Abstract
Staphylococcus aureus clonal complex 75 (herein referred to as S. argenteus) lacks the carotenoid pigment operon, crtOPQMN, responsible for production of the putative virulence factor, staphyloxanthin. Although a common cause of community-onset skin infections among Indigenous populations in northern Australia, this clone is infrequently isolated from hospital-based patients with either bacteremic or nonbacteremic infections. We hypothesized that S. argenteus would have attenuated virulence compared to other S. aureus strains due to its staphyloxanthin “deficiency.” Compared to prototypical S. aureus strains, S. argenteus was more susceptible to oxidative stress and neutrophil killing in vitro and had reduced virulence in murine sepsis and skin infection models. Transformation with pTX-crtOPQMN resulted in staphyloxanthin expression and increased resistance to oxidative stress in vitro. However, neither resistance to neutrophil killing nor in vivo virulence was increased. Thus, reduced virulence of S. argenteus in these models is due to mechanisms unrelated to lack of staphyloxanthin production.
Keywords: Staphylococcus aureus, staphyloxanthin, virulence, Australia, carotenoid pigment
Staphylococcus aureus is a very successful human commensal and pathogen. Although there are multiple lineages within the species, the core genome is well conserved, with <2% nucleotide divergence between lineages. Recently, an early branching and divergent lineage has been described that has approximately 10% nucleotide divergence compared to other S. aureus lineages [1–4]. A representative strain, MSHR1132, was found to lack the crtOPQMN gene operon encoding the carotenoid pigment, staphyloxanthin; all other tested strains from this lineage were also nonpigmented. On the basis of the nucleotide divergence and nonpigmented phenotype, this lineage, previously known as clonal complex 75, has been provisionally named S. argenteus [1].
S. argenteus is a highly prevalent and successful community-associated lineage in Indigenous populations in Australia and French Guiana [3, 5]. We have previously hypothesized that it would be less virulent than S. aureus, reflecting its ancestral phylogenetic status and apparent predilection for minor skin infections [1]. Furthermore, because staphyloxanthin confers resistance against oxidative stress and neutrophil killing [6], and inhibition of staphyloxanthin synthesis has been demonstrated to attenuate virulence in S. aureus [7], an intriguing possibility is that the virulence properties of S. argenteus differ from that of S. aureus as a result of a lack of staphyloxanthin production.
The purposes of the current study were (i) to compare the prevalence of S. argenteus in 3 distinct clinical populations of S. aureus infections (community-based impetigo, and hospital-based patients with either bacteremic or nonbacteremic infections (“hospital-based bacteremia” and “hospital-based clinical infections,” respectively); all patients were from northern Australia where S. argenteus is endemic; and (ii) to test the hypothesis that S. argenteus is less virulent than other S. aureus lineages and, if so, whether this is due to a lack of staphyloxanthin production.
METHODS
Genotyping from S. aureus Strain Collections
Three S. aureus strain collections from the “Top End” region of northern Australia were examined for the prevalence of S. argenteus: impetigo lesion isolates from children across 5 remote Australian Indigenous communities recruited from 2 previous studies between 2003 and 2007 [3, 8]; nonbacteremic isolates from the Royal Darwin Hospital (RDH) from 2006 to 2007 [4, 9]; and bloodstream isolates from the RDH from 2007 to 2010 (identified through hospital microbiology databases). Isolates were genotyped using either an allele-specific polymerase chain reaction (PCR) or high-resolution melting analysis of multilocus sequence type (ST)-derived single-nucleotide polymorphisms [3, 10]. Patient demographics and strain descriptions related to the first 2 collection cohorts above have been reported elsewhere [3, 4, 8].
Bacterial Strains and Plasmid-Transformation of crtOPQMN
S. aureus cultures were grown overnight in trypticase soy broth (TSB), diluted to an optical density (OD600) of 0.1, and grown to either exponential phase (OD600, 0.75–1.0) or stationary phase (40 hours). Cells were washed twice with phosphate buffered saline (PBS) and resuspended in 20% glycerol from which colony-forming units (CFUs) were enumerated on trypticase soy agar (TSA). Aliquots of the suspension were frozen at −80°C until further use, at which time they were individually thawed and CFUs reenumerated.
