheim et al. 1993; Yokobori et al. 1999) and the presence of a tunicate-specific trnM(UAU) gene, probably acting as a tRNA-Met elongator (Gissi et al. 2004; Yokobori et al. 2005; Iannelli, Griggio, et al. 2007; Singh et al. 2009). In our assembled mt genomes, the tRNA content was found to range from 23 to 26 genes (discussed later).
Gene Order Variability
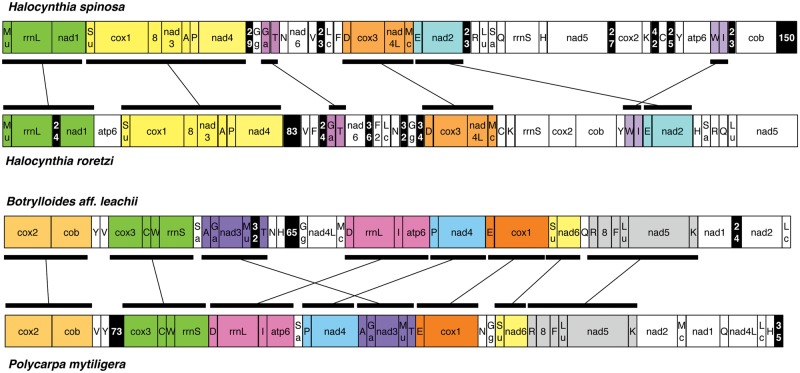
All genes are encoded by the same strand in all six sequenced mt genomes, a feature conserved in all other tunicates studied so far (Singh et al. 2009; Gissi et al. 2010). In contrast, gene order is extremely rearranged in the newly sequenced mt genomes. Among the new mt genomes, the most similar gene orders are observed between the two congeneric Halocynthia species (H. roretzi and H. spinosa) and in the Styelidae pair P. mytiligera and B. aff. leachii (fig. 3). The two Halocynthia mt genomes share six identical gene blocks (51% of the gene adjacencies are shared between the two mt genomes), with the largest of these blocks consisting of seven genes (fig. 3). Similarly, P. mytiligera and B. aff. leachii share eight identical gene blocks (69% of all mt genes), with the largest block consisting of six genes (fig. 3). The quantification of these gene order differences using the normalized breakpoint distances (BDn) shows that the gene order is more dissimilar between the congeneric Halocynthia than between the two Styelidae species (BDn values of 0.62 and 0.51, respectively). Additionally, these two pairs of species show almost identical sequence divergence (uncorrected pairwise distances in protein-coding genes: 0.388 vs. 0.379 at the amino-acid level and 0.346 vs. 0.356 at the nucleotide level). Our results thus indicate that the gene order distance is not always shorter for congeneric species than for species belonging to the same family, and thus can be unrelated to sequence divergence.
Fig. 3.—
Comparison of mt gene order between closely related species. Syntenic regions within each pair of species are marked by the same color and indicated by connected rectangles. Noncoding (NC) regions > 20 bp are marked by a black background, with numbers corresponding to the NC size (in bp). Gene abbreviations: 8, atp6: subunits 8 and 6 of the F0 ATPase; cox1-3: cytochrome c oxidase subunits 1-3; cob: cytochrome b; nad1-6 and nad4L: NADH dehydrogenase subunits 1-6 and 4L; rrnS and rrnL: small and large subunit rRNAs. tRNA genes are indicated by the one-letter code of the transported amino acid, except for: F2: duplicated trnF gene; Ga, Gly(AGR); Gg, Gly(GGN); Lu, Leu(UUR); Lc, Leu(CUN); Mc, Met(CAU); Mu, Met(UAU); Sa, Ser(AGY); Su, Ser(UCN).
