Abstract
DNA priming improves the response to inactivated influenza A(H5N1) vaccination. We compared the immunogenicity of an H5 DNA prime (using strain A/Indonesia/5/2005) followed by an H5N1 monovalent inactivated vaccine boost at 4, 8, 12, 16, or 24 weeks to that of 2 doses of H5N1 monovalent inactivated vaccine in adults. Antibody epitope repertoires were elucidated by genome-fragment phage-display library analysis, and antibody avidities for HA1 and HA2 domains were measured by surface plasmon resonance. H5 DNA priming expanded the H5-specific antibody epitope repertoire and enhanced antibody avidity to the HA1 (but not the HA2) domain in an interval-dependent manner. Enhanced HA1 binding and avidity after an interval of ≥12 weeks between prime and boost correlated with improved neutralization of homologous and heterologous H5N1 strains.
Clinical trials registration NCT01086657.
Keywords: H5N1, influenza, vaccine, antibody, affinity, Phage display, epitope, hemagglutinin, SPR, immune response, virus
(See the Brief Report by Ledgerwood et al on pages 418–22.)
The most effective way to curtail influenza pandemics is by mass vaccination. Constructing new vaccines involves reassortment between the internal genes of a donor virus such as A/PR/8/34 with the hemagglutinin (HA) and neuraminidase of the newly circulating viruses, which is performed in chicken eggs or in cell cultures by using reverse genetics techniques. The lengthy process currently used for manufacturing seasonal influenza vaccines poses a clear impediment to initiation of a global vaccination campaign against pandemic influenza.
Previously, DNA vaccines against seasonal influenza were shown to be immunogenic in mice and to elicit cross-protective antibodies against viral challenge [1]. In humans, H5 DNA priming was shown to improve the antibody response to boosting with inactivated influenza vaccine when the boost interval was 24 weeks but not 4 weeks [2].
We previously described the use of influenza virus genome–fragment phage-display library technology to decipher the epitope repertoires of polyclonal antibodies after influenza virus infection or vaccination [3–5]. Furthermore, analysis of polyclonal antibody affinity in human sera was conducted by calculating antibody off-rate constants in surface plasmon resonance (SPR)–based real-time kinetics assays [4].
Here, we investigated the effects of H5 DNA priming on the magnitude, breadth, and affinity of antibody responses to an influenza A(H5N1) monovalent inactivated vaccine (MIV) boost, with prime-boost intervals of 4, 8, 12, 16, or 24 weeks, using genome-fragment phage-display library (GFPDL) and SPR technologies. We observed significant changes in the quality of the polyclonal antibody responses (ie, epitope repertoire expansion and affinity maturation) to H5 HA after intervals of ≥12 weeks between H5 DNA priming and MIV boosting, which correlated strongly with increased homologous and heterologous H5N1 neutralization. Hemagglutination inhibition titers are presented in an article by Ledgerwood et al in this issue of the Journal [6].
METHODS
Vaccine Research Center (VRC) 310 Study Design
VRC 310 was a single-site, open-label, randomized clinical trial conducted at the National Institute of Health Clinical Center by the National Institute of Allergy and Infectious Diseases (NIAID) VRC (Bethesda, MD; clinical trials registration NCT01086657) [2]. VRC 310 was designed to further evaluate the safety, tolerability, and immunogenicity of an investigational influenza H5 DNA vaccine boosted with A(H5N1) MIV at intervals of 4, 8, 12, 16, or 24 weeks , compared with that of 2 doses of MIV, in healthy adults aged 18–60 years. Sixty-four subjects were randomly assigned to 1 of 6 groups (Supplementary Table 1), of whom 62 completed 24 weeks of follow-up. Complete details and results of the clinical trial are described by Ledgerwood et al [6] elsewhere in this issue.
H5N1 Vaccines
The H5 DNA vaccine (VRC-AVIDNA036-00-VP) was manufactured at the VRC/NIAID/Vaccine Pilot Plant operated by SAIC (Frederick, MD) and consists of a single closed-circular plasmid DNA macromolecule (VRC-9123) expressing the HA sequence derived from a human isolate of A/Indonesia/5/2005. Subvirion H5N1 MIV (A/Indonesia/5/2005; 90 µg/0.5 mL) was produced by Sanofi Pasteur (Swiftwater, PA).
