Abstract
In animal and laboratory models, cancer-associated stroma, or elements of the supporting tissue surrounding a primary tumor, has been shown to be necessary for tumor evolution and progression. However, little is understood or studied regarding the properties of intact stroma in human cancer in vivo. In addition, for breast cancer patients, the optimal volume of local tissue to treat surrounding a primary tumor is not clear. Here, we performed an interdisciplinary study of normal-appearing breast tissue using breast magnetic resonance imaging (MRI), correlative histology and array comparative genomic hybridization to identify a cancer-associated stroma in humans. Using a novel technique for segmenting breast fibroglandular tissue, quantifiable topographic percent enhancement mapping of the stroma surrounding invasive breast cancer was found to be significantly elevated within 2 cm of the tumor edge. This region was also found to harbor increased microvessel density, and genomic changes that were closely associated with host normal breast tissue. These findings indicate that a cancer-associated stroma may be identified and characterized in human breast cancer using non-invasive imaging techniques. Identification of a cancer-associated stroma may be further developed to help guide local therapy to reduce recurrence and morbidity in breast cancer patients.
Introduction
Invasive breast cancer is known to begin in the breast, and untreated, progresses by local invasion and eventually distant metastasis. For most early stage disease, local treatment involves surgical resection of the tumor and radiation therapy. Traditional radiation fields encompass the entire breast, although this paradigm is being challenged with current partial breast radiation treatment approaches. The optimal volume of “normal” tissue to treat surrounding a primary tumor remains controversial despite advances in imaging technologies. Studies in animal and laboratory models of human cancer have demonstrated the critical role of cancer-associated stroma (CAS), or the normal-appearing tissue surrounding a tumor, in tumor progression. Despite this, the CAS has not been well studied in humans to date.
The role of CAS has been shown functionally by studies demonstrating that supportive cells from the CAS are necessary for tumorigenesis in vivo,1 and that the role of CAS in tumor progression may be emulated by wounding,2 cytokine overproduction3 or low doses of ionizing radiation.4 Studies of genomic changes in the stroma have indicated that changes do exist.5 However their nature remains controversial based on the technique used to identify them; whether or not genomic changes in the CAS co-evolve with the primary tumor is a debated question.6,7 In addition, specific signals emanating from the extracellular matrix may be targeted to induce malignant cells to revert to their normal phenotype.8 Thus, identifying, characterizing and further understanding the role of CAS in breast cancers may lead to better local therapy and improved targets for treatment.
We have previously shown that stromal enhancement may have predictive value in patients receiving neoadjuvant chemotherapy for locally advanced breast cancer.9 Here, we report a novel integrated approach to identifying and characterizing a CAS phenotype in patients with small primary breast cancers using advanced MRI techniques and correlative histopathologic and genomic studies from the same patients.
Experimental
Study design and patient population
This study is a dynamic contrast enhanced MRI (DCE-MRI) and pathological tissue analysis of a retrospective cohort of women with biopsy proven breast cancers less than 3 cm in the longest diameter that had a research DCE-MRI prior to treatment (surgery, chemotherapy or radiation) and core biopsies at our institution between 1995–2002. All patients included in the cohort were candidates for breast-conserving surgery and no patient received neoadjuvant chemotherapy. An institutional review board approved the study protocol, and all patients gave informed consent. Of 49 patients that met the initial eligibility criteria, 20 were excluded due to insufficient amounts of fibroglandular breast tissue for analysis, which was defined as having less than 2 cm of radiographically normal fibroglandular breast tissue directly adjacent to the visibly enhancing tumor. The presented DCE-MRI stromal enhancement methods cannot be applied to these patients.
Four patients were excluded due to movement during the MRI image acquisition and 2 patients were excluded because their histology was not consistent with invasive breast carcinoma (IBC). The remaining 23 patients comprised the study group, Table 1.
Table 1.