Within the Ascidiidae family, the normalized breakpoint distances between A. aspersa and the two available Phallusia genomes are very close to the distance expected for random permutations (0.89–0.92). Indeed, A. aspersa shares only three gene pairs with Phallusia (trnM-cox1 and trnI-rrnS with both P. fumigata and P. mammillata; trnY-nad5 only with P. fumigata, and trnQ-trnK only with P. mammillata). This high variability in gene order within Ascidiidae is in accordance with the extensive mt genome rearrangements already observed in congeneric Phallusia species (BDn: 0.71; Iannelli, Griggio, et al. 2007).
The tremendous variability in gene order already noticed in the tunicate mt genomes is confirmed here by the enlarged data set of 19 tunicates. In this data set, the search for the largest syntenic gene blocks shared by the highest number of tunicates identifies only two very small blocks, that is, two gene pairs: the previously described cox2-cob block, found only in 11 of the 19 available species (mostly in Phlebobranchia, Stolidobranchia, and Thaliacea, but not in Aplousobranchia), and the trnP-nad4 block, found only in five Stolidobranchia and two Aplousobranchia (two different families for each main lineage). The conservation of the cox2-cob pair is suggested to be due to transcriptional constraints (revealed by ORF overlap; discussed earlier). In contrast, trnP and nad4 do not overlap but are perfectly abutted or located only 1–4 bp apart in all ascidians. Moreover, the taxonomic distribution of the trnP-nad4 gene pair covers phylogenetically distant species. Thus, based on the current data, we hypothesize the accidental conservation or appearance of this gene block in the mt genome during the frequent gene order rearrangements. The breakpoint analyses further confirm the remarkable gene order variability of Ascidiacea, as the BDn is very high and quite close to the value expected for random permutations (0.99) in almost all pairwise comparisons. This observation holds even when tRNA genes are not considered in the BDn calculations (data not shown), suggesting that ascidian tRNAs have a mobility similar to that of other genes. Considering all mt genes, nonrandom normalized breakpoint distances are observed only in the Styelidae pair P. mytiligera and B. aff. leachii (BDn: 0.51) and in congeneric comparisons (BDn: 0.08–0.41 in Ciona; 0.62 in Halocynthia; 0.71 in Phallusia).
Transfer RNA Copy Number and Secondary Structures
Among the newly sequenced mt genomes, the 4 stolidobranchs contain the canonical tunicate set of 24 tRNA genes, while the 2 phlebobranchs contain an unusual tRNA set (see tRNAs marked in red in fig. 2). Specifically, trnR is lost in R. turcicum (Corellidae), whereas 2 extra tRNAs, trnP-2 and trnQ-2, have been identified in A. aspersa (Ascidiidae) (gene coordinates: 8843–8914 and 9021–9095, respectively). As estimated by phylogenetic analyses of the entire tunicate tRNA data set (see Materials and Methods), the trnP-2 and trnQ-2 genes show no significant similarity with the trnP and trnQ of A. aspersa and other tunicates, as well as with other tRNA categories. Therefore, their origin by gene duplication or tRNA gene recruitment cannot be determined. Finally, the cloverleaf secondary structure of both trnP-2 and trnQ-2 is compatible with the tRNA functionality, although trnP-2 shows a surprisingly large overlap (14 bp) with the downstream trnI gene (supplementary table S6, Supplementary Material online).
As summarized in supplementary table S7, Supplementary Material online, losses and gains of tRNA genes are not exceptional in tunicates and, based on current data, seem to have occurred frequently in three ascidian families: Ascidiidae, Corellidae, and Pyuridae. Indeed, Phallusia mt genomes have been found to lack the trnD gene (Iannelli, Griggio, et al. 2007). Similarly, gains of extra trnI, trnH, and trnF have been found in the genomes of P. fumigata, M. sulcatus, and H. roretzi, respectively (supplementary table S7, Supplementary Material online; Iannelli, Griggio, et al. 2007; Gissi, et al. 2010). Moreover, in two of these families even congeneric species have a different tRNA gene content: for example, in the Halocynthia genus, a duplicated trnF gene is present in H. roretzi (Gissi and Pesole 2003) but not in H. spinosa (supplementary table S7, Supplementary Material online).
