Abstract
Polyunsaturated (PUFA) long-chain fatty acids (LCFAs) are more potent in eliciting molecular and tissue functional changes in monogastrics than saturated LCFA. From −21 through 10 days relative to parturition dairy cows were fed no supplemental LCFA (control), saturated LCFA (SFAT; mainly 16:0 and 18:0), or fish oil (FISH; high-PUFA). Twenty-seven genes were measured via quantitative RT-PCR in liver tissue on day −14 and day 10. Expression of nuclear receptor co-activators (CARM1, MED1), LCFA metabolism (ACSL1, SCD, ACOX1), and inflammation (IL6, TBK1, IKBKE) genes was lower with SFAT than control on day −14. Expression of SCD, however, was markedly lower with FISH than control or SFAT on both −14 and 10 days. FISH led to further decreases in expression on day 10 of LCFA metabolism (CD36, PLIN2, ACSL1, ACOX1), intracellular energy (UCP2, STK11, PRKAA1), de novo cholesterol synthesis (SREBF2), inflammation (IL6, TBK1, IKBKE), and nuclear receptor signaling genes (PPARD, MED1, NRIP1). No change in expression was observed for PPARA and RXRA. The increase of DGAT2, PLIN2, ACSL1, and ACOX1 on day 10 versus −14 in cows fed SFAT suggested upregulation of both beta-oxidation and lipid droplet (LD) formation. However, liver triacylglycerol concentration was similar among treatments. The hepatokine FGF21 and the gluconeogenic genes PC and PCK1 increased markedly on day 10 versus −14 only in controls. At the levels supplemented, the change in the profile of metabolic genes after parturition in cows fed saturated fat suggested a greater capacity for uptake of fatty acids and intracellular handling without excessive storage of LD.
Keywords: dairy cows, fat supplementation, hepatic gene network
Introduction
The liver plays a pivotal role in whole-body lipid hemostasis and responds rapidly to changes in dietary fat composition in both rodents1 and ruminants.2 Previous data with peripartal dairy cattle have underscored the potential for dietary lipid supplementation as a useful nutritional strategy to prepare3 and facilitate the hepatic metabolism of non-esterified fatty acids (NEFA)4 released in high amounts from adipose tissue during early lactation.5 However, there have been contrasting results with respect to type of dietary lipid and the response in blood hydroxybutyrate (BHBA), liver lipid content, dry matter intake (DMI), milk yield, and changes in body weight and body condition score.2,4,6,7 It is likely that differences across studies are partly due to the specific type (eg, saturated vs. unsaturated) and quantity of lipid supplemented.
In rodent liver, long-chain fatty acids (LCFAs) can bind directly to various nuclear receptors (PPAR, LXR) leading to changes in their transcriptional activity, which alters the function of pathways in proportion to changes in the mRNA expression of target genes.8,9 Pathways associated with LCFA oxidation, inflammation, and ketogenesis were recently evaluated in bovine cells leading to the recognition that saturated LCFA, eg, 16:0 and 18:0, and the fish oil-enriched 20:5n-3 were the most-potent at upregulating genes associated with PPARα activity.10,11 The response to saturated LCFA likely is a ruminant evolutionary adaptation of PPAR to metabolize the saturated LCFA which are found in large amounts in the circulation due to extensive ruminal hydrogenation.10,11
There is in vivo and in vitro evidence suggesting that the PPARα gene network might be responsive to dietary lipids and could be used to monitor the functional changes that might occur in the liver during the transition period.4,10,12,13 From that perspective, it is important to evaluate if those responses also are observed during the transition period in cows receiving supplemental lipid enriched in one or more of the main dietary LCFA. The objective of the current study was to evaluate the expression of several genes associated with the PPARα transcriptional network in liver tissue harvested from cows fed supplemental saturated fat or fish oil during the transition period.
Methods
Experimental design
The present study involved the same subset of multiparous Holstein cows used by Schmitt et al.14 Briefly, a completely random subset of 5 multiparous cows (2nd and 3rd lactation) that were fed no supplemental LCFA (control, n = 12 cows total) or supplemental LCFA from either Energy Booster (SFAT; Milk Specialties Co., Dundee, IL, USA; n = 15) or fish oil (FISH; Omega Proteins, Houston, TX, USA; n = 15) were used for hepatic phospholipid (PL) and triacylglycerol (TAG) LCFA analysis and gene expression profiling. The cows from the control, FISH, and SFAT diets were fed the respective diets from −25 (±4), −24 (±3), and −26 (±7) d, respectively, until 10 days post partum.
Biopsies
The liver tissue was harvested via percutaneous biopsy.15 The average gap between the start of feeding and the first biopsy was 11 ± 3 days. Prepartal liver biopsies were harvested at −14 ± 4, −13 ± 3, and −16 ± 6 days in cows fed the control, FISH, and SFAT diets, respectively. Postpartal biopsies were harvested at 7 ± 4 days in all the groups. Biopsied tissue (1 to 2 g) was weighed and stored in liquid N2 prior to RNA extraction. A portion of liver tissue collected (0.8 to 1.0 g) was used for analysis of PL and TAG fatty acid concentrations as described by Ballou et al.15
RNA extraction and real-time quantitative PCR (qPCR)
Complete details of the procedure for RNA extraction can be found in Schmitt et al.14 Briefly, approximately 0.2–0.3 g of liver tissue was weighted and immediately placed in ice-cold TRIzol reagent for homogenization. Genomic DNA was removed from RNA with RNase-free DNase, using RNeasy Mini Kit columns. RNA concentration was measured with NanoDrop ND-1000 spectrophotometer, while the RNA quality was assessed using the Agilent Bio-analyzer system (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). The average RNA integrity number (RIN) value for liver samples was 8 ± 0.4. Protocols for primer design, testing, selection of internal control genes (ICG) for normalization were as previously described.14 Briefly, genes selected as suitable ICG based on geNorm analysis included EDC4, SHPRH, EIF3K, UXT, ACTB, and MRPL39. The geometric mean of these genes was used to normalize gene expression data in the present study.
Genes selected for transcript profiling in this study are associated with fatty acid uptake and transport (FABP1, CD36), esterification, desaturation and lipid droplet (LD) formation (DGAT2, SCD, PLIN2, PLIN4), fatty acid oxidation (ACSL1, ACOX1, CPT1A, FGF21), gluconeogenesis (PC, PCK1), and intracellular energy (UCP2, PRKAA1, STK11). Of particular importance was the study of transcription regulators (PPARA, RXRA, PPARD, SREBF2), nuclear receptor co-activators (CARM1, MED1), nuclear receptor co-repressors (NCOR2, NRIP1), and inflammation related (IL6, TBK1, IKBKE) and apoptosis/signaling related genes (CIDEB, STK1). Primer pairs for target genes and ICG and sequencing results of primer products not shown in Tables 1 and 2 were reported previously.14,16
Table 1.
GenBank accession number, gene symbol, hybridization position, sequence and amplicon size of primers.
| Accession # | Gene | Primersa | Primers (5′-3′)b | bpc |
|---|---|---|---|---|
| XM_002695200.1 | FGF21 | F.223 R.328 | CAGAGCCCCGAAAGTCTCTTGAAAGTGCAGCGATCCGTACAG | 106 |
| NM_001034036.1 | PPARA | F.729 R.830 | CATAACGCGATTCGTTTTGGACGCGGTTTCGGAATCTTCT | 102 |
| NM_001083636.1 | PPARD | F.460 R.559 | TGTGGCAGCCTCAATATGGAGACGGAAGAAGCCCTTGCA | 100 |
| NM_001035289.2 | ACOX1 | F.180 R.279 | ACCCAGACTTCCAGCATGAGATTCCTCATCTTCTGCACCATGA | 100 |
| NM_173980.2 | PLIN2 | F.1607 R.1706 | TTTATGGCCTCATGCTTTTGCCTCAGAGCAGACCCCAATTCA | 100 |
| FJ415874.1 | CPT1A | F.141 R.240 | TCGCGATGGACTTGCTGTATACGGTCCAGTTTGCGTCTGTA | 100 |
| BC111622 | FABP1 | F.183 R.283 | GTTCATCATCACCGCTGGCTCCACTGCCTTGATCTTCTCCC | 101 |
Notes:
Primer direction (F—forward; R—reverse) and hybridization position on the sequence;
exon-exon junctions are underlined;
amplicon size in base pairs (bp).
Table 2.
Sequencing results of PCR primer products.
| Gene | Sequence |
|---|---|
| FGF21 | CGAGATCTGAAGCAAATTGAGGCAGAAATCCTTACGTGTGAGCATGACCTAGAAGATTCCGAAACCGCGA |
| PPARA | CGAGATCTGAAGCAAATTGAGGCAGAAATCCTTACGTGTGAGCATGACCTAGAAGATTCCGAAACCGCGA |
| PPARD | GCATGGGGACGGCGTCGGGCTCACTACGGCGTTCACGCTTGTGAGGGATGCAAGGGCTTCTTCCGTCC ACAAA |
| ACOX1 | ATCCTCGTATCCGCGTTCAGGGTGCGTTTAAGAAGAGTGCCATCATGGTGCAGAAGATGAGGAAATCCCC |
| PLIN2 | ACGTGCGTCGTCGTTCGTATAAAACACCTTCATGTAGGCTGTTGTATGAATTGGGGTCCGCTCTGAGAC |
| CPT1A | GGACTATGAAGGTAAACCAGGCCCGGGACGCCCTTCGTACAGGCCTCTCGCTCCAGCTGGCTCATTACA AGGGACCA |
| FABP1 | GAGGGAGGAGTGTGAGATGGAGTTCATGACTGGGAGAGAAGATCAAGGCAGTGGA |
Note: Best hits using BLASTN (http://www.ncbi.nlm.nih.gov) are shown.