Patient and tumor characteristics
| Characteristic | Value |
|---|---|
| Mean patient age | 54 years (32–77) |
| Mean pre-treatment tumor size | 1.3 cm (0.2–2.5) |
| Pathological tumor type | |
| Invasive ductal | 18 |
| Invasive lobular | 2 |
| Invasive ductal and lobular | 2 |
| Mucinous carcinoma | 1 |
| Scharff-Bloom-Richardson grade | |
| 1 | 7 |
| 2 | 10 |
| 3 | 4 |
| Unknown | 2 |
| DCIS in Ipsilateral breast? | |
| Yes | 14 |
| No | 6 |
| Unknown | 3 |
| Nodal status | |
| Positive | 8 |
| Negative | 12 |
| Unknown | 3 |
| Estrogen receptor status | |
| Positive | 20 |
| Negative | 2 |
| Unknown | 1 |
| Progesterone receptor status | |
| Positive | 19 |
| Negative | 3 |
| Unknown | 1 |
| HER2/neu Status | |
| Positive | 6 |
| Negative | 10 |
| Unknown | 7 |
Values are expressed as number of patients unless otherwise indicated. Numbers in parentheses indicate value range.
MRI Acquisition
For each of the 23 patients studied, contrast enhanced MRI using Gadopentetate dimeglumine (Magnevist, Schering, Berlin Germany) at a dose of 0.1 mmol kg–1 of body weight was performed on a 1.5 T Signa system (General Electric Medical Systems, Milwaukee, WI) with a dedicated breast coil and a high-resolution fat suppressed T1-weighted 3D fast gradient echo sequence. Scan parameters were TR/TE = 8/4.2, 20 cm field of view, 2 mm slice thickness and 256 × 192 acquisition matrix, resulting in an in-plane resolution of approximately 0.78 × 0.78 mm. Sixty slices were acquired in the sagittal orientation giving full coverage of the diseased breast in a scan time of approximately 5 min. Data were acquired prior to and immediately following contrast injection, resulting in an effective post-contrast acquisition time of 2.5 min.
MR enhancement in the normal-appearing breast tissue with proximity to the invasive breast cancer (IBC)
We first segmented the fibroglandular breast tissue from the adipose tissue, using software developed by our lab10 (Fig. 1). This semiautomated iterative segmentation method is based on a soft fuzzy c-means technique that labels MRI data depending on their adipose and fibroglandular tissue content within the entire breast. The fibroglandular tissue was further segmented into IBC tissue and normal-appearing breast tissue using a percent enhancement (PE ) map, where the PE value of a voxel, defined by (S1 – S0)/S0 × 100, is the percent signal intensity change of a voxel between the pre-contrast (S0) and post-contrast (S1) volume (Fig. 1). Voxels were classified as invasive if they were part of clusters of at least 3 connected voxels each exceeding 70% PE. This empirical threshold was shown to discriminate invasive from non-invasive lesions in more than 2000 breast MRIs obtained at our institution since 1995.
Fig. 1.
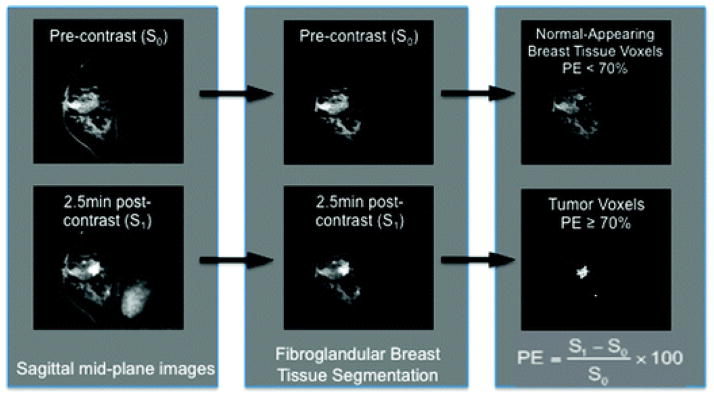
Tumor voxels are separated from normal-appearing breast following segmentation of fibroglandular breast tissue. (Left) Mid-sagittal cuts from pre- and post- contrast images are shown from a malignant tumor-bearing breast. (Middle) A fuzzy c-means based algorithm is applied to segment the fibroglandular breast tissue compartment. (Right) Voxels that represent malignant tumor are further segmented from normal-appearing fibroglandular breast tissue by percent enhancement threshold of >70%.
A tumor “proximity map” for the normal-appearing breast tissue was created which quantified the 3-dimensional distance from each normal-appearing breast tissue voxel to the nearest tumor voxel. Values of enhancement of each voxel were then recorded at specific distances around the tumor voxels. All voxels within 6 cm of a malignant voxel were analyzed. Pair-wise comparisons with Tukey’s adjusted p-values were made between voxels the edge of the tumor and those at farther distance intervals.