In addition to differences in the tRNA gene content, many ascidians also show a remarkable variability in tRNA secondary structure. In particular, in several ascidian species, trnC, trnN, and trnS(AGY) exhibit unusual cloverleaf structures, which are described in detail in supplementary table S7, Supplementary Material online, and in supplementary file S3, Supplementary Material online. Furthermore, unusual tRNA secondary structures have been sporadically found only in one tRNA of a single species (i.e., loss of the D-arm in trnA of R. turcicum) or in most tRNAs of a species (e.g., large loops up to 27 nucleotides in several tRNA of A. aspersa; large loops up to 18 nucleotides in R. turcicum; supplementary table S7, Supplementary Material online). In conclusion, all these observations point to a fast evolutionary trend of the ascidian tRNAs, concerning on the one hand the secondary structure and sequence, and on the other hand the variability in tRNA gene number.
Phylogenetic Reconstruction
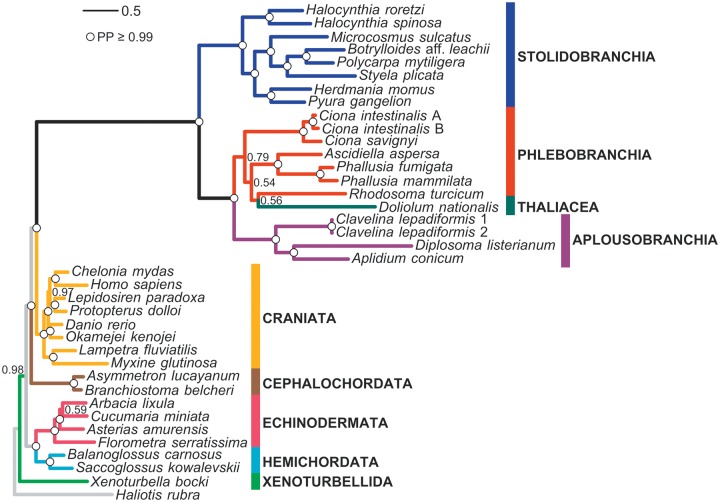
Bayesian inference on the complete mt genomes at the amino acid level provided a well-resolved deuterostome phylogeny (fig. 4). Indeed, all nodes are strongly supported (posterior probability [PP] > 0.99), except for three nodes in Tunicata and two in the remaining deuterostomes. The respective monophyly of the following major deuterostome groups is recovered: Ambulacraria (Echinodermata–Hemichordata), Chordata, and Olfactores (i.e., Craniata–Tunicata). It is worth noting that the branch lengths leading to the most recent common ancestor of Tunicata as well as those observed within this clade are strikingly long as compared to those leading to other deuterostomes.
Fig. 4.—
Phylogeny of Deuterosomes inferred from the concatenation of the 13 mitochondrial proteins. Bayesian consensus tree of 4 independent MCMC runs obtained using the CAT + GTR + Γ mixture model (38 taxa and 3,038 amino acid sites). Values at nodes correspond to Bayesian PP. Circles indicate strongly supported nodes with PP ≥ 0.99.
Within tunicates, the stolidobranch and the aplousobranch species sampled here are monophyletic (PP = 1.0). Phlebobranchs appear to be paraphyletic due to the branching of the thaliacean Doliolum with the corellid Rhodosoma (PP = 0.56). These two species are nested within a group containing all other phlebobranchs (including the monophyletic families Cionidae and Ascidiidae, PP = 1). However, the deepest nodes of the Phebobranchia–Thaliacea clade are weakly supported (PP ranging from 0.54 to 0.79; fig. 4). In contrast, Phlebobranchia–Thaliacea and Aplousobranchia form a well-defined clade (PP = 1.0). It should be noted that the particular position of the thaliacean Doliolum also supports the paraphyly of the Ascidiacea class. Within stolidobranchs, the Styelidae family (Botrylloides, Polycarpa, and Styela) is monophyletic (PP = 1.0), but is nested within a paraphyletic Pyuridae family to which all other sampled stolidobranch species belong. Within this paraphyletic group, the two Halocynthia species diverged first (PP = 1.0), then Herdmania strongly groups with Pyura (PP = 1.0), and Microcosmus unambiguously clusters with Styelidae (PP = 1.0).