As controls for our investigations, we chose well-characterized community-associated S. aureus strains to represent different S. aureus lineages: LAC (ST8; USA300) [11], MSSA476 (ST1) [12], and JKD6159 (ST93) [13]. We also included a S. epidermidis strain, RP62a. We transformed a S. argenteus strain, MSHR1132, with a plasmid containing the crtOPQMN gene operon (kindly provided by Dr F. Gotz; Tubingen, Germany). The plasmid construct pTXcrtOPQMN [14] was purified using a Wizard Plus kit from Promega, Inc. (Madison, WI). Electroporation of the plasmid DNA into MSHR1132 strain was performed as per Schenk and Laddaga [15] and transformants selected on TSA containing tetracycline (5 µg/mL).
Quantification of Carotenoids
We visually determined pigmentation status and also quantitatively compared the overall carotenoid content of the parental MSHR1132 and transformant strains, with the LAC strain used as a pigment-positive control. In brief, S. aureus cells were grown in brain heart infusion (BHI) broth to stationary phase (24 hours) at 37°C, then harvested, washed, and pelleted in PBS by centrifugation. Excess liquid was removed from the final pellets by inversion for at least 5 minutes. The S. aureus cells were then subjected to methanol extraction, and the carotenoid content spectrophotometrically determined at 450 nm wavelength [6, 16]. A minimum of 3 independent experiments was performed for all strains on different days.
Oxidative Stress Studies
Stationary-phase S. aureus cells (2 × 108 CFU) were incubated with 0, 0.375%, 0.75%, and 1.5% final concentrations of hydrogen peroxide (H2O2) in 1 mL PBS at 37°C for 1 hour; 1000 IU/mL of catalase (Sigma-Aldrich, St Louis, MO) was then added to quench residual H2O2. An aliquot of the assay was plated on TSA, and CFUs were enumerated the following day. For the singlet oxygen assay, stationary-phase S. aureus cells (5 × 108 CFUs) were incubated with 1 μg/mL methylene blue in 1 mL PBS at 37°C and 10 cm from a 100-watt light bulb for 1 hour as described elsewhere [6]. CFUs were enumerated as described above. Cell counts from control cultures containing no methylene blue or methylene blue + foil (to shield from light) demonstrated no bacterial killing. Three replicates of each experiment were performed.
Host Defense Peptides (HDPs) Susceptibilities
Although our studies above focused on the impact of the carotenoid locus on the oxidative limb of innate host defense against staphylococci, it was important to also examine the nonoxidative limb in parallel. Since cationic HDPs are very important in the nonoxidative innate immune response to infection, including skin and soft tissue infections, we analyzed the susceptibility profiles of our parental and transformant strains to 2 prototypical and relevant HDPs. Both HDPs that were used (LL-37; and hNP-1) are present in neutrophils, although stored in distinct granules (specific granules; and azurophilic granules, respectively) [17]. In addition, LL-37 is found constitutively in skin epidermal cells. Both LL-37 and hNP-1 were purchased from Peptide International (Louisville, KY). Stationary phase cells were used in these 2-hour timed-kill assays, which were carried out in the appropriate buffers as described elsewhere [16] (and see below). The concentrations of peptides used in the killing assays were 5 and 10 µg/mL for LL-37 and 10 and 20 µg/mL for hNP-1. These peptide concentrations represent levels that did not rapidly kill the parental strain in this assay, based on extensive pilot experiments.
In brief, stationary phase S. aureus cells were diluted into the peptide solutions and adjusted to desired final inoculum (103 CFU/mL) and peptide concentrations, then incubated at 37°C. After 2 hours of exposure, samples were obtained and processed for quantitative culture to evaluate the extent of killing by each HDP [16]. Final data were expressed as mean (± SD) percent surviving CFU/mL. Since there is no bona fide “resistance” breakpoint for HDPs, we statistically compared only the mean percent survivability (± SD) in the parental vs transformant strains. A minimum of 3 experimental runs on separate days was performed for each peptide.
