Abstract
Despite playing a crucial role in germline-soma differentiation, the evolutionary significance of developmentally regulated genome rearrangements (DRGRs) has received scant attention. An example of DRGR is DNA splicing, a process that removes segments of DNA interrupting genic and/or intergenic sequences. Perhaps, best known for shaping immune-system genes in vertebrates, DNA splicing plays a central role in the life of ciliated protozoa, where thousands of germline DNA segments are eliminated after sexual reproduction to regenerate a functional somatic genome. Here, we identify and chronicle the properties of 5,286 sequences that putatively undergo DNA splicing (i.e., internal eliminated sequences [IESs]) across the genomes of three closely related species of the ciliate Paramecium (P. tetraurelia, P. biaurelia, and P. sexaurelia). The study reveals that these putative IESs share several physical characteristics. Although our results are consistent with excision events being largely conserved between species, episodes of differential IES retention/excision occur, may have a recent origin, and frequently involve coding regions. Our findings indicate interconversion between somatic—often coding—DNA sequences and noncoding IESs, and provide insights into the role of DNA splicing in creating potentially functional genetic innovation.
Keywords: ciliated protozoa, developmentally regulated genome rearrangements, DNA splicing, internal eliminated sequences, genome evolution
Introduction
A fundamental goal in evolutionary biology is to identify and understand the processes that, by shaping genome content and architecture, give rise to evolutionary novelties. Developmentally regulated genome rearrangements (DRGRs) are excellent candidates for generating evolutionary innovation. DRGRs, such as chromatin/chromosome elimination and chromatin diminution, take place during germline-soma differentiation and can create vast diversity in genome architecture, often producing significant changes in the fate of the affected cells (Kloc and Zagrodzinska 2001; Zufall et al. 2005).
An intriguing example of DRGR is DNA splicing, a process that removes segments of DNA interrupting genic and/or intergenic sequences. Perhaps, best known for initiating the rearrangement of immunoglobulin and T-cell receptor genes in vertebrates during the differentiation of lymphocytes (Fugmann et al. 2000), DNA splicing also mediates a more dramatic example of chromatin diminution in ciliated protozoa. In these single-celled organisms, the whole genome is subject to extensive elimination of noncoding internal eliminated sequences (IESs) (Mochizuki and Gorovsky 2005; Kowalczyk et al. 2006; Gratias et al. 2008; Lepere et al. 2008; Kapusta et al. 2011). Such processing follows sexual reproduction and is possible because of the binucleate nature of ciliate cells. Briefly, the diploid, germline micronucleus contains the entire genome and is transcriptionally silent; after meiosis and syngamy, the micronuclear DNA regenerates a new somatic macronucleus, where all gene expression occurs, while the old, maternal macronucleus degrades. Together with major events including chromosome fragmentation and genome amplification, IES elimination crucially contributes to the development of the new macronuclear DNA.
In the ciliate Paramecium, IESs are commonly short, AT-rich, and flanked by 5′-TA-3′ dinucleotide repeats that are in turn part of larger 8-bp imperfect terminal inverted repeats (Betermier 2004). Initially observed for a limited number of IESs isolated from the micronuclear genome, these characteristics were later corroborated by a study where the isolation of micronuclear DNA, a laborious and problematic step, was elegantly overcome (Duret et al. 2008). Specifically, because IES excision in Paramecium is not entirely faithful and is preceded by a polyploidization stage (Betermier et al. 2000), Duret et al. (2008) were able to detect hundreds of TA-flanked sequences that are imperfectly excised from a few copies of the polyploid macronuclear genome of a P. tetraurelia clone. The vast majority (93%) of these TA-indels consists of true IESs, as shown by the recently published P. tetraurelia germline genome sequence (Arnaiz et al. 2012). Thus, while IES splicing in Paramecium is typically precise, events of incomplete excision occur. This excision error is of a nontrivial magnitude, as it can amount to more than 1 in 10 IESs (Duret et al. 2008).
Because unfaithful IES excision generates distinct DNA isoforms, one may ask: do some of these newly generated DNA isoforms acquire biological significance? In a hypothetical but verifiable evolutionary scenario, some events of imperfect IES excision would become established within the macronucleus of an individual. When nonlethal and heritable, these IES-retaining somatic alleles can spread through the sexual population with some probability of fixation. This is plausible in the light of our current understanding of the IES excision mechanisms as maternally determined. Specifically, in addition to being mediated by cis-acting signals located at the IESs’ 3′- and 5'-termini, IES excision in Paramecium is also epigenetically regulated. The presence of an IES in the maternal (prezygotic) macronucleus—which gradually disappears from the cytoplasm after sexual reproduction—can inhibit its own excision from the next generation’s macronuclear genome (Duharcourt et al. 1995). This homology-dependent maternal inhibition may be limited to a subset of maternally controlled IESs (Duharcourt et al. 1998). Thus, a sufficiently high number of imperfect IES excision events in the maternal macronucleus can lead to IES retention in the next generation’s macronuclear genomes (Duharcourt et al. 1995).
In this study, we build on the findings of Duret et al. (2008) in P. tetraurelia, to extend the identification of putative IESs to two additional moderately divergent P. aurelia species (P. biaurelia and P. sexaurelia) (Catania et al. 2009) whose somatic genomes have been sequenced (currently unpublished). In addition to determining physical characteristics shared by the three species-specific sets of TA-indels, we identify elements that are differentially excised between (and within) species, and provide examples of putative conversion of macronuclear-destined sequences into IESs. The existence of variability in the pattern of IES excision among closely related species of ciliated protozoa that is indicated by these results provides new insights into the roles that DRGRs—DNA splicing in particular—have in the evolution of genome architecture and gene structure in ciliates.