Selection of samples for immunohistochemistry (IHC)
Of the 23 cases analyzed by DCE-MRI, 15 had tissue available for normal stroma analysis. The remaining 8 cases did not have the corresponding tissue, or patients had lumpectomy only and normal stroma was not available for evaluation. These fifteen cases were analyzed for histologic evidence of an abnormal stromal phenotype that correlated with findings on DCE-MRI using post-surgical pathological tissue specimens. An experienced breast pathologist, Y.Y.C., reviewed all the pathology slides independent of the DCE-MRI readings. Post-surgically, tissue sections were fixed in 10% buffered formalin, routinely processed, and stained with hematoxylin and eosin (H&E). Two paraffin blocks for each case were selected using corresponding H&E stained slides: normal-appearing breast tissue less than 2 cm from the tumor edge and normal-appearing breast tissue greater than 2 cm from the tumor edge. Unstained tissue specimens corresponding to the 2 identified samples were cut from the blocks into 5 μm sections and placed on positively charged slides. All sections were subjected to routine deparaffinization using xylene and rehydrated through decreasing concentrations of ethanol before treatment with a hydrogen peroxide solution to block endogenous peroxidase activity. Antigen retrieval was performed in 10 mM citrate buffer at pH 6.0 and non-specific binding was blocked with normal horse serum in phosphate buffered saline. Tissue sections were incubated overnight at 4 °C with a primary antibody and then incubated for 30 min with a biotinylated secondary antibody. Next, the avidin–biotin complex was applied for 30 min. Subsequently, diaminobenzadine was used as the chromogen. Sections were then counterstained with hematoxylin, dehydrated through increasing concentrations of ethanol, cleared in xylene, and mounted. Primary anti-CD31 antibodies (clone JC70A, Dako, 1 : 50 dilution) and the appropriate positive and negative controls were run for each batch of slides.
Microvessel density quantification
As outlined in previous literature,11 we quantified CD31 microvessel density (MVD) for each sample under light microscopy. Three non-overlapping areas per sample considered to have the highest density of stained endothelial cells under lower magnification (100×) were identified as vascular hotspots. Each hotspot was scored for MVD under higher magnification (200×) and the average of the three hotspot measurements for each sample was used as the overall MVD. Any brown-staining endothelial cell or endothelial cell cluster that was clearly separate from adjacent microvessels was scored as a single countable microvessel. A lumen was not necessary for the structure to be identified as a microvessel and areas of necrosis or hemorrhage were avoided.
Microdissection and extraction of DNA
Post-surgical paraffin blocks for three patients were selected for microdissection. Tissue blocks from each patient included tumor (T), CAS (<2 cm from the tumor margin) and normal stroma (>2 cm from the tumor margin).
Eight consecutive 10 micron sections from formalin fixed paraffin embedded blocks were cut and placed onto standard slides. The microdissection slides were stained with methyl green. Slides were deparaffinized in xylene washes (2 × 3 min each) followed by rehydration for 2 min each through a decreasing ethanol gradient (100%, 95%, 70%, 50%, dH2O). Slides were then stained in 0.1% methyl green for 15 s, washed for 5 min in dH2O and air-dried.
Breast pathologist, Y.Y.C., reviewed and marked representative H&E stained slides for areas of tumor, cancer-associated non-epithelial stroma, cancer-associated epithelium, normal epithelium and normal non-epithelial stroma. Microdissection was performed manually using #15 and #11 scalpel blades under direct visualization using a Leica dissecting microscope. For each slide, the non-epithelial stromal components were dissected and collected first. Epithelial components were collected next into separate tubes. Finally, tumor cells were collected into separate tubes. Blades were changed for the dissection of each cell type.
Proteinase K (Sigma) was added to a final concentration of 0.4 μg μL–1 in sample buffer and the samples were incubated at 55 °C in a shaking water bath. Samples underwent digestion for 3–4 days with daily addition of fresh proteinase K until the pellets were dissolved. The proteinase K was inactivated by heating the samples to 95 °C for 10 min prior to DNA purification.