Construction of H5N1 GFPDLs and Panning of H5 GFPDLs With Polyclonal Human Sera Obtained After H5N1 Vaccine Receipt
Complementary DNA corresponding to the HA gene segment of the A/Indonesia/5/2005 strain was generated from RNA isolated from egg-grown virus. GFPDLs with the HA gene segment of the A/Indonesia/5/2005 were constructed as previously described [3, 5]. GFPDL selection was performed in solution (with protein A/G beads). Inserts of bound phages were amplified by polymerase chain reaction and sequenced.
Affinity Measurements by SPR
Steady-state equilibrium binding of human sera obtained after H5N1 vaccine receipt was monitored at 25°C, using a ProteOn SPR biosensor (BioRad). The HA1-His6 for the respective influenza virus strains was coupled to a GLC sensor chip with amine coupling with 500 resonance units (RU) in the test flow cells [4]. Sixty-microliter serum samples at 10-fold and 100-fold dilutions were injected at a flow rate of 30 µL/minute (120 seconds of contact time) for association, and dissociation was performed over a 600-second interval (at a flow rate of 30 µL/minute). Responses from the protein surface were corrected for the response from a mock surface and for responses from a separate, buffer-only injection. Calculation of binding kinetics for the human vaccine sera and analysis of data were performed with BioRad ProteOn manager software (version 2.0.1). Antibody off-rate constants, which describe the fraction of complexes that decay per second, were determined directly from the serum/plasma sample interaction with rHA1 or rHA2 protein, using SPR in the dissociation phase (as described above), and were calculated using the BioRad ProteOn manager software for the heterogeneous sample model. Off-rate constants were determined from 2 independent SPR runs.
Neutralization Assay
Virus-neutralizing activity was analyzed by a microneutralization assay based on the methods of the pandemic influenza reference laboratories of the Centers for Disease Control and Prevention (CDC). Low-pathogenicity H5N1 viruses generated by reverse genetics were obtained from the CDC.
Statistical Analyses
Differences between groups were examined for statistical significance, using the Student t test. An unadjusted P value of < .05 was considered to be significant.
RESULTS
GFPDL Analysis Identified a Broader Antibody Epitope Repertoire Against H5 After an Interval of 24 Weeks, Compared With an Interval of 4 Weeks, Between H5 DNA Prime and H5N1 MIV Boost
VRC 310 is described in the companion article by Ledgerwood et al [6] and in Supplementary Table 1. The microneutralization titers against the homologous A/Indonesia/5/2005(H5N1) clade 2.1 strain and the heterologous A/Vietnam/1203/2004(H5N1) clade 1 strain were measured 2 weeks after the H5N1 MIV boost (Supplementary Table 2). Patients in group 1 (an MIV prime followed by an MIV boost at 24 weeks) elicited very modest neutralizing antibody titers and achieved a seroconversion rate of 66.7%. H5 DNA priming followed by an H5N1 MIV boost ≥12 weeks later (groups 4–6) generated more-robust homologous H5N1 neutralization titers. In group 6 (H5 DNA priming followed by an MIV boost after 24 weeks), 100% seroconversion was achieved, along with significant neutralization of heterologous H5N1 clades, including A/Vietnam/1203/2004 (Supplementary Table 2; data not shown).
To better understand the role of H5 DNA priming on the antibody epitope repertoire following vaccination, GFPDL analysis was used to identify the HA epitopes recognized by postprime and postboost polyclonal antibodies from subjects in group 1, compared with subjects in group 6. For this analysis, sera were pooled from 5 responders per group who had similar microneutralization titers (Figure 1). The total number and distribution of phage inserts captured after the primary vaccination (with MIV or H5 DNA) were low and were predominated by HA2 inserts, and none of these sera had neutralizing titers against the homologous A/Indonesia/5/2005 strain. Following MIV boosting, the number of bound phages expressing HA peptides in both group 1 and group 6 increased. GFPDL analysis revealed a significantly greater number of total inserts captured by sera from group 6, compared with sera from group 1 (Figure 1B). More importantly, the number and diversity of antibody-selected phages with epitope sequences spanning the HA1 domain (including the receptor-binding domain) were greater in group 6, compared with group 1 (Figure 1A and 1B). Also, much larger HA inserts representative of conformation-dependent epitopes were selected in group 6, compared with group 1. These data suggested that heterologous (ie, H5 DNA and MIV) prime-boost H5N1 vaccination with a 24-week interval was beneficial for B-cell recruitment and diversification of the polyclonal antibody epitope repertoire.