Materials and Methods
Treatment of Sequence Reads, Mapping Procedure, and Extraction of TA-Indel Sequences
Assembled macronuclear genome sequences and corresponding raw sequence reads and quality data were collected from three Paramecium species, P. tetraurelia, P. biaurelia, and P. sexaurelia, all belonging to the P. aurelia species complex (Sonneborn 1975). The data files were downloaded from ParameciumDB (Arnaiz et al. 2007; Arnaiz and Sperling 2011) for P. tetraurelia, and generated at Indiana University for P. biaurelia and P. sexaurelia (McGrath CL, Doak TG, Lynch M, unpublished data). Cultures of P. biaurelia and P. sexaurelia (stock V1-4 and AZ8-4, respectively, from the collection of A. Potekhin) were grown to saturation in Wheat Grass Powder (Pines International, Lawrence, KS) medium previously inoculated with Aerobacter aerogenes. Relatively pure macronuclei (free of bacterial, mitochondrial, and micronuclear DNA contamination) were obtained by the method of Aury et al. (2006). An almost complete absence of reads for the mitochondrial genome supports the efficacy of the macronuclear purification. Macronuclear DNA was further purified by the CTAB method (Ausubel et al. 1994). Libraries for Illumina and 454 (8 kb inserts) sequencing were generated at the Center for Genomics and Bioinformatics (Indiana University). The average genomic coverage for P. biaurelia and P. sexaurelia is approximately 45× and 42×, respectively.
To identify putative IESs for each of the three Paramecium species, we designed a bioinformatics pipeline that screens the collections of macronuclear sequence reads for incorrect excision events, an approach used in a recent study by Duret et al. (2008). Specifically, raw sequence read ends were trimmed until the terminal sites (grouped in windows of 30 nucleotides) showed an average quality score higher than 10. Nucleotides in sequence reads were masked when their quality score was less than 10 or when they matched the P. tetraurelia telomeric repeat, that is, three repeats of the hexanucleotide CCC[CA]AA, with at most one mismatch. Trimmed and masked sequence reads were subsequently mapped to the corresponding macronuclear genome assembly using BLAT (Kent 2002). The alignment quality of retained BLAT hits was improved using the MUSCLE program (Edgar 2004) and indels were treated as bona fide IESs (hereafter referred to simply as IESs) if: 1) the length of the hit genomic region covers ≥50% of the read length (masked nucleotides not included); 2) the hit genomic region contains an insertion-deletion (indel) whose size falls between 10 and 5,000 bp; 3) the sequence read maps to a unique location; 4) the indel is immediately flanked by a TA dinucleotide—which is known to flank IESs in several Paramecium species—and preceded or followed by at least 30 ungapped and unmasked sites; 5) the indel is included in an alignment that contains at most two ≥10-nt gaps and where the overall level of sequence identity is ≥95%. A summary of the key bioinformatics steps, including the count of sequence reads after the filtering step and the total number of putative IESs collected after the realignment step is presented in supplementary table S1, Supplementary Material online.
We additionally used BLAT to extract the total number of gapped and ungapped sequence reads that map uniquely onto the detected IES loci. For this analysis, merged IES-flanking regions (40 nucleotides [nt] from each end) and IES-flanking regions (40 nt from each end) plus the intervening IES were used to query the collection of reads from the corresponding species.
Characterization of IESs
As in Duret et al. (2008) our approach detected two types of TA-flanked sequences: 1) present in the genome assembly but excised from one or more sister reads (we call these sequences cryptic IESs), and 2) absent from the genome assembly but inserted in one or more reads (we call these sequences incompletely excised IESs).
Note that some incompletely excised IESs may be spurious because of reads arising from contaminating micronuclear DNA. Although incompletely excised IESs and IESs as micronuclear DNA contaminants cannot be distinguished in our study, we expect the latter type of IESs to be uncommon for at least two reasons: 1) macronuclear DNA enrichment treatments were adopted before genome sequencing for each of the three species surveyed (as a result, the sequence reads of the three Paramecium species rarely match mitochondrial DNA), and 2) in P. aurelia cells, the macronuclear DNA (∼800 haploid copies) is approximately 200 times more abundant than the micronuclear DNA (four haploid copies). In fact, for P. tetraurelia, micronuclear DNA contamination has been suggested not to exceed 50 in 106 reads (Duret et al. 2008).
Although most characterization used custom perl scripts, the degree of intraspecies conservation of 8-bp sequences at the IES termini and the 11-bp regions immediately flanking the IESs was analyzed using RNA Structure Logo (Schneider and Stephens 1990; Gorodkin et al. 1997). The logo generation process takes into account the nucleotide distribution in each of the three P. aurelia species’ IES data set. In the resulting logo, the height of each nucleotide is proportional to its observed frequency relative to the expected frequency—nucleotides whose frequency is less than expected are displayed upside down. The following options were used in RNA Structure Logo: plain sequence, no field assignment, and logo type 2.
Reciprocal Mapping of IESs among Species and Retrieval of Orthologs and Paralogs
After collecting and characterizing physical characteristics of the IESs for each of the three P. aurelia species, we examined the location of the IESs. As the P. tetraurelia genome has been annotated, the location of IESs in this species was inferred by comparing the scaffold coordinates of the IESs with those of the annotated genes. To infer the location of the IESs detected for the remaining two species, whose genomes have not yet been annotated, we took advantage of the annotation of the P. tetraurelia genome. Specifically, we used the BLAST program (Altschul et al. 1997) to map the sequence reads associated with the IESs in P. biaurelia and P. sexaurelia to the P. tetraurelia genome. All regions sharing significant sequence similarity between species (E value = 10−5) were retrieved but only BLAST best hits were used to infer IES location. In cases where BLAST best hits only partially covered the sequence reads and did not encompass the IES, the coordinates of the mapped sequence were used to extrapolate the IES position.
BLAST was also employed to determine the absence (or presence) of the IESs detected in one P. aurelia species in the macronuclear DNA of the remaining species. Three independent BLAST input sequences were used: 1) merged IES-flanking regions (40 nt from each end); 2) IES-flanking regions (40 nt from each end) plus the intervening IES; and 3) IES sequences only. BLAST hits produced by the first set of input sequences reveal the absence of IESs in the macronuclear genome of a given P. aurelia species. Hits were only accepted if their coverage extends for more than 3/4 the length of the query sequence (i.e., >60 bp). BLAST hits produced by the second and third sets of input sequences uncover events of IES retention. These BLAST hits are expected to partially overlap. Hits produced by the second set of input sequences were accepted (and visually examined) if at least a 20-nt tract of the IES detected in one species matched the macronuclear genome sequence of either of the other two species. Finally, the BLAST hits obtained for the first and second set of input sequences were used to calculate the overall average fraction of differentially excised IESs between the species.