DNA was purified from the samples by water extraction using Millipore Microcon Ultracel YM-30 columns. Double-deionized water (ddH20) was added to the columns and centrifuged at 12 000 × g at room temperature to wet the columns. DNA samples were applied to the columns and centrifuged at 12 000 × g at room temperature to load the DNA into the columns. Two washes were performed with ddH2O and centrifugation at 12 000 × g at room temperature. The columns were inverted into collecting tubes and centrifuged at 13 000 × g to elute the DNA in water.
Quantitative real-time PCR (Taqman) was performed on 1 μL of the samples diluted serially up to 1 : 2000 in nuclease free water. Threshold cycles were compared to known DNA concentration standards in order to quantify sample DNA concentrations.
Comparative genomic hybridization (CGH)
The three cases that were chosen for CGH analysis had both MRI and CD-31 staining performed; they each had ample tumor, and near and far stroma tissues for microdissection. Limitations in resources precluded us from performing CGH analysis of the entire set. Fifty ng of DNA from each sample were used for whole genome amplification using the GenomePlex Complete Whole Genome Amplification Kit (Sigma WGA2). Two μg of the resulting amplified DNA were labeled with the Agilent Genomic Enzymatic Labeling Kit (Agilent 5190-0449) per manufacturer protocol. Normal diploid human male DNA (Promega) was amplified in the same fashion and counter-labeled for use as reference. Dye switching was performed on three samples to evaluate for dye bias. The labeled DNA samples were hybridized to Agilent G3 Human Catalog 4 × 180k CGH microarrays representing over 170 000 coding and noncoding human DNA sequences.
Statistical analysis
A mixed random effects model was used to evaluate the percent enhancement in normal breast tissue voxels as a function of distance from tumors. Patient identity was the random effect to control for intra-patient correlation among tissue voxels from the same patients. Tukey’s adjusted p-values were used to control for multiple comparisons for differences between distance categories. The paired t-test was used to evaluate microvessel density differences between tissues from near or far distance from tumor.
Two statistical analyses on array CGH data were performed. The first consisted of identifying gains and losses for each sample. To facilitate this, circular binary segmentation was used to segment the log ratios from each into regions of estimated equal copy number.12 Segments greater than 0.25 standard deviations (SDs) from the median estimated copy number were considered gains, and those less than 0.25 SDs from the same were considered losses. The second analysis was pair-wise, but otherwise similar to the first. The log ratios of the second were subtracted from the log ratios of the first, after which the same segmentation and calling of gains and losses was performed. Because the subtraction considerably reduced the variability, the cutoff for alteration was increased to 0.5 SDs.
Results
A 3-dimensional topographic map of percent-enhancement shows elevated enhancement at the tumor periphery in normal appearing breast fibroglandular tissue
We previously showed that the mean percent enhancement of breast stroma in patients receiving neoadjuvant chemotherapy for advanced breast cancer was correlated with disease-free survival.9 Subsequently, we developed automated techniques which allow segmentation of the fibroglandular tissue in volumetric shells that conform to the architecture of the tumor edge. This allowed us to evaluate the 3-dimensional fibroglandular tissue in its entirety, and to quantify enhancement levels, voxel-by-voxel, around the tumor. In the present study, each breast MRI was processed for segmentation of fibroglandular tissue from tumor, Fig. 1. The percent-enhancement for each non-tumor voxel was quantitated and a topographic map was created according to proximity to the tumor, Fig. 2A. We found that for all patients, enhancement levels in normal appearing breast tissue within 2 cm of the tumor were significantly higher than all more distant regions, Fig. 2B. Specifically, the mean enhancement at 0–1 cm (p = <0.0001) and at 1–2 cm (p = 0.006) from the edge of the tumor was significantly higher than farther distance intervals.
Fig. 2.
