Abstract
The past decade has seen tremendous advances in our understanding of the genetic factors influencing response to a variety of drugs, including those targeted at treatment of cardiovascular diseases. In the case of clopidogrel, warfarin, and statins, the literature has become sufficiently strong that guidelines are now available describing the use of genetic information to guide treatment with these therapies, and some health centers are using this information in the care of their patients. There are many challenges in moving from research data to translation to practice; we discuss some of these barriers and the approaches some health systems are taking to overcome them. The body of literature that has led to the clinical implementation of CYP2C19 genotyping for clopidogrel, VKORC1, CYP2C9; and CYP4F2 for warfarin; and SLCO1B1 for statins is comprehensively described. We also provide clarity for other genes that have been extensively studied relative to these drugs, but for which the data are conflicting. Finally, we comment briefly on pharmacogenetics of other cardiovascular drugs and highlight β-blockers as the drug class with strong data that has not yet seen clinical implementation. It is anticipated that genetic information will increasingly be available on patients, and it is important to identify those examples where the evidence is sufficiently robust and predictive to use genetic information to guide clinical decisions. The review herein provides several examples of the accumulation of evidence and eventual clinical translation in cardiovascular pharmacogenetics.
I. Introduction
A. Pharmacogenetics
There is significant interpatient variability in drug response, much of which has a genetic basis. Specifically, genotype can influence drug metabolism, drug transport, and a person’s sensitivity to a drug. Pharmacogenetics involves applying DNA sequence data to predict drug response and to inform drug discovery and development. The term pharmacogenomics is generally considered a broader term referring to multiple genes affecting drug response, whereas pharmacogenetics refers to a more limited set of genes. However, the difference between the two is largely arbitrary, and for the purposes of this review and because in many cases we are discussing a single gene, pharmacogenetics will be the sole term used. To date, cardiovascular pharmacogenetics has predominately focused on genetic variants with implications for existing therapies. However, examples of pharmacogenetic application in cardiovascular drug discovery and development are beginning to emerge. The recent creation of the Encyclopedia of DNA Elements (ENCODE), which describes the biochemical functions of 80% of the components within the human genome, is expected to further our understanding of genetic contributions to cardiovascular drug pharmacokinetics and pharmacodynamics and lead to novel targets for drug development.
Among the earliest evidence of genetic influences of cardiovascular drug response in humans was the discovery that an inherited deficiency in protein C contributes to coumarin-induced skin necrosis (McGehee et al., 1984). Over the past decade, scientists have made significant strides toward uncovering genetic determinants of response to a number of cardiovascular agents, and we are beginning to see genetic information enter the clinical arena to guide cardiovascular therapy decisions. The evidence supporting the clinical utility of genotype-guided therapy for clopidogrel and warfarin are farthest along at this point. However, the pharmacogenetic evidence is accumulating with statins, β-blockers, and other drugs.
B. Candidate Gene versus Nonbiased Approaches to Discovery in Pharmacogenetics
Candidate gene approaches to genetic discovery have been applied for many years, although in disease genetics, this approach has largely been unsuccessful, with the exception of certain metabolic traits, e.g., lipids, uric acid. Rather, the unbiased approaches to discovery, e.g., those used in genome-wide association studies (GWAS) have been much more successful in identifying genetic variants that can be replicated across studies and populations. In contrast, candidate gene approaches have been considerably more successful in pharmacogenetics, particularly when the candidate gene encodes a major drug metabolizing enzyme, drug transporter, or protein target for the drug. Unlike diseases where the exact mechanisms are not known in most cases, the major (or sole) protein responsible for a drug’s action and the major drug-metabolizing enzymes and transporters are typically known and likely explain the greater success with candidate gene approaches in pharmacogenetics. In fact, the examples we discuss herein, CYP2C19 for clopidogrel, CYP2C9 and VKORC1 for warfarin, and SLCO1B1 for statins, were all first studied as candidate genes, with GWAS studies later confirming in all cases that these were the major genetic signatures for those respective drug responses. Although genetic variability in the major protein target or drug-metabolizing enzyme is not always associated with drug response, these are the protein categories in which there has been greatest success with a candidate gene approach in pharmacogenetics. It is also clear that GWAS can reveal novel discoveries (Tantisira et al., 2011) in pharmacogenetics, and moving forward, most discoveries in pharmacogenetics are likely to come from this non-biased approach.
C. Opportunities for Personalized Medicine
On the basis of the results of large, randomized clinical trials, important advances have been made in recent years to define appropriate treatments for certain diseases. On the basis of results of such trials, expert consensus panels then develop consensus guidelines for the treatment of a given disease. Such guidelines exist for nearly all the major cardiovascular diseases (Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults, 2001; Chobanian et al., 2003; Mansia et al., 2007; Hunt et al., 2009; Kushner et al., 2009; Anderson et al., 2011; Catapano et al., 2011; Hamm et al., 2011; McMurray et al., 2012). Since these guidelines are often based on data from large clinical trials, the same therapy is often recommended for all persons with a given disease, regardless of individual characteristics. Although these treatments are efficacious in overall clinical trial populations, there is no guarantee they will be safe or effective for an individual patient.
Personalized medicine takes into account the evidence base from clinical trials while also focusing on an individualized treatment approach, taking into account a variety of factors that are specific to the patient, including age, family history of disease, concomitant diseases, concomitant medications, lifestyle factors (e.g., smoking), among others. And increasingly, the term personalized medicine has been used to include the use of genetic information in arriving at an individualized or personalized treatment plan.
Pharmacogenetics offers the opportunity to predict drug response based on an individual’s DNA and treat accordingly, or personalized medicine. This is happening with increasing frequency in guiding treatment of a variety of cancers, but such approaches are also possible within cardiovascular disease. For example, genotyping for cytochrome P450 (P450) 2C19 variants may inform the most effective antiplatelet therapy for an individual undergoing coronary artery stent placement. Genotyping may also be applied to predict risk for drug toxicity, such as risk for myopathy with statins. Eventually, it may be possible to apply pharmacogenetics on a broader scale to choose the best combination of drugs to treat complex diseases, such as heart failure; however, pharmacogenetics is still in its infancy in this regard. This review will focus on pharmacogenetic data that are currently being used to guide treatment in the clinical setting within the area of cardiovascular disease.
II. Implementing Pharmacogenetics in the Clinical Setting
A. Drug Labeling and Pharmacogenetics
Over the past decade there has been increasing focus from the Food and Drug Administration (FDA), European Medicines Agency (EMA), Pharmaceuticals and Medical Devices Agency in Japan (PMDA), and other regulatory agencies on the impact of pharmacogenetics on drug efficacy, safety, and pharmacokinetics (Lesko and Zineh, 2010; Zineh and Pacanowski, 2011). Those at the FDA have sought to be leaders in the United States in this area by highlighting the potential clinical implications of genetic information while also hoping to advance use of genomics in drug development to help curb the attrition of drugs during late clinical trial phases. The FDA’s efforts in pharmacogenetics began in the early 2000s, with requests to the pharmaceutical industry to submit genomic information (under a voluntary genomic data submissions program) and through evaluation of approved drugs that might warrant a label revision based on pharmacogenetic data. There are now over 110 drugs for which there is pharmacogenetic information in the FDA-approved label, with the majority discussing germline polymorphisms and the remainder focused on somatic mutations and responses to anticancer drugs. An updated listing of drugs with pharmacogenetic labeling can be found on the FDA website (www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm) and a quick reference to information on the FDA label, along with easy access to the FDA-approved label, can be found in the Drug Labels section of PharmGKB (www.pharmgkb.org).
Various types of pharmacogenetic information are included in the FDA labels, ranging from information about the impact of genetic polymorphisms on pharmacokinetics of the drug to boxed warnings, the highest level of warning in the FDA label. Cardiovascular drugs are well represented on this list and include atorvastatin, carvedilol, clopidogrel, hydralazine, isosorbide dinitrate, metoprolol, prasugrel, pravastatin, propranolol, ticagrelor, timolol, and warfarin. As with other drugs, there is a range of pharmacogenetic information provided for these drugs, from a boxed warning on the clopidogrel label, informing clinicians of the effect of CYP2C19 genotype on efficacy, to the prasugrel label, which indicates that genetic variation in a variety of CYP enzymes, including CYP2C19, does not affect prasugrel’s pharmacokinetics or its efficacy. The cardiovascular drugs also include those that had pharmacogenetic label revisions postapproval, including the very old drug warfarin, and those where the pharmacogenetic information was part of the new drug application and so the pharmacogenetic information was part of the original label at the time of approval (e.g., prasugrel, ticagrelor). Pharmacogenetic label revisions by the FDA have created controversy in some quarters [such as arguments that the FDA has been premature in their label revisions (Nissen, 2011)] but have also helped to increase awareness of potential clinical implications of pharmacogenetics, thus advancing the movement toward clinical implementation for certain examples.
The EMA has also engaged in several activities related to pharmacogenetics, including formation of a Pharmacogenomics Working Party and publication of guidelines for pharmaceutical companies regarding pharmacogenetic methodologies for the evaluation of medicine products. Similar to the FDA, drug developers may seek advice related to pharmacogenetics through the Pharmacogenomics Working Party. The EMA also provides European Public Assessment Reports (EPARs), and some of these, including clopidogrel, contain pharmacogenetic information. Unlike the FDA, the EMA does not currently maintain a list of EPARs containing pharmacogenetic information. However, excerpts from some of the EPARs with pharmacogenetic information as well as access to the complete EPAR with the pharmacogenetic information highlighted are available through the Pharmacogenomic Knowledgebase (http://pharmgkb.org). The EMA labels appear alongside the FDA labels on the Pharmacogenomic Knowledgebase for an easy comparison between the two. Of note, many older drugs, such as warfarin, were never evaluated by the EMA, and as a result, drug labeling may vary by nation.
The PMDA in Japan has introduced a program that involves consultation with drug manufacturers to identify strategies for integrating pharmacogenetic biomarkers in the drug development process (Ichimaru et al., 2010). Pharmacogenetic information has been added to the labeling for many drugs marketed in Japan. In fact, the majority of drugs with FDA-approved pharmacogenetic labeling also contain pharmacogenetic information in their PMDA-approved package insert (Otsubo et al., 2012). However, in fewer instances is this information included in the warning or contraindication section of label. Cardiovascular agents comprise 9% of the agents in PMDA-approved pharmacogenetic labeling. There are also examples of cardiovascular drugs that have been relabeled to include genetic information in other Asian countries; however, pharmacogenetics in most Asian countries does not appear to have risen to a similar regulatory level as in Japan.
B. General Overview of Personalized Medicine through Pharmacogenetics
In the last one to two years there has been an increase in the number of institutional programs focused on clinical use of pharmacogenetic information to individualize (personalize) treatment regimens, but such an approach is far from new. For example, at St. Jude Children’s Research Hospital, TPMT genotyping has been used to guide treatment with 6-mercaptopurine in patients with acute lymphoblastic leukemia for over a decade (Crews et al., 2011). Many academic medical centers, including Mayo Clinic, have used CYP2D6 genotyping to guide treatment with antidepressants and antipsychotics since 2004 when the Amplichip received FDA approval and provided a clinically approved means for CYP2D6 genotyping (Mrazek 2010). However, despite isolated examples like these, there generally has not been broad acceptance of pharmacogenetic testing in clinical practice. Many thought the addition of pharmacogenetic information to FDA-approved labels of certain drugs would spur the clinical use but this did not occur in a broad way. As a result, pharmacogenetics researchers began to investigate reasons behind lack of clinical implementation of pharmacogenetics, and there is now general agreement about many of the barriers to such implementation, which are summarized in Table 1 (Lesko and Zineh, 2010; Crews et al., 2011; Johnson et al., 2012a; O'Donnell et al., 2012; Pulley et al., 2012). These barriers include lack of knowledge among clinicians about pharmacogenetic data or uncertainty about how to interpret and act on pharmacogenetic information. There are also financial and logistical barriers, which include costs for genotyping and potential lack of reimbursement, remembering when to order the test, and logistical challenges if the turnaround time results in the genetic information being delivered after a decision about drug therapy or dose has been made.
TABLE 1.
Potential barriers to clinical implementation of pharmacogenetics
| Knowledge barriers |
| • Lack of awareness of the pharmacogenetic data |
| • Uncertainty about how to interpret a pharmacogenetic test result |
| • Uncertainly about what action to take based on a pharmacogenetic test result |
| Logistical/financial barriers |
| • Remembering when to order a pharmacogenetic test in a busy clinical practice |
| • Turnaround time for pharmacogenetic test |
| • Cost of pharmacogenetic test |
| • Concerns about lack of reimbursement for pharmacogenetic test |
| Evidence barriers |
| • Lack of randomized controlled clinical trial data documenting benefit of pharmacogenetic guided treatment approach |
| • “Genetic exceptionalism” for genetic and pharmacogenetic tests |
Some also argue that the level of evidence in pharmacogenetics is not sufficient to warrant clinical implementation, based on the lack of robust randomized controlled trials documenting improved outcomes with a pharmacogenetic approach. Contrarily, there are others that argue there is a problem with “genetic exceptionalism,” whereby genetic and pharmacogenetic test results are held to a higher standard than other diagnostic tests (Relling et al., 2010; Altman 2011). In recognition of these barriers, investigators and clinicians at a variety of institutions have worked to overcome as many of them as possible (Crews et al., 2011; Johnson et al., 2012a; O'Connor et al., 2012a; O'Donnell et al., 2012; Pulley et al., 2012). Overcoming barriers of knowledge requires education of physicians and other clinicians on pharmacogenetics, but it seems clear that this alone will not be sufficient. Incorporation of genetic information into clinical practice will also require electronic clinical decision support tools that help the busy clinician interpret the genetic test information and provide recommendations about what to do with the pharmacogenetic information. Many recent personalized medicine programs have adopted this facilitative approach (Crews et al., 2011; Johnson et al., 2012a; O'Donnell et al., 2012; Pulley et al., 2012).
To help address issues of turnaround time and cost, many are also advocating a multigene, chip-based approach, such that genetic information is generated at a point of care and the genetic information remains available to guide drug therapy decisions over the course of the patient’s life span (Crews et al., 2011; Johnson et al., 2012a; O'Donnell et al., 2012; Pulley et al., 2012). Such an approach avoids the issue of turnaround time, because the data are available preemptively and it is more cost-effective than genotyping one single nucleotide polymorphism (SNP) or one gene at a time.
Evidentiary barriers are harder to address if the expectation is that pharmacogenetic information cannot be used in clinical practice unless there are randomized controlled trial data documenting the benefit of pharmacogenetic guided approaches. For clopidogrel and warfarin, such studies are ongoing, and the data should be available by the middle of this decade. However, many argue that if this high level of evidence is required in all cases, it will be impossible to use pharmacogenetics in all but a limited number of scenarios, because it is highly unlikely that such costly studies will be done routinely. In fact, many argue that such an approach is unnecessary, and rather the evidentiary bar should be that there is little likelihood for harm from a pharmacogenetic approach with some potential for benefit (Relling et al., 2010; Altman, 2011). More specifically, it has been suggested that pharmacogenetics should be viewed as a tool to guide therapy, not dissimilar from use of serum creatinine to guide dosing of renally cleared drugs. Particularly in scenarios where there are acceptable alternatives or the genetic information is being used to guide dose, it has been argued that noninferiority should be sufficient (Altman, 2011). In contrast, for situations where withholding therapy would be the outcome of the genetic test, then the level of evidence needs to be substantially higher. We will discuss examples from each of these below.