The detection of variably excised IESs was followed by the (BLAST-mediated) retrieval of all the detectable DNA regions that are orthologous or paralogous to the region encompassing the polymorphic IES across the three P. aurelia species. The DIALIGN sequence alignment program (Morgenstern 1999) and manual editing were used to generate accurate sequence alignments, which were visualized with BioEdit (Hall 1999). The Maximum Composite Likelihood model implemented in MEGA 4.0 and bootstrap statistics (1,000 replicates) were applied to all the sites of the examined regions to build Neighbor-Joining trees (Tamura et al. 2007).
Results
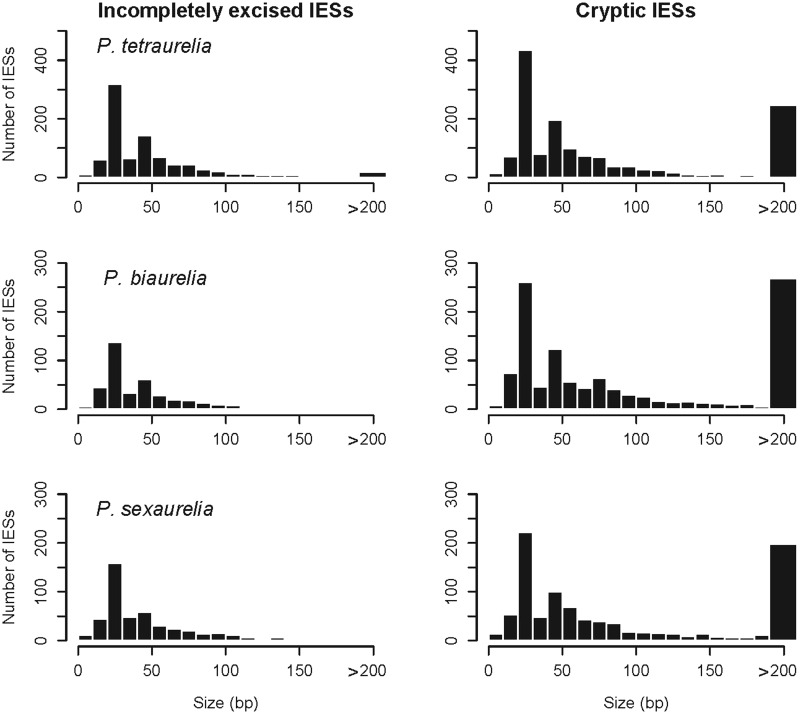
By mapping sequence reads of each Paramecium species onto the corresponding genome assembly, we identified 2,345 putative IESs in P. tetraurelia, 1,538 in P. biaurelia and 1,403 in P. sexaurelia (table 1). The putative IESs that we detected in this study (hereafter referred to as simply IESs) can be differentiated into two classes: 1) incompletely excised IESs (i.e., sequences absent from the somatic genome assembly but found in one or more reads) and 2) cryptic IESs (i.e., sequences present in the somatic genome assembly but excised from one or more sister reads). Using the recently published P. tetraurelia germline DNA sequence (Arnaiz et al. 2012), we could verify that approximately 90% of the incompletely excised IESs are either full-length (73%) or fragmented (17%) IESs. The identification of about half as many putative elements in P. biaurelia and P. sexaurelia as in P. tetraurelia is likely due to the much shorter average read length produced by the technology employed to sequence the P. biaurelia and P. sexaurelia somatic genomes (supplementary table S1, Supplementary Material online). The surveyed IESs are nonoverlapping and single-copy. The AT-richness in these sequences is 4–5% higher than the average AT-content of the macronuclear genome (Aury et al. 2006; McGrath CL, Doak TG, Lynch M, unpublished data). Also, the average AT-content of P. biaurelia and P. sexaurelia IESs is 4–5% higher than that of P. tetraurelia IESs (supplementary tables S2A and S2B, Supplementary Material online). The IES size distribution is similar to that reported for IESs in P. tetraurelia (Gratias and Betermier 2001; Arnaiz et al. 2012) and comparable across the three species, with one major mode at approximately 27 bp and a minor mode at approximately 44 bp followed by a long tail (fig. 1). Based on the published P. tetraurelia somatic genome annotation (for the remaining two genomes the process of annotation is currently ongoing), we conclude that all three species-specific sets of IESs are located both in coding and noncoding regions (table 1).
Table 1.
Summary of IES Counts, Types, AT-Contents, and Genomic Locations
|
Paramecium tetraurelia |
P. biaurelia |
P. sexaurelia |
||||
|---|---|---|---|---|---|---|
| Incompletely Excised | Cryptic | Incompletely Excised | Cryptic | Incompletely Excised | Cryptic | |
| Nonoverlapping, single-copy IESs | 875 | 1,470 | 398 | 1,140 | 462 | 941 |
| AT richness | 0.76 ± 0.08 | 0.78 ± 0.08 | 0.81 ± 0.07 | 0.79 ± 0.08 | 0.81 ± 0.07 | 0.80 ± 0.07 |
| Number (percentage) of IESs with size multiple of 3 (i.e., 3n) | 276 (32%) | 471 (32%) | 117 (29%) | 378 (33%) | 137 (30%) | 297 (32%) |
| Location in the genomea | All IESs (3n IESs) | |||||
| Coding exon | 464 (141) | 370 (99) | 194 (67) | 325 (111) | 226 (63) | 180 (47) |
| UTR | 9 (5) | 8 (2) | 9 (1) | 7 (1) | 10 (3) | 2 (0) |
| Intron | 29 (5) | 4 (1) | 12 (3) | 1 (0) | 21 (8) | 5 (2) |
| Intergenic | 373 (125) | 788 (274) | 45 (8) | 186 (47) | 45 (17) | 111 (40) |
aBased on the P. tetraurelia genome annotation. Estimates for cryptic IESs refer to elements that overlap with only one of the four classes of DNA sequence.
Fig. 1.—
IES size distribution. Size distribution (in base pairs) of the two sets of putative IESs (i.e., imperfectly excised and cryptic IESs) detected for Paramecium tetraurelia, P. biaurelia, and P. sexaurelia.