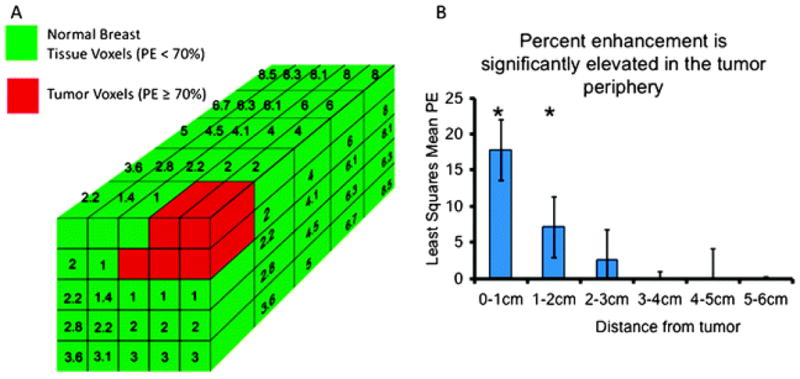
A 3-dimensional topographic map was created based on the proximity of each normal breast tissue voxel from its nearest neighboring tumor voxel. (A) Tumor voxels, identified by PE > 70%, are shown in red, while normal breast tissue voxels, identified by PE < 70%, are shown in green. Numbers written in each green box in the schema show the distance in millimetres of the green voxel to its closest red voxel. (B) Mean percent enhancement was significantly higher at the tumor edge, and decreased with distance through the fibroglandular breast tissue. Pairwise comparisons with Tukey’s adjusted p-values indicated that the mean enhancement at 0–1 cm (p = <0.0001) and at 1–2 cm (p = 0.006) from the edge of the MBL was significantly higher than farther distance intervals.
Microvessel density is significantly increased in areas of increased enhancement at the tumor periphery
The basis for increased enhancement in tissue is an increased rate of gadolinium extravasation from the vascular compartment. We now know that neoangiogenesis from the stromal compartment is necessary for tumor progression, and has been found to occur even in the setting of in situ carcinoma.13,14 Thus, we hypothesized that the increased enhancement detected in the normal-appearing tissue in the tumor periphery on MRI was due to increased vascularity around the tumor. To validate this finding, we measured microvessel density (MVD), detected by IHC labeling of CD31 antigen, and quantified the number of microvessels visualized per microscopic field in 15 of the 23 cases. The mean MVD for breast tissue <2 cm from the IBC (80 vessels mm 2, 95% CI : 61–98) was found to be significantly higher than the mean MVD in breast tissue >2 cm from the IBC (54 vessels mm 2, 95% CI : 36–72, p = 0.04 for two-sided t-test) (Fig. 3C).
Fig. 3.
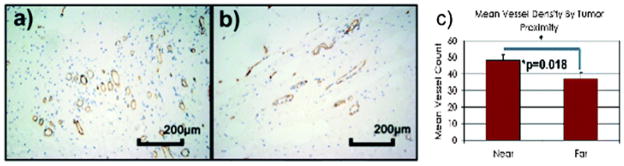
Microvessel density quantified by CD31 immunohistochemistry was compared between tissue sections close (within 2 cm) and far (>2 cm) of the tumor edge. Micrograph showing CD 31 immunohistochemistry in (a) near and (b) far tissue sections. (c) The mean MVD for breast tissue <2 cm from the IBC (80 vessels mm−2, 95% CI : 61–98) was found to be significantly higher than the mean MVD in breast tissue >2 cm from the IBC (54 vessels mm−2, 95% CI : 36–72, p = 0.04 for two-sided t-test).
Comparisons of genomic changes in microdissected tumor, cancer-associated stroma and normal stroma using array CGH show that cancer-associated stroma closely resembles normal stroma
We and others have found phenotypic changes in the periphery of tumors, such as increased angiogenesis, that support the hypothesis that a transformed stroma is necessary for malignant tumor progression. In the present study, to investigate whether genome-wide changes in the tumor are represented in the cancer-associated stroma or vice versa, we chose to perform array CGH from 3 separate cases. DNA from microdissected specimens was isolated from the tumor, CAS and normal stroma (NS). Epithelial and non-epithelial compartments were isolated separately from the stromal tissue (Fig. 4).
Fig. 4.