Neutrophil Bactericidal Activity Assays
Human neutrophils were isolated from heparinized venous blood of healthy donors using a standard method [18]. All work was performed in accordance with protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases, National Institutes of Health. All human blood donors provided written informed consent prior to participation in the study.
S. aureus strains were cultured to midexponential phase of growth as described above, washed in PBS, and then opsonized in 50% normal human serum for 30 minutes. Bacteria were washed in PBS and resuspended in Roswell Park Memorial Institute (RPMI) 1640 buffered with 10 mM Hepes (RPMI/H) and at 107 CFU/mL. In total, 106 bacteria were combined with 106 neutrophils (600 μL final volume) and rotated gently for 2 hours at 37°C. Based on extensive pilot studies, experience in the literature, and our bacteria:neutrophil ratios, we chose 2 hours as an optimal time point for these assays. An aliquot of the assay was removed at 30 minutes for subsequent analysis by light microscopy. At the end of the 2-hour time period, saponin (0.1% final concentration) was added to permeabilize neutrophils, and assay tubes were chilled on ice for 15 minutes. An aliquot of the assay was plated on Luria-Bertani agar, and CFUs were enumerated the following day. Percentage of S. aureus survival was calculated with the equation (CFUPMN+S. aureus/CFUS. aureus) × 100.
For analysis of samples by light microscopy, samples were prepared on slides by using a Cytospin 4 and Wright-Giemsa stain. In total, 250 neutrophils from 5 fields of view were scored to determine the percentage of neutrophils with bound and/or ingested S. aureus (total associated). Nine independent experiments were performed.
Murine Infection Studies
Male BALB/c mice (the Jackson Laboratory, Bar Harbor, ME) aged 4–6 weeks were housed in a centralized research animal facility at Duke University, North Carolina, and provided with food and water ad libitum. All animal experiments were performed in accordance to National Institutes of Health (NIH) guidelines, the Animal Welfare Act, and US federal law, and were approved by Duke University's Institutional Animal Care and Use Committee (IACUC).
For the skin abscess model [13], mice were anesthetized with intraperitoneal ketamine/xylazine, had their right flank shaved and then subcutaneously inoculated with 1 × 107 CFU of exponential phase bacteria in 50 μL PBS. Key experiments were repeated using stationary phase bacteria as specified in the results. Paracetamol was added to the drinking water to a concentration of 1 mg/mL for analgesia as prescribed by the IACUC. Mice were assessed daily, with lesion size measured using calipers, and the infection area calculated by length × width. A subset of mice was killed at day 3 at which time photographs were taken, skin and muscle harvested for histology, and tissue homogenized for enumeration of CFUs. Histopathology changes were assessed by a dermatopathologist who was blinded to the infecting strains. Changes were defined as mild inflammation for inflammatory cells in the subcutaneous layer and muscle; moderate inflammation for inflammatory cells in subcutaneous layer, muscle and soft tissue; and severe inflammation for severe necrosis in epidermis, dermis, subcutaneous layer, and deep muscle.
For the sepsis model [19], mice were intraperitoneally inoculated with 1 × 109 CFU of exponential phase bacteria in 200 μL PBS and observed every 24 hours for 5 days. Moribund mice were killed.
Statistical Analysis
Categorical data are presented as proportions and compared using χ2 or Fisher exact test. Continuous data are presented as means with standard errors of the mean and compared using the Mann–Whitney U test or ANOVA and Tukey post-test. Survival analysis is presented as Kaplan–Meier plots and compared using the log rank test. Two-sided P values < .05 were considered significant. Analyses were performed using Stata 11 (StataCorp, TX) and GraphPad Prism 5.01 (GraphPad Software, Inc, CA).