Inverted Repeats and Conserved Flanking Motifs of the Detected IESs
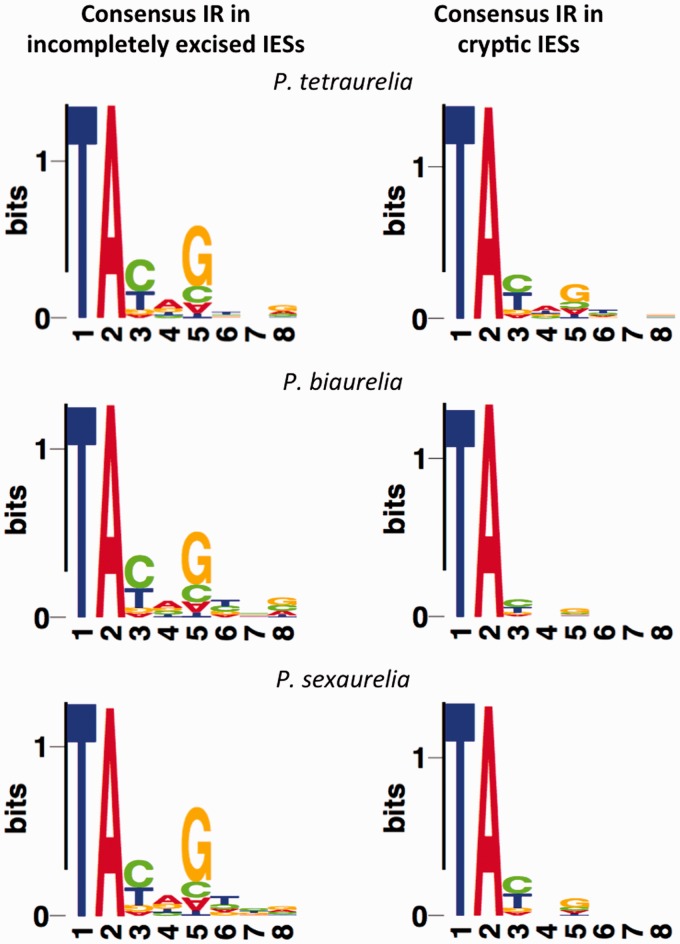
The consensus of the 5′- and 3′-ends of IESs in Paramecium (5′-TAYAGYNR-3′) resembles the termini of Tc1/mariner transposons (Klobutcher and Herrick 1995). As in previous work, a large fraction of the extremities of IESs identified in this study produce a similar consensus (fig. 2). When the two classes of IESs are considered, incompletely excised IESs share the canonical consensus, whereas the termini of cryptic IESs display a much lower level of conservation (fig. 2; supplementary tables S2A and S2B, Supplementary Material online). The enrichment (or deficit) of particular nucleotides in the IES termini is comparable across the three species and mainly involves positions 3, 4, and 5 (after the conserved TA dinucleotide). Also, when the two ends of IESs are compared, positions 3, 4, and 5 are significantly more conserved than positions 6, 7, and 8 (supplementary table S3, Supplementary Material online). This conserved pattern of nucleotide enrichment suggests that IES termini—particularly the nucleotides at positions 3, 4, and 5—play a function associated with presumably equivalent mechanisms of IES recognition/excision across P. aurelia species.
Fig. 2.—
Nucleotide composition and frequency of IES termini consensus sequences. Nucleotide composition and frequency of the consensus sequences of the 8-bp imperfect terminal inverted repeat (IR) of IESs detected for Paramecium tetraurelia, P. biaurelia, and P. sexaurelia. Nucleotide frequency is expressed as the proportion between the observed frequency relative to the expected frequency (the expected frequency is calculated on the basis of the average nucleotide composition of the species-specific set of IESs).
An exploratory search for further cis-regulatory excision signals—in addition to the crucial TA dinucleotide (Mayer and Forney 1999; Matsuda et al. 2004)—revealed a distinct and recurrent (about twice expected) 5′-GG|CC-3′ motif (where | indicates the intervening IES) flanking incompletely excised IESs in each of the three P. aurelia species (supplementary fig. S1A, Supplementary Material online). Although less pronounced, an enrichment of Gs and Cs is apparent around cryptic IESs as well (supplementary fig. S1B, Supplementary Material online). A similar observation had been reported for a limited number of IESs in P. tetraurelia (Gratias and Betermier 2003) and suggests that these cis sites too may be involved in the process of IES definition or excision. A hypothesis to explain such involvement—other than possibly facilitating the generation of excision-associated loop structures—is that an overrepresentation of guanines and cytosines around IESs in an AT-rich genome signals the presence of an IES to the excision machinery.
Constraints on Distribution of IESs within Genes
Analyses of spliced RNA and DNA sequences have detected selective constraints on the evolution of coding regions (Duret et al. 2008; Jaillon et al. 2008). Briefly, both spliceosomal introns and IESs in coding sequences appear to share patterns of selection to provide either in-frame stop codons (premature translation termination codons or PTCs) or frame shifts (length ≠ a multiple of 3 (3n), which will quickly result in translation termination). The most likely hypothesis to explain such selection is that in the event of (accidental) retention in transcripts, noncoding sequences without these features would be invisible to the cellular surveillance systems and can generate toxic products.
We verified the existence of similar selective constraints in our data set. To this end, we studied the overall number of 3n IESs and the relative abundance of PTC-free and PTC-containing IESs in the genome of P. tetraurelia. As previously reported (Duret et al. 2008), we found that fewer 3n (cryptic) IESs than expected by chance map to coding sequences (χ2 = 6.47, df = 1, P = 0.011), and fewer 3n PTC-free (incompletely excised) IESs than expected map to coding sequences (χ2 = 4.74, df = 1, PPTC-free-3n = 0.029; χ2 = 1.34, df = 1, PPTC-containing-3n = 0.246) (supplementary table S4, Supplementary Material online). We detected no deviation from expected frequencies within intergenic regions (χ2 = 0.049, df = 1, Pimperfectly excised IESs = 0.826; χ2 = 1.125, df = 1, Pcryptic IESs = 0.289).