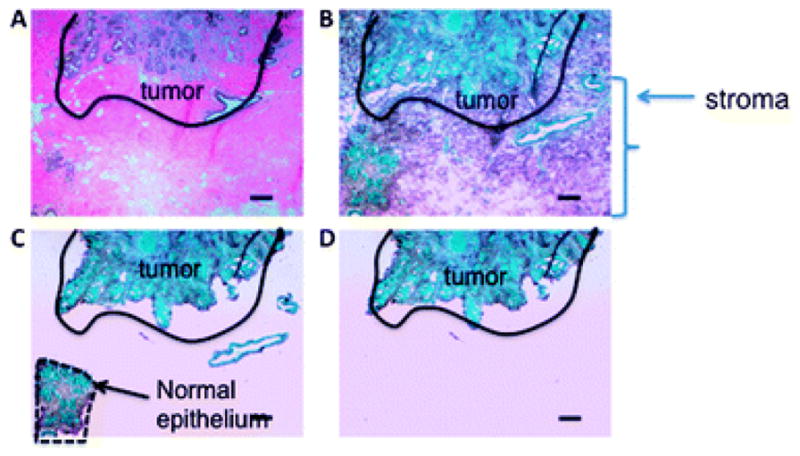
Thin sections from near and far tissue blocks were microdissected for genomic array analysis. (A) A hematoxylin and eosin stained section shows tumor (black outline) and surrounding stroma. (B) Cresyl green stain showing stromal elements. (C) Non-epithelial normal tissue was microdissected. (D) Normal epithelium was isolated from all other stromal elements.
We found that the percentage of the genome altered, either gained or lost, was substantially higher in two of the three tumor samples than it was in any of the other samples. It was 42.7% altered in tumor from case “A” and 34.7% altered in tumor from case “B”. In the third case “C”, tumor was only 5.9% altered. The non-tumor samples were altered between 0.1% and 2.9% for all three cases, and therefore most alterations in the tumors were not replicated elsewhere, representative case shown in Fig. 5.
Fig. 5.
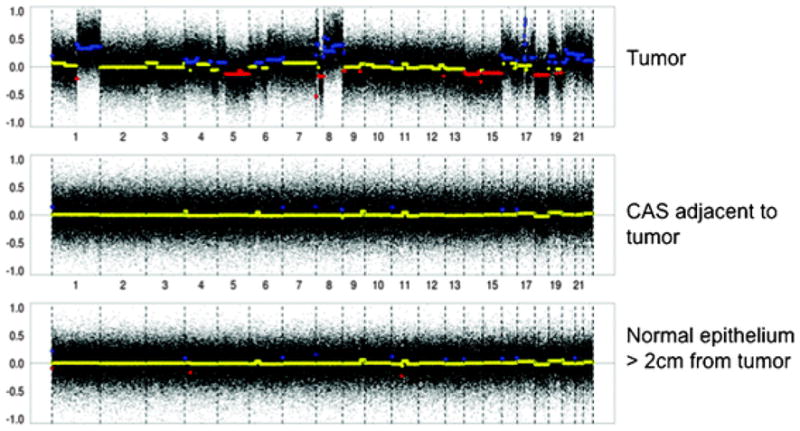
Array comparative genomic hybridization was performed on tumor, adjacent stroma and stroma far from the tumor in 3 cases. Representative genome wide changes are depicted with mean values in yellow, gains in blue and losses in red. Compared to tumor, stroma adjacent to and far from the tumor harbored significantly less chromosomal changes than that in the tumor.
We also examined pair-wise differences between CAS and NS samples. To do this, we compared CAS epithelium to NS epithelium and CAS non-epithelial stroma to NS non-epithelial stroma. Overall, we found that the percentage of the genome differentially altered was minimal, ranging from 0.0% to 0.6%. We also analyzed pair-wise differences between non-tumor epithelium and stroma. Specifically, we compared CAS non-tumor epithelium to CAS stroma and NS non-tumor epithelium to NS stroma. Here the percentage of the genome differentially altered was never more than 0.1%. The lower rates of alterations in the pair-wise analyses suggested that alterations in the non-tumor samples, whatever the causes, were repeated across the different types of samples.
Discussion
The emergence of the malignant phenotype relies both on initiated epithelial cells and an activated or permissive stroma in order for cancer progression to occur.1,8,15,16 Studies of cancer stroma in laboratory animals and in culture have shown the critical and specific role of stroma, and revealed novel approaches to targeting the dependent relationship between a growing tumor and its supporting stroma.1,4,17 Active investigation is ongoing addressing how the cancerous epithelial cells co-opt stromal cells, and vice versa. However, in humans, the role of cancer-associated stroma in clinical practice has been limited. Here, we report an MRI-based tool to characterize a cancer-associated phenotype based on topographically mapping enhancement levels in normal-appearing tissue surrounding invasive tumors. This region is histologically associated with increased microvessel density, although genomically resembles its normal tissue counterpart. These findings offer new insight to cancer-associated stroma in humans, and potentially a novel tool to help guide local therapy.