Discussion
Efficient Assembly of Complete Mitochondrial Genomes
Our results confirm that complete mt genome sequences of different species can be successfully assembled from raw data obtained by NGS of a library of mixed total DNA without barcodes. A striking advantage of our approach is that it neither depends on specific primers nor on the availability of fragments from closely related species for enrichment. This advantage is manifested in the fact that long-range PCR failed to amplify the complete mt sequences of two of the sampled species (R. turcicum, and P. mytiligera).
A crucial conclusion is that for the data and methodologies used in this work, de novo genome assemblers fail to properly assemble mt genomes. If uniform coverage of reads across the genome is assumed by a de novo genome assembler, highly covered regions may, as a result, be excluded from the assembly process. For assembling a nuclear genome, this is a sensible assumption, as it excludes repetitive regions that may otherwise cause assembly artifacts (Miller et al. 2010). However, when it comes to mt genomes that are present in variable copy numbers compared to the nuclear genome, this is disadvantageous. In agreement with this, the mt genome with the highest coverage, that is, that of P. gangelion, was not completely assembled by SOAPdenovo (tables 1 and 2). Concerning ABySS, the developers of the program indicate that high coverage sequences will assemble better at higher k-mer lengths and vice versa (Robertson et al. 2010), thus a different k-mer length might have allowed ABySS to correctly assemble the mt genome of Polycarpa. We reasoned that de novo transcriptome assemblers, which anticipate differential coverage, may provide a solution to this problem and indeed our results confirmed this intuition. We did not exhaustively explore the parameter space of de novo genome assemblers, for instance by scanning a wide range of k-mer lengths, since an exhaustive study is impractical unless vast dedicated computational resources are available (i.e., multiple high RAM servers). Thus, although our conclusions on the effectiveness of de novo genome assemblers for the problem at hand are not fully comprehensive, our data clearly indicate that de novo transcriptome assemblers provide an efficient means to successfully assemble mt genomes from mixed DNA libraries, especially when computational resources are limited.
From the point of view of manipulations at the bench, it should be noted that our strategy to sequence a mixture of total DNA is fast, relatively cheap, and does not require great effort. It is also notable that our approach allows us to obtain the complete mt genome of the degraded Rhodosoma sample (see also Groenenberg et al. [2012], who sequenced the degraded DNA of a museum specimen preserved in 70% ethanol). Approaches based on long PCR amplifications (Pollock et al. 2000; McComish et al. 2010; Timmermans et al. 2010) or on mt DNA enrichment (Dettai et al. 2012) require well-preserved biological samples, containing complete or almost complete mitochondrial molecules. Our approach, however, is dependent on the tissue used for total DNA extraction. Indeed, the genome projects of C. intestinalis and C. savignyi both produced high coverage and nearly complete mt genome scaffolds (Iannelli, Pesole, et al. 2007). In contrast, the genome project of the planktonic tunicate Oikopleura dioica (Larvacea) did not produce mt scaffolds (they were instead partially predicted from transcript sequences) since it was carried out starting from sperm (Denoeud et al. 2010), where mitochondria are present in very low numbers. These data indicate that a considered choice of the DNA extraction tissue is of great importance for the success of our strategy: the ovary is a mitochondria-rich tissue and is thus the best candidate, provided that it can be easily identified and dissected in the target tunicate species. Otherwise, muscle tissue or, for small-sized tunicates, the entire organism has to be used.