C. Cardiovascular Pharmacogenetics and Personalized Medicine
As noted above, consensus guidelines are important tools for establishing standards for treatment of disease, and cardiovascular diseases are considered by many to be the disease group with the largest body of evidence to guide treatment decisions (Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults, 2001; Chobanian et al., 2003; Hunt et al., 2009; Kushner et al., 2009; Anderson et al., 2011). However, the “one size fits all” approach that can result from consensus guidelines fails to take into account the substantial interpatient variability that exists in responses to medications. In fact, there is significant interpatient variability in response to cardiovascular agents, including warfarin, clopidogrel, statins, and β-blockers, among others, with some patients deriving no benefit and other patients experiencing intolerable or serious adverse effects with these agents. In the absence of genetic data, it is often difficult to predict how a patient will respond to a certain cardiovascular agent, and this is where pharmacogenetics presents potential opportunities for individualizing care. In some cases, nongenetic biomarkers are useful to predict drug response and select therapy, as is the case with plasma renin activity measurement in the setting of resistant hypertension (Olson et al., 2012). However, for other diseases, including thrombotic disease, there is no reliable biomarker to predict drug response prior to drug administration. Furthermore, neurohormonal biomarkers may exhibit diurnal variation or changes with body position. Thus, there are advantages to using a biomarker that is not subject to such variation, such as genotype.
Guidelines are helpful tools for assisting clinicians in interpreting the literature, and to address the absence of guidelines around clinical implementation of pharmacogenetics, the Clinical Pharmacogenetics Implementation Consortium (CPIC) was established in 2009 (www.pharmgkb.org/page/cpic). The objective of CPIC is not to recommend whether genetic testing should be conducted relative to a certain drug, but whether the data are sufficiently robust to use genetic information to guide therapy if the genetic data are available (Relling and Klein, 2011). The working premise for this group is that increasingly large amounts of genetic information will be available on patients, and soon the question will no longer be “should I order a pharmacogenetic test” but rather “I have the genetic data; should I use it to influence what I do clinically.” The CPIC guidelines are one line of evidence that some of the most robust cases for clinical implementation of pharmacogenetics come from the cardiovascular drugs. As of October 2012, CPIC has published six sets of pharmacogenetics guidelines, and three of these are for cardiovascular drugs (clopidogrel, warfarin, and simvastatin). This review focuses on the pharmacogenetic data for these three cardiovascular agents that are currently considered by CPIC to be clinically actionable. We also highlight examples of commercially available pharmacogenetic testing for which the data supporting its use are questionable.
III. Clopidogrel Pharmacogenetics
A. Clopidogrel Metabolism
Clopidogrel is a thienopyridine antiplatelet drug used in patients after an acute coronary syndrome or percutaneous coronary intervention to prevent future cardiovascular events. It works by binding to the platelet purinergic P2Y12 receptor and irreversibly inhibiting adenosine diphosphate-mediated platelet activation and aggregation for the life of the platelet (~10 days) (Fig. 1). In the United States in 2011, clopidogrel was the number two selling drug among all prescription drugs by dollars ($6.8 billion) and number six by prescriptions (28 million), making it a major drug in the treatment of cardiovascular disease.
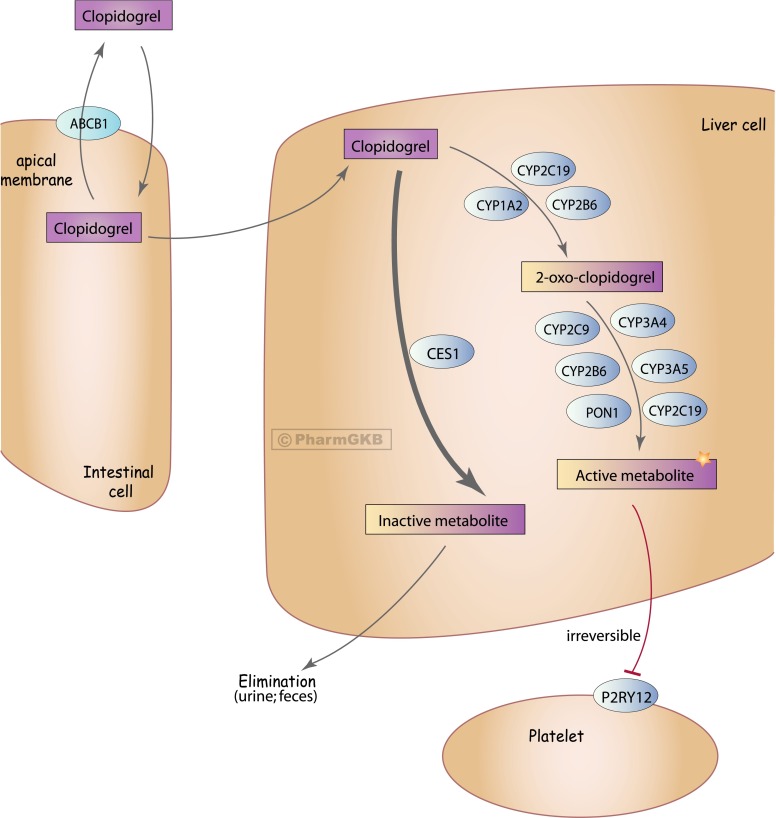
Fig. 1.
Clopidogrel pharmacokinetic and target pathway. ABCB1, ATP-binding cassette, subfamily B, member 1, encodes P-glycoprotein; CES1, carboxylesterase 1; CYPXXX, gene name and encoded proteins for the various cytochrome P450 enyzmes involved in metabolism of clopidogrel; P2RY12, purinergic receptor P2Y12; PON1, paroxonase 1. Copyright to the Pharmacogenomics Knowledge Base (PharmGKB), with permission given by PharmGKB and Stanford University to reproduce this figure.
Clopidogrel is a prodrug and undergoes rapid metabolism primarily (approximately 85%) by carboxylesterase 1 to an inactive metabolite. The activation of clopidogrel is largely mediated by various cytochrome P450 enzymes, as shown in Fig. 1, in a two-step enzymatic activation process. The first activation step is thought to be mediated largely by CYP2C19, CYP1A2, and CYP2B6, generating the intermediate metabolite 2-oxo-clopidogrel. This is followed by metabolism to the active metabolite, for which CYP3A, CYP2C19, CYP2B6, and CYP2C9 are involved (Kazui et al., 2010; Sangkuhl et al., 2010; Floyd et al., 2012). Genetic studies suggested a role for paroxonase 1 in the metabolic activation, but more recent studies have called this into question (see details below).
B. CYP2C19 Genetic Polymorphisms
That genetic polymorphisms contribute to interpatient variability in drug metabolism via CYP2C19 was first recognized in 1994 (de Morais et al., 1994). That the common loss of function polymorphism (*2) for CYP2C19 was an important determinant of clopidogrel effect was not recognized until 2006 (Hulot et al., 2006), nearly a decade after its approval by the FDA. It was not until these and other data were published that the critical role of CYP2C19 in the bioactivation of clopidogrel was appreciated.
CYP2C19 contains a common loss of function polymorphism, called *2 (rs4244285, c.681G>A, p.P227P), which creates a cryptic splice site and premature stop codon 20 amino acids later (de Morais et al., 1994), resulting in loss of function. Allele frequencies for this single nucleotide polymorphism (SNP) are approximately 0.12, 0.15, and up to 0.35 in those of European, African, and Asian ancestry, respectively. This means that 25–30% of those of European and African ancestry and up to 60% of Asians carry at least one loss of function *2 allele, which significantly impacts their ability to metabolize drugs via the CYP2C19 enzyme (Scott et al., 2012). Other less frequent loss of function alleles include *3, *4, *5, *6, *7, and *8. Among these, *3 and *8 are the most frequent. Details of these and other genetic polymorphisms that influence metabolism via CYP2C19 are comprehensively reviewed in the PharmGKB very important pharmacogene CYP2C19 summary (Scott et al., 2012). Collectively, these polymorphisms result in approximately 2–5% of European and African ancestry individuals and 15% of Asians being homozygous for a loss of function allele, categorized as poor metabolizers for CYP2C19, with an additional approximately 25–35% of whites and blacks and 45–50% of Asians who are heterozygous for loss of function alleles or intermediate metabolizers. Thus a large portion of the population has impaired capacity to metabolize via CYP2C19, which can have clinically important implications for certain drugs, including clopidogrel.
CYP2C19 also contains a promoter region polymorphism, called *17, which is also common in the population, with minor allele frequencies in European, African, and Asian populations of approximately 0.21, 0.16, and 0.03, respectively (Scott et al., 2012). This SNP is located in a transcription factor binding site and has been associated with increased metabolism of a variety of CYP2C19 substrates (Sim et al., 2006; Baldwin et al., 2008; Rudberg et al., 2008). As such it is considered a gain of function polymorphism. Although this polymorphism has detectable effects on pharmacokinetics of CYP2C19 substrates, the magnitude of effect is smaller than for the loss of function alleles, and the clinical implications of the *17 allele remain unclear (Li-Wan-Po et al., 2010).
C. CYP2C19 and Clopidogrel Pharmacogenetics
1. Pharmacokinetics and Pharmacodynamics (Ex Vivo Platelet Reactivity) Data
The relationships between CYP2C19 genotype and pharmacokinetics, antiplatelet effect, and cardiovascular outcomes are comprehensively summarized in the clopidogrel CYP2C19 CPIC guidelines (Scott et al., 2012). In 2006, the first evidence of the link between CYP2C19 genotype and clopidogrel efficacy emerged, with the publication of a small study in healthy volunteers that identified that CYP2C19 variant carriers (*2 or *3) had significantly less antiplatelet effect from clopidogrel as assessed by ex vivo platelet reactivity studies (Hulot et al., 2006). Specifically, the individuals carrying a loss of function allele had significantly higher on-treatment platelet reactivity. This was followed by a series of additional healthy volunteer studies that confirmed the impact of CYP2C19 genotype on the antiplatelet effect and provided evidence for loss of function variant carriers having significantly lower plasma concentrations of the clopidogrel active metabolite(Brandt et al., 2007; Kim et al., 2008; Umemura et al., 2008). Numerous studies in patients with cardiovascular disease further documented that CYP2C19 loss of function carriers have reduced concentrations of active metabolite and less antiplatelet effect (Mega et al., 2009; Varenhorst et al., 2009; Collet et al., 2011; Hulot et al., 2011; Gong et al., 2012; Price et al., 2012). As a result, there is little to no discrepancy in the literature regarding the impact of the CYP2C19 loss of function alleles on clopidogrel active metabolite concentrations or on-treatment platelet reactivity.
Whether the effect of genotype on active metabolite concentrations can be overcome with increased clopidogrel dosing has been the subject of several studies. The most informative of these was a study by Mega et al. (2011) in which they tested the effect of double, triple, and quadruple doses (150, 225, 300 mg daily, respectively) compared with usual doses (75 mg daily) in *1/*2 and *2/*2 individuals. Individuals with the *1/*1 genotype were also included as the comparator group and treated with 75 mg and 150 mg daily, and all assessments were based on on-treatment platelet reactivity. The data from this study show that in all genotype groups, each dose increment leads to reduced platelet reactivity, yet for *2/*2 individuals even fourfold higher doses cannot generate an antiplatelet effect that matches what is achieved in *1/*1 individuals on 75 mg daily. This suggests that one cannot overcome the homozygous loss of function genotypes with increased dose. In contrast, for the heterozygotes (*1/*2), they found that 225 mg or 300 mg daily provided an antiplatelet effect similar to that achieved in *1/*1 individuals at the normal 75-mg dose. On the other hand, 150 mg daily was not enough to overcome the effect of the single loss of function allele. This is supported by other studies that suggest that 150 mg daily does not produce a sufficient antiplatelet effect in *2 carriers (Price et al., 2012). Clinically, this means that tripling the dose for clopidogrel in those carrying a single loss of function allele may be a viable option. However, the data do not support dose escalation as a means of achieving sufficient platelet inhibition in homozygotes for loss of function alleles.
There are fewer data on the impact of CYP2C19*17 on active drug concentrations, although existing data supporting that *17 is associated with higher concentrations of active metabolites (Tiroch et al., 2010). That the *17 allele is associated with increased CYP2C19 activity is further supported by a number of studies showing a gene-dose effect, with *17/*17 patients having the greatest antiplatelet effect (lowest on-treatment platelet reactivity), with intermediate effects for *1/*17 relative to *1/*1 (Sibbing et al., 2010; Harmsze et al., 2012; Li et al., 2012). A recent meta-analysis of *17 found that only 38% of *17 carriers have high on-treatment platelet reactivity compared with 51% of noncarriers (P = 0.0003) (Li et al., 2012).
Overall, the data strongly support that CYP2C19 loss of function alleles lead to significantly lower active metabolite concentrations and reduced antiplatelet efficacy, whereas the *17 allele has the opposite effect, with higher active metabolite concentrations and greater antiplatelet effect. One area of debate is the effect when an individual is *2*17 or carries both a loss of function and gain of function allele. Two clopidogrel studies provide insight into this question based on platelet reactivity data. These studies both suggest that *2 has a much greater impact than *17, and so in individuals carrying one of each allele, the *2 allele predominates, such that the patient’s antiplatelet response is likely to be similar to that of a patient who is *1*2 (Sibbing et al., 2010; Harmsze et al., 2012).
2. Clinical Outcomes Data
That CYP2C19 genotype might impact cardiovascular outcomes in patients treated with clopidogrel became evident in late 2008 with the simultaneous publication of two articles in the New England Journal of Medicine and one in Lancet (Collet et al., 2009; Mega et al., 2009; Simon et al., 2009). These articles indicated risk of major adverse cardiovascular events (MACE) (e.g., stent thrombosis, recurrent myocardial infarction, stroke, death) based on CYP2C19 loss of function genotype, with two of the articles suggesting risk in carriers of a single *2 (or other) allele, and the third suggesting the risk was confined to *2 homozygotes. A variety of additional articles also suggested marked risk for stent thrombosis in patients under percutaneous coronary intervention (PCI), with hazard ratios for stent thrombosis in many of the studies exceeding 3.0 (Sibbing et al., 2009; Collet et al., 2009; Mega et al., 2009; Simon et al., 2009). Later in 2009, Shuldiner et al. (2009) published the first genome-wide association study (GWAS) with clopidogrel. They found that only the CYP2C19 genomic region achieved a genome-wide significant association with antiplatelet response. They further documented an association of CYP2C19 genotype with cardiovascular outcomes in patients treated with clopidogrel. On the basis of accumulating evidence relating CYP2C19 genotype to cardiovascular outcomes from these and other studies, in March 2010 the FDA issued a boxed warning about reduced clopidogrel efficacy based on CYP2C19 genotype, discussed in detail below.