We previously showed that global enhancement levels in the entire breast could have predictive value in patients receiving neoadjuvant chemotherapy for locally advanced breast cancer.9 In this exploratory study, we addressed patients with small tumors to determine whether enhancement in the normal appearing breast tissue could be informative. The patterns or “steps” of how the stroma becomes supportive for cancer growth are not well understood. However, studies indicate that in situ cancers are able to elicit angiogenesis, even without an invasive component, and that cancer cells can elaborate and induce extensive stromal remodeling.13 Although initial studies indicate that stromal gene expression can be a potent predictor of outcomes, the extent of remodeling has not been correlated with clinical breast cancer behavior.7 The ability to detect the extent of abnormal stroma that could increase risk for recurrence is potentially an important clinical tool. Breast cancer is often approached using surgery and radiation in order to preserve the breast, and avoid mastectomy. However, the degree of surgical excision, and targeted radiation, either as a boost or in the context of partial breast radiotherapy, are variable in practice. The optimal amount of normal tissue to treat surrounding a tumor is debatable and varies among institutions. Therefore, a tool that could guide local therapy to optimally reduce cancer recurrence risk could potentially avoid re-excision surgeries that are often necessary, and decrease unneeded treatment.
Our results also give insight into the underlying genomic underpinnings of the cancer-associated stroma. Whether the stroma co-evolves genomically with the tumor, or whether its involvement is predominantly epigenetic is not well understood.7 Several studies have shown that loss of heterozygosity, and specific mutations including p53 and PTEN, are identifiable in the stroma of breast cancers.18,19 However, others have not found significant genetic alterations in stroma.6 Array CGH revealed that genomic changes in the cancer-associated stroma were congruent with that of normal tissue, and not the tumor. Thus, phenotypic changes such as increased angiogenesis and evidence for stromal remodeling, may be primarily driven through epigenetic mechanisms, and may be targetable. Further investigations along these lines would be important.
Finally, we use an empirically derived 70% PE threshold to identify invasive lesions visualized on MRI. Although there is no direct radiology–pathology correlation, we have found excellent correlation between invasive tumor size measured by MRI using the 70% threshold, and pathologic tumor size.20 In fact, MRI has been shown to overestimate the size of tumor, not underestimate;21–23 thus, it is unlikely that the stromal measurements (<70% PE ) are within the tumor. In the cases selected for the present study, patients did receive surgery soon after MRI, and all cases were carefully reviewed for size and extent of DCIS surrounding the tumor periphery.
We would like to acknowledge the limitations of this retrospective study. One, a pathologic correlation of the size of lesions seen on MRI has not been established. Although others have shown that MRI tends to overestimate tumor size,21–23 our empiric approach to identifying tumor boundaries has not been validated pathologically. In addition, the small numbers in our study overall may limit whether our results can be generalized. Despite this, to our knowledge, our study is the first to identify and characterize a cancer-associated stroma in breast cancer patients using evaluation of enhancement patterns on MRI. We have integrated a biological concept with novel application of imaging tools and corresponding pathologic and genomic tissue analysis. Future studies are necessary to further evaluate and validate the clinical utility of these findings.
Insight, innovation, integration.
We describe a novel approach to evaluating cancer-associated stroma in patients with breast cancer. Using an innovative MRI technique to assess normal appearing breast tissue, elevated enhancement was quantified in relationship to proximity to the tumor. On pathologic evaluation of the corresponding tissue, this region was found to harbor significantly increased microvessel density. Despite changes in phenotype, and elevated enhancement on MRI, there were minimal genomic changes detected by array CGH. Our study is the first to integrate molecular biology, pathology and imaging techniques to lend insight into cancer-associated stroma in patients.
Acknowledgments
We thank Hui Zhang for expert technical assistance and Dylan Smart for assistance with the figures. This work is supported by NIH grant 1R01CA124891 (to CP) and by the American Cancer Society RSG-07-1110-01-CCE (to CP); Doris Duke Charitable Foundation (N.N.), California Breast Cancer Research Program CBCRP 13IB-0171 (C.K.) and NIH Grant R01 CA 069587 to N.H.