Fatty acid analysis
Details of these procedures have already been published by Ballou et al.15 Briefly, a 100 mg liver sample was used to separate both PL and TAG via thin-layer chromatography using hexane-diethyl etheracetic acid (90:30:1, vol/vol/vol) as the elution phase. Methyl esters of fatty acids (FA) were prepared by incubation with 2 M potassium hydroxide in methanol for 15 min at room temperature. The ester mixture was separated using a Hewlett Packard 5890 gas chromatograph (Hewlett Packard, Avondale, PA, USA) equipped with a flame-ionization detector and a Supelco 2560 100-m capillary column (Supelco, Bellefonte, PA, USA).15
Statistical analysis
After normalization with the geometric mean of the ICG, the qPCR data from all treatments (prepartum and postpartum) were log-2 transformed prior to statistical analysis.14 A repeated measures model was fitted to gene expression, FA, NEFA, and DMI data using Proc MIXED in SAS. The model consisted of time, treatment, and time × treatment interaction as fixed effects, as well as cow as the random effect. An autoregressive covariate structure was used. All means were compared using the PDIFF statement of SAS.
Results
Dry matter intake and blood NEFA
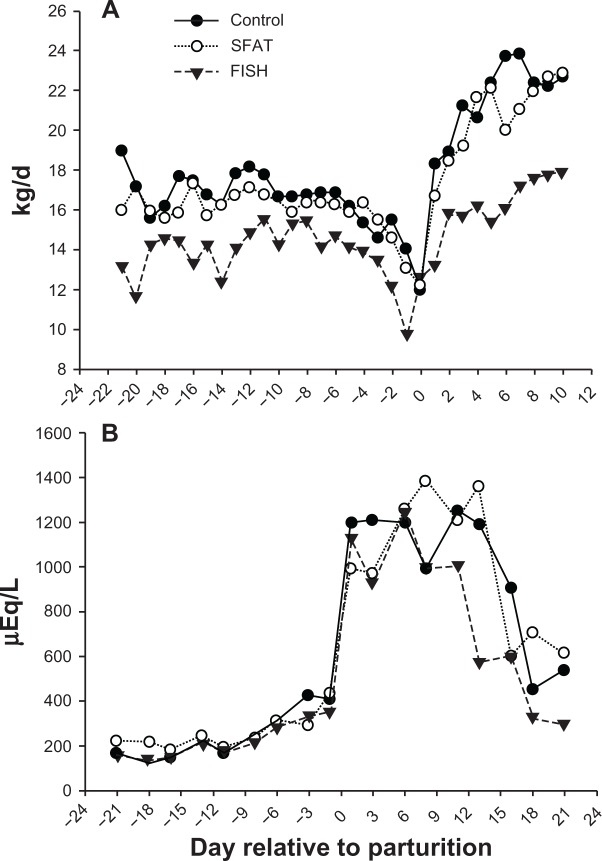
The DMI was significantly (P < 0.05) affected by the interaction of diet and day. Supplementing FISH led to lower peripartal DMI compared to SFAT and control cows; the maximum decrease was observed close to parturition. A postpartal increase in DMI was observed in all the groups but cows supplemented with FISH still had lower DMI than the other two groups by the end of the supplementation period (Fig. 1A). The plasma NEFA concentration was not affected significantly by diet (P < 0.05). Irrespective of lipid type, NEFA increased at calving and remained elevated for the subsequent 10 days (Fig. 1B). Cows supplemented with FISH had comparatively lower postpartal NEFA relative to controls or SFAT, which agreed with the lower DMI during that time-frame.
Figure 1.
Dry matter intake (A) and blood NEFA concentration (B) during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT). There was a significant (P < 0.05) Diet × Time effect for dry matter intake; whereas, for NEFA only Time was significant (P < 0.05).
Hepatic fatty acid composition
Phospholipids
Mean concentration of palmitic acid (16:0) increased (P < 0.05) after parturition in all groups including control. The hepatic PL fatty acid content of stearic acid (18:0) decreased (diet × day P < 0.05) after parturition in control and SFAT but increased with FISH and remained elevated by day 11 (Table 3). Supplementing FISH increased (diet × day P < 0.05) the proportion of trans-18:1 isomers, with a maximal concentration on day −10 followed by a gradual decrease by day 11 (Tables 1 and S1). Concomitantly, FISH led to lower (diet × day P < 0.05) prepartal linoleic acid (18:2n-6) content at −10 days followed by a steady increase after parturition. By day 11, there were no differences in 18:2n-6 among diets. Overall concentration of 18:1trans11 was greatest (P < 0.05) in FISH compared with control or SFAT from −10 through 11 days. In contrast, conjugated linoleic acid (CLA) concentration was lower (diet P < 0.05) in lipid-supplemented cows than control cows. However, CLA concentration peaked in all the groups (day P < 0.05) at parturition (1 day) followed by a gradual decrease by day 11. The concentrations of other 18:1trans isomers were little affected by supplemental lipid (Table S1).
Table 3.
Percentage of long-chain fatty acids in liver phospholipids during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT).
| Fatty acid† | Treatment | Day | SEM† | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| −21 | −10 | 1 | 11 | Diet | Time | D × T‡ | |||
| 16:0 | CON | 9.47 | 11.28 | 12.86 | 12.90 | 0.96 | 0.34 | <0.01 | 0.39 |
| SFAT | 10.58 | 10.98 | 12.55 | 12.12 | |||||
| FISH | 12.55 | 10.32 | 13.25 | 13.33 | |||||
| 18:0 | CON | 30.58α | 30.07a,α | 27.34a,b,β | 27.01γ | 1.11 | 0.62 | <0.01 | <0.01 |
| SFAT | 31.08α | 31.01a,α | 26.80a,β | 27.65β | |||||
| FISH | 30.03α | 25.59b,β | 29.68b,α,γ | 28.44α,γ | |||||
| 18:1cis9 | CON | 0.57 | 0.82 | 0.90 | 0.78 | 0.11 | 0.21 | <0.01 | 0.73 |
| SFAT | 0.75 | 0.81 | 0.88 | 0.88 | |||||
| FISH | 0.43 | 0.70 | 0.83 | 0.84 | |||||
| 18:1trans11 | CON | 0.76a | 0.91a | 0.87a | 0.79a,b | 0.14 | <0.01 | <0.01 | <0.01 |
| SFAT | 0.74a | 0.56a | 0.86a | 0.54a | |||||
| FISH | 0.37b,α | 4.95b,β | 1.50b,γ | 0.97b,δ | |||||
| 18:2c9t11 | CON | 0.16 | 0.20 | 0.27 | 0.22 | 0.02 | <0.01 | <0.01 | 0.23 |
| SFAT | 0.14 | 0.13 | 0.28 | 0.16 | |||||
| FISH | 0.07 | 0.13 | 0.21 | 0.17 | |||||
| 18:2n-6 | CON | 9.64α | 10.35a,α | 13.16a,β | 13.15β | 0.88 | <0.01 | <0.01 | 0.03 |
| SFAT | 9.62α | 9.87a,α | 13.25a,β | 13.45β | |||||
| FISH | 10.29α | 7.09b,β | 9.46b,α | 13.16γ | |||||
| 18:3n-3 | CON | 0.93b,α | 0.95α | 1.37a,b,β | 1.24α | 0.12 | 0.49 | <0.01 | 0.03 |
| SFAT | 1.12a,b,α | 0.89α | 1.64a,β | 1.17α | |||||
| FISH | 1.26a,α | 0.79β | 1.14b,α | 1.28α | |||||
| 20:4n-6 | CON | 11.76α,β | 10.60α | 10.92b,α | 12.29a,β,γ | 0.53 | <0.01 | <0.01 | 0.02 |
| SFAT | 11.29α | 10.45α,β | 9.87a,b,β | 11.42a,α | |||||
| FISH | 11.24α | 10.34α,β | 9.01a,β,γ | 8.82b,γ | |||||
| 20:5n-3 | CON | 1.43 | 1.43a | 1.44a | 1.69a | 0.21 | <0.01 | <0.01 | <0.01 |
| SFAT | 1.59α | 1.51a,α | 1.52a,α | 2.21a,β | |||||
| FISH | 1.30α | 4.03b,β | 4.33b,β | 3.94b,β | |||||
| 22:5n-3 | CON | 0.71 | 0.68 | 0.43 | 0.36 | 0.07 | 0.01 | <0.01 | 0.10 |
| SFAT | 0.64 | 0.65 | 0.41 | 0.28 | |||||
| FISH | 0.70 | 0.93 | 0.65 | 0.27 | |||||
| 22:6n-3 | CON | 0.94 | 0.92a | 0.92a | 0.89a | 0.29 | <0.01 | <0.01 | <0.01 |
| SFAT | 0.97 | 0.84a | 0.83a | 0.76a | |||||
| FISH | 0.90α | 7.72b,β | 7.97b,β | 6.58b,γ | |||||
Notes:
Standard error of the mean;
diet × time interaction.
Difference (P < 0.05) between diets on the same day.
Significant interactions (P < 0.05) within a diet and between days.
Table S1.