After the publication of a relatively long string of articles that fairly consistently linked CYP2C19 genotype and cardiovascular outcomes, pharmacogenetic analyses of two clopidogrel clinical trials raised questions about the previous associations (Paré et al., 2010).These analyses arose from the CURE and ACTIVE-A trials. CURE was a study of clopidogrel added to standard therapy in patients with non-ST segment elevation myocardial infarction (NSTEMI), in which the vast majority (80%) did not undergo PCI. Clopidogrel produced a 20% reduction in risk of MACE and has since become standard therapy in this population (Yusuf et al., 2001). In the genetic subset from CURE, no evidence of an association between CYP2C19 genotype and cardiovascular outcomes was found (Paré et al., 2010). ACTIVE-A was a study testing the efficacy of clopidogrel plus aspirin versus aspirin alone in patients with atrial fibrillation who could not tolerate warfarin (Connolly et al., 2006). Superiority of warfarin compared with clopidogrel plus aspirin had been previously documented (Connolly et al., 2006). In the primary trial, clopidogrel plus aspirin exhibited modest benefit (11% risk reduction) over aspirin and increased risk of bleeding (Connolly et al., 2009), although clopidogrel is not currently indicated in treatment of atrial fibrillation. In the genetic substudy, there was no evidence of an association between CYP2C19 genotype and cardiovascular outcomes (Paré et al., 2010). The results from these studies challenged the earlier publications suggesting risk for reduced protection from MACE with clopidogrel based on genotype.
3. Meta-Analyses of CYP2C19 and Clopidogrel
a. CYP2C19 loss of function alleles
Accumulating evidence for CYP2C19 and clopidogrel, and some inconsistencies in study results, led to a series of meta-analyses since 2010. Meta-analyses published since 2010 are summarized in Table 2 (Hulot et al., 2010; Mega et al., 2010; Bauer et al., 2011; Holmes et al., 2011; Liu et al., 2011; Sofi et al., 2011; Li et al., 2012; Zabalza et al., 2012). As is evident based on the samples sizes for the various meta-analyses, a variety of approaches have been taken in selection of studies to be included in the analysis. Some studies have focused on the published literature, whereas others have also used data in abstracts and public databases. Some have been focused on specific patient populations, whereas others have been broader in their inclusion. Additionally, different meta-analytic approaches have been taken.
TABLE 2.
Meta-analyses of CYP2C19—clopidogrel outcomes studies
| Citation | Pt Population in Meta-analysis (Sample Sizea) | CYP2C19*2c Genotype Relative Risk for MACE (95% CI) | CYP2C19*17 Genotype Relative Risk for MACE, Stent Thrombosis (ST), and/or Bleeding (95% CI) | CYP2C19*2c Genotype Relative Risk for Stent Thrombosis (95% CI) |
|---|---|---|---|---|
| Hulot et al., 2010 | Established CAD (n = 11,959) | *2 carrier: 1.29 (1.12–1.49) | NA | n = 4905 |
| *1*2: 1.59 (0.88–2.88) | *2 carrier: 3.45 (2.14–5.57) | |||
| *2*2: 2.05 (1.15–3.63) | *1*2: 3.34 (1.84–5.93) | |||
| *2*2: 4.68 (1.55–14.11) | ||||
| Sofi et al., 2011d | Established CAD (n = 8043) | *2 carrier: 1.96 (1.14–3.37) | NA | n = 4.975 |
| *2 carrier: 3.82 (2.23–6.54) | ||||
| Mega et al., 2010 | Aggressively managed CAD patients (n = 9685) | *2 carrier: 1.57 (1.13–2.16) | NA | n = 6094 |
| *1*2: 1.55 (1.11–2.17) | *2 carrier: 2.81 (1.81–4.37) | |||
| *2*2: 1.76 (1.24–2.50) | *1*2: 2.67 (1.69–4.22) | |||
| *2*2: 3.97 (1.75–9.02) | ||||
| Bauer et al., 2011 | Established CAD (n = 18,529) | *2 carrier: 1.11 (0.89–1.39) | *17 carrier (n = 9128) | n = 19,328 |
| MACE: 0.93 (0.75–1.14) | *2 carrier: 1.77 (1.31–2.40) | |||
| ST: 0.99 (0.60–1.62) | ||||
| Liu et al., 2011 | Established CAD (n = 24,120) | *2 carrier: 1.26 (1.06–1.50) | *17 carrier (n = NR) | ST n = NR |
| MACE: 0.82 (0.69–0.98) | *2 carrier: 2.58 (1.77–3.77) | |||
| Holmes et al., 2011 | Unselectedb (n = 26,251) | *2 carrier: 1.18 (1.09–1.28) | NA | n = 16,008 |
| *2 carrier: 1.75 (1.50–2.03) | ||||
| Li et al., 2012 | Established CAD (n = 9428) | NA | *17 carrier: (n = 9428) MACE: 0.82 (0.72–0.94) | NA |
| Bleeding: 1.25 (n = 12,228)(1.07–1.47) | ||||
| Zabalza et al., 2012d | Established CAD (n = 16,360) | 1.23 (0.97–1.55) | *17 carrier (n = 7660): | n = 8686 |
| MACE: 0.75 (0.66–0.87) | *2 carrier: 2.24 (1.52–3.30) | |||
| Bleeding: 1.26 (1.05–1.50) |
NA, not assessed; NR, not reported; ST, stent thrombosis.
Indicates largest sample size in the analysis; usually for *2 carrier status.
This meta-analysis included all studies of clopidogrel pharmacogenetics, independent of patient disease status or indication for clopidogrel; excluded studies with stent thrombosis as primary endpoint. Data reported are treatment only analysis with fixed effects model.
*2 refers to any loss of function allele, among which *2 represents the vast majority.
Meta-analyses are listed by online publication year. In some cases the final print manuscript was published the following year.
In general the meta-analyses summarized in Table 2 provide insight in four areas: the impact of the *2 and other loss of function alleles on MACE, and on stent thrombosis, and the impact of the gain of functional *17 allele on MACE and bleeding.
The one area for which the data are highly consistent and considered indisputable is the risk for stent thrombosis in *2 carriers. Seven meta-analyses have analyzed stent thrombosis risk after PCI and all found a significant increased risk in *2 carriers, with hazard ratios ranging from 1.75 to 3.82, with a median hazard ratio of 2.58. Two of the meta-analyses provide data on *1*2 and *2*2 also, and in both of these cases, the risk in the homozygous loss of function patients is higher than in the heterozygotes, but the heterozygotes are also at significant risk. Collectively there appears to be little debate regarding the risk of stent thrombosis, an event associated with significant morbidity and mortality, in carriers of a CYP2C19 loss of function allele treated with clopidogrel. This suggests the potential clinical value for genotyping patients undergoing a PCI and considering alternative antiplatelet therapy in those carrying a *2 allele.
There is much greater heterogeneity across studies and the meta-analyses related to MACE in patients treated with clopidogrel. Among the seven meta-analyses summarized in Table 2, all report point estimates greater than 1, ranging from 1.11 to 1.96 among *2 carriers, with a median of 1.26. However, in two of these the association was not statistically significant and many have argued that even if statistically significant, these effect sizes are not large enough to be of clinical relevance. Many of the meta-analyses report significant heterogeneity between the studies and/or suggest a small study bias whereby the smaller studies, mostly published earlier, revealed greater effect sizes than the larger studies (Hulot et al., 2010; Bauer et al., 2011; Holmes et al., 2011; Sofi et al., 2011; Zabalza et al., 2012). This is in contrast to the analyses on stent thrombosis where in many analyses heterogeneity and small study bias appear to be absent. Thus, the interpretation by some is that there is no detectable impact of CYP2C19 genotype on cardiovascular outcomes (Nissen 2011). Others, including many pharmacologists, have a slightly different interpretation of these data (Johnson et al., 2012b). This interpretation relies on our understanding of the pharmacology and bioactivation of clopidogrel and magnitude of benefit. Specifically, there are clear data on the role of CYP2C19 on bioactivation of clopidogrel (vide supra), and if there are reduced concentrations of the active metabolite, then it follows that efficacy will be reduced. However, the magnitude of benefit of clopidogrel is variable across populations, and thus very large sample sizes would be needed to detect the impact of loss of function polymorphisms in a population where the magnitude of benefit is small. Specifically, clinical trial data suggest the greatest benefit of clopidogrel is in patients undergoing PCI, where such treatment is associated with 35–80% reductions in risk. In contrast, in lower risk CAD patients, such as those in the CURE trial described above, and where PCI was uncommon, clopidogrel was associated with only 20% risk reduction compared with placebo (Yusuf et al., 2001). The atrial fibrillation population from ACTIVE-A obtained an 11% risk reduction with clopidogrel (Connolly et al., 2009). Thus, it is not surprising that a reduction in clopidogrel active metabolite might have a difficult-to-detect effect in patient populations where the magnitude of benefit is small, but is more evident for the populations where the benefit is greatest. The greatest benefit of clopidogrel is likely during the first month after PCI, when risk for stent thrombosis is highest; thus it is not surprising that the impact of the loss of function polymorphisms is most evident for this outcome.
On the basis of the above assessment, we have argued that the clinical relevance of CYP2C19 pharmacogenetics with clopidogrel is indication specific, meaning that the greatest clinical value of genetic testing is in patients undergoing PCI (Johnson et al., 2012b). It is these patients who obtain the greatest benefit from treatment, who are at risk for stent thrombosis, and thus are at greatest risk from loss of CYP2C19 function. The consistency of the data for risk of stent thrombosis across all meta-analyses supports this view. In addition, if patients who are post-PCI are considered those of greatest clinical relevance for clopidogrel pharmacogenetics, the meta-analysis by Mega et al. (2010) is most useful. This is the only meta-analysis that focused on high-risk, aggressively managed patients, 91% of whom underwent a PCI. On the basis of the data from this meta-analysis, the risk of stent thrombosis with *2 is substantial, as in the other studies. However, the risk for MACE is also substantially increased among heterozygotes (hazard ratio of 1.55) and homozygotes (HR of 1.76) for carriers of a loss of function allele.
Collectively, the outcomes data for CYP2C19 loss of function alleles suggest there is substantial and clinically relevant risk for stent thrombosis and MACE after PCI. The clinical impact of the loss of function alleles is less evident and likely of smaller magnitude in those patients treated with clopidogrel for other indications.
b. CYP2C19 *17 (gain of function) allele
There is a smaller literature base for the *17 gain of function polymorphism than for the loss of function alleles, although it too has been evaluated in a number of studies and subject to meta-analyses. The *17 allele potentially represents the opposite of the loss of function alleles, namely the potential for greater generation of active metabolite, greater efficacy, and perhaps increased bleeding risk as a result of higher levels of active metabolite. As summarized in Table 2, there are four meta-analyses focused on the *17 allele, with sample sizes in the 8000 to 12,000 range. Among these meta-analyses, three of the four suggest significantly more benefit in *17 than in non-*17 carriers, with average risk reduction of approximately 18%. Two of the meta-analyses evaluated bleeding risk, and both found approximately 25% increased risk of bleeding in *17 carriers.
Collectively, the data suggest statistically different efficacy and bleeding risk in carriers of the *17 allele. Whether these differences are clinically relevant could be debated. It is also important to note that attributable to linkage disequilibrium, the *17 and *2 variants reside on different alleles. As a result, it is possible that the literature on the *17 polymorphism is to some degree simply reflecting the impact of the absence of a *2 allele. Because the analyses to date for *17 do not control for the *2 allele, it is difficult to have clarity on this issue. It will be critical moving forward that analyses focused on these two polymorphisms are done in a way that the alternate is a covariate in the analysis, so that the precise role, particularly for *17, is clearer.
D. Other Genes Associated with Clopidogrel Efficacy or Safety
1. PON1
In 2011, a high profile manuscript suggested that CYP enzymes did not play a major role in the bioactivation of clopidogrel, but rather the esterase paroxonase 1 (encoded by PON1) was the rate-limiting enzyme responsible for the conversion of 2-oxo-clopidogrel to the active metabolite (Bouman et al., 2011). They additionally identified 41 cases of nonfatal stent thrombosis and 71 stent thrombosis-free controls from a large cohort of patients treated with PCI and found the functional, nonsynonymous Gln192Arg polymorphism was associated with clopidogrel-associated stent thrombosis, with the patients with a Gln192Gln polymorphism having a hazard ratio of 12.9 for risk of stent thrombosis versus patients with an Arg192Arg polymorphism. They further supported the clinical association in an independent cohort, where the hazard ratio for stent thrombosis was 10.6 for Gln192Gln patients. Taken together with previous findings, these data suggested perhaps there were two major genes influencing efficacy of clopidogrel.
However, numerous follow-up studies have provided essentially no support for this original publication. Specifically, numerous studies found no relationship between PON1 genotype and on-treatment platelet reactivity, including a meta-analysis that included 17 studies (Campo et al., 2011; Lewis et al., 2011; Trenk et al., 2011; Reny et al., 2012). Likewise, numerous additional studies have found no link between stent thrombosis or cardiovascular events during clopidogrel treatment based on PON1 genotype, including a meta-analysis of 11 studies (Lewis et al., 2011; Simon et al., 2011; Trenk et al., 2011; Delaney et al., 2012; Pare et al., 2012; Reny et al., 2012). Although it is difficult to explain the original compelling findings in the context of the multiple subsequent studies, the totality of the data does not support a role for the PON1 Gln192Arg polymorphism on the antiplatelet effect or cardiovascular outcomes associated with clopidogrel therapy.
2. ABCB1
ABCB1 encodes P-glycoprotein, an ATP-binding cassette (ABC) efflux transporter that plays an important role in the transport of many drugs. A synonymous polymorphism in the cytoplasmic loop of the transporter (c.3435 C>T) has been commonly studied, with noted, but inconsistent associations for many drugs. Early studies of this polymorphism in patients treated with clopidogrel suggested the T allele (particularly TT homozygotes) had lower active metabolite concentrations (Taubert et al., 2006) and an approximately 75% increased risk of MACE (Simon et al., 2009; Mega et al., 2010). The influence of this genotype has therefore been tested in a variety of other studies and a recently published meta-analysis that focused on patients treated with clopidogrel for documented CAD, many of whom underwent PCI (Su et al., 2012). On the basis of data from four studies with 358 patients, there was no association between high on-treatment platelet reactivity and ABCB1 genotype (OR 1.01; 95% CI 0.51–1.97). There was also no relationship with long-term MACE (after >1 year of treatment), with an OR 1.09 (95% CI 0.77–1.54). Evaluation of early cardiovascular events (within approximately 1 month) suggested potential increased risk in T allele carriers, with an OR of 1.48 (95% CI 1.06–2.06). However, stent thrombosis, myocardial infarction, stroke, all-cause mortality, and bleeding were not different by ABCB1 genotype. Although the number of studies and patients in each analysis was relatively small, overall the data do not provide a compelling argument for the influence of ABCB1 genotype on clopidogrel efficacy.
E. Clinical Implementation of Clopidogrel Pharmacogenetics
1. Drug Label Warnings
In March 2010, after the publication of a variety of studies suggesting risk based on CYP2C19 genotype, the FDA issued a boxed warning indicating potential for reduced efficacy (increased adverse cardiovascular outcomes) based on CYP2C19 genotype. Boxed warnings are the highest level of warning in the FDA-approved product label and are typically used to draw special attention of clinicians to issues of serious concern for the drug. The text of the clopidogrel boxed warning is shown in Table 3. The warning is limited to “poor metabolizers” (i.e., patients homozygous for loss of function alleles, e.g., *2*2) and focuses on post-acute coronary syndrome and patients undergoing PCI. The boxed warning is silent on heterozygous loss of function carriers (e.g., *1*2), but as noted above, additional data published since the boxed warning was issued suggest risk for patients with this genotype post-PCI. The boxed warning provides guidance on the data, suggesting risk based on CYP2C19, but does not mandate genetic testing, is not specific on the exact patients who should be targeted for genetic testing, and is vague on alternative treatment approaches in poor metabolizers.