References
- 1.Cunha GR, Hayward SW, Wang YZ. Role of stroma in carcinogenesis of the prostate. Differentiation. 2002;70:473–485. doi: 10.1046/j.1432-0436.2002.700902.x. [DOI] [PubMed] [Google Scholar]
- 2.Dolberg DS, Hollingsworth R, Hertle M, Bissell MJ. Wounding and its role in RSV-mediated tumor formation. Science. 1985;230:676–678. doi: 10.1126/science.2996144. [DOI] [PubMed] [Google Scholar]
- 3.Sieweke MH, Thompson NL, Sporn MB, Bissell MJ. Mediation of wound-related Rous sarcoma virus tumorigenesis by TGF-beta. Science. 1990;248:1656–1660. doi: 10.1126/science.2163544. [DOI] [PubMed] [Google Scholar]
- 4.Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- 5.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 6.Campbell I, Polyak K, Haviv I. Clonal mutations in the cancer-associated fibroblasts: the case against genetic coevolution. Cancer Res. 2009;69:6765–6768. doi: 10.1158/0008-5472.CAN-08-4253. discussion 6769. [DOI] [PubMed] [Google Scholar]
- 7.Eng C, Leone G, Orloff MS, Ostrowski MC. Genomic alterations in tumor stroma. Cancer Res. 2009;69:6759–6764. doi: 10.1158/0008-5472.CAN-09-0985. [DOI] [PubMed] [Google Scholar]
- 8.Weaver VM, et al. Reversion of the malignant phenotype of human breast cell s in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattangadi J, et al. Breast stromal enhancement on MRI is associated with response to neoadjuvant chemotherapy. AJR, Am J Roentgenol. 2008;190:1630–1636. doi: 10.2214/AJR.07.2533. [DOI] [PubMed] [Google Scholar]
- 10.Klifa C, et al. Quantification of breast tissue index from MR data using fuzzy clustering. Conf Proc IEEE Eng Med Biol Soc. 2004;3:1667–1670. doi: 10.1109/IEMBS.2004.1403503. [DOI] [PubMed] [Google Scholar]
- 11.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis—correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 12.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–66. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 13.Brown LF, et al. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin Cancer Res. 1999;5:1041–1056. [PubMed] [Google Scholar]
- 14.Guidi AJ, et al. Vascular permeability factor (vascular endothelial growth factor) expression and angiogenesis in patients with ductal carcinoma in situ of the breast. Cancer. 1997;80:1945–1953. doi: 10.1002/(sici)1097-0142(19971115)80:10<1945::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 15.Bissell MJ, Labarge MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunha GR, Hayward SW, Wang YZ, Ricke WA. Role of the stromal microenvironment in carcinogenesis of the prostate. Int J Cancer. 2003;107:1–10. doi: 10.1002/ijc.11335. [DOI] [PubMed] [Google Scholar]
- 18.Moinfar F, et al. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60:2562–2566. [PubMed] [Google Scholar]
- 19.Patocs A, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 20.Esserman L, Hylton N, George T, Weidner N. Contrast-enhanced magnetic resonance imaging to assess tumor histopathology and angiogenesis in breast carcinoma. Breast J. 1999;5:13–21. doi: 10.1046/j.1524-4741.1999.005001013.x. [DOI] [PubMed] [Google Scholar]
- 21.Behjatnia B, Sim J, Bassett LW, Moatamed NA, Apple SK. Does size matter? Comparison study between MRI, gross, and microscopic tumor sizes in breast cancer in lumpectomy specimens. Int J Clin Exp Pathol. 3:303–309. [PMC free article] [PubMed] [Google Scholar]
- 22.Grimsby GM, et al. Is there concordance of invasive breast cancer pathologic tumor size with magnetic resonance imaging? Am J Surg. 2009;198:500–504. doi: 10.1016/j.amjsurg.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 23.Onesti JK, Mangus BE, Helmer SD, Osland JS. Breast cancer tumor size: correlation between magnetic resonance imaging and pathology measurements. Am J Surg. 2008;196:844–848. doi: 10.1016/j.amjsurg.2008.07.028. discussion 849–850. [DOI] [PubMed] [Google Scholar]