Figure 1.
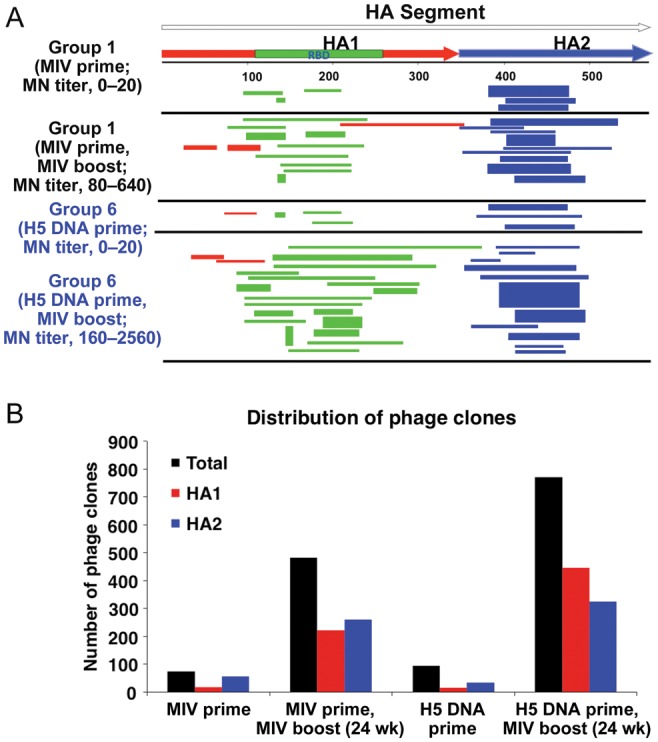
H5 hemagglutinin (HA) DNA priming significantly expands the antibody epitope repertoire in HA1 generated following receipt of an A/Indonesia/5/2005(H5N1) monovalent inactivated vaccine (MIV) boost. A, Schematic alignment of the peptides recognized by sera obtained after H5 DNA priming and H5N1 MIV boosting, as identified by panning of H5 genome-fragment phage-display libraries with A/Indonesia/5/2005(H5N1). The amino acid designation is based on the A/Indonesia/5/2005(H5N1) HA protein sequence (Supplementary Figure 1). Bars indicate identified inserts in HA1 (red bars) and HA2 (blue bars). Phage displaying peptides from sequences in the receptor-binding domain are depicted with green bars within HA1 segment. The thickness of each bar represent the frequencies of repetitively isolated phage inserts (only clones with a frequency of ≥2 are shown). B, Distribution of phage clones in different HA domains after affinity selection with sera obtained from adults after MIV-prime, MIV-MIV boost, DNA-prime, and DNA-MIV boost is shown. Abbreviation: MN, microneutralization.
Intervals of ≥12 Weeks Between H5 DNA Priming and H5N1 MIV Boosting Elicited Higher-Affinity Antibodies (With Slower Antigen-Antibody Complex Dissociation Rates) Specific to the H5N1 HA1 Globular Head
We previously described the expression of properly folded HA1 and HA2 domains from pandemic influenza virus strains (ie, 2009 pandemic influenza A[H1N1] and A[H5N1]) in a bacterial system and their use in SPR for measurements of domain-specific binding and kinetics analysis [4, 7–9]. In the current study, individual samples from all subjects were used for measurements of total polyclonal serum antibody binding to chips coated with the A/Indonesia/5/2005 HA1 and HA2 domains, using an SPR-based real-time kinetics assay (Supplementary Figure 1). The total antibody-binding titers (maximum RU values) were higher in sera from groups 2–6, compared with sera from group 1, reaching statistical significance for group 5 and group 6 (16- and 24-week intervals, respectively, between prime and boost; Supplementary Figure 1A).