TABLE 3.
FDA-boxed warning on clopidogrel (Plavix) product label
| WARNING: DIMINISHED EFFECTIVENESS IN POOR METABOLIZERS |
|---|
| • Effectiveness of Plavix depends on activation to an active metabolite by the cytochrome P450 (CYP) system, principally CYP2C19. |
| • Poor metabolizers treated with Plavix at recommended doses exhibit higher cardiovascular event rates after acute coronary syndrome (ACS) or percutaneous coronary intervention (PCI) than patients with normal CYP2C19 function. |
| • Tests are available to identify a patient's CYP2C19 genotype and can be used as an aid in determining therapeutic strategy. |
| • Consider alternative treatment or treatment strategies in patients identified as CYP2C19 poor metabolizers. |
The EMA and PMDA also approved pharmacogenetic-related labeling for clopidogrel. The EMA EPAR for clopidogrel states that CYP2C19 poor metabolizer status is associated with lower systemic exposure to the active metabolite of clopidogrel and a diminished antiplatelet response, which may manifest as higher cardiovascular event rates after myocardial infarction compared with the extensive metabolizer phenotype. The information in the PMDA-approved clopidogrel labeling is graded as level 5, meaning that it is for reference information only, compared with the level 1 (e.g., boxed warfarin) information in the FDA-approved label.
2. Consensus Statements on CYP2C19 Genotyping and Clopidogrel
Shortly after release of the boxed warning on the clopidogrel label, the American Heart Association/American College of Cardiology Foundation issued a clinical alert to provide physicians with guidance on the necessity of genotyping for CYP2C19 in clopidogrel-treated patients (Holmes et al., 2010). They expressed concerns about the lack of outcomes data documenting the benefit of routine genotyping and concluded the available literature did not support CYP2C19 genotyping for all patients being prescribed clopidogrel. They did, however, acknowledge the potential benefit for genotyping in patients at high risk for poor outcomes after PCI, such as patients with diabetes and patients with complex atherosclerotic disease. When CYP2C19 genotype is determined, alternative therapy is recommended in those with loss of function genotypes. They also suggested the potential role for platelet reactivity testing but acknowledge the role for platelet reactivity testing and genotyping, alone or in combination, requires further study.
Recent treatment guidelines for patients with acute coronary syndrome, those undergoing PCI, and those with non-ST segment elevation myocardial infarction (NSTEMI) or unstable angina have all commented on the role of CYP2C19 genotyping. In general, their recommendations are consistent with the original American Heart Association/American College of Cardiology Foundation Clinical Alert in that they recommend against mandatory or routine genetic testing in all patients being considered for treatment with clopidogrel (Anderson et al., 2011; Levine et al., 2011; Wright et al., 2011). The PCI guidelines suggest genotyping may be appropriate in patients at high risk for whom inadequate platelet inhibition with clopidogrel would increase the likelihood for poor clinical outcomes. Consistent with the literature, the NSTEMI/unstable angina guidelines are more negative regarding potential use of genotyping in this group of patients.
The CPIC and Dutch Pharmacogenetics Working Group (DPWG) have also published guidelines on clopidogrel pharmacogenetics. The guidelines above focus primarily on whether or when pharmacogenetic testing should be ordered and what decisions might be made based on the genotype. CPIC and DPWG guidelines have a slightly different focus; they do not specifically comment on whether the pharmacogenetic test should be ordered, but rather comment on whether the level of evidence is sufficiently strong to let the genetic information, if available, guide the approach to treatment. The CPIC guidelines recommend that in patients with acute coronary syndromes or PCI, CYP2C19 genotype can be used to guide therapy, with alternative antiplatelet therapy recommended as a strong recommendation in loss of function homozygotes and a moderate recommendation for heterozygotes. They suggest that if one is considering whether to genotype, consideration could be given to testing all patients undergoing PCI or focusing only on those at high risk for poor outcomes post-PCI. Similarly, the DPWG recommends considering an alternative drug, such as prasugrel, for individuals with a CYP2C19*2 or *3 allele.
Collectively the guidelines all suggest that it is reasonable to consider CYP2C19 genotyping in patients at high risk for poor outcomes after PCI and that alternative antiplatelet therapy should be considered in those for whom the genotype poses potential risk for reduced antiplatelet efficacy. There is generally agreement that, at present, genotyping should not be mandated and cannot be recommended for routine use in patients at lower risk, such as those without PCI. This approach reduces the population size in whom clopidogrel can be guided by genotype but balances the use of these data in the populations likely to derive the greatest benefit from genotype data against the lack of evidence to support such an approach in patients at lower risk. Ongoing clinical trials should help further clarify the precise patient populations in whom CYP2C19 genotyping is most appropriate to guide treatment decisions with clopidogrel.
3. Examples of CYP2C19 Clopidogrel Pharmacogenetic Clinical Implementation
On the basis of the available data, some centers have elected to begin genotyping CYP2C19 in the clinical setting. Scripps Health may have been the first medical center to make CYP2C19 genotyping routinely available for those patients undergoing PCI when they launched their initiative in October 2009 (http://www.scripps.org/news_items/3521-scripps-becomes-first-in-u-s-to-offer-genetic-tests-to-stent-patients). The genotype test was made available as a clinical laboratory test that could be ordered by physicians.
In September 2010, the Vanderbilt University Medical Center initiated their Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment program, which is a pre-emptive, chip-based pharmacogenetics program initially targeted at patients in the cardiac catheterization laboratory who may require clopidogrel therapy (Pulley et al., 2012). As of November 2012, over 10,000 patients had been genotyped on the preemptive genotyping chip (D. Roden, personal communication), some of whom had their clopidogrel therapy altered based on being heterozygous or homozygous for loss of function CYP2C19 alleles.
The University of Florida initiated a preemptive chip-based program similar to that at Vanderbilt in June 2012 (Johnson et al., 2012a), with the early focus on patients in the cardiac catheterization laboratory who might require clopidogrel. The pharmacogenetic test is part of the standard order set for patients undergoing cardiac catheterization, indicating the physician decision to consider CYP2C19 genotyping a standard approach to these patients.
These examples highlight the clinical implementation of clopidogrel pharmacogenetics, which are consistent with the guidelines in focusing on those with clopidogrel treatment post-PCI. Other institutions are adopting similar approaches, and at some centers CYP2C19 genotyping for post-PCI clopidogrel is becoming a standard of care.
IV. Warfarin Pharmacogenetics
A. Warfarin Shortcomings
Even with the recent approval of newer agents, warfarin remains the mainstay therapy for oral anticoagulation, accounting for an estimated 1.6 million treatment visits and over $144 million in expenditures in the United States in the fourth quarter of 2011 (Kirley et al., 2012). Warfarin also remains one of the most challenging medications to manage despite over 60 years of experience with the drug. In fact, warfarin currently ranks as the leading drug-related cause of serious adverse events leading to hospitalization in the United States (Budnitz et al., 2011). Challenges with warfarin stem from its narrow therapeutic index, wide interpatient variability in the dose required to achieve optimal anticoagulation, and numerous drug and food interactions.
Warfarin is dosed to achieve an international normalized ratio (INR) of 2 to 3 for most indications. The dose required to obtain therapeutic anticoagulation varies among patients from as little as 0.5 mg/day to as much as 10 mg/day or higher (Wadelius et al., 2009). Inappropriate dosing increases the risk for warfarin-related adverse sequelae. In particular, when the INR exceeds 4 or falls below 1.5, the patient is at increased risk for bleeding and thromboembolic complications, respectively. These risks are highest during the initial months of warfarin therapy (Hylek et al., 2003, 2007). Although clinical factors contribute to warfarin response, they account for only about 15 to 20% of the interpatient variability in dose requirements and are usually insufficient in themselves to accurately predict the therapeutic dose (Gage et al., 2008).
B. Genetic Contributions to Warfarin Response
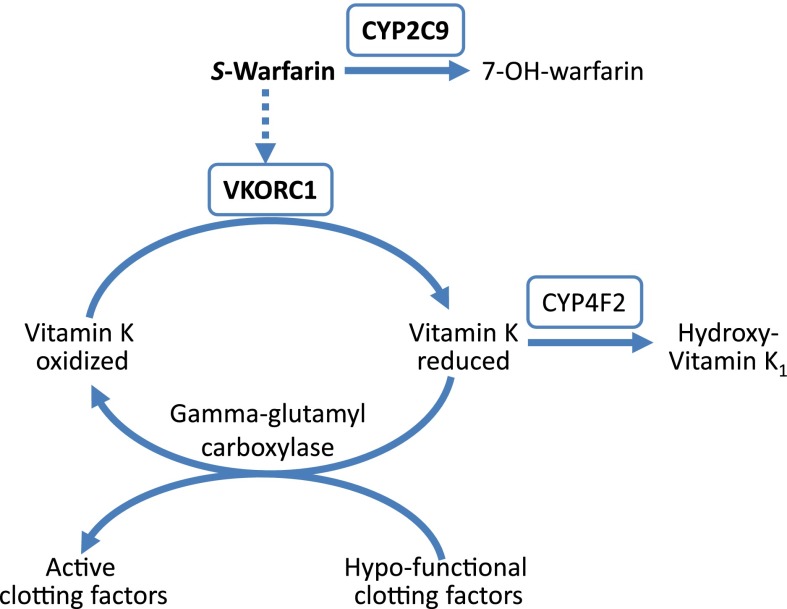
Both candidate gene and GWAS data over the past 10 years have clearly shown that genotype contributes to the interpatient variability in warfarin dose requirements (Aithal et al., 1999; Scordo et al., 2002; Rieder et al., 2005; Aquilante et al., 2006; Cooper et al., 2008; Takeuchi et al., 2009; Cha et al., 2010). CYP2C9 and VKORC1, which encode for vitamin K epoxide reductase complex 1, are the major genes influencing warfarin pharmacokinetics and pharmacodynamics, respectively (Fig. 2). The CYP2C9 enzyme metabolizes the more potent S-enantiomer of warfarin to the inactive 7-hydroxy warfarin protein, whereas VKORC1 is the target protein of warfarin, responsible for converting vitamin K epoxide to its reduced form, which is an essential cofactor in carboxylation and activation of clotting factors II, VII, IX, and X. The CYP4F2 gene, which encodes a vitamin K oxidase, also influences warfarin pharmacodynamics and dose requirements, but to a lesser extent. Although the majority of warfarin pharmacogenetic data is in the adult population, recent evidence supports a major influence of CYP2C9 and VKORC1 genotypes on warfarin dose requirements in children as well (Biss et al., 2012).
Fig. 2.
Warfarin pharmacokinetic and pharmacodynamic pathway. CYP2C9, cytochrome P450 2C9, metabolizes the more potent S-enantiomer of warfarin; VKORC1, vitamin K epoxide reductase complex 1, target site for warfarin; CYP4F2, cytochrome P450 4F2, metabolizes vitamin K. The genes for proteins shown in boxes are the primary genes influencing warfarin dose requirements.
1. CYP2C9
There are over 35 CYP2C9 alleles, of which the CYP2C9*2 and *3 alleles are most extensively studied and result from nonsynonymous SNPs in the gene’s coding region that are important for enzyme activity (Table 4) (Crespi and Miller, 1997; Ieiri et al., 2000). S-warfarin clearance is reduced by approximately 40% with CYPC9*2 and 75% with CYP2C9*3 (Takahashi et al., 1998; Scordo et al., 2002). Accordingly, warfarin dose requirements are approximately 20% lower with the CYP2C9*1/*2 genotype and 35% lower with the CYP2C9*1/*3 genotype compared with the CYP2C9*1/*1 genotype (Lindh et al., 2009). Doses of 1 mg/d or lower may be necessary in patients with the CYP2C9*3/*3 genotype to prevent over-anticoagulation and bleeding.
TABLE 4.
Reported prevalence of CYP2C9 and VKORC1 gene polymorphisms by ancestry (Limdi et al., 2008a,b; Cavallari et al., 2010; Limdi et al., 2010; Chan et al., 2012a)
| Allele | Prevalence* |
||
|---|---|---|---|
| European | African | Asian | |
| % | |||
| CYP2C9*2 (R144C) | 24 | 3-4 | <1 |
| CYP2C9*3 (I359L) | 12 | 1-3 | 6-8 |
| CYP2C9*5 (D360E) | <1 | 1-2 | <1 |
| CYP2C9*6 (10601delA) | <1 | 1 | <1 |
| CYP2C9*8 (R150H) | NR | 12 | <1 |
| CYP2C9*11 (R335W) | <1 | 3 | <1 |
| VKORC1 -1639G>A | 61 | 20 | 99 |
| CYP4F2 V433M | 40 | 14 | 40–42 |
Percentage of individuals who have the variant.
CYP2C9 allele frequencies differ by ancestry, with the CYP2C9*2 and *3 alleles occurring much more commonly among those of European versus African descent (Table 4). The CYP2C9*5, *6, *8, and *11 alleles predominate among those of African ancestry, with the *8 allele being the most common (Scott et al., 2009; Perera et al., 2011). Decreased S-warfarin clearance and lower warfarin dose requirements have been reported with these alleles (Dickmann et al., 2001; Tai et al., 2005; Limdi et al., 2008a; Perera et al., 2011; Liu et al., 2012). Overall, CYP2C9 genotype explains approximately 7 to 10% of the interpatient variability in warfarin dose requirements (Aquilante et al., 2006; Cavallari et al., 2010).
CYP2C9 variant alleles are also associated with an increased risk for over-anticoagulation and bleeding during warfarin therapy (Aithal et al., 1999; Limdi et al., 2008b). The risk for bleeding attributable to CYP2C9 polymorphism is highest during the initial months of warfarin therapy. However, there is evidence that it persists during chronic therapy, suggesting a need for close monitoring for signs and symptoms of bleeding throughout warfarin therapy for carriers of a variant CYP2C9 allele (Limdi et al., 2008b).