Concentration of fatty acids in hepatic phospholipids during the peripartal period.
| Fatty acid | Diet† | Day | SEM‡ | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| −21 | −10 | 1 | 11 | Diet | Time | D × T¶ | |||
| 14:0 | CON | 0.25 | 0.35 | 0.20 | 0.20 | 0.09 | 0.71 | <0.01 | 0.22 |
| SFAT | 0.53 | 0.31 | 0.18 | 0.16 | |||||
| FISH | 0.52 | 0.19 | 0.20 | 0.17 | |||||
| 14:1trans | CON | <0.01 | 0.02 | <0.01 | <0.01 | 0.01 | 0.60 | 0.17 | 0.82 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | <0.01 | 0.01 | <0.01 | <0.01 | |||||
| 14:1cis | CON | 0 | <0.01 | <0.01 | <0.01 | <0.01 | 0.43 | 0.61 | 0.19 |
| SFAT | 0 | 0 | <0.01 | <0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 15:0 | CON | 0.34 | 0.34 | 0.22 | 0.22 | 0.04 | 0.76 | <0.01 | 0.45 |
| SFAT | 0.39 | 0.32 | 0.19 | 0.18 | |||||
| FISH | 0.43 | 0.25 | 0.19 | 0.18 | |||||
| 15:1trans | CON | <0.01 | 0.04 | <0.01 | <0.01 | 0.01 | 0.44 | 0.15 | 0.78 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | <0.01 | 0.02 | <0.01 | <0.01 | |||||
| 16:1trans | CON | 0.29 | 0.28 | 0.18 | 0.22 | 0.03 | 0.03 | <0.01 | 0.09 |
| SFAT | 0.27 | 0.24 | 0.18 | 0.16 | |||||
| FISH | 0.28 | 0.39 | 0.22 | 0.17 | |||||
| 16:1cis | CON | 0.62 | 0.66 | 0.70 | 0.65 | 0.06 | <0.01 | 0.52 | 0.85 |
| SFAT | 0.68 | 0.67 | 0.73 | 0.66 | |||||
| FISH | 0.63 | 0.55 | 0.59 | 0.53 | |||||
| 17:0 | CON | 1.35 | 1.27 | 0.83 | 0.82 | 0.10 | 0.95 | <0.01 | 0.37 |
| SFAT | 1.39 | 1.30 | 0.75 | 0.81 | |||||
| FISH | 1.12 | 1.43 | 0.84 | 0.80 | |||||
| 17:1trans | CON | 0.09 | 0.12 | 0.04 | 0.05 | 0.02 | 0.01 | <0.01 | 0.67 |
| SFAT | 0.11 | 0.09 | 0.06 | 0.04 | |||||
| FISH | 0.04 | 0.09 | 0.01 | <0.01 | |||||
| 18:1trans5 | CON | <0.01 | 0 | <0.01 | <0.01 | 0.03 | 0.10 | 0.56 | 0.55 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | 0.10 | 0.06 | 0.01 | <0.01 | |||||
| 18:1trans7 | CON | <0.01 | <0.01a | <0.01 | <0.01 | <0.01 | 0.21 | 0.09 | <0.01 |
| SFAT | 0.01 | <0.01a | <0.01 | 0.01 | |||||
| FISH | <0.01α | 0.03b,β | <0.01α | <0.01α | |||||
| 18:1trans8 | CON | 0.08b,α | <0.01c,β | <0.01β | <0.01b,c,β | 0.03 | 0.78 | 0.25 | <0.01 |
| SFAT | <0.01a,α | <0.01a,b,α | <0.01α | 0.11a,β | |||||
| FISH | <0.01a,α | 0.07b,β | <0.01α | <0.01b,α | |||||
| 18:1trans9 | CON | 0.09 | 0.11 | 0.09 | 0.08 | 0.05 | 0.84 | 0.16 | 0.39 |
| SFAT | 0.09 | 0.10 | 0.16 | 0.03 | |||||
| FISH | 0.09 | 0.17 | 0.03 | 0.02 | |||||
| 18:1trans10 | CON | 0.17 | <0.01 | <0.01 | 0.02 | 0.07 | 0.91 | 0.59 | 0.12 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | <0.01 | 0.16 | <0.01 | 0.04 | |||||
| 18:1trans12 | CON | 0.39α,β | 0.17a,α | 0.53a,b,α,β | 0.63β | 0.15 | <0.01 | 0.01 | <0.01 |
| SFAT | 0.35 | 0.38a | 0.50a | 0.49 | |||||
| FISH | 0.18α | 1.24b,β | 0.90b,β,γ | 0.71γ | |||||
| 18:1t13-14 | CON | <0.01 | <0.01 | <0.01 | 2.86 | 0.93 | 0.39 | 0.41 | 0.45 |
| SFAT | <0.01 | 0 | 0 | <0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 18:1cis11 | CON | 0.57 | 0.82 | 0.90 | 0.78 | 0.11 | 0.21 | <0.01 | 0.73 |
| SFAT | 0.75 | 0.81 | 0.88 | 0.88 | |||||
| FISH | 0.43 | 0.70 | 0.83 | 0.84 | |||||
| 18:1cis12 | CON | 0.29 | 0.19 | 0.26 | 0.24 | 0.07 | 0.74 | 0.99 | 0.22 |
| SFAT | 0.23 | 0.22 | 0.26 | 0.13 | |||||
| FISH | 0.14 | 0.29 | 0.17 | 0.30 | |||||
| 18:1cis13 | CON | 0.05 | 0.01 | 0.04 | 0.04 | 0.02 | 0.57 | 0.63 | 0.42 |
| SFAT | <0.01 | 0.01 | 0.04 | 0.02 | |||||
| FISH | 0 | 0.06 | 0.03 | 0.03 | |||||
| 18:1cis16 | CON | 0.04 | 0.10 | 0.14 | 0.13 | 0.03 | 0.38 | 0.01 | 0.11 |
| SFAT | 0.09 | 0.09 | 0.13 | 0.06 | |||||
| FISH | 0.05 | 0.16 | 0.12 | 0.16 | |||||
| 20:0 | CON | 0.08 | 0.10 | 0.06 | 0.06 | 0.03 | 0.02 | <0.01 | 0.11 |
| SFAT | 0.08 | 0.07 | 0.05 | 0.04 | |||||
| FISH | 0.21 | 0.12 | 0.07 | 0.05 | |||||
| 18:3n-6 | CON | 0.40 | 0.42a | 0.39a | 0.38a | 0.08 | <0.01 | 0.34 | 0.04 |
| SFAT | 0.34α,β | 0.48a,α,β | 0.33a,α | 0.53a,β | |||||
| FISH | 0.39α | 0.08b,β | 0.11b,β | 0.15b,β | |||||
| 18:2alltrans | CON | 0.13 | 0.14 | 0.07 | 0.06 | 0.02 | <0.01 | <0.01 | 0.38 |
| SFAT | 0.14 | 0.16 | 0.08 | 0.10 | |||||
| FISH | 0.08 | 0.03 | 0.01 | 0.01 | |||||
| 20:3n-3 | CON | 6.55 | 6.75 | 4.53 | 4.27 | 0.59 | <0.01 | <0.01 | 0.14 |
| SFAT | 5.92 | 6.86 | 4.00 | 4.38 | |||||
| FISH | 5.54 | 3.26 | 2.14 | 2.38 | |||||
| 22:1 | CON | 0.01 | 0.01 | 0.01 | 0.02 | 0.10 | 0.39 | 0.46 | 0.48 |
| SFAT | <0.01 | 0.01 | 0.01 | 0.01 | |||||
| FISH | 0.30 | 0.03 | <0.01 | <0.01 | |||||
| 22:2n-6 | CON | 0.01 | 0.01 | <0.01 | <0.01 | 0.12 | 0.07 | 0.59 | 0.75 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | 0.35 | 0.14 | 0.07 | 0.04 | |||||
| 22:3n-3 | CON | <0.01 | 0.01 | 0 | 0.11 | 0.07 | 0.92 | 0.72 | 0.25 |
| SFAT | 0.02 | 0.03 | 0.09 | 0.01 | |||||
| FISH | 0.18 | <0.01 | <0.01 | <0.01 | |||||
| 22:4n-6 | CON | 2.71 | 2.58 | 1.60 | 1.27 | 0.22 | <0.01 | <0.01 | 0.08 |
| SFAT | 2.36 | 2.45 | 1.59 | 1.10 | |||||
| FISH | 2.07 | 0.93 | 0.36 | 0.26 | |||||
| Unknown | CON | 5.33 | 3.89 | 2.04 | 2.60 | 0.80 | 0.92 | <0.01 | 0.10 |
| SFAT | 3.93 | 3.81 | 1.70 | 2.74 | |||||
| FISH | 3.30 | 6.00 | 1.71 | 2.26 | |||||
Notes:
Difference (P < 0.05) between different diet at same day.
Denote significant interactions (P < 0.05) within a same diet at different days.
CON = control diet containing no supplemental lipid: SFAT = CON supplemented with Energy Booster: FISH = CON supplemented with fish oil;
standard error of the mean;
diet by time interaction.
SFAT and FISH-supplemented cows had greater (diet × day P < 0.05) α-linolenic acid (18:3n-3) content at −21 days compared with controls, while a lower concentration was observed at −10 days in the same groups (Table 3). As compared with SFAT and control, the FISH-supplemented cows had a lower (diet × day P < 0.05) proportion of γ-linolenic acid (18:3n-6) at −10 days but a gradual increase was observed at 1 and 11 days (Table S1). As expected, the concentration of eicosapentaenoic acid (20:5n-3) and docosahexaenoic acid (22:6n-3) was greatest with FISH compared with control or SFAT from −10 days through 11 days. However, arachidonic acid (20:4n-6) decreased markedly with FISH and SFAT after parturition as compared with control (Table 3).