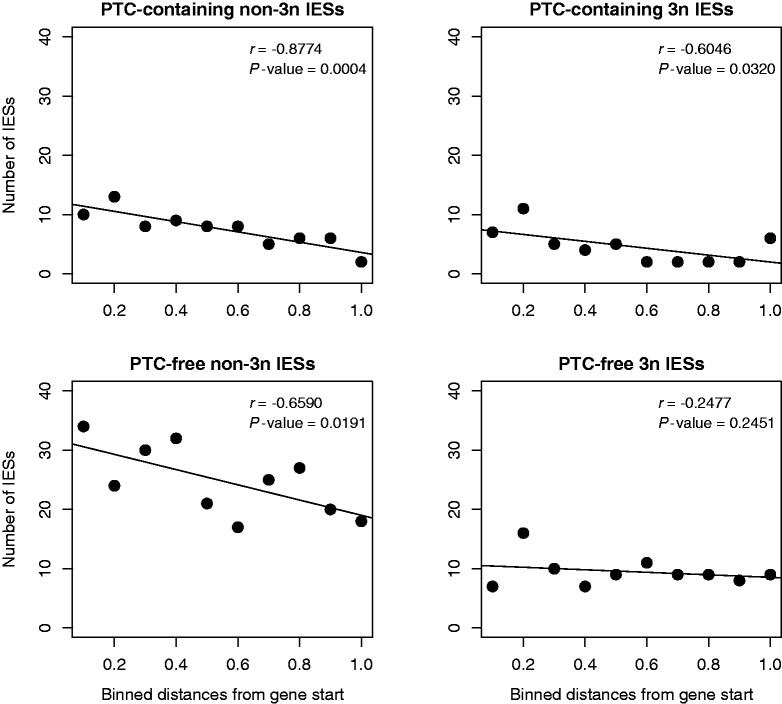
In addition, we asked: how is a selective response triggered by imperfect IES excision events? Current observations suggest that imperfect IES excisions typically involve only a small fraction (typically ∼1/13) of the highly polyploid (∼800n) somatic genome. It is unlikely that this limited fraction of IES-retaining somatic DNA copies has major effects on the fitness of the carrier. Thus, we must conclude that imperfect excisions may occasionally involve relatively large fractions of the somatic DNA copies: fractions that are large enough to impinge on fitness and trigger an adequate selective response. Under this selective scenario, imperfectly excised IESs that reside within genes may exhibit a positional trend that favors their elimination. Specifically, a PTC erroneously introduced in a transcript and proximal to the translation start site leads to a more rapid interruption of the expensive process of protein synthesis and would be most efficiently recognized by the nonsense-mediated decay (NMD) system (van Hoof and Green 1996; Silva et al. 2006; Longman et al. 2007). Although the NMD efficiency of PTC recognition has not yet been investigated in Paramecium, if NMD operates in Paramecium as it does in other eukaryotic systems we may expect to observe the following: 1) PTC-free non-3n and PTC-containing IESs accumulating at the 5′-ends of genes and 2) no positional bias for stopless 3n IESs, as their retention is invisible to NMD.
In our data set, the location of IESs within P. tetraurelia genes is indeed biased toward the 5′-end: there is a 5′-end bias for both 3n and non-3n IESs (3n IESs: r = −0.5270, P value = 0.0588; non-3n IESs: r = −0.8544, P value = 0.0008), whereas a separate analysis of 3n vs. non-3n IESs reveals that the preferential location near a gene’s 5′-end involves PTC-containing IESs and PTC-free non-3n IESs. No significant trend is observed, as expected, for PTC-free 3n IESs (fig. 3).
Fig. 3.—
Position of IESs in coding sequences. Correlation analyses between number of exon-mapping IESs (partitioned according to size and presence of PTCs) and IES distance from the gene start in Paramecium tetraurelia. Distance values are allocated in 10 bins and are equal to the ratio between the IES distance from the gene start and the total gene length.
Overall, these results are consistent with a scenario in which 1) the per-locus fraction of inaccurate IES excisions occasionally involves a number of somatic DNA copies that is sufficiently large to affect individual fitness and 2) transcripts that incorporate IESs are efficiently degraded. Despite this indication of efficient counter-selection, our results suggest that the incorporation of IESs into coding sequences occasionally takes place. These infrequent events, which could have substantial evolutionary implications, are examined below.
Most Putative (Imperfectly Excised) IESs in One P. aurelia Species Are Absent from the Macronuclear Genome of the Remaining Species
The availability of the macronuclear genome sequences of three closely related species of ciliates in tandem with a relevant collection of IESs provides an excellent opportunity for investigating the contribution of IESs (i.e., germline-specific sequences) to somatic genome content. Specifically, as a result of mutations in excision signals, some IESs could become integral parts of the somatic DNA of an individual and, when nonlethal and heritable, the IES-containing alleles could spread and fix in the species. Under this scenario, the alignment of homologous somatic DNA sequences of closely related species would display insertion–deletion polymorphisms associated with IES retention.
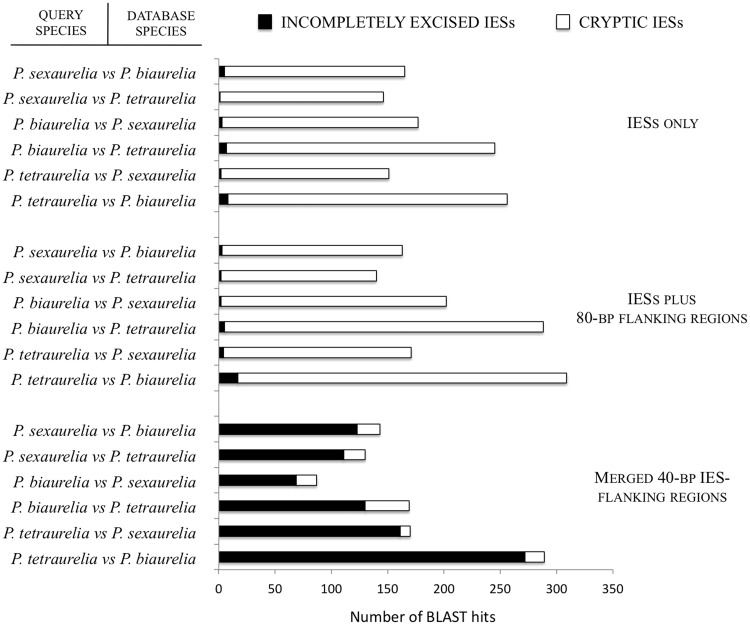
To gain insights into this hypothetical scenario, we asked if imperfectly excised IESs in one P. aurelia species are absent from macronuclei of the other P. aurelia species. This analysis showed that the majority of incompletely excised IESs in a given P. aurelia species are indeed absent from the putative orthologous (or least divergent homologous) loci of the other two species (fig. 4; supplementary table S5, Supplementary Material online). IESs can be absent from the somatic genome assemblies for two reasons: they are excised from the corresponding germline DNA, or they are absent from the germline DNA. In the former case, one should be able to detect the signature of this excision, the 5′-TA-3′ dinucleotide, known to remain in the somatic DNA after IES removal. Indeed, in 97–100% of the cases, a 5′-TA-3′ is conserved at the expected position in between-species comparisons. Although it is difficult to draw firm quantitative conclusions from these observations, these results are consistent with most IES excision events being conserved across the three P. aurelia species.