RESULTS
The Epidemiological Niche of S. argenteus Differs From that of Other S. aureus Lineages
Large, representative S. aureus strain collections recovered from 3 distinct clinical populations from the same geographical region of northern Australia were used to compare the prevalence of S. argenteus across these cohorts (Figure 1). S. argenteus made up 8% (36 of 431) of methicillin-susceptible Staphylococcus aureus (MSSA) and 69% (62 of 90) of methicillin-resistant Staphylococcus aureus (MRSA) recovered from impetigenous lesions in children from remote Australian Indigenous communities. However, this clone accounted for only 1% (3 of 226) and 9% (21 of 229) of MSSA and MRSA nonbacteremic hospital-infection isolates and 1% (3 of 220) and 0% (0 of 47) of MSSA and MRSA hospital bacteremias at the RDH (P < .001 for a comparison of proportions across the 3 cohorts for both MSSA and MRSA). Among the nonbacteremic hospital-infection MRSA isolates, S. argenteus was more commonly recovered from Indigenous patients (17 of 126 [13%]) than from non-Indigenous patients (4 of 103 [4%]; P = .02).
Figure 1.

Proportion of Staphylococcus argenteus among Staphylococcus aureus isolates in 3 distinct clinical cohorts from northern Australia. The clinical cohorts are community-based impetigo, hospital-based clinical disease, and hospital-based bacteremia. S. argenteus is represented by solid red bars; other S. aureus lineages by striped blue bars. A, Methicillin-susceptible S. aureus (MSSA). B, Methicillin-resistant S. aureus (MRSA).
Quantification of Carotenoids
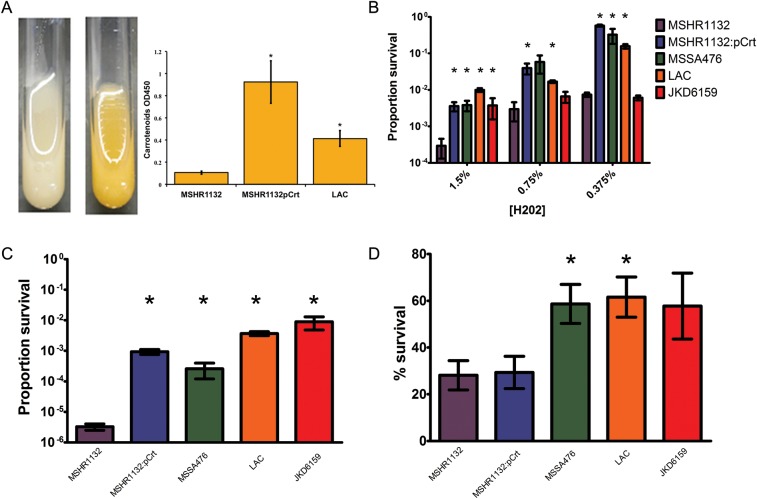
Transformant construct MSHR1132 containing pTX-crtOPQMN displayed a typical golden pigmented phenotype (Figure 2A). As expected, the quantity of carotenoid content of the transformant strain was significantly higher than its parental isolate, which was at the lower limit of detection by this assay (Figure 2A). The LAC control strain, which is representative of the widespread USA300 clone, also produced significantly more carotenoid than the parental strain.
Figure 2.
Plasmid transformation and oxidative stress and neutrophil survival assays. A, Pigment production. Following transformation with the plasmid pTX-crtOPQMN a yellow phenotype consistent with expression of staphyloxanthin is demonstrated (MSHR1132 on left, MSHR1132:Pcrt on right). Both the transformant strain and LAC (USA300) strain produced significantly more carotenoid pigment than the parental MSHR1132 strain (both P < .02). B, H2O2 mediated killing. Survival following incubation in H2O2 1.5%, 0.75% and 0.375%. MSHR1132 is more susceptible to H2O2 killing compared to MSHR1132:Pcrt and other Staphylococcus aureus strains. C, Singlet-oxygen mediated killing. MSHR1132 is more susceptible to singlet-oxygen mediated killing compared to MSHR1132:Pcrt and other S. aureus strains. D, Phagocytic interaction with neutrophils. MSHR1132 has similar survival compared to MSHR1132:Pcrt but has reduced survival compared to MSSA476 and LAC. Carotenoid pigment quantification and oxidative stress assays were performed in triplicate and neutrophil survival assays from 9 blood donors. Columns represent means with standard deviation (SD) for the carotenoid pigment assays and with standard errors of the mean (SEM) for all other assays. * indicates P < .05 for comparison with MSHR1132.