Triacylglycerol
Concentration of 16:0 was greatest (P < 0.05) overall in cows fed SFAT, but 18:0 was not affected by diet (Table 4). A postpartal increase (day P < 0.05) in 16:0 and a decrease (day P < 0.05) in stearic acid was observed (Table 4). Similar to the PL fatty acid fraction, FISH-supplemented cows had a tendency (diet × day P = 0.06) for a postpartal increase in TAG concentration of trans-vaccenic acid (18:1trans11) as compared with control and SFAT (Table 4). Overall, however, concentration of 18:1trans11 was greatest (P < 0.05) for cows fed FISH than SFAT or control. Similar to PL, the CLA concentration of hepatic TAG also increased soon after calving and remained elevated compared with prepartal concentrations. A postpartal increase (day P < 0.05) was observed in the concentration of 18:2n-6 regardless of diet. The concentrations of other 18:1trans isomers were little affected by supplemental lipid (Table S1).
Table 4.
Percentage of long-chain fatty acids in liver triacylglycerol during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT).
| Fatty acid | Treatment | Day | SEM† | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| −21 | −10 | 1 | 11 | Diet | Time | D = T‡ | |||
| 16:0 | CON | 32.21 | 33.23 | 38.76 | 39.37 | 1.80 | 0.03 | <0.01 | 0.87 |
| SFAT | 35.10 | 36.70 | 39.14 | 40.42 | |||||
| FISH | 33.05 | 33.03 | 35.46 | 37.10 | |||||
| 18:0 | CON | 20.59 | 18.00 | 6.87 | 7.56 | 1.93 | 0.18 | <0.01 | 0.82 |
| SFAT | 18.11 | 18.87 | 9.37 | 6.27 | |||||
| FISH | 16.86 | 15.56 | 6.59 | 5.77 | |||||
| 18:1cis9 | CON | 14.28 | 13.82 | 20.90 | 20.72 | 2.63 | 0.92 | <0.01 | 0.21 |
| SFAT | 14.35 | 12.81 | 21.16 | 19.57 | |||||
| FISH | 20.04 | 14.00 | 14.92 | 21.69 | |||||
| 18:1trans11 | CON | 0.85 | 0.99 | 1.07 | 0.72 | 0.24 | 0.04 | 0.01 | 0.06 |
| SFAT | 0.84 | 0.66 | 1.22 | 0.86 | |||||
| FISH | 0.41 | 1.47 | 1.76 | 1.43 | |||||
| 18:2c9t11 | CON | 0.07 | 0.12 | 0.26 | 0.26 | 0.04 | 0.85 | <0.01 | 0.33 |
| SFAT | 0.12 | 0.08 | 0.26 | 0.20 | |||||
| FISH | 0.11 | 0.04 | 0.29 | 0.28 | |||||
| 18:2n-6 | CON | 4.48 | 4.88 | 6.45 | 7.75 | 0.60 | 0.14 | <0.01 | 0.80 |
| SFAT | 4.29 | 4.36 | 5.52 | 6.42 | |||||
| FISH | 3.68 | 4.01 | 6.22 | 7.31 | |||||
| 18:3n-3 | CON | 0.50 | 0.73 | 1.36 | 1.34 | 0.20 | 0.33 | <0.01 | 0.19 |
| SFAT | 0.89 | 0.46 | 1.54 | 1.16 | |||||
| FISH | 0.62 | 0.54 | 1.74 | 1.74 | |||||
| 20:4n-6 | CON | 2.24 | 2.77 | 0.60 | 0.81 | 0.59 | 0.90 | <0.01 | 0.44 |
| SFAT | 2.92 | 2.42 | 0.45 | 0.51 | |||||
| FISH | 1.81 | 3.93 | 0.66 | 0.57 | |||||
| 20:5n-3 | CON | 0.06α | 0.24β | 0.07a,α | 0.11a,α,β | 0.06 | <0.01 | 0.23 | 0.04 |
| SFAT | 0.15 | 0.11 | 0.06a | 0.10a | |||||
| FISH | 0.09α | 0.25α,β | 0.36b,β,γ | 0.29b,β,γ | |||||
| 22:5n-3 | CON | 0.62α | 1.30b,β | 0.62a,α | 0.61a,α | 0.20 | <0.01 | 0.02 | <0.01 |
| SFAT | 0.52 | 0.69a | 0.61a | 0.56a | |||||
| FISH | 0.47α | 0.84a,b,α | 1.61b,β | 1.57b,β | |||||
| 22:6n-3 | CON | 0.04b | 0.27a,b | 0.04a | 0.06a | 0.10 | <0.01 | <0.01 | <0.01 |
| SFAT | 0.35a,α | 0.04a,β | 0.06a,β | 0.04a,β | |||||
| FISH | 0.09a,b,α | 0.46b,β | 1.26b,γ | 1.01b,γ | |||||
Notes:
Standard error of the mean;
diet × time interaction.
Difference (P < 0.05) between diets on the same day.
Significant interactions (P < 0.05) within a diet and between days.
Compared with the PL fraction, supplementing lipid was not effective (diet P > 0.05) in altering the 18:3n-3 and 20:4n-6 concentration of the hepatic TAG fraction. However, a postpartal increase (day P < 0.05) in 18:3n-3 and a concomitant decrease in 20:4n-6 concentration was observed regardless of diet (Table 4). Similar to PL, there was an increase (diet × day P < 0.05) in the proportion of all n-3 fatty acids in the TAG fraction of FISH-supplemented cows; 20:5n-3, 22:5n-3, and 22:6n-3 had a steady increase in concentration by day 1 followed by still greater concentrations relative to the prepartum levels. Overall, the postpartal concentration of 20:5n-3, 22:5n-3, and 22:6n-3 was markedly lower in SFAT and control than FISH (Table 4). A postpartal increase (diet × day P < 0.05) was observed in the proportion of 18:3n-6, namely due to the response in the control group. The overall increase, however, was of a lower magnitude in FISH as compared with SFAT (Table S2).
Hepatic gene expression
Fatty acid transport and LD formation
The expression of genes involved in fatty acid uptake/transport, storage, and oxidation changed in a different fashion with lipid supplementation as compared to the control cows. The expression of the LCFA-uptake protein CD36 in the prepartal period was lower (diet × day P < 0.05) with SFAT than FISH and control, and was markedly down regulated after parturition in response to FISH. In contrast, an increase (diet × day P < 0.05) in FABP1 expression postpartum was observed in cows fed SFAT and the controls, at which point expression was greater in those cows compared with FISH (Table 5).
Table 5.
Relative expression (treatment means, log-2 scale) of genes involved in fatty acid uptake, esterification, desaturation, lipid droplet formation, fatty acid oxidation, gluconeogenesis and cellular energy during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT).
| Gene | Prepartum | Postpartum | SEM† | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Control | FISH | SFAT | Control | FISH | SFAT | Diet | Time | D × T‡ | ||
| Fatty acid uptake/transport | ||||||||||
| CD36 | −0.02a | −0.59a,b,* | −1.48b | 0.12a | −1.91b,* | −1.07a,b | 0.43 | 0.06 | 0.18 | <0.01 |
| FABP1 | 1.34* | 1.39 | 0.99* | 1.99a,* | 0.99b | 1.50a,b,* | 0.24 | 0.20 | 0.13 | <0.01 |
| Esterification, desaturation, lipid droplet formation | ||||||||||
| DGAT2 | 1.49a | 0.96a,b | 0.33b,* | 2.09a | 1.00b | 1.52a,b,* | 0.34 | 0.13 | <0.01 | <0.05 |
| SCD | −0.43a,* | −4.11c | −2.02b | −1.69a,* | −4.21b | −1.72a | 0.38 | <0.01 | 0.22 | 0.09 |
| PLIN2 | 2.59 | 1.97* | 1.42* | 3.21a | 0.97b,* | 2.29a,* | 0.41 | <0.05 | 0.43 | <0.01 |
| PLIN4 | −2.57 | −2.38 | −2.71 | −1.90 | −3.38 | −3.12 | 0.51 | 0.45 | 0.33 | 0.09 |
| Fatty acid oxidation | ||||||||||
| ACSL1 | 3.95a,* | 2.88a,b,* | 2.72b,* | 4.76a,* | 2.12c,* | 3.40b,* | 0.41 | <0.01 | 0.21 | 0.02 |
| ACOX1 | 2.71a | 2.24a,* | 1.22b,* | 2.24a,b | 1.36b,* | 2.48a,* | 0.33 | 0.10 | 0.9 | <0.01 |
| CPT1A | 3.08 | 2.92 | 2.20 | 3.54 | 1.76 | 2.20 | 0.71 | 0.20 | 0.54 | 0.14 |
| FGF21 | −0.51* | −0.06 | 1.07 | 5.55a,* | 2.10b | 1.08b | 1.09 | 0.33 | <0.01 | <0.01 |
| Gluconeogenesis | ||||||||||
| PC | 3.57* | 3.25 | 2.83 | 5.73a,* | 2.95b | 3.54b | 0.53 | 0.06 | <0.01 | <0.01 |
| PCK1 | 3.76* | 3.72 | 3.49 | 5.13a,* | 3.10b | 3.21b | 0.51 | 0.17 | 0.56 | 0.04 |
| Intracellular energy | ||||||||||
| UCP2 | 2.98 | 2.76* | 1.60 | 1.96 | 0.76* | 1.62 | 0.67 | 0.33 | 0.04 | 0.04 |
| PRKAA1 | 3.80 | 3.59* | 2.90 | 4.19a,b | 2.36b,* | 2.90a,b | 0.50 | 0.22 | 0.16 | 0.01 |
| STK11 | 4.09 | 3.78* | 2.96 | 3.57 | 2.39* | 2.81 | 0.44 | 0.21 | <0.01 | 0.04 |
Notes:
Standard error of the mean;
diet × time interaction.