Correlation analysis of SPR-derived total anti-HA1–binding antibodies (maximum RU values) and H5N1 microneutralization titers of all study participants showed a very strong correlation not only for the homologous vaccine strain, A/Indonesia/5/2005 (R2 = 0.715), but also for several heterologous H5N1 strains: A/Vietnam/1203/2004 (clade 1, R2 = 0.735), A/Egypt/3072/2010 (clade 2.2.1, R2 = 0.578), A/Anhui/1/2005 (clade 2.3.4, R2 = 0.619), and A/Turkey/1/2005 (clade 2.2, R2 = 0.587) (Supplementary Figure 1B–F).
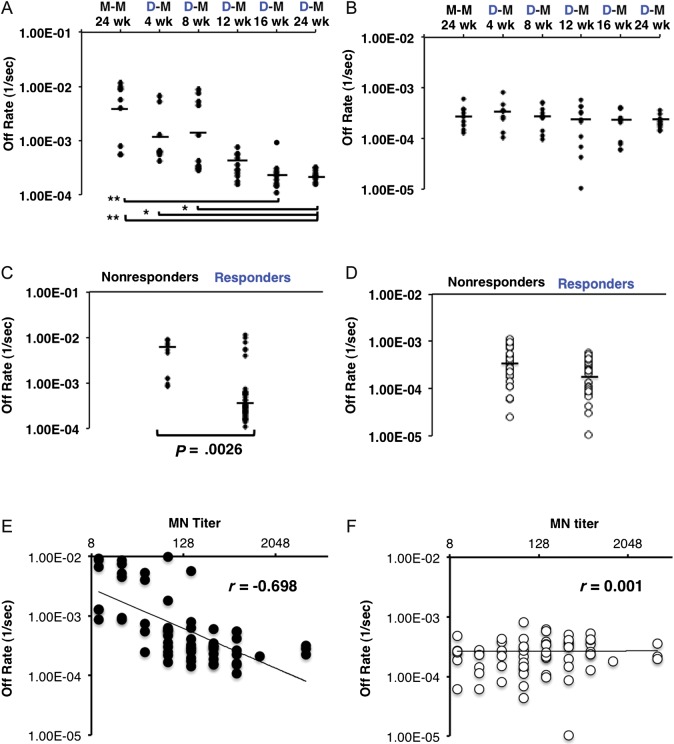
Individual sera from H5N1-vaccinated subjects were analyzed by SPR, and antibody off-rate constants that characterize the stability of the antigen-antibody complex (ie, the fraction of antigen-antibody complexes that decay per second) were calculated using BioRad ProteOn manager software for the heterogeneous sample model. The polyclonal antibody–binding off rates of individual postboost sera (10–11 individuals per group) were calculated separately for the H5N1 HA1 and HA2 domains (Figure 2A and 2B). Significantly slower dissociation rates (≥1 log change) were observed for the polyclonal antibody binding to HA1-coated chips with sera from groups 4, 5, and 6 (12-, 16-, and 24-week intervals, respectively), indicating increasing antibody affinity to the HA1 domain in the groups with longer intervals between H5 DNA prime and MIV boost (Figure 2A). This was further confirmed when data were analyzed separately for responders (microneutralization titer, ≥40) and nonresponders (microneutralization titer, <40). The average serum anti-HA1 antibody off rates were significantly lower in the responders (Figure 2C). Additionally, the cluster of sera with faster anti-HA1 antibody dissociation rates (weaker antibody affinity) in the responder groups were all from groups 1–3. In contrast, no clear difference in serum antibody–binding dissociation rates to the HA2 domain was observed among any of the 6 vaccinated groups or between responders and nonresponders (Figure 2B and 2D). The HA2-binding off rates (10−3–10−5 per second) tended to be slower than the HA1-binding off rates (10−2–10−4 per second), suggesting the presence of affinity-matured cross-reactive anti-HA2 antibodies in adults, probably due to the high conservation between the H5N1 HA2 domain and the seasonal A(H1N1) HA2 domain, as previously described [5].
Figure 2.