Oxidative Stress Resistance Is Augmented by Staphyloxanthin
We compared resistance to oxidative stress in vitro using H2O2 and singlet oxygen assays (Figure 2B and 2C). MSHR1132 was significantly more susceptible to H2O2 compared to MSSA476 and LAC and more susceptible to singlet oxygen than all other S. aureus strains. The transformant MSHR1132 (containing pTX-crtOPQMN) strain was significantly more resistant than MSHR1132 in these assays (P < .05 for both assays), confirming the ability of staphyloxanthin to modulate the antibacterial effects of H2O2 and singlet oxygen.
Susceptibility to Host Defense Peptides (HDPs)
The transformant strain consistently appeared to be more susceptible than the parental strain to both LL-37 and hNP-1. However, these data differences did not reach statistical significance (Table 1).
Table 1.
In vitro Susceptibilities of Study Strains to Two Distinct Host Defense Peptides
| Strains | LL-37 (5 µg/mL) | LL-37 (10 µg/mL) | hNP-1 (10 µg/mL) | hNP-1 (20 µg/mL) |
|---|---|---|---|---|
| MSHR1132 | 96 ± 16 | 38 ± 19 | 76 ± 14 | 68 ± 25 |
| MSHR1132:pCrt | 86 ± 21 | 26 ± 21 | 56 ± 11 | 33 ± 12 |
Per cent survival (mean ± SD) after 2 hours exposure to two concentrations of host defense peptides
Survival of MSHR1132 Following Phagocytic Interaction With Human Neutrophils Is Not Augmented by Staphyloxanthin
To determine if the noted in vitro differences in resistance to oxidative stress among the strains translated into differential survival following interaction with host phagocytic leukocytes, we tested bacterial survival during co-culture with human neutrophils (Figure 2D). The binding/uptake of bacteria by neutrophils was similar for all bacterial strains tested (data not shown). LAC and MSSA476 were more resistant to killing by neutrophils than MSHR1132 (both comparisons P < .05). Surprisingly, MSHR1132 containing pTX-crtOPQMN was not more resistant to neutrophil killing that the parent strain MSHR1132. Thus, although expression of staphyloxanthin conferred increased resistance to oxidative stress on MSHR1132, this did not translate into improved survival following phagocytic interaction with neutrophils.
In vivo Virulence in Murine Skin Infection and Sepsis Models
We next compared the different community-associated S. aureus lineages in both skin abscess and systemic sepsis models (Figure 3). In the skin abscess model with exponential phase bacteria, JKD6159 (ST93) was clearly the most virulent strain and MSHR1132 the least virulent strain, as assessed by the measured area of infected lesion, CFUs recovered from skin abscesses, and histologically determined inflammation and necrosis. MSHR1132 caused larger lesions than RP62a (S. epidermidis). The JKD6159 strain was also the most virulent and strain MSHR1132 the least virulent, in the murine sepsis model (Figure 3D). Of note, there was no difference between MSHR1132 and transformant MSHR1132 containing pTX-crtOPQMN in any outcome metric in either infection model. In addition, a mean of 89% (±4.5%) of CFUs of the transformant strain displayed a pigmented phenotype at day 3 following inoculation in the skin abscess model, thus demonstrating excellent plasmid stability.
Figure 3.