Difference (D × T, P < 0.05) between diets at prepartum or postpartum times.
Difference (D × T, P < 0.05) prepartum vs. postpartum within diet.
Feeding SFAT led to lower (diet × day P < 0.05) DGAT2 prepartum when compared to control or FISH, however that diet resulted in an increase in expression of DGAT2 postpartum, at which point expression of DGAT2 was similar between control and FISH. Quite remarkably, the expression of SCD was lowest (diet P < 0.05) for SFAT and FISH than controls during the prepartal period. Its expression, however, decreased markedly after parturition in control cows but not in lipid-supplemented cows. The LD-associated proteins PLIN2 (formerly known as adipophilin) and PLIN4 (formerly known as S3-12) did not differ in expression among groups in the prepartal period. However, there was a decrease in expression of PLIN2 in cows fed FISH while an increase was observed in cows fed SFAT (diet × day P < 0.05). The overall result was that PLIN2 expression postpartum was similar between SFAT and controls and lower with FISH.
Fatty acid oxidation
The expression of ACSL1 and ACOX1 in the prepartal period was greater in cows fed FISH than SFAT. After parturition, however, cows fed FISH had lower expression and cows fed SFAT had greater expression of both genes resulting in an overall interaction effect (diet × day P < 0.05). The control cows also had an increase in ACSL1 expression after parturition, actually resulting in greater overall expression than SFAT and FISH. Whereas expression of the PPARα target CPT1A did not differ due to treatment or day, the expression of FGF21 which is another PPARα target increased to the greatest extent (diet × day P < 0.05) postpartum in the controls. Thus, in the postpartal period FGF21 expression was greatest in controls relative to FISH and SFAT.
Genes involved in gluconeogenesis and intracellular energy sensing
There was no interaction (diet × day P > 0.05) in the prepartal period for the expression of PC and PCK1; however, expression of both genes increased (diet × day P < 0.05) markedly postpartum in the control cows, at which point cows fed FISH and SFAT had lower overall expression largely due to a lack of change in expression from the prepartal period. In a similar fashion, there was no significant prepartal interaction effect on the expression of UCP2, PRKAA1, and STK11, which are involved in intracellular sensing of ATP and signaling via the AMPK pathway. However, cows fed FISH had a marked decrease (diet × day P < 0.05) in expression of these three genes in the postpartal period leading to a lower overall PRKAA1 expression postpartum.
Transcription regulators and nuclear co-activators and repressors
No obvious pre- and postpartal differences were observed in the expression of PPARA, while RXRA had an overall downregulation after parturition (time P = 0.06), due primarily to feeding control and FISH (Table 6). Feeding SFAT resulted in lower (diet × day P < 0.05) SREBF2 prepartum compared with controls, but unlike FISH it did not elicit a decrease in expression in the postpartum period. Thus, overall SREBF2 expression postpartum was similar in controls and SFAT and lower in FISH. Similar to SREBF2, feeding FISH led to lower (diet × day P < 0.05) expression of PPARD between the pre and postpartal period at which point cows fed both FISH and SFAT had lower (diet × day P < 0.05) expression than controls.
Table 6.
Relative expression (treatment means, log-2 scale) of genes encoding transcription regulators, nuclear receptor co-activators and co-repressors during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT).
| Gene | Prepartum | Postpartum | SEM† | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Control | FISH | SFAT | Control | FISH | SFAT | Diet | Time | D × T‡ | ||
| Transcription regulators | ||||||||||
| PPARA | 0.41 | 0.63 | 0.26 | 0.38 | 0.49 | 0.14 | 0.23 | 0.54 | 0.31 | 0.88 |
| RXRA | 3.17 | 2.85 | 2.31 | 2.60 | 1.36 | 2.24 | 0.57 | 0.30 | 0.06 | 0.77 |
| PPARD | 3.18 | 2.86* | 1.93 | 3.72a | 1.19b,* | 2.22b | 0.45 | 0.04 | 0.18 | <0.01 |
| SREBF2 | 4.43a | 3.74a,b,* | 3.18b | 3.72a | 2.59b,* | 3.74a | 0.36 | 0.01 | 0.66 | 0.02 |
| Nuclear receptor co-activators | ||||||||||
| CARM1 | 4.20a | 3.48b,* | 2.84b | 4.04a | 2.10b,* | 3.24a,b | 0.37 | 0.03 | 0.08 | <0.01 |
| MED1 | 4.30a | 3.57b,* | 2.89b | 4.21a | 2.15c,* | 3.37b | 0.38 | 0.02 | 0.12 | <0.01 |
| Nuclear receptor co-repressors | ||||||||||
| NCOR2 | 3.67a | 3.00a,b,* | 2.35b | 3.69a | 1.91b,* | 2.73b | 0.33 | 0.01 | 0.30 | 0.04 |
| NRIP1 | 3.60a | 2.78b,* | 2.40b | 3.48a | 1.64b,* | 2.72b | 0.36 | 0.03 | 0.14 | 0.03 |
Notes:
Standard error of the mean;
diet × time interaction.
Difference (D × T, P < 0.05) between diets at the prepartum or postpartum times.
Difference (D × T, P < 0.05) prepartum vs. postpartum within diet.
Feeding FISH and SFAT vs. control led to lower expression in the prepartal period of the nuclear receptor co-regulators CARM1, NCOR2, MED1, and RIP1 (diet × day P < 0.05). Expression of all these genes decreased (diet × day P < 0.05) in the postpartum period exclusively in cows fed FISH such that control cows still had greater overall expression followed by cows fed SFAT.
Inflammation and apoptosis
The expression of IL6, TBK1, and IKBKE in the pre-partal period was lowest (diet × day P < 0.05) in cows fed SFAT in comparison to the control or FISH cows (Table 7). Interestingly, cows fed FISH experienced a decrease (diet × day P < 0.05) in postpartal expression of these genes while expression in SFAT-fed or control cows remained unchanged. Except for TBK1 and IKBKE, expression of IL6 was lower (diet × day P < 0.05) overall postpartum in cows fed SFAT and FISH. Cows fed FISH also had a decrease (diet × day P < 0.05) in CIDEB expression postpartum at which point expression was lower compared with controls and SFAT.
Table 7.
Relative expression (treatment means, log-2 scale) of genes involved in inflammation and cell growth during the peripartal period in cows fed (n = 5/treatment) control, fish oil (FISH), or saturated lipid (SFAT).
| Gene | Prepartum | Postpartum | SEM† | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Control | FISH | SFAT | Control | FISH | SFAT | Diet | Time | D × T‡ | ||
| Inflammation related genes | ||||||||||
| IL6 | 3.04a | 2.11a,* | 1.06b | 2.32a | 0.90b,* | 1.48b | 0.35 | 0.01 | 0.03 | 0.02 |
| TBK1 | 4.37a | 3.68a,* | 2.78b | 4.04a | 1.98b,* | 3.44a | 0.38 | 0.03 | 0.05 | <0.01 |
| IKBKE | 3.58a | 3.28a,* | 2.17b | 3.34a | 1.39b,* | 2.76a | 0.37 | 0.05 | 0.03 | <0.01 |
| Apoptosis | ||||||||||
| CIDEB | 5.38 | 4.40* | 4.18 | 5.63a | 3.52b,* | 4.82a | 0.40 | 0.02 | 0.99 | 0.04 |
Notes:
Standard error of the mean;
diet × time interaction.
Difference (D × T, P < 0.05) between diets at the prepartum or postpartum times.
Difference (D × T, P < 0.05) prepartum vs. postpartum within diet.