Similar to another NGS strategy of mt genomes (McComish et al. 2010), our approach has two specific potential pitfalls: 1) chimeric assemblies combining mt sequences from different species and 2) chimeric assemblies combining mt sequences and, if existing, nuclear-mitochondrial sequences (Numt, i.e., nuclear copies of mitochondrial DNA; Hazkani-Covo et al. 2010). The first problem can be mitigated by mixing DNA from divergent species, and by using a longer k-mer value, which allows increasing the overlap in the assembly process. Moreover, the use of a paired-end library can reduce the chance of sequences from different species being chimerically assembled. Regarding Numt-mtDNA chimeras, because the sequenced mt and nuclear DNA vary significantly in their copy numbers, the extremely high sequencing coverage obtained with the Illumina or other NGS platforms virtually guarantees that the assembly process will not combine mt and Numt reads. In support of this view, Maricic et al (2010) estimated that less than 0.1% of the reads similar to human mtDNA are of Numt origin. In addition, in our case the paired-end sequencing approach considered should minimize the chances of Numt-mtDNA chimeras.
Extreme Plasticity of Tunicate Mitochondrial Gene Order
Our increase in taxon sampling emphasizes the existence of extreme gene rearrangements in ascidians (Gissi et al. 2010). The few gene blocks that were thought to be conserved among ascidian orders now appear to be artifacts resulting from an insufficient sampling of species diversity. As a case in point, the trnL(UUR)-nad5 block, which was observed to be conserved among the three previously available Stolidobranchia (Gissi et al. 2010), was found to be absent in two of our newly analyzed species (H. spinosa, and P. gangelion) and in Herdmania momus (Singh et al. 2009). Similarly, the cox2-cob block appears to be less conserved than previously thought (Gissi et al. 2010). Our results show that no gene order is conserved at the ordinal level among ascidians and support a general saturation of the gene order rearrangements, except for phylogenetically closely related species (congeneric or intrafamily). This leads us to conclude that gene order cannot be reliably used for reconstructing ascidian phylogenetic relationships, starting from the family and up to higher taxonomic levels. The ascidians mt genomes also show a remarkable variability in their tRNA gene content and in tRNA secondary structures. In general, the origin of additional mt tRNAs can either result from gene duplication or tRNA gene recruitment (Lavrov and Lang 2005; Belinky et al. 2008). In the latter case, a tRNA gene is duplicated, but the anticodon and the acceptor site (the amino acid binding site) of one of the duplicated tRNAs undergoes substitutions, resulting in a tRNA that recognizes a different codon. In ascidians, gene duplication is clearly the phenomenon at the origin of the two trnI of P. fumigata since both tRNAs are nearly identical (Iannelli, Griggio, et al. 2007). Similarly, the two trnF of H. roretzi cluster together in phylogenetic analyses of the whole ascidian tRNA data set (data not shown). However, in the case of the extra trnP and trnQ tRNAs of A. aspersa, these extra genes appear to strongly differ from their homologs in both sequence and length of their secondary structure elements. This suggests that tRNA gene recruitment might exist in tunicates although, due to the high sequence divergence, we cannot clearly determine its existence, unlike what was achieved for the slower-evolving mitogenomes of sponges (Lavrov and Lang 2005; Belinky et al. 2008). Regarding the evolution of tRNA secondary structures, additional mt genomic sequences are required to evaluate the level of homoplasy of tRNA characters such as D-arm loss.
Phylogenetic Signal of Mitogenomic Data for Tunicate Phylogeny
As previously demonstrated (Singh et al. 2009), the mitogenomic approach (using only protein-coding genes) proved useful for inferring deep-level phylogenetic relationships within Deuterostomia. All clades are well resolved and, with few exceptions, posterior probabilities are higher or equal to 0.99. Despite their fast evolutionary rate, mt genomes contain a phylogenetic signal, that can be efficiently recovered provided that analyses are conducted at the amino acid level, with a reasonable taxon sampling, and using a site-heterogeneous mixture model of protein evolution (Singh et al. 2009).