2. VKORC1
Nonsynonymous variants in the VKORC1 coding region lead to warfarin resistance, where very high doses (e.g., >20 mg/day) are necessary to obtain therapeutic anticoagulation (Rost et al., 2004). Warfarin resistance variants are rare in most populations, with the exception of the Ashkenazi Jewish population, in whom the p.Asp36Try variant occurs at a prevalence of approximately 8% (Scott et al., 2008). There are also two common SNPs located in the gene regulatory regions, c.−1639G>A and c.1173C>T, that contribute to the interpatient variability in dose observed in the general population of warfarin-treated patients. It is unclear if just one or both of these SNPs is functional (Wang et al., 2008). However, −1639G>A and 1173C>T occur in near complete linkage disequilibrium across populations, and thus, either may be considered for warfarin dose prediction (Limdi et al., 2010). The −1639AA, AG, and GG genotypes (or 1173TT, CT, and CC genotypes) are associated with high, intermediate, and low sensitivity to warfarin, respectively. Thus, compared with the −1639AG genotype, higher warfarin doses are needed with the GG genotype to effectively inhibit vitamin K reduction and subsequent clotting factor activation, whereas lower doses are needed with the AA genotype. The −1639A allele frequency varies by ancestry, as shown in Table 4, which largely explains differences in warfarin dose requirements between African, European, and Asian populations. Specifically, African populations have a higher frequency of the low sensitivity GG genotype and generally require higher doses than those of European descent, who have a high frequency of the intermediate sensitivity AG genotype. Asians have a high frequency of the high sensitivity AA genotype and usually require very low doses of warfarin. The VKORC1 −1639G>A variant explains approximately 20 to 25% of the dose variability in Europeans and Asians, but only 5 to 7% in African Americans (Aquilante et al., 2006; Cavallari et al., 2010; Limdi et al., 2010). The lesser variability explained in African Americans is primarily attributable to the lower −1639A allele frequency in this group (Limdi et al., 2010). Nonetheless, persons of African descent derive a similar benefit from genotype-guided warfarin dosing as others. Recent evidence suggests that an additional VKORC1 variant may further contribute to warfarin dose variability in African Americans (Perera et al., 2011).
3. CYP4F2
CYP4F2 metabolizes vitamin K to hydroxyl-vitamin K, resulting in less vitamin K available for clotting factor activation. The activity of CYP4F2 is reduced in individuals with the CYP4F2 p.Val433Met SNP (rs2108622, c.1297G>A), resulting in reduced vitamin K metabolism and greater vitamin K availability (McDonald et al., 2009). Studies in both Caucasians and Asians have shown higher warfarin dose requirements with the Met433Met genotype (Caldwell et al., 2008; Cha et al., 2010; Sagreiya et al., 2010; Gong et al., 2011; Wei et al., 2012). In a GWAS done in European and Asian populations, CYP4F2 emerges as a predictor of warfarin dose after controlling for CYP2C9 and VKORC1, explaining an additional 1 to 3% of the overall variability in warfarin dose requirements (Takeuchi et al., 2009; Cha et al., 2010).
C. Clinical Utility Data
Early clinical trials evaluating outcomes with genotype-guided warfarin dosing yielded inconsistent results (Anderson et al., 2007; Caraco et al., 2008; Burmester et al., 2011). However, these trials were limited by their small sample size, with each including fewer than 250 patients. Several larger multicenter, randomized, clinical trials assessing whether genotype-guided warfarin dosing is superior to traditional dosing are underway in the United States and Europe. Perhaps the most anticipated of these is the Clarification of Optimal Anticoagulation through Genetics (COAG) trial, in which over 1200 patients are being randomized to either a genotype-guided or clinical-guided warfarin dosing strategy (French et al., 2010). The study is planned for completion in 2013.
While we await the results of ongoing clinical trials, data from two large comparative effectiveness studies are available to support genotype-guided warfarin therapy. In the first study, nearly 900 patients starting warfarin therapy were offered free CYP2C9 and VKORC1 genotyping, with results provided to their physician with an interpretive report, and outcomes were compared with those from 2688 historical controls (Epstein et al., 2010). During the initial 6 months of warfarin therapy, patients who underwent genotyping had 31% fewer hospitalizations for any cause and 28% fewer hospitalizations for bleeding or thromboembolism compared with controls. More recently, the CoumaGen-II study compared genotype-guided warfarin dosing using one of two pharmacogenetic dosing algorithms in 504 total patients to standard dosing in a parallel control group (n = 1911) (Anderson et al., 2012). Patients in the genotype-guided arm were rapidly genotyped for CYP2C9 and VKORC1 variants, with results available to inform the first dose. The investigators reported that genotype-guided therapy, regardless of the algorithm used, was superior to standard dosing in reducing the percent of out-of-range INRs and the percent of INRs ≥ 4 or ≤ 1.5 at 3 months. The number of INR measurements was similar between groups, suggesting that the improvement in anticoagulation control in the genotype-guided group was not secondary to more intensive monitoring. There were also fewer serious adverse events, including hemorrhagic events, thromboembolic events, and death, at 3 months with genotype-guided therapy.
D. Clinical Implementation of Warfarin Pharmacogenetics
1. Warfarin Labeling
In August 2007, the FDA approved the addition of pharmacogenetic data to the warfarin labeling. The label states that lower doses “should be considered for patients with certain genetic variations in CYP2C9 and VKORC1 enzymes.” The warfarin labeling was further revised in January 2010 to include a dosing table based on CYP2C9 and VKORC1 genotypes (Table 5). The table may be used as a quick guide for clinicians to dose warfarin when genotype is available, realizing that clinical factors still need to be taken into account. Pharmacogenetic information is also included in the warfarin labeling approved by the PMDA in Japan and by regulatory body in Taiwan.
TABLE 5.
Recommended warfarin starting dose in mg/day according to VKORC1 and CYP2C9 genotypes per the FDA-approved warfarin labeling
| VKORC1 -1639 | CYP2C9 |
||||
|---|---|---|---|---|---|
| *1/*1 | *1/*2 | *1/*3 or *2/*2 | *2/*3 | *3/*3 | |
| GG | 5–7 | 5–7 | 3–4 | 3–4 | 0.5–2 |
| AG | 5–7 | 3–4 | 3–4 | 0.5–2 | 0.5–2 |
| AA | 3–4 | 3–4 | 0.5–2 | 0.5–2 | 0.5–2 |
2. CPIC Guidelines
Guidelines from expert consensus groups, including the American College of Chest Physicians and the American College of Medical Genetics, currently recommended against routine use of genetic testing to guide warfarin dosing (Flockhart et al., 2008; Guyatt et al., 2012). These groups traditionally rely on evidence from randomized, controlled clinical trials to guide therapy, and thus, their stances on warfarin genetic testing are not especially surprising. While we await results from ongoing clinical trials, CPIC guidelines are available to assist with genotype-guided warfarin dosing (Johnson et al., 2011b). These guidelines do not address when or who to genotype, leaving this to the discretion of the clinician. Rather they strongly endorse the use of genetic information to guide warfarin dosing when such information is available and provide guidance on how to interpret and apply such information. The guidelines recommend using one of two published dosing algorithms (Gage et al., 2008; Klein et al., 2009) to guide dosing decisions. Both algorithms were derived from large patient populations, account for both clinical and genetic factors, and are freely available at www.warfarindosing.org. Furthermore, both algorithms were shown to more accurately predict warfarin dose than other methods, including use of the table in the warfarin labeling (Finkelman, Gage et al., 2011). The algorithm by Gage et al. (2008) allows for refinement of dose estimation based on INR response to initial warfarin doses. In the event that computer access is unavailable, the pharmacogenetic table in the FDA-approved warfarin label may serve as an alternative guide to dosing.
3. Implementation in Clinical Practice
Although translation of warfarin pharmacogenetics into clinical practice has been slow to arrive, examples of such are beginning to emerge. The University of Illinois Hospital and Health Sciences Center recently announced efforts to routinely genotype all patients newly starting warfarin during hospitalization for CYP2C9 and VKORC1 variants (Hostettler, 2012). Genotypes are targeted to be available within 24 hours of the initial warfarin dose so that they may inform subsequent dosing. Residency-trained pharmacy fellows provide warfarin dosing recommendations, with dosing oversight provided by more senior clinical pharmacists with expertise in warfarin pharmacogenetics, and medical oversight provided by attending physicians from either the cardiology or hematology service.
As part of the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment program (Lieb et al., 2012), Vanderbilt University recently began utilizing chip-based genotype data on relevant warfarin pharmacogenetic genes to guide warfarin dosing in their patients. Likewise, the University of Florida is genotyping patients who provide consent for a broad array of variants across the genome that may be used to inform use of a number of drugs, including warfarin, in the future. CYP2C9, VKORC1, and CYP4F2 are included on their personalized medicine custom array (Johnson et al., 2012a) and expect to clinically implement warfarin pharmacogenetics in the near future.
V. Statin Pharmacogenetics
A. SLCO1B1 and Statin-Induced Myopathy
A number of large randomized controlled trials have demonstrated significant reductions in coronary events and stroke with statin therapy for both primary and secondary prevention of cardiovascular disease (Baigent et al., 2005). Thus, statins are among the most widely prescribed medications and are generally safe and well tolerated. Myopathy is the most common side effect, with symptoms ranging from mild myalgias without creatine kinase (CK) elevation to life-threatening rhabdomyolysis with markedly elevated CK levels, muscle damage, and acute renal injury. Although mild myalgias may not result in any physical harm, they threaten patient adherence to statin therapy. The incidence of statin-associated myopathy was reportedly between 1 and 5% in clinical trials, with a higher incidence observed in clinical practice (Thompson et al., 2003; Bruckert et al., 2005).
The risk for statin-induced myopathy is greater with higher statin doses or inhibition of statin metabolism or clearance secondary to drug interactions or decreased hepatic or renal function (Thompson et al., 2003).There is also a heritable component to the risk for statin-induced myopathy. The strongest data in this regard exist for the solute carrier organic anion transporter family, member 1B1 (SLCO1B1) gene. This gene encodes the organic anion transporting polypeptide 1B1, which transports most statins, with the exception of fluvastatin, to the liver. The SLCO1B1 genotype was identified as a risk factor for statin-induced myopathy in a GWAS of DNA samples from the Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine trial, in which participants received low (20 mg/day) or high (80 mg/day) simvastatin after myocardial infarction (Link et al., 2008). Of more than 300,000 variants interrogated in 85 patients with myopathy confirmed via evidence of CK elevation, and in 90 patients without myopathy during high-dose simvastatin therapy, the only variant to reach genome-wide significance for association with statin-induced myopathy was the noncoding rs4363657 variant on chromosome 12. This SNP is in near complete linkage disequilibrium with the c.521T>C (rs4149056, p.Val174Ala) SNP. The odds of myopathy were 4.5-fold greater with a single 521C allele and nearly 17-fold greater with the CC versus TT genotype. In a replication cohort consisting of patients treated with simvastatin 40 mg/day as part of the Heart Protection Study, the 521T > CSNP remained associated with statin-induced myopathy, with a relative risk of 2.6 per C allele. The heterozygous 521CT genotype is present in 11 to 36% of individuals, whereas the CC genotype occurs in up to 6% of individuals.
The SLCO1B1 521C allele is contained within the SLCO1B1*5 haplotype, which has been associated with reduced organic anion transporting polypeptide 1B1 activity and increased statin plasma concentrations (Kameyama et al., 2005; Pasanen et al., 2007). Consistent with previous data, among over 500 patients receiving atorvastatin, simvastatin, or pravastatin, the SLCO1B1*5 allele was associated with an increased risk for adverse statin-induced effects, defined as statin discontinuation for any side effect, myalgia, or CK greater than three times the upper limit of normal (Voora et al., 2009). Additional studies have confirmed the association between the SLCO1B1*5 allele and statin-induced myopathy, with the effects conferred by SLCO1B1 genotype appearing to be greatest with simvastatin (Donnelly et al., 2011; Brunham et al., 2012). The effect of SLCO1B1 genotype on statin-induced LDL-cholesterol reduction has also been examined, and although associated with cholesterol reduction with simvastatin and rosuvastatin the effect size was small (Akao et al., 2012; Chasman et al., 2012; Hopewell et al., 2013). Results from an additional study suggest that the effect size with the SLCO1B1 genotype may be greater among older persons (Akao et al., 2012). Nonetheless, the clinical utility of the SLCO1B1 genotype is limited to predicting myopathy risk at this time.
With the goal of reducing myopathy and optimizing patient adherence, the CPIC recently published guidelines addressing statin use when SLCO1B1 genotype is known (Wilke et al., 2012). These recommendations are limited to simvastatin, for which the strongest data are available. Similar to other CPIC guidelines, they do not make recommendation for when or who to genotype, but rather provide guidance on how to act on available genotype information. Consistent with the FDA-approved labeling, the guidelines recommend avoiding the 80 mg/day simvastatin dose regardless of genotype. An exception to this may be a patient with the 521TT genotype and a history of tolerating the 80 mg/day dose. Otherwise, in patients with the 521TT genotype, a maximum dose of 40 mg daily is recommended. The data suggest that the relative risk for myopathy with the 40 mg/day dose is approximately 2.5 per C allele (Link et al., 2008). Thus, for patients with the 521CT and 521CC genotypes, the risk of myopathy is such that the guidelines recommend no greater than a 20 mg daily dose or an alternative statin. If the cholesterol goals are not achieved with the genotype-guided maximum recommended simvastatin, alternative therapy is recommended. The guidelines also recommend considering use of routine CK surveillance in patients with the CT or CC genotype at the 20-mg and particularly 40-mg dose to screen for serious myopathies.
Vanderbilt University is an example of an institution where SLCO1B1 genotyping has been implemented into clinical practice (Lieb et al., 2012). Patients with risk factors for cardiovascular disease who may need statin therapy in the future are genotyped for SLCO1B1 521T>C preemptively, and results are placed in the electronic health record. An electronic order for simvastatin will trigger an electronic alert to warn about the increased risk for myopathy in patients with the CC or CT genotype.
B. Controversy with KIF6 Genotype
In contrast to SLCO1B1, which has primarily been examined in relation to statin safety, the gene for kinesin-like protein 6 (KIF6), a protein involved in intracellular transport, has been extensively assessed in regard to statin efficacy. Initial genetic substudies of two large, randomized, placebo-controlled trials with statins found an association between the KIF6 p.Trp719Arg (rs20455, c.2155T>C) polymorphism and clinical outcomes with statin therapy, with carriers of the 719Arg allele having greater reduction in clinical events compared with noncarriers. Specifically, among participants receiving placebo in the secondary prevention Cholesterol and Recurrent Events trial and the primary prevention West of Scotland Coronary Prevention Study, the 719Arg allele was associated with an increased risk for cardiovascular events, defined as recurrent myocardial infarction in Cholesterol and Recurrent Events and coronary heart disease in West of Scotland Coronary Prevention Study (Iakoubova et al., 2008b). Carriers of the 719Arg allele appeared to derive greater benefit from pravastatin compared with noncarriers. Similar findings were observed in a separate trial of patients with vascular disease (Iakoubova et al., 2010). In a third genetic substudy, investigators reported a greater reduction in coronary events with intensive statin therapy (atorvastatin 80 mg/day) in 719Arg carriers compared with noncarriers (Iakoubova et al., 2008a). On the basis of these data, a commercially available KIF6 genotyping assay was developed and marketed to predict risk for cardiac events and response to statins.