Skin infection and sepsis models using BALB/c mice and exponential phase bacteria. A, Area of skin abscess lesions demonstrating the marked virulence of JKD6159 (P < .01 at all time points compared to other strains). For greater clarity, B presents the same data without JKD6159. MSHR1132 caused smaller lesions than MSSA476 and LAC at all time points (except days 9–10) and there was no difference between MSHR1132 and MSHR1132:Pcrt (except days 8–9). For each Staphylococcus aureus strain, there were at least 20 mice for days 1–3 and 10 mice for days 4–12. Points represent mean area, and bars represent the standard errors of the mean (SEM). C, CFUs recovered from abscess lesions. There were fewer colony-forming units (CFUs) for MSHR1132 compared to MSSA476, LAC and JKD6159; but no difference between MSHR1132 and MSHR1132:Pcrt. JKD6159 has more CFUs than all other strains. For each S. aureus strain there were 8 mice. Bars represent mean CFUs and SEM, and * if P < .05 compared to MSHR1132. D, Intraperitoneal sepsis. Survival curve following intraperitoneal inoculation of 1 × 109 CFUs. Survival for MSHR1132 was significantly greater compared to MSSA476, LAC, and JKD6159 (*P < .05) but was not different compared to MSHR1132:Pcrt. Survival for JKD6159 was similar compared to LAC at 1 × 109 CFUs but significantly worse using an inoculation of 5 × 108 (P < .01, data not shown). At least 10 mice were used for each S. aureus strain. E–N, Photos and histological sections of representative lesions for strains MSHR1132 (E and J), MSHR1132:Pcrt (F and K), MSSA476 (G and L), LAC (H and M) and JKD6159 (I and N). There was minimal and mild inflammation for MSHR1132 (J) and MSHR1132:Pcrt (K), respectively; mild inflammation for MSSA476 (L) with inflammatory cells in the subcutaneous layer and muscle; moderate inflammation for LAC (M) with inflammatory cells in sub-cutaneous layer, muscle and soft tissue; and severe inflammation for JKD6159 (N) with severe necrosis in epidermis, dermis, subcutaneous layer, and deep muscle.
Given that stationary phase bacteria may produce greater amounts of staphyloxanthin as compared to log phase bacteria [6], we repeated the in vivo murine skin infection studies using stationary phase bacteria (Figure 4) for MSHR1132 (n = 14) and MSHR1132:Pcrt (n = 14). Again, no difference was seen with regard to size of skin lesions at any time point, nor was there a difference in CFUs at day 3 (n = 3 for each strain). Following inoculation with stationary phase MSSA476 (n = 15), the size of skin lesions and CFUs at day 3 were consistent with greater virulence for MSSA476 compared to MSHR1132. Thus, similar to the findings in the neutrophil bactericidal activity assays, staphyloxanthin-mediated resistance to oxidative stress did not translate to increased strain virulence.
Figure 4.

Skin infection model using BALB/c mice and stationary phase bacteria. A, Area of skin abscess lesions. MSHR1132 caused smaller lesions than MSSA476 at all time points and there was no difference between MSHR1132 and MSHR1132:Pcrt. For each Staphylococcus aureus strain, there were at least 14 mice for days 1–3 and 10 mice for days 4–12. Points represent mean area, and bars represent the standard errors of the mean (SEM). B, Colony-forming units (CFUs) recovered from abscess lesions. There were fewer CFUs for MSHR1132 compared to MSSA476; but no difference between MSHR1132 and MSHR1132:Pcrt. For each S. aureus strain there were 3 mice. Bars represent mean CFUs and SEM, and * if P < .05 compared to MSHR1132.
DISCUSSION
Several key observations emerged from this study. First, the phylogenetically divergent S. argenteus lineage occupies a distinct epidemiological niche as compared to other S. aureus strains in our geographic locale-of-interest. We observed that S. argenteus was less likely to cause hospital-based clinical disease but rather appears well adapted to cause superficial skin lesions. Given its prevalence in remote Indigenous populations with poor levels of housing and hygiene in northern Australia and French Guiana [3, 5], and the rarity of S. argenteus within the overall global population sampling of S. aureus (notwithstanding the bias toward sampling from clinical disease and hospital settings), it would appear that S. argenteus is less successful than other S. aureus in human populations with access to modern-day hygiene and healthcare facilities.