Fig. 4.—
Conservation and variability of IES excision between Paramecium aurelia species. The BLAST program is employed to determine whether imperfectly excised IESs and cryptic IESs detected in one P. aurelia species (query species) are present or absent from the macronuclear genome assembly of the remaining species (database species). We screen the genome assembly of the database species using: 1) merged IES-flanking regions (40 nt from each end); 2) IES-flanking regions (40 nt from each end) plus the intervening IES; and 3) IES sequences only. The graph represents the cumulative number of hits in the genome assembly of the database species.
Most cryptic IESs, on the contrary, are detected in the macronuclear DNA of the other two species (fig. 4; supplementary table S5, Supplementary Material online). This observation is consistent with the majority of cryptic IESs being the product of erroneous excision, presumably resulting from a partial resemblance of nearby sequences to canonical recognition or excision signals (fig. 2; supplementary fig. S1B and table S2B, Supplementary Material online).
IES Excision Variability between P. aurelia Species
We also detected some cases of variability in IES excision among the three P. aurelia species (fig. 4; supplementary table S5, Supplementary Material online). For example, when the 875 P. tetraurelia incompletely excised IESs were used (with or without additional flanking regions) to query the genome of the two remaining species, 14 and 4 of these IESs were found to match significantly (E value = 10−5) and uniquely with the P. biaurelia and P. sexaurelia assembled macronuclear genomes, respectively. This suggests that a fraction of the IESs detected in a given P. aurelia species may no longer be (or may have never been) excised from the macronuclear genome of other species of P. aurelia.
Cases of IES presence–absence polymorphisms involve cryptic IESs as well. For example, when 40-bp regions immediately flanking the 1470 P. tetraurelia cryptic IESs were merged and used to query the other species’ genome sequences, 10 and 9 have hits in the P. biaurelia and the P. sexaurelia genomes, respectively. This latter observation is intriguing, suggesting that regions rarely excised from the macronuclear DNA of one P. aurelia species can be faithfully excised from homologous loci in closely related species.
Could the observed IES excision polymorphisms reflect misassembly errors due to micronuclear contamination? In principle, if a collection of reads mapping onto a genomic region consists of both gapped and un-gapped sequences (e.g., macronuclear and micronuclear sequences, respectively), erroneous information could be incorporated during the assembly process, and, as a result, a comparison between two or more genomes would exhibit artificial insertion/deletion polymorphisms. These sequence misincorporation events would be expected to be more common for genomic regions that are supported by only a few reads. Also, the likelihood of DNA region misincorporation would be expected to be the highest when the ratio between gapped and ungapped reads approaches 50%. To reduce this type of inaccuracy, we filtered our data set for genomic regions covered by at least eight reads and whose current configuration is supported by a fraction of reads higher than 70%. In addition, we only included loci that can be reliably aligned between all three P. aurelia species. Under these arbitrary thresholds, we retrieved nine IES-associated regions that are differentially excised between P. aurelia species (supplementary table S6, Supplementary Material online). These regions contain two imperfectly excised IESs, and seven cryptic IESs, amounting to 0.12% of all the imperfectly excised IESs and 0.20% of all the cryptic IESs detected in our study.
Finally, we asked how many of the approximately 45,000 germline-specific sequences in P. tetraurelia are differentially excised between the surveyed species. We found that at least 71 and 15 of these IESs are found in the macronuclear genome assembly of P. biaurelia and P. sexaurelia, respectively (supplementary table S7, Supplementary Material online). Remarkably, approximately 50% of these differentially excised IESs reside within coding sequences (supplementary table S8, Supplementary Material online). It is worth noting that as the orthology relationships between the examined species are currently under investigation, the (BLAST-mediated) identification of differential IES excision events in our study relies on the levels of between-species sequence conservation of IES-flanking regions.
Preliminary Characterization of IESs That Are Differentially Excised between P. aurelia Species
The nine IES loci that appear as differentially excised between P. aurelia species correspond to regions that in P. tetraurelia are annotated as exonic (n = 5), intronic (n = 1), and intergenic (n = 3) (supplementary table S6, Supplementary Material online). Although the exact structure of the genes in P. biaurelia and P. sexaurelia is currently unknown, the location of variably excised IESs hints at the occurrence of between-species changes in protein primary structure and, perhaps, in levels of gene expression. With respect to possible changes in gene primary structure, it is worth noting that only one of the IESs mapping to coding exons has a size that is a multiple of 3. This implies that the retention/excision of the remaining non-3n IESs would lead to a shift in reading frame (as defined in P. tetraurelia), unless gene structure differs between species and/or additional indel mutations maintain the reading frame.
When compared with a family of homologous sequences within and across species (i.e., paralogous and orthologous loci), the insertion–deletion IES polymorphisms are typically restricted to only one member of the family, which suggests that these events are recent. In one set of sequence alignments a differential IES retention/excision event between paralogous regions was jointly detected for two species (supplementary fig. S2, Supplementary Material online). It is worth noting that absence of a putative IES from a genomic locus may indicate 1) that the IES is excised or 2) that the missing sequence is entirely absent from the micronuclear DNA. With our current data, we cannot discriminate between these two possibilities.