H5 DNA priming enhance antibody affinity (slower off rates) to H5N1 HA1 (but not HA2) with an interval of 12–24 weeks between H5 DNA prime and monovalent inactivated vaccine (MIV) boost. Surface plasmon resonance (SPR) analysis of human sera obtained after vaccination from 6 prime-boost groups (10–11 individuals per group) in the vaccine trial was performed with properly folded functional oligomeric H5N1 HA1 (A) and H5N1 HA2 (B) domains from the A/Indonesia/5/2005 strain. Antibody off-rate constants that describe the fraction of antibody-antigen complexes decaying per second were determined directly from the serum sample interaction with rHA1 (1–330) and rHA2 (331–480), using SPR in the dissociation phase. Serum antibody off-rate constants (each symbol represents 1 individual) were determined as described in Materials and Methods. The off-rate constants were determined from two independent SPR runs. *P < .05 and **P < .005, by the Student t test, for differences between mean off-rate constants for human sera obtained after vaccination. D-M, H5 DNA prime, MIV boost; M-M, MIV prime, MIV boost. C and D, A/Indonesia/5/2005(H5N1) responders (microneutralization [MN] titer, ≥ 40) have significantly higher antibody affinity to rHA1 (but not rHA2) than nonresponders (MN titer, < 40). Antibody off-rate constants from the serum sample interaction with rHA1 protein (C) and rHA2 (D), determined using SPR analysis, for all of the individuals in 6 prime-boost groups following prime-boost vaccination were analyzed for A/Indonesia/5/2005(H5N1) responders (MN titer, ≥ 40) and compared to the antibody affinity to rHA1 for nonresponders (MN titer, < 40). The mean off-rate constants of human sera obtained after vaccination were statistically significant between responders and nonresponders only in the HA1 domain, not in the HA2 domain (P < .05, by the Student t test). The off-rate constants were determined from 2 independent SPR runs. E and F, Serum antibody off rates for H5 A/Indonesia/5/2005(H5N1) rHA1 but not HA2 after prime-boost are strongly correlated with the in vitro neutralizing capacity against the homologous A/Indonesia/5/2005(H5N1) strain. Antibody off-rate constants were determined directly from the plasma sample interaction with rHA1 (1–330) or rHA2, using SPR in the dissociation phase. SPR analysis of human sera obtained after boosting from all vaccine groups included in the vaccine trial was performed with rHA1 (E) or rHA2 (F) of the A/Indonesia/5/2005(H5N1) strain. Each symbol represents 1 individual. Antibody affinity of human sera obtained after H5N1 vaccination against HA1 of A/Indonesia/5/2005(H5N1) correlated with homologous A/Indonesia/5/2005(H5N1) MN titers.
The contribution of the antibody affinity of sera obtained after H5N1 vaccination to virus neutralization was evaluated by correlation plots of the antibody off rates for A/Indonesia/5/2005 HA1 and HA2 domains versus microneutralization titers. A negative correlation was observed between the anti-HA1 antibody dissociation rates and serum microneutralization titers (r = −0.698; Figure 2E) but not between the anti-HA2 serum antibody off rates and virus neutralization (Figure 2F).
Together, these data suggest that the H5 DNA and MIV prime-boost regimen with intervals of ≥12 weeks between the prime and boost generated antibodies with higher diversity against the HA1 receptor-binding domain and increased affinity for the globular head, which correlated with better neutralization of the H5N1 vaccine strain and stronger cross-neutralization of heterologous H5N1 from diverse clades.
DISCUSSION
DNA priming with H5 HA improves the antibody response elicited by boosting with A/Indonesia/5/2005(H5N1) MIV [2]. The current study demonstrated that H5 DNA and MIV prime-boost regimen was superior to the MIV-MIV prime-boost approach in terms of the diversity of the antibody epitope repertoire and antibody affinity maturation against the HA1 (but not HA2) domain. The most significant changes were observed with a ≥12-week interval between prime and boost. The increase in antibody affinity for the HA1 domain strongly correlated with the neutralization of the A/Indonesia/5/2005(H5N1) vaccine strain (clade 2.1), as well as several heterologous H5N1 strains.