A striking pattern evidenced in our phylogram (fig. 4) is the high evolutionary rate of tunicates with respect to other deuterostomes. This acceleration seems to have occurred back along the Tunicata ancestral branch, and has been maintained in all the extant clades (Singh et al. 2009). Mitochondrial protein-coding genes also point to Aplousobranchia as the fastest-evolving tunicates, although the mt taxon sampling of Aplousobranchia is still very poor and, within them, the family Clavelinidae evolves at a slower rate. This Aplousobranchia rate pattern has also been documented for the nuclear 18S rRNA gene (Tsagkogeorga et al. 2009).
Within Tunicata, the Ascidiacea class forms a paraphyletic group, because the thaliacean Doliolum is nested within ascidians. Moreover, we found a monophyletic assemblage of Phlebobranchia, Thaliacea, and Aplousobranchia as the sister group of a monophyletic Stolidobranchia. This is in agreement with the tunicate phylogeny based on the nuclear 18S rRNA gene published by Tsagkogeorga et al. (2009). The phylogenetic relationships are unresolved concerning the branching pattern among Thaliacea and Phlebobranchia, but suggest a paraphyly of the Phlebobranchia with respect to Thaliacea, with Doliolum appearing as the sister taxon of Rhodosoma (Corellidae). Although the Phlebobranchia paraphyly has weak statistical support (fig. 4), it should be noted that in other phylogenetic studies too it has always been observed with low support (Swalla et al. 2000; Yokobori et al. 2005; Zeng and Swalla 2005; Tsagkogeorga et al. 2009; Stach et al. 2010). Clearly, an improved taxon sampling of thaliaceans is required to better identify their evolutionary affinities with respect to phlebobranchs and aplousobranchs (Govindarajan et al. 2011).
Within Stolidobranchia, the mitogenomic data strongly support the paraphyly of Pyuridae with respect to Styelidae, as also suggested by 18S rRNA (Tsagkogeorga et al. 2009). In particular, these molecular data strongly support the branching of Microcosmus with Styelidae (Styela, Polycarpa, and Botrylloides) in a clade that is the sister group of Herdmania-Pyura, and identify the genus Halocynthia as the earliest diverging lineage within Stolidobranchia. This branching order is in agreement with previous studies either focusing on the overall phylogenetic relationships within Tunicata (Tsagkogeorga et al. 2009; Stach et al. 2010) or more specifically on the relationships between Styelidae and Pyuridae (Pérez-Portela et al. 2009).
Conclusions
Our work demonstrates that mt genomes can be reliably assembled from NGS data derived from pooled total DNA extractions coming from different species, using de novo transcriptome assemblers. The novel strategy proposed here provides an affordable approach to obtain complete mt sequences for taxa, such as tunicates, where either scarce availability of mt sequences or fast substitution and rearrangement rates render use of the standard mtDNA sequencing strategies by long PCR products and “universal” primers labor intensive and nearly impracticable.