However, subsequent substudies of other large, randomized, placebo-controlled trials with atorvastatin, rosuvastatin, and simvastatin revealed no association between KIF6 genotype and either risk for coronary events or response to statins, leading clinicians and scientists to question the appropriateness of using KIF6 genotype to guide statin therapy (Hopewell et al., 2011; Ridker et al., 2011; Arsenault et al., 2012). For example, in a large primary prevention study of subjects with a baseline LDL cholesterol <130 mg/dl but elevated high-sensitivity C-reactive protein (a purported marker for increased cardiovascular disease), KIF6 genotype had no impact on the reduction in risk for major vascular events with rosuvastatin (Ridker et al., 2011). Similarly, KIF6 genotype had no effect on outcomes with high dose atorvastatin in patients with stable cardiovascular disease (Arsenault et al., 2012) or with simvastatin in patients with or at risk for vascular disease (Hopewell et al., 2011). Inconsistent findings among studies have generated considerable controversy regarding the clinical utility of KIF6 testing. For all the examples discussed in this review, the functional basis and biologic relevance of the encoded protein and the SNP are well understood. This is not the case for KIF6. The questionable biologic plausibility for the association between KIF6 and statin response in the face of similar reductions in LDL cholesterol with statin therapy in 719Arg carriers and noncarriers have contributed to this controversy. Similar to other large meta-analyses, a recent meta-analysis of 37 case-control, prospective cohort or randomized studies including data from over 144,000 patients found no association between KIF6 genotype and risk for cardiovascular disease (Ference et al., 2011). However, among patients with higher baseline LDL cholesterol, the 719Arg allele reportedly conferred both an increased risk for cardiovascular disease and greater reduction in cardiovascular disease risk per unit reduction in LDL cholesterol with statin therapy. The investigators concluded that the 719Arg allele may increase vulnerability to LDL cholesterol, thus influencing clinical benefit with LDL reduction. Nonetheless, until this is confirmed, the clinical utility of using KIF6 genotype as a marker of statin efficacy remains questionable.
C. Other Genes Linked to Statin Response
A number of plausible candidate genes have been examined for their role in mediating statin response. These include the genes encoding 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the target protein of statins; apolipoprotein E (APOE), which transports cholesterol through the bloodstream; and the LDLR gene encoding the LDL receptor. In addition, the ABCG2, which is involved in the disposition of rosuvastatin, has been linked to LDL-cholesterol response to rosuvastatin (Tomlinson et al., 2010; Chasman et al., 2012). The loci for the HMGCR, APOE, and LDLR genes were associated with LDL-cholesterol levels in a GWAS (Trompet et al., 2011), and additional candidate gene or whole genome studies have shown all three genes to influence statin-induced reductions in LDL cholesterol (Andriani et al., 1990; Willer et al., 2008; Thompson et al., 2009; Hopewell et al., 2013). However, effect sizes were small, and in one GWAS, associations could not be replicated (Hopewell et al., 2013). The APOE gene was also associated with clinical outcomes with simvastatin in myocardial infarction survivors (Gerdes et al., 2000); however, the gene’s association with statin-mediated outcomes has not been consistently replicated (Shiffman et al., 2012). A recent GWAS of samples from three large randomized placebo-controlled trials of pravastatin therapy revealed a SNP in the DnaJ homolog subfamily C member 5B (DNAJC5B) that was associated with reduction in coronary heart disease events with pravastatin (Shiffman et al., 2012). The functional role of DnaJ in modulating outcomes with statin therapy remains to be determined, and its association with statin response requires validation before it can be considered as a clinical marker of statin response. Although these data are interesting, they require further replication and evaluation to know if they will be of clinical utility in the future. That the effect sizes with these associations tend to be relatively small and that LDL cholesterol response can easily be measured may limit clinical utility of most markers associated with LDL cholesterol lowering with statins.
VI. Other Examples in Cardiovascular Pharmacogenetics
A. β-Blockers
The data on clopidogrel, warfarin, and simvastatin have been judged to be sufficiently robust, well replicated, and clinically meaningful that there are now examples of clinical utilization of pharmacogenetic data to guide therapy for all of these drugs. In this review we focused on those examples with clinical utility. The other cardiovascular drug class that may be closest to clinical utility of pharmacogenetics is the β-blockers. As with the examples above, the strongest data on β-blocker pharmacogenetics arise from known functional polymorphisms.
The two of the most commonly used β-blockers in heart failure, metoprolol and carvedilol, both undergo substantial metabolism by the highly polymorphic CYP2D6 enzyme, whose gene contains loss of function, deletion, and duplication polymorphisms. The pharmacokinetics of these drugs are clearly affected based on CYP2D6 genotype; however, there is less evidence for differences in efficacy or side effects (Fukami et al., 2004; Terra et al., 2005b; Dorn, 2009; Shin and Johnson 2010; Lu 2011; Chan et al., 2012b). This is most likely attributed to the fact that patients on β-blockers typically undergo dose titration to a heart rate <60 beats/min, thus minimizing the impact of pharmacokinetic differences.
There are also a number of studies suggesting functional polymorphisms in adrenergic receptor signaling genes are associated with differential response to β-blockers, particularly in hypertension and heart failure. The genes with the strongest data are ADRB1¸ ADRA2C, GRK5, and GRK4 (Johnson et al., 2003; Mialet Perez et al., 2003; Terra et al., 2005a; Liggett et al., 2006; Lobmeyer et al., 2007; Pacanowski et al., 2008; O'Connor et al., 2012b; Vandell et al., 2012). These data suggest differential responses to β-blockers by genotype that include blood pressure response, improvement in left ventricular ejection fraction, and survival differences in hypertension and heart failure. These data were reviewed in detail recently, and the reader is referred there for more information (Johnson and Liggett, 2011). It is of interest to note that based on ADRB1 genotype associations with outcomes in heart failure with the β-blocker bucindolol, a pharmaceutical company is currently undertaking a genotype-targeted clinical trial, enrolling only those patients with ADRB1 Arg389Arg genotype in a comparative trial with controlled/extended release metoprolol (http://phx.corporate-ir.net/phoenix.zhtml?c=109749&p=irol-newsArticle&ID=1427666&highlight=; Retrieved 9 March 2011).
Thus, although the pharmacogenetic data for β-blockers have not yet arisen to the point of clinical utility, they are almost certainly the next closest examples among the cardiovascular drugs.
B. Other Cardiovascular Drugs
Other cardiovascular drugs have been studied, including angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, antiarrhythmic drugs, aspirin, and drugs causing long QT syndrome, but in general the associations have only been tested in a single study or there have been inconsistencies across studies (Sasaki, 1996; Johnson et al., 2011a; Lu, 2011; Roden et al., 2011; Chan et al., 2012b; Talameh et al., 2012). It remains unclear whether genetics contributes less to the response to these drugs or whether the right studies, or sufficiently large studies, have just not been conducted.
Insights might also arise from pharmacogenetics studies for new cardiovascular drugs. For example, data were recently published for the new oral anticoagulant dabigatran, which showed that a polymorphism in CES1 was significantly associated with bleeding risk (Paré et al., 2013). These data arose from a GWAS and highlight that we may continue to uncover new understanding based on pharmacogenetic data, and these data might someday be of clinical utility.
VII. Summary
At the turn of the new millennium there were very little to almost no data on the pharmacogenetics of cardiovascular drugs, and with the exception of the first warfarin pharmacogenetics study, all the data highlighted above for clopidogrel, warfarin, statins, and β-blockers have all arisen since 2000. In that time frame, the data for clopidogrel, warfarin, and simvastatin have become sufficiently robust that there is now limited utilization of genetic information to guide their use clinically. Many barriers yet remain to the clinical implementation of pharmacogenetics for cardiovascular drugs; yet if one considers the knowledge advances that have been made in the last 10–12 years, it is exciting to think about where we will be in the coming decade. It is also apparent that the genome might just be the tip of the iceberg for understanding variable drug response, because epigenetics, transcriptomics, and metabolomics also are likely important and are more strongly affected by environmental and other factors that can change over a person’s life span. That there is much more to learn and apply to personalized approaches to care is clear, but genetics represents a tool that in certain situations, such as the ones described above, can be used now.
Costs of genotyping have plummeted, such that it is now possible to sequence a human genome for under $1000. On the basis of these technological advances, it is expected that genetic data will increasingly be available on patients to guide their clinical care. There are many additional barriers to this within the healthcare system and for society, but for such advances to occur, we must continue to work to understand the genetic influences on drug response and to define those examples where the genetic associations are sufficiently robust and clinically meaningful to warrant their use to guide treatment decisions.
Abbreviations
- ABC
ATP-binding cassette
- CAD
coronary artery disease
- CI
confidence interval
- CK
creatine kinase
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- DPWG
Dutch Pharmacogenetics Working Group
- EMA
European Medicines Agency
- EPARs
European Public Assessment Reports
- FDA
Food and Drug Administration
- GWAS
genome-wide association study
- INR
international normalized ratio
- KIF6
kinesin-like protein 6
- LDL
low-density lipoprotein
- MACE
major adverse cardiovascular event
- NSTIMI
non-ST segment elevation myocardial infarction
- OR
odds ratio
- PCI
percutaneous coronary intervention
- PMDA
Pharmaceuticals and Medical Devices Agency in Japan
- SNP
single nucleotide polymorphism
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Johnson, Cavallari.
Footnotes
This work was supported in part by the National Institutes of Health [Grants U01 GM074492 and R01 NS073346] (to J.A.J.); and the American Heart Association Midwest Affiliate [Grant 10GRNT3750024] (to L.H.C.).
References
- Aithal GP, Day CP, Kesteven PJ, Daly AK. (1999) Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 353:717–719 [DOI] [PubMed] [Google Scholar]
- Akao H, Polisecki E, Kajinami K, Trompet S, Robertson M, Ford I, Jukema JW, de Craen AJ, Westendorp RG, Shepherd J, et al. (2012) Genetic variation at the SLCO1B1 gene locus and low density lipoprotein cholesterol lowering response to pravastatin in the elderly. Atherosclerosis 220:413–417 [DOI] [PubMed] [Google Scholar]
- Altman RB. (2011) Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin Pharmacol Ther 89:348–350 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Adams CD, Antman EM, Bridges CR, Califf RM, Casey DE, Jr, Chavey WE, 2nd, Fesmire FM, Hochman JS, Levin TN, et al. 2011 WRITING GROUP MEMBERS. ACCF/AHA TASK FORCE MEMBERS (2011) 2011 ACCF/AHA Focused Update Incorporated Into the ACC/AHA 2007 Guidelines for the Management of Patients With Unstable Angina/Non-ST-Elevation Myocardial Infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 123:e426–e579 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, Kahn SF, May HT, Samuelson KM, Muhlestein JB, et al. Couma-Gen Investigators (2007) Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation 116:2563–2570 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Horne BD, Stevens SM, Woller SC, Samuelson KM, Mansfield JW, Robinson M, Barton S, Brunisholz K, Mower CP, et al. (2012) A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II). Circulation 125:1997–2005 [DOI] [PubMed] [Google Scholar]
- Andriani A, Mosiello G, Chiappetta MG, Gregori G, Rinaldi P. (1990) [Clinico-hematological evaluation of 57 patients undergoing antiaggregant treatment with indobufen (in Italian)]. Recenti Prog Med 81:49–53 [PubMed] [Google Scholar]
- Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, Gaston KL, Waddell CD, Chirico MJ, Johnson JA. (2006) Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther 79:291–302 [DOI] [PubMed] [Google Scholar]
- Arsenault BJ, Boekholdt SM, Hovingh GK, Hyde CL, DeMicco DA, Chatterjee A, Barter P, Deedwania P, Waters DD, LaRosa J, et al. TNT and IDEAL Investigators (2012) The 719Arg variant of KIF6 and cardiovascular outcomes in statin-treated, stable coronary patients of the treating to new targets and incremental decrease in end points through aggressive lipid-lowering prospective studies. Circ Cardiovasc Genet 5:51–57 [DOI] [PubMed] [Google Scholar]
- Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, et al. Cholesterol Treatment Trialists’ (CTT) Collaborators (2005) Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366:1267–1278 [DOI] [PubMed] [Google Scholar]
- Baldwin RM, Ohlsson S, Pedersen RS, Mwinyi J, Ingelman-Sundberg M, Eliasson E, Bertilsson L. (2008) Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol 65:767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer T, Bouman HJ, van Werkum JW, Ford NF, ten Berg JM, Taubert D. (2011) Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ 343:d4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biss TT, Avery PJ, Brandão LR, Chalmers EA, Williams MD, Grainger JD, Leathart JB, Hanley JP, Daly AK, Kamali F. (2012) VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood 119:868–873 [DOI] [PubMed] [Google Scholar]
- Bouman HJ, Schömig E, van Werkum JW, Velder J, Hackeng CM, Hirschhäuser C, Waldmann C, Schmalz HG, ten Berg JM, Taubert D. (2011) Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med 17:110–116 [DOI] [PubMed] [Google Scholar]
- Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, 2nd, Lachno DR, Salazar D, Winters KJ. (2007) Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost 5:2429–2436 [DOI] [PubMed] [Google Scholar]
- Bruckert E, Hayem G, Dejager S, Yau C, Bégaud B. (2005) Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients—the PRIMO study. Cardiovasc Drugs Ther 19:403–414 [DOI] [PubMed] [Google Scholar]
- Brunham LR, Lansberg PJ, Zhang L, Miao F, Carter C, Hovingh GK, Visscher H, Jukema JW, Stalenhoef AF, Ross CJ, et al. (2012) Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J 12:233–237 [DOI] [PubMed] [Google Scholar]
- Budnitz DS, Lovegrove MC, Shehab N, Richards CL. (2011) Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med 365:2002–2012 [DOI] [PubMed] [Google Scholar]
- Burmester JK, Berg RL, Yale SH, Rottscheit CM, Glurich IE, Schmelzer JR, Caldwell MD. (2011) A randomized controlled trial of genotype-based Coumadin initiation. Genet Med 13:509–518 [DOI] [PubMed] [Google Scholar]
- Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, et al. (2008) CYP4F2 genetic variant alters required warfarin dose. Blood 111:4106–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo G, Ferraresi P, Marchesini J, Bernardi F, Valgimigli M. (2011) Relationship between paraoxonase Q192R gene polymorphism and on-clopidogrel platelet reactivity over time in patients treated with percutaneous coronary intervention. J Thromb Haemost 9:2106–2108 [DOI] [PubMed] [Google Scholar]
- Caraco Y, Blotnick S, Muszkat M. (2008) CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther 83:460–470 [DOI] [PubMed] [Google Scholar]
- Catapano AL, Reiner Z, De Backer G, Graham I, Taskinen MR, Wiklund O, Agewall S, Alegria E, Chapman M, Durrington P, et al. European Society of Cardiology (ESC) European Atherosclerosis Society (EAS) (2011) ESC/EAS Guidelines for the management of dyslipidaemias The Task Force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and the European Atherosclerosis Society (EAS). Atherosclerosis 217:3–46 [DOI] [PubMed] [Google Scholar]
- Cavallari LH, Langaee TY, Momary KM, Shapiro NL, Nutescu EA, Coty WA, Viana MA, Patel SR, Johnson JA. (2010) Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther 87:459–464 [DOI] [PubMed] [Google Scholar]