The findings in the current study on the superiority of H5 DNA and MIV prime-boost regimen over the homologous MIV prime-boost protocol were similar to findings with oil-in-water adjuvanted pandemic influenza vaccines [4, 5]. The significant impact on the diversity and quality of the antibody responses observed in the different vaccination approaches suggests an important role for induction of early robust T-helper responses [10]. Antigen presentation of DNA vaccines differs from that of other vaccines, possibly increasing the number and diversity of CD4 clones recruited [11], which may lead to a greater expansion of antigen-specific B cells.
The importance of antibody avidity for protection against infection and disease severity has been demonstrated recently for several viral diseases, including infection with swine-like 2009 pandemic H1N1 [12, 13], and loss of protection against mumps 20 years after measles, mumps, and rubella vaccination [14]. Additionally, antibody avidity correlated with heterologous protection against foot-and-mouth disease nonvaccine serotypes [15].
DNA priming provides an approach that may improve the magnitude, quality, and breadth of antibody responses to conventional vaccine products and is not dependent on isolation and construction of vaccine virus.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Maryna Eichelberger and Marina Zaitseva, for their thorough review of the manuscript, and the VRC 310 Study Team.
Disclaimer. The funders had no role in conducting this research.
Financial support. This work was supported by the Food and Drug Administration and the National Institutes of Health.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
References
- 1.Wei CJ, Boyington JC, McTamney PM, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–4. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 2.Ledgerwood JE, Wei CJ, Hu Z, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–24. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khurana S, Suguitan AL, Jr., Rivera Y, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6:e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khurana S, Verma N, Yewdell JW, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khurana S, Chearwae W, Castellino F, et al. Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci Transl Med. 2010;2:15ra5. doi: 10.1126/scitranslmed.3000624. [DOI] [PubMed] [Google Scholar]
- 6.Ledgerwood JE, Zephir K, Hu Z, et al. Boost interval matters: a randomized phase 1 study to identify the minimum interval to observe the H5 DNA influenza vaccine priming effect. J Infect Dis. 2013;208:418–22. doi: 10.1093/infdis/jit180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khurana S, Verma S, Verma N, et al. Properly folded bacterially expressed H1N1 hemagglutinin globular head and ectodomain vaccines protect ferrets against H1N1 pandemic influenza virus. PLoS One. 2010;5:e11548. doi: 10.1371/journal.pone.0011548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Khurana S, Larkin C, Verma S, et al. Recombinant HA1 produced in E. coli forms functional oligomers and generates strain-specific SRID potency antibodies for pandemic influenza vaccines. Vaccine. 2011;29:5657–65. doi: 10.1016/j.vaccine.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khurana S, Verma S, Verma N, et al. Bacterial HA1 vaccine against pandemic H5N1 influenza virus: evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets. J Virol. 2011;85:1246–56. doi: 10.1128/JVI.02107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galli G, Medini D, Borgogni E, et al. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc Natl Acad Sci U S A. 2009;106:3877–82. doi: 10.1073/pnas.0813390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Kong WP, Nabel GJ. Enhanced breadth of CD4 T-cell immunity by DNA prime and adenovirus boost immunization to human immunodeficiency virus Env and Gag immunogens. J Virol. 2005;79:8024–31. doi: 10.1128/JVI.79.13.8024-8031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monsalvo AC, Batalle JP, Lopez MF, et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat Med. 2010;17:195–9. doi: 10.1038/nm.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma N, Dimitrova M, Carter DM, et al. Influenza virus H1N1pdm09 infections in the young and old: evidence of greater antibody diversity and affinity for the hemagglutinin globular head domain (HA1 domain) in the elderly than in young adults and children. J Virol. 2012;86:5515–22. doi: 10.1128/JVI.07085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontio M, Jokinen S, Paunio M, Peltola H, Davidkin I. Waning antibody levels and avidity: implications for MMR vaccine-induced protection. J Infect Dis. 2012;206:1542–8. doi: 10.1093/infdis/jis568. [DOI] [PubMed] [Google Scholar]
- 15.Lavoria MA, Di-Giacomo S, Bucafusco D, Franco-Mahecha OL, Perez-Filgueira DM, Capozzo AV. Avidity and subtyping of specific antibodies applied to the indirect assessment of heterologous protection against foot-and-mouth disease virus in cattle. Vaccine. 2012;30:6845–50. doi: 10.1016/j.vaccine.2012.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.