Discussion
Hepatic PL and TAG fatty acid composition
Changes observed in the hepatic fatty acid profiles of PL and TAG were likely driven by a combination of (1) alterations in LCFA composition of adipose tissue induced by feeding, (2) mobilization of LCFA postpartum in response to lipolytic signals, and (3) biohydrogenation of PUFA in the rumen leading to increases in trans fatty acids. The contribution of adipose tissue LCFA to the PL and TAG fraction of lipids in dairy cattle liver has been demonstrated previously.2,17 Schmitt et al14 using the same cows from this study, reported that all groups had an increase in liver TAG after parturition, which is a typical response observed due to the increase in circulating NEFA (ie, from mobilization of adipose tissue) (Fig. 1B). Although not measured in these cows, it is likely that the pool of LCFA stored in adipose tissue TAG and also in liver PL during the prepartal period in particular, contributed to the observed changes in gene expression profiles. This idea is supported by the fact that both FISH and SFAT upregulated several genes associated with lipid metabolism and the PPAR pathway namely in the prepartal period.14
Dietary fat supplementation often produces a pronounced effect on hepatic PL composition.2 In the present study, the postpartal decrease of 18:0 in the concentration and the increase in 18:1trans isomers (particularly trans11) with FISH was consistent with greater biohydrogenation of dietary PUFA.15 The post-partal increase in 16:0 and 18:2n-6 regardless of diet likely was associated with increased adipose tissue lipolysis and subsequent transport and storage in the liver.17 The observed decrease in the PL concentration of γ linolenic acid (18:3n-6) might have been due to allosteric inhibition or transcriptional regulation of the elongases as well as the Δ5- and Δ6-desaturases or to competition between n-6 and n-3 substrates for incorporation into PL.18
The observed increase in the hepatic PL and TAG content of α linolenic acid (18:3n-3) likely accounted for the increase in content of 20:5n-3, 22:5n-3, and 22:6n-3 at calving2; α linoleic acid can be desaturated and elongated to 20:5n-3 and 22:6n-3. The decrease in dihomo-γ-linoleic acid (20:3n-6) and arachidonic acid (20:4n-6) with FISH and SFAT after parturition as compared with the control (Table 3) might have been due to a low baseline concentration of linoleic acid (18:2n-6) in hepatic PL15 as linoleic acid is desaturated and elongated to 20:3n-6 and subsequently to 20:4n-6.2 Another possibility is that greater supply of 18:2 resulted in competition and displacement of other long chain PUFAs for esterification into PL.2 Similarly, increased tissue content may inhibit elongation and desaturation of 18:2 and 18:3 as observed in calves.19 The lower level of γ linolenic acid in FISH could have affected the concentration of arachidonic acid as γ linolenic acid is the intermediate in the conversion of linoleic acid to arachidonic acid.20
Intracellular metabolism and LCFAs in bovine
The metabolism of specific LCFAs has been studied previously in vitro using hepatocytes isolated from neonatal calves.21,22 Despite the inherent difficulties in comparing in vitro and in vivo studies, in the context of our study, data generated with incubations of 16:0 alone or in combination with 18:0, 18:1, 20:5n-3, or 22:6n-3 are of interest. For instance, greater total oxidation (CO2 and acid-soluble products) of 14C-16:0 were observed with incubations of 1 mM 16:0 plus 1 mM 20:5n-3 and 18:1 when compared with incubation of 1 mM 16:0 alone.21 In contrast, the incubation containing 20:5n-3 did not increase use of 14C-palmitic acid for cellular TAG synthesis, while 18:1 did. Palmitate plus 22:6n-3 did not affect total oxidation of 14C-palmitic acid or its esterification to TAG.21 Results from that initial study clearly underscored the differential utilization of LCFA for esterification or fatty acid oxidation in liver cells. Although there were no data on mRNA or protein expression of lipid metabolism enzymes, it would not be unreasonable to suspect (based on data from model organisms) that changes in expression would have been partly responsible for the changes observed. For instance, 18:1 either from exogenous sources or synthesized from 18:0 via SCD, is central for cellular TAG synthesis and LD formation,23 and is probably an essential step in very-low-density lipoprotein (VLDL) synthesis in the liver.
A second study confirmed the response of 18:1 only in terms of enhancing the use of 16:0 for esterification, but did not confirm the effect of 20:5n-3 on ketone body formation from 16:0.22 Furthermore, there was an increase in cellular TAG with incubations of 22:6n-3. However, contrary to the first study incubation of 1 mM 16:0 plus 1 mM 18:0 resulted in greater BHBA concentration in culture media as a result of greater oxidation rate of 14C-16:0.22 Although we did not detect statistical differences in blood BHBA due to lipid supplementation,14 the likely fluctuations in BHBA metabolism during lactation, when compared with in vitro systems, would make it difficult to use this marker to discern physiological effects in vivo.
Additional analyses from this second study included rate of gluconeogenesis and concentration of cellular glycogen. Linolenic acid plus 16:0 resulted in the highest rates of gluconeogenesis from 14C-propionic acid and greatest amounts of intracellular glycogen, along with reduced TAG production.22 An interesting finding was that 22:6n-3 plus 16:0 incubation, or either plus 20:5n-3, increased cellular TAG content and incorporation of 14C-palmitic acid into cellular TAG. In addition, 22:6n-3 plus 16:0 decreased metabolism of 14C-propionic acid to glucose or to cellular glycogen in the medium.22
The in vitro work with hepatocytes has, by necessity, been short-term compared with the longer-term feeding studies. Furthermore, the above studies used supra-physiological concentrations of each LCFA. Their expected concentration in the circulation of the cow after parturition is unlikely to reach the 1 mM level. In fact, unless the cow is under a ketotic situation, the peak total NEFA concentration after parturition is rarely greater than 1.5 mM.24 Although the resulting data from these in vitro studies have helped expand our knowledge of the metabolic effects of specific LCFA, the observed results are challenging to place in the in vivo context. Thus, gene expression data obtained over a longer time-frame from cows in field conditions may provide a more physiologically-relevant view of the likely mechanistic effects of enriching diets with specific LCFA.
Hepatic gene expression
Fatty acid transport and metabolism
The expression of FABP1 in dairy cows has been previously characterized as increasing between parturition and 14 days postpartum, a response which was proposed to denote downstream activation of PPARα via NEFA metabolism.5 However, in the present study, even with the increase in NEFA postpartum (Fig. 1B), the FABP1 expression remained stable in cows fed SFAT and FISH while it was upregulated in controls. That response suggests that irrespective of the type of fat supplemented, this intra-cellular transporter could be modulated without the influence of the rising post-partum NEFA. On the other hand, the pronounced down-regulation of CD36 after parturition in cows fed FISH (Table 5) suggested a possible biological role of PUFA in the control of the intracellular flux of LCFA. Such a response would be contradictory with non-ruminant data (ie, upregulation of CD36 with PUFA) indicating an intra-species difference in the response of liver cells to supplementation with PUFA, as already proposed in bovine kidney cells.10
The contrasting response between liver tissue in the present study and bovine kidney cells10 in the same species is not surprising and obviously is likely related with the different types of cells studied in addition to the endocrine and metabolic environment. The complexity of the LCFA pool that hepatocytes are exposed to also has shown to influence the degree of PPAR gene network activation in rodent liver.25–27 As stated before, feeding FISH led to a large difference in the concentration of 18:1trans11, 18:1trans12, 22:6n-3, and 20:5n-3, all of which could potentially play a role in the activation of the PPARα gene network at least judging from in vitro data.28
Esterification, desaturation, and LD formation
The enzyme SCD is responsible for biosynthesis of monounsaturated fatty acids 18:1 and 16:1 from 16:0 and 18:0, which are substrates for de novo synthesis of PL, cholesterol esters, and TAG.29,30 Our results with SCD agree with reports from other animal species where both omega-3 and omega-6 PUFA decreased SCD expression partly by decreasing mRNA stability.30–34 On the other hand, the SCD down-regulation with SFAT is contrary to in vitro reports with bovine kidney cells10 in which SCD was upregulated by exogenous 16:0 and 18:0, the main LCFA in the blood of ruminants. However, the downregulation of CD36 could partly explain the lower SCD if in fact there was less 18:0 and 16:0 taken up from blood, leading to reduced substrate availability.
In rodents, SCD and DGAT2 are located adjacently in the outer endoplasmic reticulum membrane.35 It has been proposed that SCD indirectly influences the activity of DGAT2 by supplying substrates (16:1 and 18:1) for synthesis of TAG.29 Our data revealed a similar pattern of expression response for SCD and DGAT2 across diets, namely postpartum. The DGAT2 enzyme plays a key role in the cytosolic accumulation of TAG,36–38 which could in the post-absorptive state provide LCFA for lipoprotein synthesis. The lower expression of this gene postpartum with the FISH group than with the control and the lack of difference between FISH and SFAT could be taken as an indication of decreased use of LCFA for TAG synthesis. Such response would have channeled LCFA towards PL and/or cholesterol ester synthesis, VLDL synthesis, or oxidation.23
The lower SCD expression also could have been associated with the response observed for PLIN2, another enzyme linked with liver TAG accumulation and size of LDs.39 Studies in vitro with different non-ruminant cell lines40,41 demonstrated that ADFP is upregulated by PUFA, which was obviously not induced by FISH. The increase of PLIN2 expression postpartum we observed in the SFAT group agrees with previous data from rodent hepatocytes40 and underscores the fact that the increase in NEFA alone is not the sole mechanism for activating transcription of this gene. Hepatic TAG accumulation typically follows the rise in NEFA postpartum.14,24 As an integral LD protein, PLIN2 may serve more critical roles in managing the turnover of neutral lipid stores to facilitate the coordinated release of LCFA into lipoproteins in response to changes in metabolic state.23
Unlike PLIN2, PLIN4 is an exchangeable cytosolic LD protein that facilitates rapid protein association with the immature LD.5,42 Generally in the liver, the PLIN4 is associated with smaller and peripheral LD and is stimulated by high concentrations of LCFA while PLIN2 is associated with bigger and more mature LD.43 The observed changes postpartum for PLIN2 in cows fed SFAT (upregulation) and FISH (downregulation) seem to suggest a difference in potency of LCFA, or alternatively, as in the case of the FISH group, a protective mechanism of the cell to buffer from excessive LCFA influx.
Fatty acid oxidation
In the liver, ACSL1 is central for the synthesis of LCFA-acyl-CoA, which can then be channeled towards β-oxidation.44 The regulation of expression of this enzyme by PPARα activation in rodents is well-established,44–46 and recent work demonstrated a similar response in bovines.10,47 Previous work with peripartal dairy cows fed diets without supplemental lipids revealed that ACSL1 increases expression between −14 to 1 day postpartum after which it remains unchanged through 14 days. This response could help in the metabolism of incoming NEFA, thus channeling them towards β-oxidation.5,48 We observed a similar response in the control cows and those fed SFAT. Thus, the down regulation of ACSL1 in cows fed with FISH, along with CD36 in particular, would agree with the concept of different LCFA eliciting opposite effects on transcription of ASCL1 and CD36. It could be envisioned that such a response would prevent excessive influx of LCFA but also serve to control their use for esterification or oxidation.