- Cha PC, Mushiroda T, Takahashi A, Kubo M, Minami S, Kamatani N, Nakamura Y. (2010) Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet 19:4735–4744 [DOI] [PubMed] [Google Scholar]
- Chan SL, Suo C, Lee SC, Goh BC, Chia KS, Teo YY. (2012a) Translational aspects of genetic factors in the prediction of drug response variability: a case study of warfarin pharmacogenomics in a multi-ethnic cohort from Asia. Pharmacogenomics J 12:312–318 [DOI] [PubMed] [Google Scholar]
- Chan SW, Hu M, Tomlinson B. (2012b) The pharmacogenetics of β-adrenergic receptor antagonists in the treatment of hypertension and heart failure. Expert Opin Drug Metab Toxicol 8:767–790 [DOI] [PubMed] [Google Scholar]
- Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. (2012). Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: the Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet 5:257–264 [DOI] [PubMed] [Google Scholar]
- Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, et al. National Heart, Lung, and Blood Institute Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. National High Blood Pressure Education Program Coordinating Committee (2003) The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA 289:2560–2572 [DOI] [PubMed] [Google Scholar]
- Collet JP, Hulot JS, Anzaha G, Pena A, Chastre T, Caron C, Silvain J, Cayla G, Bellemain-Appaix A, Vignalou JB, et al. CLOVIS-2 Investigators (2011) High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS-2 (Clopidogrel and Response Variability Investigation Study 2). JACC Cardiovasc Interv 4:392–402 [DOI] [PubMed] [Google Scholar]
- Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F, et al. (2009) Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 373:309–317 [DOI] [PubMed] [Google Scholar]
- Connolly S, Pogue J, Hart R, Pfeffer M, Hohnloser S, Chrolavicius S, Pfeffer M, Hohnloser S, Yusuf S, ACTIVE Writing Group of the ACTIVE Investigators (2006) Clopidogrel plus aspirin versus oral anticoagulation for atrial fibrillation in the Atrial fibrillation Clopidogrel Trial with Irbesartan for prevention of Vascular Events (ACTIVE W): a randomised controlled trial. Lancet 367:1903–1912 [DOI] [PubMed] [Google Scholar]
- Connolly SJ, Pogue J, Hart RG, Hohnloser SH, Pfeffer M, Chrolavicius S, Yusuf S, ACTIVE Investigators (2009) Effect of clopidogrel added to aspirin in patients with atrial fibrillation. N Engl J Med 360:2066–2078 [DOI] [PubMed] [Google Scholar]
- Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, et al. (2008) A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112:1022–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi CL, Miller VP. (1997) The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH:cytochrome P450 oxidoreductase. Pharmacogenetics 7:203–210 [DOI] [PubMed] [Google Scholar]
- Crews KR, Cross SJ, McCormick JN, Baker DK, Molinelli AR, Mullins R, Relling MV, Hoffman JM. (2011) Development and implementation of a pharmacist-managed clinical pharmacogenetics service.Am J Health Syst Pharm 68:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. (1994) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269:15419–15422 [PubMed] [Google Scholar]
- Delaney JT, Ramirez AH, Bowton E, Pulley JM, Basford MA, Schildcrout JS, Shi Y, Zink R, Oetjens M, Xu H, et al. (2012) Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther 91:257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmann LJ, Rettie AE, Kneller MB, Kim RB, Wood AJ, Stein CM, Wilkinson GR, Schwarz UI. (2001) Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol 60:382–387 [DOI] [PubMed] [Google Scholar]
- Donnelly LA, Doney AS, Tavendale R, Lang CC, Pearson ER, Colhoun HM, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. (2011) Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther 89:210–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW., 2nd (2009) Pharmacogenetic profiling in the treatment of heart disease. Transl Res 154:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein RS, Moyer TP, Aubert RE, O Kane DJ, Xia F, Verbrugge RR, Gage BF, Teagarden JR. (2010) Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study). J Am Coll Cardiol 55:2804–2812 [DOI] [PubMed] [Google Scholar]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (2001) Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 285:2486–2497 [DOI] [PubMed] [Google Scholar]
- Ference BA, Yoo W, Flack JM, Clarke M. (2011) A common KIF6 polymorphism increases vulnerability to low-density lipoprotein cholesterol: two meta-analyses and a meta-regression analysis. PLoS ONE 6:e28834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman BS, Gage BF, Johnson JA, Brensinger CM, Kimmel SE. (2011) Genetic warfarin dosing: tables versus algorithms. J Am Coll Cardiol 57:612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart DA, O’Kane D, Williams MS, Watson MS, Flockhart DA, Gage B, Gandolfi R, King R, Lyon E, Nussbaum R, et al. ACMG Working Group on Pharmacogenetic Testing of CYP2C9, VKORC1 Alleles for Warfarin Use (2008) Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med 10:139–150 [DOI] [PubMed] [Google Scholar]
- Floyd CN, Passacquale G, Ferro A. (2012) Comparative pharmacokinetics and pharmacodynamics of platelet adenosine diphosphate receptor antagonists and their clinical implications. Clin Pharmacokinet 51:429–442 [DOI] [PubMed] [Google Scholar]
- French B, Joo J, Geller NL, Kimmel SE, Rosenberg Y, Anderson JL, Gage BF, Johnson JA, Ellenberg JH, COAG (Clarification of Optimal Anticoagulation through Genetics) Investigators (2010) Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials 11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T, Nakajima M, Yoshida R, Tsuchiya Y, Fujiki Y, Katoh M, McLeod HL, Yokoi T. (2004) A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin Pharmacol Ther 76:519–527 [DOI] [PubMed] [Google Scholar]
- Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, et al. (2008) Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther 84:326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Gerdes C, Kervinen K, Savolainen M, Klausen IC, Hansen PS, Kesäniemi YA, Faergeman O. (2000) The apolipoprotein epsilon4 allele determines prognosis and the effect on prognosis of simvastatin in survivors of myocardial infarction : a substudy of the Scandinavian simvastatin survival study. Circulation 101:1366–1371 [DOI] [PubMed] [Google Scholar]
- Gong IY, Crown N, Suen CM, Schwarz UI, Dresser GK, Knauer MJ, Sugiyama D, Degorter MK, Woolsey S, Tirona RG, et al. (2012) Clarifying the importance of CYP2C19 and PON1 in the mechanism of clopidogrel bioactivation and in vivo antiplatelet response. Eur Heart J 33:2856–2464a [DOI] [PubMed] [Google Scholar]
- Gong IY, Schwarz UI, Crown N, Dresser GK, Lazo-Langner A, Zou G, Roden DM, Stein CM, Rodger M, Wells PS, et al. (2011) Clinical and genetic determinants of warfarin pharmacokinetics and pharmacodynamics during treatment initiation. PLoS ONE 6:e27808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt GH, Akl EA, Crowther MGutterman DD, Schuünemann HJ, American College of Chest Physicians Antithrombotic Therapy and Prevention of Thrombosis Panel (2012). Executive summary: antithrombotic therapy and prevention of thrombosis, 9th ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 141(Suppl 2): 7S–47S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm CW, Bassand JP, Agewall S, Bax J, Boersma E, Bueno H, Caso P, Dudek D, Gielen S, Huber K, et al. ESC Committee for Practice Guidelines (2011) ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: The Task Force for the management of acute coronary syndromes (ACS) in patients presenting without persistent ST-segment elevation of the European Society of Cardiology (ESC). Eur Heart J 32:2999–3054 [DOI] [PubMed] [Google Scholar]
- Harmsze AM, van Werkum JW, Hackeng CM, Ruven HJ, Kelder JC, Bouman HJ, Breet NJ, Ten Berg JM, Klungel OH, de Boer A, et al. (2012) The influence of CYP2C19*2 and *17 on on-treatment platelet reactivity and bleeding events in patients undergoing elective coronary stenting. Pharmacogenet Genomics 22:169–175 [DOI] [PubMed] [Google Scholar]
- Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. (2010) ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol 56:321–341 [DOI] [PubMed] [Google Scholar]
- Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. (2011) CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA 306:2704–2714 [DOI] [PubMed] [Google Scholar]
- Hopewell JC, Parish S, Clarke R, Armitage J, Bowman L, Hager J, Lathrop M, Collins R, MRC/BHF Heart Protection Study Collaborative Group (2011) No impact of KIF6 genotype on vascular risk and statin response among 18,348 randomized patients in the heart protection study. J Am Coll Cardiol 57:2000–2007 [DOI] [PubMed] [Google Scholar]
- Hopewell JC, Parish S, Offer A, Link E, Clarke R, Lathrop M, Armitage J, Collins R, MRC/BHF Heart Protection Study Collaborative Group (2013) Impact of common genetic variation on response to simvastatin therapy among 18 705 participants in the Heart Protection Study. Eur Heart J 34:982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettler S. (2012) Genetic Test Will Help Dose Blood Thinner, UIC News, Chicago [Google Scholar]
- Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, Aiach M, Lechat P, Gaussem P. (2006) Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 108:2244–2247 [DOI] [PubMed] [Google Scholar]
- Hulot JS, Collet JP, Cayla G, Silvain J, Allanic F, Bellemain-Appaix A, Scott SA, Montalescot G. (2011) CYP2C19 but not PON1 genetic variants influence clopidogrel pharmacokinetics, pharmacodynamics, and clinical efficacy in post-myocardial infarction patients. Circ Cardiovasc Interv 4:422–428 [DOI] [PubMed] [Google Scholar]
- Hulot JS, Collet JP, Silvain J, Pena A, Bellemain-Appaix A, Barthélémy O, Cayla G, Beygui F, Montalescot G. (2010) Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol 56:134–143 [DOI] [PubMed] [Google Scholar]
- Hunt SA, Abraham WT, Chin MH, Feldman AM, Francis GS, Ganiats TG, Jessup M, Konstam MA, Mancini DM, Michl K, et al. American College of Cardiology Foundation. American Heart Association (2009) 2009 Focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines Developed in Collaboration With the International Society for Heart and Lung Transplantation. J Am Coll Cardiol 53:e1–e90 [DOI] [PubMed] [Google Scholar]
- Hylek EM, Evans-Molina C, Shea C, Henault LE, Regan S. (2007) Major hemorrhage and tolerability of warfarin in the first year of therapy among elderly patients with atrial fibrillation. Circulation 115:2689–2696 [DOI] [PubMed] [Google Scholar]
- Hylek EM, Go AS, Chang Y, Jensvold NG, Henault LE, Selby JV, Singer DE. (2003) Effect of intensity of oral anticoagulation on stroke severity and mortality in atrial fibrillation. N Engl J Med 349:1019–1026 [DOI] [PubMed] [Google Scholar]
- Iakoubova OA, Robertson M, Tong CH, Rowland CM, Catanese JJ, Blauw GJ, Jukema JW, Murphy MB, Devlin JJ, Ford I, et al. (2010) KIF6 Trp719Arg polymorphism and the effect of statin therapy in elderly patients: results from the PROSPER study. Eur J Cardiovasc Prev Rehabil 17:455–461 [DOI] [PubMed] [Google Scholar]
- Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, Simonsen KL, Kirchgessner TG, Cannon CP, Devlin JJ, et al. (2008a) Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. J Am Coll Cardiol 51:449–455 [DOI] [PubMed] [Google Scholar]
- Iakoubova OA, Tong CH, Rowland CM, Kirchgessner TG, Young BA, Arellano AR, Shiffman D, Sabatine MS, Campos H, Packard CJ, et al. (2008b) Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. J Am Coll Cardiol 51:435–443 [DOI] [PubMed] [Google Scholar]
- Ichimaru K, Toyoshima S, Uyama Y. (2010) PMDA’s challenge to accelerate clinical development and review of new drugs in Japan. Clin Pharmacol Ther 88:454–457 [DOI] [PubMed] [Google Scholar]
- Ieiri I, Tainaka H, Morita T, Hadama A, Mamiya K, Hayashibara M, Ninomiya H, Ohmori S, Kitada M, Tashiro N, et al. (2000) Catalytic activity of three variants (Ile, Leu, and Thr) at amino acid residue 359 in human CYP2C9 gene and simultaneous detection using single-strand conformation polymorphism analysis. Ther Drug Monit 22:237–244 [DOI] [PubMed] [Google Scholar]
- Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. (2012a) Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther 92:437–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Cavallari LH, Beitelshees AL, Lewis JP, Shuldiner AR, Roden DM. (2011a) Pharmacogenomics: application to the management of cardiovascular disease. Clin Pharmacol Ther 90:519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, Anderson JL, Kimmel SE, Lee MT, Pirmohamed M, et al. Clinical Pharmacogenetics Implementation Consortium (2011b) Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther 90:625–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Liggett SB. (2011) Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin Pharmacol Ther 89:366–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Roden DM, Lesko LJ, Ashley E, Klein TE, Shuldiner AR. (2012b) Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther 91:774–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. (2003) Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther 74:44–52 [DOI] [PubMed] [Google Scholar]
- Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. (2005) Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 15:513–522 [DOI] [PubMed] [Google Scholar]
- Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, Ikeda T, Kurihara A. (2010) Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 38:92–99 [DOI] [PubMed] [Google Scholar]
- Kim KA, Park PW, Hong SJ, Park JY. (2008) The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther 84:236–242 [DOI] [PubMed] [Google Scholar]
- Kirley K, Qato DM, Kornfield RStafford RS, Alexander GC. (2012) National trends in oral anticoagulant use in the United States, 2007 to 2011. Circ Cardiovasc Qual Outcomes 5:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, et al. International Warfarin Pharmacogenetics Consortium (2009) Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 360:753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner FG, Hand M, Smith SC, Jr, King SB, 3rd, Anderson JL, Antman EM, Bailey SR, Bates ER, Blankenship JC, Casey DE, Jr, et al. (2009) 2009 focused updates: ACC/AHA guidelines for the management of patients with ST-elevation myocardial infarction (updating the 2004 guideline and 2007 focused update) and ACC/AHA/SCAI guidelines on percutaneous coronary intervention (updating the 2005 guideline and 2007 focused update) a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 54:2205–2241 [DOI] [PubMed] [Google Scholar]
- Lesko LJ, Zineh I. (2010) DNA, drugs and chariots: on a decade of pharmacogenomics at the US FDA. Pharmacogenomics 11:507–512 [DOI] [PubMed] [Google Scholar]
- Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM, et al. (2011) 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation 124:e574–e651 [DOI] [PubMed] [Google Scholar]
- Lewis JP, Fisch AS, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, Shen H, Tanner K, Horenstein RB, Pakzy R, et al. (2011) Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. Clin Pharmacol Ther 90:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. (2010) Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol 69:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tang HL, Hu YF, Xie HG. (2012) The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost 10:199–206 [DOI] [PubMed] [Google Scholar]
- Lieb W, Völzke H, Pulley JM, Roden DM, Kroemer HK. (2012) Strategies for personalized medicine-based research and implementation in the clinical workflow. Clin Pharmacol Ther 92:443–445 [DOI] [PubMed] [Google Scholar]
- Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, et al. (2006) A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA 103:11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, Acton RT. (2008a) Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics 9:511–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, Baird MF, Acton RT. (2008b) Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther 83:312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limdi NA, Wadelius M, Cavallari L, Eriksson N, Crawford DC, Lee MT, Chen CH, Motsinger-Reif A, Sagreiya H, Liu N, et al. International Warfarin Pharmacogenetics Consortium (2010) Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 115:3827–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh JD, Holm L, Andersson ML, Rane A. (2009) Influence of CYP2C9 genotype on warfarin dose requirements—a systematic review and meta-analysis. Eur J Clin Pharmacol 65:365–375 [DOI] [PubMed] [Google Scholar]
- Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R, SEARCH Collaborative Group (2008) SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 359:789–799 [DOI] [PubMed] [Google Scholar]