Second, we hypothesized that the lack of staphyloxanthin production might be associated with the triad of increased susceptibility to oxidative killing, enhanced neutrophil-mediated killing, plus reduced virulence in multiple animal models. Although the in vivo virulence of wild-type S. argenteus was indeed less than that of other S. aureus strains in murine infection models, neither the resistance to killing by neutrophils nor in vivo virulence was augmented by the expression of staphyloxanthin. Our findings suggest that S. argenteus is intrinsically more susceptible to killing by neutrophils by mechanisms unrelated to a lack of staphyloxanthin production. This stands in contrast to previous findings where an isogenic S. aureus Δcrt mutant that did not express staphyloxanthin demonstrated increased susceptibility to oxidative stress and killing by neutrophils compared to the wild-type strain, as well as reduced virulence in a murine skin infection model [6].
Given the phylogenetic divergence of S. argenteus from other S. aureus, it is possible that S. argenteus lacks the regulatory pathways to appropriately use staphyloxanthin in response to stress. However, 2 known regulators of staphyloxanthin—the RNA polymerase sigma factor, SigB, and its sensing module, RsbU [20]—are both present in S. argenteus and have a 99% amino acid identity to these proteins found in other S. aureus strains. Furthermore, MSHR1132 containing pTX-crtOPQMN clearly demonstrated an ability to use crtOPQMN in response to oxidative stress to maintain survivability.
In investigating the nonoxidative limb of host defense against staphylococci we noted that MSHR1132 containing pTX-crtOPQMN is likely to be more susceptible than the parental wild-type strain to in vitro killing by 2 HDPs relevant to neutrophil- and epidermal-related killing of S. aureus (although these data did not reach statistical significance due to high standard deviations as is common in these assays). This may be a strain-specific finding as we have previously demonstrated in other S. aureus strains that carotenoid overexpression often makes cells more “resistant” to HDPs [16, 21]. Nonetheless, an increased susceptibility to HDPs of the transformant strain could partly explain why increased resistance to oxidative killing did not translate to increased resistance to neutrophil killing or increased virulence in the in vivo mice models. In other words, the gain in resistance against oxidative stress may have been counterbalanced by a reduction in resistance to HDPs.
The targeted inhibition of staphyloxanthin has been shown to attenuate the virulence of selected S. aureus strains. Liu et al [7] elegantly demonstrated that inhibition of staphyloxanthin production using a cholesterol biosynthesis inhibitor resulted in impaired survival of S. aureus against oxidative stress and killing in whole blood. Furthermore, bacterial counts in the kidney in an intraperitoneal sepsis model were significantly lower in mice treated with the staphyloxanthin inhibitor as compared to control mice. While such therapies appear promising, our results demonstrate that these pathways may not be of importance for virulence in all S. aureus or related strains. The fact that EMRSA-16 (clonal complex 30, ST36) has proven to be a successful hospital lineage despite a nonsense mutation in crtM and a nonpigmented phenotype [22] also supports the notion that staphyloxanthin is not essential for success as a lineage of S. aureus. It may be that targeted therapies, such as those directed against biosynthesis of staphyloxanthin, will prove to be more effective against certain lineages of S. aureus than others.
Third, the findings presented here and elsewhere [13] regarding the ST93 lineage (as represented by JKD6159) highlight the virulence of this successful Australian clone. ST93 has become the dominant community-associated MRSA lineage in Australia [13], as well as representing up to 20% of MSSA strains in some parts of Australia [4]. The reasons for the high virulence of ST93, especially in skin and soft tissue infections, remain to be elucidated. However, our findings demonstrate that this is not as a direct result of increased resistance to oxidative stress and/or increased resistance to killing by neutrophils. The molecular basis for the high virulence phenotype of ST93 merits further investigation.
Notes
Acknowledgments. We thank Christopher Montgomery and Kyra Chua for advice regarding the murine skin infection model and Benjamin Howden for supplying strain JKD6159.