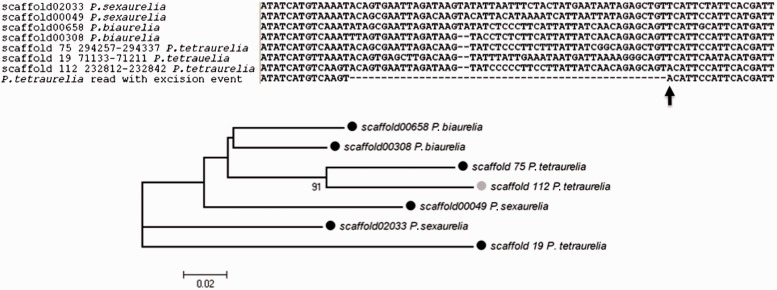
Because of the central role played by the flanking TA dinucleotides in IES excision, the mutational loss of TA dinucleotides around an IES is expected to prevent excision. Alternatively, the creation of sufficiently distant TA dinucleotides in an appropriate sequence context may lead to the erroneous recognition of the intervening sequence as an IES. As a putative example of this, we detected a TA-flanked IES that aligns with TT-flanked macronuclear DNA sequences, within and across P. aurelia species (fig. 5). These TT-flanked macronuclear DNA sequences align with ungapped reads only. A parsimonious interpretation for this finding is that a macronuclear DNA region of P. tetraurelia has undergone excision as a result of a mutational change (T|T → T|A) not found in other paralogs and orthologs. This macronuclear sequence has a size that is multiple of 3, and is part of an expressed P. tetraurelia gene (GSPATG00029756001) coding for a protein with unknown function.
Fig. 5.—
Putative example of a mutational event triggering erroneous excision of a macronuclear DNA region. A T → A mutational change (indicated by the arrow) presumably triggers the excision of a macronuclear-destined sequence (part of the gene model GSPATG00029756001) in Paramecium tetraurelia. The alignment includes all detectable homologous sequences in the surveyed Paramecium species (BLAST E value = 10−5). The unrooted NJ-tree is based on regions that immediately flank the IES under examination (114 sites). Bootstrap values lower than 75% are not shown. Estimates of sequencing coverage are provided for each of the scaffold regions that are shown in the alignment: 13× (11 ungapped and 2 gapped reads) for P. tetraurelia scaffold 112; 10× and 19× (all ungapped reads) for P. tetraurelia scaffold 19 and scaffold 75, respectively; 6× and 12× (all ungapped reads) for P. biaurelia scaffold00658 and scaffold00308, respectively; 10× and 17× (all ungapped reads) for P. sexaurelia scaffold2033 and scaffold00049, respectively.
Discussion
DNA splicing is one of the most intensively studied DRGRs, and particular attention has been devoted to the mechanisms in ciliates that effect the excision of IESs (Mochizuki and Gorovsky 2005; Kowalczyk et al. 2006; Gratias et al. 2008; Lepere et al. 2008; Kapusta et al. 2011). Far less consideration, however, has been given to the evolutionary significance of this biological process.
The availability of macronuclear genome sequences of closely related species of the ciliate Paramecium (Aury et al. 2006) (McGrath CL, Doak TG, Lynch M, unpublished data) provides an excellent opportunity for a comparative study, both to identify regions potentially involved in the regulation of DNA splicing and to explore the contribution of this process to the reshaping of a species’ genome content.
In this study, we identify a total of 5,286 putative IESs across three moderately divergent species of the P. aurelia species complex, P. tetraurelia, P. biaurelia, and P. sexaurelia (Sonneborn 1975; Catania et al. 2009). The thorough macronuclear DNA purification procedure applied here, as in Aury et al. (2006), suggests that the vast majority of the IESs isolated from the surveyed macronuclear DNA sequences result from less-than-perfect IES excision, which takes place during the regeneration of a new macronuclear genome at each sexual generation (Duret et al. 2008).
We find that the imperfectly excised sequences share several similar features: 1) they are typically short (<100 bp) (fig. 1) and AT-rich (≥76%) (table 1); 2) they include imperfect inverted terminal repeats whose nucleotide frequencies and compositions are mostly conserved across species (fig. 2); 3) they are immediately flanked by an excess of Gs and Cs (supplementary fig. S1, Supplementary Material online); and 4) they are located in intergenic, intronic, and coding regions (table 1).
The effect of natural selection on the evolution of these sequences is apparent for IESs that reside in genes. In particular, fewer than expected 3n IESs are located within coding regions in P. tetraurelia, the only species whose genome is currently annotated. This deficit extends to the subset of IESs that are 3n in size and do not contain PTCs. A hypothesis for the latter observation maintains that PTC-free 3n IESs incorporated into mature mRNAs would unsafely elude cellular surveillance systems. Thus, selection appears to operate on IESs that interrupt coding regions, eliminating those IESs that do not provide premature translation termination in the case of inaccurate excision (Duret et al. 2008). This scenario is both plausible and consistent with an additional observation of our study: IESs that introduce a PTC as a result of imperfect excision accumulate at the 5′-end of gene transcripts (fig. 3). This preferential location of IESs corresponds to the region of the transcript where in other eukaryotes NMD most efficiently identifies aberrant mRNA transcripts (van Hoof and Green 1996; Silva et al. 2006; Longman et al. 2007) and more rapidly promotes termination of protein synthesis. Thus, our in silico observations suggest that the way the NMD pathway operates in Paramecium and in other multicellular eukaryotes is similar.
The finding that the majority of IESs detected as imperfectly excised in one species are absent from the macronuclear genome assembly of other species is compatible with the hypothesis that the IES excision profile is largely conserved between closely related P. aurelia species. A nonmutually exclusive explanation for our observations, however, is that an IES detected in one species is not observed in other species simply because it is absent from their micronuclear genome. In other words, it is possible that many IESs are species specific. A formal test for this hypothesis requires a better understanding of the IES loss/gain dynamics within species and the nontrivial inference of orthologous genomic regions between the surveyed Paramecium species.
We detected IES excision variability between and within P. aurelia species (fig. 4, supplementary table S5, Supplementary Material online). Our results indicate that a subset (0.20%) of the sequences that are incorrectly excised from only a few copies of the polyploid macronuclear genome of one particular P. aurelia species (i.e., cryptic IESs) may be faithfully excised—or lost—from the macronuclear genome of other P. aurelia species. In the same way, IESs that are incorrectly retained in only a few copies of the macronuclear genome of a P. aurelia species (i.e., imperfectly excised IESs) appear to be an integral part of the assembled macronuclear genome of other P. aurelia species (0.12%). The existence of differential events of IES excision/retention between P. aurelia species is consistent with the results of an additional analysis: when we query the entire set of approximately 45,000 IESs in the P. tetraurelia genome, we find that at least 86 IESs are differentially excised between species. It is worth noting that a more accurate quantification of differential IES excision events between species requires the availability of higher sequencing coverages (and thus a larger number of IESs) and completion of the P. biaurelia and P. sexaurelia assemblies.