- Liu Y, Jeong H, Takahashi H, Drozda K, Patel SR, Shapiro NL, Nutescu EA, Cavallari LH. (2012) Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther 91:660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Hao PP, Zhang MX, Zhang C, Gao F, Zhang Y, Chen YG. (2011) Association of genetic variants in CYP2C19 and adverse clinical outcomes after treatment with clopidogrel: an updated meta-analysis. Thromb Res 128:593–594 [DOI] [PubMed] [Google Scholar]
- Lobmeyer MT, Gong Y, Terra SG, Beitelshees AL, Langaee TY, Pauly DF, Schofield RS, Hamilton KK, Herbert Patterson J, Adams KF, Jr, et al. (2007) Synergistic polymorphisms of beta1 and alpha2C-adrenergic receptors and the influence on left ventricular ejection fraction response to beta-blocker therapy in heart failure. Pharmacogenet Genomics 17:277–282 [DOI] [PubMed] [Google Scholar]
- Lu GW. (2011) [A slight experience gained from life course and academic career (in Chinese)]. Sheng Li Ke Xue Jin Zhan 42:321–328 Progress in physiology [PubMed] [Google Scholar]
- Mansia G, De Backer G, Dominiczak A, Cifkova R, Fagard R, Germano G, Grassi G, Heagerty AM, Kjeldsen SE, Laurent S, et al. European Society of Hypertension. European Society of Cardiology (2007) 2007 ESH-ESC Guidelines for the management of arterial hypertension: the task force for the management of arterial hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). Blood Press 16:135–232 [DOI] [PubMed] [Google Scholar]
- McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. (2009) CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol 75:1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGehee WG, Klotz TA, Epstein DJ, Rapaport SI. (1984) Coumarin necrosis associated with hereditary protein C deficiency. Ann Intern Med 101:59–60 [DOI] [PubMed] [Google Scholar]
- McMurray JJ, Adamopoulos S, Anker SD, Auricchio A, Böhm M, Dickstein K, Falk V, Filippatos G, Fonseca C, Gomez-Sanchez MA, et al. Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. ESC Committee for Practice Guidelines (2012) ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail 14:803–869 [DOI] [PubMed] [Google Scholar]
- Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, et al. (2009) Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 360:354–362 [DOI] [PubMed] [Google Scholar]
- Mega JL, Hochholzer W, Frelinger AL, 3rd, Kluk MJ, Angiolillo DJ, Kereiakes DJ, Isserman S, Rogers WJ, Ruff CT, Contant C, et al. (2011) Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA 306:2221–2228 [DOI] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, et al. (2010) Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, Dorn GW, Liggett SB. (2003) Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med 9:1300–1305 [DOI] [PubMed] [Google Scholar]
- Mrazek DA. (2010) Psychiatric pharmacogenomic testing in clinical practice. Dialogues Clin Neurosci 12:69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen SE. (2011) Pharmacogenomics and clopidogrel: irrational exuberance? JAMA 306:2727–2728 [DOI] [PubMed] [Google Scholar]
- O’Connor SK, Ferreri SP, Michaels NM, Greco AJ, Viera AJ, Faruki H, McLeod HL, Roederer MW. (2012a) Exploratory planning and implementation of a pilot pharmacogenetic program in a community pharmacy. Pharmacogenomics 13:955–962 [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Fiuzat M, Carson PE, Anand IS, Plehn JF, Gottlieb SS, Silver MA, Lindenfeld J, Miller AB, White M, et al. (2012b) Combinatorial pharmacogenetic interactions of bucindolol and β1, α2C adrenergic receptor polymorphisms. PLoS ONE 7:e44324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, Cox NJ, Ratain MJ. (2012) The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 92:446–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson N, DeJongh B, Hough A, Parra D. (2012) Plasma renin activity-guided strategy for the management of hypertension. Pharmacotherapy 32:446–455 [DOI] [PubMed] [Google Scholar]
- Otsubo Y, Asahina Y, Noguchi A, Sato Y, Ando Y, Uyama Y. (2012) Similarities and differences between US and Japan as to pharmacogenomic biomarker information in drug labels. Drug Metab Pharmacokinet 27:142–149 [DOI] [PubMed] [Google Scholar]
- Pacanowski MA, Gong Y, Cooper-Dehoff RM, Schork NJ, Shriver MD, Langaee TY, Pepine CJ, Johnson JA, INVEST Investigators (2008) Beta-adrenergic receptor gene polymorphisms and beta-blocker treatment outcomes in hypertension. Clin Pharmacol Ther 84:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré G, Eriksson N, Lehr T, Connolly S, Eikelboom J, Ezekowitz MD, Axelsson T, Haertter S, Oldgren J, Reilly P, et al. (2013) Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation 127:1404–1412 [DOI] [PubMed] [Google Scholar]
- Paré G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, Simonsen K, Bhatt DL, Fox KA, Eikelboom JW. (2010) Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 363:1704–1714 [DOI] [PubMed] [Google Scholar]
- Pare G, Ross S, Mehta SR, Yusuf SAnand SS, Connolly SJ, Fox KA, Eikelboom JW. (2012). Effect of PON1 Q192R genetic polymorphism on clopidogrel efficacy and cardiovascular events in the Clopidogrel in the Unstable Angina to Prevent Recurrent Events trial and the Atrial Fibrillation Clopidogrel Trial with Irbesartan for Prevention of Vascular Events. Circ Cardiovascular Genet 5:250–256 [DOI] [PubMed]
- Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M. (2007) Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther 82:726–733 [DOI] [PubMed] [Google Scholar]
- Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, Nicolae D, Cox NJ. (2011) The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther 89:408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MJ, Murray SS, Angiolillo DJ, Lillie E, Smith EN, Tisch RL, Schork NJ, Teirstein PS, Topol EJ, GIFT Investigators (2012) Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol 59:1928–1937 [DOI] [PubMed] [Google Scholar]
- Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, et al. (2012) Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 92:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Altman RB, Goetz MP, Evans WE. (2010) Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol 11:507–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Klein TE. (2011) CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89:464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reny JL, Combescure C, Daali Y, Fontana P, PON1 Meta-Analysis Group (2012) Influence of the paraoxonase-1 Q192R genetic variant on clopidogrel responsiveness and recurrent cardiovascular events: a systematic review and meta-analysis. J Thromb Haemost 10:1242–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, MacFadyen JG, Glynn RJ, Chasman DI. (2011) Kinesin-like protein 6 (KIF6) polymorphism and the efficacy of rosuvastatin in primary prevention. Circ Cardiovasc Genet 4:312–317 [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. (2005) Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 352:2285–2293 [DOI] [PubMed] [Google Scholar]
- Roden DM, Johnson JA, Kimmel SE, Krauss RM, Medina MW, Shuldiner A, Wilke RA. (2011) Cardiovascular pharmacogenomics. Circ Res 109:807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hörtnagel K, Pelz HJ, Lappegard K, Seifried E, Scharrer I, Tuddenham EG, et al. (2004) Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 427:537–541 [DOI] [PubMed] [Google Scholar]
- Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. (2008) Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther 83:322–327 [DOI] [PubMed] [Google Scholar]
- Sagreiya H, Berube C, Wen A, Ramakrishnan R, Mir A, Hamilton A, Altman RB. (2010) Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenet Genomics 20:407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkuhl K, Klein TE, Altman RB. (2010) Clopidogrel pathway. Pharmacogenet Genomics 20:463–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Oki T, Iuchi A, Tabata T, Yamada H, Manabe K, Fukuda K, Abe M, Ito S. (1996) Relationship between the angiotensin converting enzyme gene polymorphism and the effects of enalapril on left ventricular hypertrophy and impaired diastolic filling in essential hypertension: M-mode and pulsed Doppler echocardiographic studies. J Hypertens 14:1403–1408 [DOI] [PubMed] [Google Scholar]
- Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. (2002) Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther 72:702–710 [DOI] [PubMed] [Google Scholar]
- Scott SA, Edelmann L, Kornreich R, Desnick RJ. (2008) Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet 82:495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. (2009) CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics 10:1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Shuldiner AR, Hulot JS, Thorn CF, Altman RB, Klein TE. (2012) PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenet Genomics 22:159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D, Trompet S, Louie JZ, Rowland CM, Catanese JJ, Iakoubova OA, Kirchgessner TG, Westendorp RG, de Craen AJ, Slagboom PE, et al. (2012) Genome-wide study of gene variants associated with differential cardiovascular event reduction by pravastatin therapy. PLoS ONE 7:e38240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Johnson JA. (2010) Beta-blocker pharmacogenetics in heart failure. Heart Fail Rev 15:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, et al. (2009) Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 302:849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbing D, Gebhard D, Koch W, Braun S, Stegherr J, Morath T, Von Beckerath N, Mehilli J, Schömig A, Schuster T, et al. (2010) Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost 8:1685–1693 [DOI] [PubMed] [Google Scholar]
- Sibbing D, Stegherr J, Latz W, Koch W, Mehilli J, Dörrler K, Morath T, Schömig A, Kastrati A, von Beckerath N. (2009) Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J 30:916–922 [DOI] [PubMed] [Google Scholar]
- Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. (2006) A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79:103–113 [DOI] [PubMed] [Google Scholar]
- Simon T, Steg PG, Becquemont L, Verstuyft C, Kotti S, Schiele F, Ferrari E, Drouet E, Grollier G, Danchin N. (2011) Effect of paraoxonase-1 polymorphism on clinical outcomes in patients treated with clopidogrel after an acute myocardial infarction. Clin Pharmacol Ther 90:561–567 [DOI] [PubMed] [Google Scholar]
- Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Méneveau N, Steg PG, Ferrières J, Danchin N, Becquemont L, French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators (2009) Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 360:363–375 [DOI] [PubMed] [Google Scholar]
- Sofi F, Giusti B, Marcucci R, Gori AM, Abbate R, Gensini GF. (2011) Cytochrome P450 2C19*2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J 11:199–206 [DOI] [PubMed] [Google Scholar]
- Su J, Xu J, Li X, Zhang H, Hu J, Fang R, Chen X. (2012) ABCB1 C3435T polymorphism and response to clopidogrel treatment in coronary artery disease (CAD) patients: a meta-analysis. PLoS ONE 7:e46366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai G, Farin F, Rieder MJ, Dreisbach AW, Veenstra DL, Verlinde CL, Rettie AE. (2005) In-vitro and in-vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenet Genomics 15:475–481 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kashima T, Nomizo Y, Muramoto N, Shimizu T, Nasu K, Kubota T, Kimura S, Echizen H. (1998) Metabolism of warfarin enantiomers in Japanese patients with heart disease having different CYP2C9 and CYP2C19 genotypes. Clin Pharmacol Ther 63:519–528 [DOI] [PubMed] [Google Scholar]
- Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, et al. (2009) A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talameh JA, McLeod HL, Adams KF, Jr, Patterson JH. (2012) Genetic tailoring of pharmacotherapy in heart failure: optimize the old, while we wait for something new. J Card Fail 18:338–349 [DOI] [PubMed] [Google Scholar]
- Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, Lange C, Lazarus R, Sylvia J, Klanderman B, et al. (2011) Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 365:1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert D, von Beckerath N, Grimberg G, Lazar A, Jung N, Goeser T, Kastrati A, Schömig A, Schömig E. (2006) Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther 80:486–501 [DOI] [PubMed] [Google Scholar]
- Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hill JA, Aranda JM, et al. (2005a) Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenet Genomics 15:227–234 [DOI] [PubMed] [Google Scholar]
- Terra SG, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hamilton KK, Aranda JM, Hill JA, et al. (2005b) beta-Adrenergic receptor polymorphisms and responses during titration of metoprolol controlled release/extended release in heart failure. Clin Pharmacol Ther 77:127–137 [DOI] [PubMed] [Google Scholar]
- Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, Hovingh GK, Kastelein JJ. (2009) Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet 2:173–181 [DOI] [PubMed] [Google Scholar]
- Thompson PD, Clarkson P, Karas RH. (2003) Statin-associated myopathy. JAMA 289:1681–1690 [DOI] [PubMed] [Google Scholar]
- Tiroch KA, Sibbing D, Koch W, Roosen-Runge T, Mehilli J, Schömig A, Kastrati A. (2010) Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J 160:506–512 [DOI] [PubMed] [Google Scholar]
- Tomlinson B, Hu M, Lee VW, Lui SS, Chu TT, Poon EW, Ko GT, Baum L, Tam LS, Li EK. (2010) ABCG2 polymorphism is associated with the low-density lipoprotein cholesterol response to rosuvastatin. Clin Pharmacol Ther 87:558–562 [DOI] [PubMed] [Google Scholar]
- Trenk D, Hochholzer W, Fromm MF, Zolk O, Valina CM, Stratz C, Neumann FJ. (2011). Paraoxonase-1 Q192R polymorphism and antiplatelet effects of clopidogrel in patients undergoing elective coronary stent placement. Circ Cardiovasc Genet 4:429–436 [DOI] [PubMed] [Google Scholar]
- Trompet S, de Craen AJ, Postmus I, Ford I, Sattar N, Caslake M, Stott DJ, Buckley BM, Sacks F, Devlin JJ, et al. PROSPER Study Group (2011) Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med Genet 12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K, Furuta T, Kondo K. (2008) The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost 6:1439–1441 [DOI] [PubMed] [Google Scholar]
- Vandell AG, Lobmeyer MT, Gawronski BE, Langaee TY, Gong Y, Gums JG, Beitelshees AL, Turner ST, Chapman AB, Cooper-DeHoff RM, et al. (2012) G protein receptor kinase 4 polymorphisms: β-blocker pharmacogenetics and treatment-related outcomes in hypertension. Hypertension 60:957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenhorst C, James S, Erlinge D, Brandt JT, Braun OO, Man M, Siegbahn A, Walker J, Wallentin L, Winters KJ, et al. (2009) Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J 30:1744–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. (2009) The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol 54:1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, Holm L, McGinnis R, Rane A, Deloukas P. (2009) The largest prospective warfarin-treated cohort supports genetic forecasting. Blood 113:784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. (2008) Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 112:1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Ye F, Xie D, Zhu Y, Zhu J, Tao Y, Yu F. (2012) A new algorithm to predict warfarin dose from polymorphisms of CYP4F2, CYP2C9 and VKORC1 and clinical variables: derivation in Han Chinese patients with non valvular atrial fibrillation. Thromb Haemost 107:1083–1091 [DOI] [PubMed] [Google Scholar]
- Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D, Krauss RM, Roden DM, Feng Q, Cooper-Dehoff RM, et al. Clinical Pharmacogenomics Implementation Consortium (CPIC) (2012) The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther 92:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RS, Anderson JL, Adams CD, Bridges CR, Casey DE, Jr, Ettinger SM, Fesmire FM, Ganiats TG, Jneid H, Lincoff AM, et al. (2011) 2011 ACCF/AHA focused update of the Guidelines for the Management of Patients with Unstable Angina/Non-ST-Elevation Myocardial Infarction (updating the 2007 guideline): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines developed in collaboration with the American College of Emergency Physicians, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol 57:1920–1959 [DOI] [PubMed] [Google Scholar]
- Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK, Clopidogrel in Unstable Angina to Prevent Recurrent Events Trial Investigators (2001) Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med 345:494–502 [DOI] [PubMed] [Google Scholar]
- Zabalza M, Subirana I, Sala J, Lluis-Ganella C, Lucas G, Tomás M, Masiá R, Marrugat J, Brugada R, Elosua R. (2012) Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart 98:100–108 [DOI] [PubMed] [Google Scholar]
- Zineh I, Pacanowski MA. (2011) Pharmacogenomics in the assessment of therapeutic risks versus benefits: inside the United States Food and Drug Administration. Pharmacotherapy 31:729–735 [DOI] [PubMed] [Google Scholar]