Financial support. This work was supported by an Australian National Health and Medical Research Council Postdoctoral Training Fellowship (grant 508829), an Australian-American Fulbright Scholarship, and a Royal Australasian College of Physicians, Bayer Medical Research Fellowship for S. Y. C. T.; grants R01-AI068804 and K24-AI093969 from the National Institutes of Health (NIH) for V. G. F.; RO1-AI039108-14 from the NIH for A. S. B.; and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. V. G. F. served as Chair of V710 Scientific Advisory Committee (Merck), has received grant support from Merck, Cerexa, Pfizer, Novartis, Advanced Liquid Logics, MedImmune, and the National Institutes of Health, has been a paid consultant for Merck, Astellas, Cubist, Cerexa, Durata, Pfizer, NovaDigm, Novartis, Medicines Company, Biosynexus, MedImmune, Galderma, and Inimex, and has received honoraria from Merck, Astellas, Cubist, Pfizer, Theravance, and Novartis. A. S. B. has current grant support from the following pharmaceutical companies: Cubist, Astellas, Trius and Polymedix; he also has grant support from the National Institutes of Health. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Holt DC, Holden MT, Tong SY, et al. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol. 2011;3:881–95. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng JW, Holt DC, Lilliebridge RA, et al. Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol. 2009;47:2295–300. doi: 10.1128/JCM.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald M, Dougall A, Holt D, et al. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote Aboriginal communities. J Clin Microbiol. 2006;44:3720–7. doi: 10.1128/JCM.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong SY, Lilliebridge RA, Bishop EJ, et al. Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of Northern Australia. J Infect Dis. 2010;202:760–9. doi: 10.1086/655396. [DOI] [PubMed] [Google Scholar]
- 5.Ruimy R, Angebault C, Djossou F, et al. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis. 2010;202:924–34. doi: 10.1086/655901. [DOI] [PubMed] [Google Scholar]
- 6.Liu GY, Essex A, Buchanan JT, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–15. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu CI, Liu GY, Song Y, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–4. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews RM, Kearns T, Connors C, et al. A regional initiative to reduce skin infections amongst aboriginal children living in remote communities of the Northern Territory, Australia. PLoS Negl Trop Dis. 2009;3:e554. doi: 10.1371/journal.pntd.0000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong SY, Bishop EJ, Lilliebridge RA, et al. Community-associated strains of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus in Indigenous northern Australia: epidemiology and outcomes. J Infect Dis. 2009;199:1461–70. doi: 10.1086/598218. [DOI] [PubMed] [Google Scholar]
- 10.Lilliebridge RA, Tong SYC, Giffard PM, Holt DC. The utility of high-resolution melting analysis of SNP nucleated PCR amplicons—a MLST based Staphylococcus aureus typing scheme. PLoS ONE. 2011;6:e19749. doi: 10.1371/journal.pone.0019749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voyich JM, Otto M, Mathema B, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–70. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 12.Holden MTG, Feil EJ, Lindsay JA, et al. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–91. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua KY, Seemann T, Harrison PF, et al. The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct. PLoS ONE. 2011;6:e25887. doi: 10.1371/journal.pone.0025887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Gotz F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem. 2005;280:32493–98. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 15.Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–8. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 16.Mishra NN, Liu GY, Yeaman MR, et al. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother. 2011;55:526–31. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kai-Larsen Y, Agerberth B. The role of the multifunctional peptide LL-37 in host defense. Front Biosci. 2008;13:3760–7. doi: 10.2741/2964. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci U S A. 2002;99:6901–6. doi: 10.1073/pnas.092148299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshmukh HS, Hamburger JB, Ahn SH, McCafferty DG, Yang SR, Fowler VG., Jr Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect Immun. 2009;77:1376–82. doi: 10.1128/IAI.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giachino P, Engelmann S, Bischoff M. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J Bacteriol. 2001;183:1843–52. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra NN, Rubio A, Nast CC, Bayer AS. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int J Microbiol. 2012;2012:683450. doi: 10.1155/2012/683450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAdam PR, Templeton KE, Edwards GF, et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109:9107–12. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]