Supplementary Material
Supplementary files S1–S3 and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Naomi Paz for editing the text, Riccardo Brunetti for confirming the identity of Botrylloides aff. leachii, Adriana Giumbo for her comments on genome annotation, and Liron Goren for taking the Botrylloides picture. Revital Ben-David-Zaslow provided invaluable help with the collections of the Zoological Museum at Tel Aviv University. They thank Moran Yassour for assembly and Trinity use advice. They also thank the Israeli Ministry of Science Culture & Sport for supporting the National Collections of Natural History at Tel Aviv University. This work was supported by the Israel Science Foundation (663/10) to D.H. and the Ministero dell’Istruzione, dell’Università e della Ricerca, Italy (PRIN-2009) to C.G. This publication is the contribution no. 2013-033 of the Institut des Sciences de l’Evolution de Montpellier (UMR 5554 – CNRS – IRD).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Belinky F, Rot C, Ilan M, Huchon D. The complete mitochondrial genome of the demosponge Negombata magnifica (Poecilosclerida) Mol Phylogenet Evol. 2008;47:1238–1243. doi: 10.1016/j.ympev.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
- Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- Binladen J, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Bourque G, Sankoff D. Breakpoint phylogenies. Genome Inform Ser Workshop Genome Inform. 1997;8:25–34. [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C, et al. Refining the Ciona intestinalis model of central nervous system regeneration. PLoS One. 2009;4: e4458. doi: 10.1371/journal.pone.0004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Denoeud F, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettai A, et al. Conveniently pre-tagged and pre-packaged: extended molecular identification and metagenomics using complete metazoan mitochondrial genomes. PLoS One. 2012;7:e51263. doi: 10.1371/journal.pone.0051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onorio de Meo P, et al. MitoZoa 2.0: a database resource and search tools for comparative and evolutionary analyses of mitochondrial genomes in Metazoa. Nucleic Acids Res. 2012;40:D1168–D1172. doi: 10.1093/nar/gkr1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrheim GA, Corfield VA, Harley EH, Ricketts MH. Nucleotide sequence of cytochrome oxidase (subunit III) from the mitochondrion of the tunicate Pyura stolonifera: evidence that AGR encodes glycine. Nucleic Acids Res. 1993;21:3587–3588. doi: 10.1093/nar/21.15.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton TM, Chunwongse J, Tanksley SD. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep. 1995;13:207–209. [Google Scholar]
- Geller JB, Darling JA, Carlton JT. Genetic perspectives on marine biological invasions. Annu Rev Mar Sci. 2010;2:367–393. doi: 10.1146/annurev.marine.010908.163745. [DOI] [PubMed] [Google Scholar]
- Gissi C, et al. Hypervariability of ascidian mitochondrial gene order: exposing the myth of deuterostome organelle genome stability. Mol Biol Evol. 2010;27:211–215. doi: 10.1093/molbev/msp234. [DOI] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Complete mtDNA of Ciona intestinalis reveals extensive gene rearrangement and the presence of an atp8 and an extra trnM gene in ascidians. J Mol Evol. 2004;58:376–389. doi: 10.1007/s00239-003-2559-6. [DOI] [PubMed] [Google Scholar]
- Gissi C, Pesole G. Transcript mapping and genome annotation of ascidian mtDNA using EST data. Genome Res. 2003;13:2203–2212. doi: 10.1101/gr.1227803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan AF, Bucklin A, Madin LP. A molecular phylogeny of the Thaliacea. J Plankton Res. 2011;33:843–853. [Google Scholar]
- Groenenberg DJS, Pirovano W, Gittenberger E, Schilthuizen M. The complete mitogenome of Cylindrus obtusus (Helicidae, Ariantinae) using Illumina next generation sequencing. BMC Genomics. 2012;13:114. doi: 10.1186/1471-2164-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J, Tischler G, Ning Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012;13:901–915. doi: 10.2217/pgs.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli F, Griggio F, Pesole G, Gissi C. The mitochondrial genome of Phallusia mammillata and Phallusia fumigata (Tunicata, Ascidiacea): high genome plasticity at intra-genus level. BMC Evol Biol. 2007;7:155. doi: 10.1186/1471-2148-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli F, Pesole G, Sordino P, Gissi C. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 2007;23:417–422. doi: 10.1016/j.tig.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Lambert G. A global overview of ascidian introductions and their possible impact on endemic fauna. In: Sawada H, Yokosawa H, Lambert CC, editors. The biology of ascidians. Tokyo (Japan): Springer-Verlag; 2001. pp. 267–269. [Google Scholar]