Manually curated analyses of a limited number of cases of putative differential IES excision/retention between the three P. aurelia species reveal that these events are 1) mostly unique (i.e., involve only one species and no other paralogs in that species); 2) flanked by a 5′-TA-3′ dinucleotide; and 3) often located within regions annotated as coding exons in the P. tetraurelia genome. These observations suggest that the majority of alternative DNA rearrangements have arisen recently, at most after the intermediary whole genome duplication, before the formation of the P. aurelia species complex (Aury et al. 2006). A compatible explanation is that genes with alternative rearrangements are frequently lost. Additionally, the location of these events implies that variable IES excision between species may, theoretically, alter both protein sequence and gene expression.
Our study suggests that interconversion between macronuclear-destined DNA sequences and IESs takes place in Paramecium. This two-way process is reminiscent of the interconversion hypothesized for spliceosomal introns and IESs in a study on spirotrichous ciliates (Chang et al. 2007) and of a model advanced for the evolutionary origin of spliceosomal introns in eukaryotes (Catania and Lynch 2008). In the intronization model, exonic and intronic sequences undergo a process of mutual conversion that includes a transient phase where sequences are neither fully exonic nor fully intronic, an intrinsic property of sequences undergoing alternative splicing (Catania and Lynch 2008). The interesting similarity between IESs and spliceosomal introns goes further, including a positional bias along genes (Catania and Lynch 2008; this study), the mechanism of selective cost (Lynch 2002; this study), and the potential role of these sequences in the generation of new genomic information (Gao and Lynch 2009; this study), which could impinge on phenotype.
All ciliates examined have IESs. Although it is not known if these IESs developed in the ancestral ciliate or evolved independently in different ciliates, it is reasonable that some of these sequences have had enough time to be co-opted for various other functions and to have developed specific selective roles in some genes in some species. For example, imperfect excision of IESs from coding sequence, when not harmful, may alter the primary structure of a gene and perhaps the function of the coded protein. Also, the retention of IESs located in noncoding regions could alter (or create de novo) regulatory elements.
Our study represents a first attempt to explore the existence of variability in the pattern of IES excision among closely related species of ciliated protozoa. Despite the limitations of our approach in inferring the presence or absence of IESs in a genome, our findings are encouraging, providing clear imperatives for future investigations, and compatible with a scenario in which DNA splicing contributes to the creation of potentially functional genetic innovation.
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank R.K. Butlin for comments on an earlier version of this manuscript. This work was supported by a Marie Curie International Incoming Fellowship IIF 254202 to F.C. and by the National Science Foundation grant NSF EF-0328516-A006 to M.L.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Cain S, Cohen J, Sperling L. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Sperling L. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 2011;39:D632–D636. doi: 10.1093/nar/gkq918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, et al. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, et al. Current protocols in molecular biology. New York: John Wiley & Sons Inc; 1994. [Google Scholar]
- Betermier M. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res Microbiol. 2004;155:399–408. doi: 10.1016/j.resmic.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Betermier M, Duharcourt S, Seitz H, Meyer E. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol Cell Biol. 2000;20:1553–1561. doi: 10.1128/mcb.20.5.1553-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Lynch M. Where do introns come from? PLoS Biol. 2008;6:e283. doi: 10.1371/journal.pbio.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Wurmser F, Potekhin AA, Przybos E, Lynch M. Genetic diversity in the Paramecium aurelia species complex. Mol Biol Evol. 2009;26:421–431. doi: 10.1093/molbev/msn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WJ, et al. Intron evolution and information processing in the DNA polymerase alpha gene in spirotrichous ciliates: a hypothesis for interconversion between DNA and RNA deletion. Biol Direct. 2007;2:6. doi: 10.1186/1745-6150-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, et al. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: a somatic view of the germline. Genome Res. 2008;18:585–596. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gao X, Lynch M. Ubiquitous internal gene duplication and intron creation in eukaryotes. Proc Natl Acad Sci U S A. 2009;106:20818–20823. doi: 10.1073/pnas.0911093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- Gratias A, Betermier M. Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie. 2001;83:1009–1022. doi: 10.1016/s0300-9084(01)01349-9. [DOI] [PubMed] [Google Scholar]
- Gratias A, Betermier M. Processing of double-strand breaks is involved in the precise excision of Paramecium internal eliminated sequences. Mol Cell Biol. 2003;23:7152–7162. doi: 10.1128/MCB.23.20.7152-7162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratias A, et al. Developmentally programmed DNA splicing in Paramecium reveals short-distance crosstalk between DNA cleavage sites. Nucleic Acids Res. 2008;36:3244–3251. doi: 10.1093/nar/gkn154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Jaillon O, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- Kapusta A, et al. Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet. 2011;7:e1002049. doi: 10.1371/journal.pgen.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Zagrodzinska B. Chromatin elimination—an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- Kowalczyk CA, Anderson AM, Arce-Larreta M, Chalker DL. The germ line limited M element of Tetrahymena is targeted for elimination from the somatic genome by a homology-dependent mechanism. Nucleic Acids Res. 2006;34:5778–5789. doi: 10.1093/nar/gkl699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Mayer KM, Forney JD. Identification of single nucleotide mutations that prevent developmentally programmed DNA elimination in Paramecium tetraurelia. J Eukaryot Microbiol. 2004;51:664–669. doi: 10.1111/j.1550-7408.2004.tb00606.x. [DOI] [PubMed] [Google Scholar]
- Mayer KM, Forney JD. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–604. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, et al. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA. 2006;12:2160–2170. doi: 10.1261/rna.201406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. The Paramecium-Aurelia complex of 14 sibling species. Trans Am Microsc Soc. 1975;94:155–178. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- Zufall RA, Robinson T, Katz LA. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol. 2005;304:448–455. doi: 10.1002/jez.b.21056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.