The lack of difference for ACOX1 expression postpartum between control and SFAT suggested that, on the one hand, enhanced dietary LCFA flux into liver did not alter peroxisomal oxidation (ie, at a greater availability of LCFA, the liver from SFAT-fed cows might have been capable of oxidizing a greater fraction) and on the other hand, that not enough dietary LCFA reached the liver to cause an effect. The latter case is likely because of the mammary gland’s preference for taking up LCFA from the circulation, and thus in a way buffering other tissues.15 The effect of fish oil on ACOX1 expression has been evaluated in several rodent studies which demonstrated that PUFA are potent activators.25,49–51 Mechanistically, such a response makes sense as it is the first and rate-limiting enzyme of the peroxisomal fatty acid β-oxidation pathway,52 which is important in peripartal liver lipid metabolism.53 The prepartal response with FISH vs. SFAT appeared to be in line with rodent data. However, the postpartal decrease in ACOX1 with FISH coupled with the increase with SFAT suggested that saturated LCFAs were more potent in activating ACOX1, which agrees with in vitro data.10 These results highlight that in peri-partal cows, feeding FISH (at least at the levels of this study) might not be effective in enhancing LCFA oxidative capacity.
In our analysis of adipose tissue gene expression from these cows, we found some evidence that lipid supplementation could influence the production of adipokines by adipose tissue, hence, influence indirectly the hepatic capacity of β-oxidation.14 It has been proposed in rodents that adiponectin binding to its hepatic receptor (ADIPOR2) leads to activation of both AMPK and PPAR signaling pathways and, consequently, ACOX1 activation.5,54 We observed that FISH led to lower postpartal expression of ADIPOR2 and ADIPOQ in the adipose, and lower ADIPOR2 in liver tissue.14 In contrast, ADIPOR2 expression in liver and ADIPOQ in adipose tissue were the same in the control and SFAT. Such responses could be mechanistically related with the lower postpartal ACOX1 and a potential overall reduction in β-oxidation when dietary FISH supplementation.
Recently, it was reported that FISH inhibits de novo lipogenesis and β-oxidation, and decreases insulin resistance in non-ruminants. The improvement in insulin sensitivity is mediated by down regulating the PPARG network, ChREBP, and SREBF1. Furthermore, FISH also increases adiponectin (powerful insulin-sensitizing agent) production in adipose tissue.55 The decrease in DMI (Fig. 1A) along with the lower milk yield,14 and the observed down regulation of SCD and DGAT2 in the present study and in adipose tissue (SCD, DGAT2 ChREBP, and SREBF1) in the study of Schmitt et al,14 suggests that FISH may actually have had a negative impact on genes involved in LCFA oxidation. Additional research seems warranted to elucidate the underlying mechanisms and their physiological relevance.
Peroxisomal oxidation results in the increased production of shorter-chain fatty acyl-CoA that generally are channeled to be completely oxidized in mitochondria.5 Although CPT1A has a key role in this process, the lack of change in expression in our study seems to confirm previous data, providing evidence that this enzyme is not strongly controlled at the transcription level even during periods of severe negative energy balance.56 Rodent CPT1A is markedly upregulated in response to undernutrition and fasting, thus underscoring additional differences between species in the control of hepatic LCFA oxidation. Despite the apparent lack of CPT1A activation (ie, it is a PPARα target in rodents) the upregulation of FGF21 (another PPARα target) postpartum provided evidence of transcriptional adaptations that could have been driven via PPARα.
In rodents, the PPARα protein is required for the normal activation of hepatic LCFA oxidation, TAG clearance, and ketogenesis.57 Despite obvious biological variation in its expression across treatments, the pattern of FGF21 expression that was observed in this study seemed to confirm that this protein is as important in coordinating hepatic adaptations to undernutrition in ruminants as in rodents. However, the lack of postpartal increase in cows fed SFAT or FISH could indicate that excess LCFA influx into liver actually might feedback-inhibit the transcriptional activation of FGF21 and more potently when feeding PUFA (eg, FISH) than saturated LCFA. From a mechanistic standpoint, the attenuation of FGF21 did not seem to curtail LCFA oxidation in cows fed SFAT (ie, ACOX1 expression increased) and blood BHBA was similar among treatments.14 Overall, the present and previous data4 suggest that FGF21 in cows is associated with negative energetic balance.
Genes involved in gluconeogenesis and intracellular energy sensing
The predominant glucose precursors in dairy cows are propionate and lactate, although the contribution of alanine and glycerol becomes quantitatively more important during conditions of propionate deficit (ie, the early postpartal period).58,59 Partitioning of lactate and alanine towards gluconeogenesis is under the control of PC, the expression of which increases sharply after parturition,13,60 feed restriction,61 and experimentally-induced glucose deficit62 to facilitate flux of alanine and lactate toward oxaloacetate rather than acetyl-CoA.
The upregulation of PC at calving is linked with increased concentrations of NEFA in plasma. This is not surprising as several metabolic reactions are regulated by LCFA at the level of the activation of genes that encode key regulatory enzymes,63,64 including gluconeogenesis and fatty acid metabolism. The observed increase in linoleic acid concentration in PL after calving in controls (and comparatively lower in FISH) seems to support in vitro data, demonstrating that linoleic acid could serve to activate PC and enhance the capacity for fatty acid oxidation and glucose synthesis.64 Furthermore, there is evidence that greater supply of linoleic acid can enhance the rate of gluconeogenesis in bovine hepatocytes.22 Cellular-membrane bound linoleic acid also can serve as a ligand for NR that regulate gene expression. For example, in hepatocyte cultures, this LCFA increased the PCK1 mRNA level in a dose-dependent manner.65
The exogenous LCFA can enhance the rates of hepatic mitochondrial oxidation by uncoupling oxidation from ATP production.5 The uncoupling proteins (UCPs) are key players in mitochondrial oxidation, the activity of which can be induced by LCFA. Monounsaturated and PUFA appear more effective than saturated LCFA in activating the liver specific iso-form UCP2.66UCP2 is a demonstrated non-ruminant PPARα target in vivo and has been proposed to play a role in lipid metabolism, insulin resistance, glucose utilization, regulation of reactive oxygen species, and macrophage-mediated immunity.67,68
Armstrong and Towle66 reported induced expression of hepatic UCP2 with high concentrations of LCFA. To some extent, that effect lends support to the observed lower postpartal UCP2 expression in cows fed FISH vs. SFAT as they had numerically-lower blood NEFA (Fig. 1B). Insulin69 and aspirin66 significantly reduced the expression of UCP2 mRNA in rodent liver. Because the high NEFA concentration postpartum seems to directly impair the ability of the pancreas to secrete insulin in postpartal dairy cows,70 it is unlikely that in the present study insulin had a mechanistic role on the expression of UCP2. In contrast, the fact that aspirin reduced UCP2 in rodent liver despite high concentrations of LCFA could indicate that in our study the prostaglandin pathway might have been physiologically relevant. This idea is supported by the lower hepatic PL concentration of 20:4n-6 (the immediate precursor of prostaglandins) in cows fed FISH when compared with those fed SFAT.
Nuclear receptor co-activators and co-repressors
The co-activator-associated arginine methyltransferase I (CARM1) is a critical component of glucose metabolism in rodent hepatocytes.71 There it physically interacts with cAMP-responsive element binding factor CREB before both being recruited to the PCK1 and glucose-6-phosphatase promoters in a cAMP- dependent manner particularly during periods of dietary glucose short-falls (eg, undernutrition, starvation, negative energy balance).72 CARM1 regulates gene expression by multiple mechanisms including methylation of histones and co-activation of steroid receptor transcription.73 The observed post-partal decrease of CARM1 with a FISH supplemented diet agrees with the lower expression of PCK1 (Table 5). In addition, it is possible that differences in intracellular cAMP concentration also might have played a role in both CARM1 and PCK1 upregulation. We speculate that because of the lower rate of milk production and DMI in response to FISH, the intra-cellular levels of cAMP (driven by the increase in glucagon after parturition)74 would have been lower in those cows. The lower DMI due to feeding FISH was clearly a long-term response which preceded parturition. Thus, cows had likely adapted by the time of parturition in a way that the lower rate of milk production was driven by a level of DMI which was appropriate to meet the energy demands of the mammary gland.
Mediator 1 (MED1) is required for high-fat diet-induced hepatic steatosis via PPARγ, and loss of MED1 protects rodents against fatty liver.73 A biologically-similar role for PPARG in ruminant liver is unlikely because this isoform is substantially lower in abundance than PPARA or PPARD (data not shown). In fact, in this study the expression of PPARD and not PPARA was affected by the onset of lactation and by the type of lipid fed. Our data seem to suggest a mechanistic link between PPARD, CARM1, and MED1, particularly postpartum when the responses due to diet for the three genes were the same. This suggestion is supported by the observed responses in the expression of the co-repressors NCOR2 and NRIP1.
Nuclear receptor co-repressor 2 (NCOR2), in tandem with specific NR and different DNA binding transcription factors, represses the transcription of target genes.75 In non-ruminants, NRIP1 seems to play dual roles in metabolic tissues but the precise mechanisms driving its co-activator role remain elusive.76 What seems evident from previous rodent studies is that NRIP1 is required for expression of genes associated with energy metabolism (eg, UCP1, CPT1A) partly under control of β-adrenergic stimulation and PPARα and PPARδ.77
Transcription regulators of lipid metabolism
In non-ruminants, sterol regulatory element binding proteins SREBF1 and SREBF2 act as a central hub to control the transcription of genes required for cholesterol, fatty acid, TAG, and PL.78,79 In the liver, SREBF1 preferentially regulates genes involved in fatty acid synthesis while SREBF2 regulates genes associated with cholesterol biosynthesis.80,81 The activation of SREBF2 in the liver is mediated by SREBF cleavage binding protein (SCAP). CIDEB controls cholesterol biosynthesis by regulating the levels of SCAP at the transcriptional level.80 In the present study, the post-partal decrease in SREBF2 with FISH (Table 6) could have been a consequence of lower CIDEB expression (Table 7), which may have decreased abundance of SCAP and, thus, reduced the transport and activation of SREBF2. From a physiological perspective, such decrease in SREBF2 expression could have resulted in lower endogenous synthesis of cholesterol, which is an important process in liver as a part of the lipoprotein synthesis pathway.