- Lambert G. Adventures of a sea squirt sleuth: unraveling the identity of Didemnum vexillum, a global ascidian invader. Aquat Invasions. 2009;4:5–28. [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Lang BF. Transfer RNA gene recruitment in mitochondrial DNA. Trends Genet. 2005;21:129–133. doi: 10.1016/j.tig.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, et al. Comparative studies of de novo assembly tools for next-generation sequencing technologies. Bioinformatics. 2011;27:2031–2037. doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Whitten M, Paabo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5: e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrototaro F, Dappiano M. New record of the non-indigenous species Microcosmus squamiger (Ascidiacea: Stolidobranchia) in the harbour of Salerno (Tyrrhenian Sea, Italy) Mar Biodiv Rec. 2008 1:2008e12. [Google Scholar]
- McComish BJ, Hills SFK, Biggs PJ, Penny D. Index-free de novo assembly and deconvolution of mixed mitochondrial genomes. Genome Biol Evol. 2010;2:410–424. doi: 10.1093/gbe/evq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monniot C, Monniot F, Laboute P. Coral reef ascidians of New Caledonia. Paris (France): ORSTOM editions; 1991. [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Iannelli F, Gissi C, Pesole G. GeneSyn: a tool for detecting conserved gene order across genomes. Bioinformatics. 2004;20:1472–1474. doi: 10.1093/bioinformatics/bth102. [DOI] [PubMed] [Google Scholar]
- Pérez-Portela R, Bishop JDD, Davis AR, Turon X. Phylogeny of the families Pyuridae and Styelidae (Stolidobranchiata, Ascidiacea) inferred from mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 2009;50:560–570. doi: 10.1016/j.ympev.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, D'Souza M. PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics. 2000;16:439–450. doi: 10.1093/bioinformatics/16.5.439. [DOI] [PubMed] [Google Scholar]
- Pollock DD, Eisen JA, Doggett NA, Cummings MP. A case for evolutionary genomics and the comprehensive examination of sequence biodiversity. Mol Biol Evol. 2000;17:1776–1788. doi: 10.1093/oxfordjournals.molbev.a026278. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One. 2011;6:e22594. doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Shenkar N, et al. World Ascidiacea Database [Internet] 2012. [cited 2013 Jun 14]. Available from: http://www.marinespecies.org/ascidiacea. [Google Scholar]
- Shenkar N, Loya Y. The solitary ascidian Herdmania momus: native (Red Sea) versus non-indigenous (Mediterranean) populations. Biol Invasions. 2008;10:1431–1439. [Google Scholar]
- Shenkar N, Swalla BJ. Global diversity of Ascidiacea. PLoS One. 2011;6:e20657. doi: 10.1371/journal.pone.0020657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, et al. Tunicate mitogenomics and phylogenetics: peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny. BMC Genomics. 2009;10:534. doi: 10.1186/1471-2164-10-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach T, Braband A, Podsiadlowski L. Erosion of phylogenetic signal in tunicate mitochondrial genomes on different levels of analysis. Mol Phylogenet Evol. 2010;55:860–870. doi: 10.1016/j.ympev.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Cameron CB, Corley LS, Garey JR. Urochordates are monophyletic within the deuterostomes. Syst Biol. 2000;49:52–64. doi: 10.1080/10635150050207384. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4b10. Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- Timmermans MJTN, et al. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 2010;38:1–4. doi: 10.1093/nar/gkq807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkogeorga G, et al. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol Biol. 2009;9:187. doi: 10.1186/1471-2148-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkogeorga G, Turon X, Galtier N, Douzery EJP, Delsuc F. Accelerated evolutionary rate of housekeeping genes in tunicates. J Mol Evol. 2010;71:153–167. doi: 10.1007/s00239-010-9372-9. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Oshima T, Wada H. Complete nucleotide sequence of the mitochondrial genome of Doliolum nationalis with implications for evolution of urochordates. Mol Phylogenet Evol. 2005;34:273–283. doi: 10.1016/j.ympev.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Yokobori S, et al. Complete DNA sequence of the mitochondrial genome of the ascidian Halocynthia roretzi (Chordata, Urochordata) Genetics. 1999;153:1851–1862. doi: 10.1093/genetics/153.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Swalla BJ. Molecular phylogeny of protochordates: chordate evolution. Can J Zool. 2005;83:24–33. [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, McEwen GK, Margulies EH, Birney E. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS One. 2009;4:e8407. doi: 10.1371/journal.pone.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.