Inflammation and apoptosis
Dietary LCFA can impact immunity through the production of cytokines and molecules involved in the regulation of immune responses.82 Omega-3 and omega-6 PUFAs are important immunomodulators.83 The transcription factors interferon regulatory factor 3 (IRF3) and NFkB are the central points of an integrated network of genes involved in the innate immune response, whereas inhibitors of kappa light polypeptide gene enhancer in B-cells, kinase epsilon (IKBKE) and TANK-binding kinase (TBK1), play a pivotal role in coordinating the activation of both those genes.84 The decrease in expression of IL6, TBK1, and IKBKE postpartum in cows fed FISH could have been associated with lower concentration of 20:4n-6 in PL. This fatty acid is the major substrate for prostaglandins and a key link between PUFA and inflammation.82,85
The observed postpartal increase in 20:5n-3 and 22:6n-3 in PL of cows fed FISH likely inhibited 20:4n-6 metabolism directly, via substrate competition, or indirectly, by altering the expression of inflammatory genes through effects on transcription factor activation.85 Feeding plant or fish oil rich in omega-3 PUFA generally reduces inflammatory reactions and production of interleukin (IL)-1, IL-6, MMP-3 (STR1), and tumor necrosis factor.86,87 Such type of effects might be less pronounced in dairy cattle because of the substantial degree of biohydroge-nation of PUFA in the rumen and also because of the substantial uptake of LCFA by the mammary gland during lactation. However, the data provides strong evidence that the enrichment of omega-3 PUFA in liver PL due to feeding FISH likely was important in lowering the expression of inflammatory genes. A possible mechanism for such an effect could have encompassed PPARδ, which was recently shown to be activated in dairy cattle liver during inflammation.88
The cell death-inducing DFFA-like effector b (CIDEB) protein has a high level of expression in non-ruminant liver, and its deficiency affects energy expenditure, plasma TAG levels,89 and also alters the expression of genes involved in various metabolic and signaling networks.80 The CIDEB protein is localized to the LD, endoplasmic reticulum, and Golgi apparatus and facilitates VLDL lipidation and maturation in the liver by interacting with ApoB.80 In the present study, the decrease in CIDEB expression with FISH may have led to reduced expression of SREBF2, de novo cholesterol biosynthesis, and potentially an augmentation of the cellular inflammatory status.
Conclusions
The present study revealed the type of dietary fatty acid which affects the hepatic fatty acid profile of PL and TAG. At the levels supplemented, the change in the profile of metabolic genes after parturition in cows fed saturated fat suggested a greater capacity for uptake of fatty acids and intracellular handling without excessive storage of LD. The lack of difference in liver TAG concentration between lipid-supplemented groups and the downregulation of metabolic genes after parturition in cows fed fish oil suggested that it might not be effective in enhancing oxidative capacity of LCFA. Such response contrasts the effect of very-long chain PUFA in monogastric species. The results highlighted that both saturated and very-long chain PUFA seemed equally effective at helping decrease inflammatory gene expression but FISH had a more potent effect after parturition. Based on the combined data from this study additional studies to better delineate the effective doses of saturated and very-long chain PUFA to feed around parturition seem warranted.
Acknowledgments
Eduardo Schmitt was supported by a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior from the Brazilian Ministry of Education.
Footnotes
Funding
Partial support for the gene expression work was provided by Section 1433 Animal Health and Disease funds under project ILLU-538-961 (National Institute of Food and Agriculture, Washington, DC).
Author Contributions
Conceived and designed the experiment: MAB, EJD. Conceived and/or performed the analyses: HA, ES, JJL. Wrote the manuscript: HA, ES, MNC, JJL. Agree with manuscript results and conclusions: HA, ES, MAB, MNC, EJD, JJL. Jointly developed the structure and arguments for the paper: HA, ES, JJL. Made critical revisions and approved final version: HA, ES, MAB, MNC, EJD, JJL. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Supplementary Tables
Table S2.
Concentration of fatty acids in hepatic triacylglycerol during the peripartal period.
| Fatty acid | Diet† | Day | SEM‡ | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| −21 | −10 | 1 | 11 | Diet | Time | D × T¶ | |||
| 14:0 | CON | 3.94 | 5.34 | 5.06 | 3.98 | 0.53 | 0.06 | 0.08 | 0.62 |
| SFAT | 5.24 | 5.30 | 5.17 | 5.15 | |||||
| FISH | 4.00 | 5.39 | 4.17 | 4.16 | |||||
| 14:1trans | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.07 | 0.11 | 0.08 | 0.04 |
| SFAT | 0.33a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | <0.01 | <0.01 | |||||
| 14:1cis | CON | 0.06a,α | 0.14α | 0.43β | 0.35β | 0.06 | 0.16 | 0.01 | 0.03 |
| SFAT | 0.05a,α | 0.08α | 0.34β | 0.34β | |||||
| FISH | 0.32b,α | <0.01β | 0.39α | 0.43α | |||||
| 15:0 | CON | 1.50 | 1.58 | 1.22 | 1.24 | 0.24 | 0.37 | 0.01 | 0.72 |
| SFAT | 2.00 | 1.50 | 1.11 | 1.19 | |||||
| FISH | 1.35 | 1.53 | 2.00 | 1.06 | |||||
| 15:1trans | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.64 | 0.11 | 0.08 | 0.04 |
| SFAT | 0.31a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | <0.01 | <0.01 | |||||
| 16:1trans | CON | 0.65a,b | 0.68 | 0.64a | 0.66 | 0.11 | 0.02 | 0.36 | 0.02 |
| SFAT | 0.90a | 0.58 | 0.74a | 0.60 | |||||
| FISH | 0.58b,α | 0.85α,β | 1.12b,β | 0.88β | |||||
| 16:1cis | CON | 1.70 | 2.01 | 4.03 | 3.30 | 0.42 | 0.05 | <0.01 | 0.11 |
| SFAT | 1.87 | 1.74 | 2.64 | 3.80 | |||||
| FISH | 2.44 | 3.03 | 3.90 | 3.41 | |||||
| 17:0 | CON | 1.87 | 1.77 | 0.84 | 1.04 | 0.25 | 0.83 | <0.01 | 0.98 |
| SFAT | 1.73 | 1.79 | 0.98 | 0.84 | |||||
| FISH | 1.59 | 1.70 | 0.91 | 0.91 | |||||
| 17:1trans | CON | <0.01 | <0.01 | 0.05 | 0.05 | 0.02 | 0.40 | <0.01 | 0.71 |
| SFAT | 0.04 | <0.01 | 0.07 | 0.09 | |||||
| FISH | 0.03 | <0.01 | 0.06 | 0.06 | |||||
| 17:1cis | CON | <0.01 | <0.01 | 0.12 | <0.01 | 0.08 | 0.58 | 0.32 | 0.55 |
| SFAT | 0.07 | <0.01 | <0.01 | 0.10 | |||||
| FISH | <0.01 | <0.01 | 0.13 | 0.20 | |||||
| 18:1trans5 | CON | <0.01 | <0.01 | <0.01 | <0.01 | 0.10 | 0.37 | 0.63 | 0.48 |
| SFAT | 0.28 | <0.01 | 0.07 | 0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | 0.09 | |||||
| 18:1trans7 | CON | <0.01 | <0.01 | <0.01 | 0.01 | 0.11 | 0.40 | 0.45 | 0.48 |
| SFAT | 0.32 | <0.01 | 0.01 | 0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 18:1trans68 | CON | 0.04 | <0.01 | <0.01 | 0.11 | 0.34 | 0.55 | <0.01 | 0.71 |
| SFAT | <0.01 | 0.04 | 0.09 | 0.11 | |||||
| FISH | <0.01 | <0.01 | 0.04 | 0.06 | |||||
| 18:1trans9 | CON | <0.01 | 0.07 | 0.12 | 0.02 | 0.05 | 0.67 | 0.03 | 0.52 |
| SFAT | <0.01 | 0.04 | 0.12 | 0.15 | |||||
| FISH | 0.03 | 0.04 | 0.09 | 0.09 | |||||
| 18:1trans10 | CON | <0.01 | <0.01 | 0.06 | 0.20 | 0.07 | 0.61 | 0.66 | 0.53 |
| SFAT | <0.01 | 0.07 | <0.01 | <0.01 | |||||
| FISH | 0.05 | 0.04 | 0.04 | 0.04 | |||||
| 18:1trans12 | CON | <0.01 | 0.04 | 0.27 | 0.21 | 0.12 | 0.36 | <0.01 | 0.89 |
| SFAT | 0.14 | <0.01 | 0.39 | 0.28 | |||||
| FISH | 0.20 | <0.01 | 0.34 | 0.40 | |||||
| 18:1t13-14 | CON | 1.21 | 1.30 | 0.49 | 0.35 | 0.38 | 0.51 | <0.01 | 0.71 |
| SFAT | 1.68 | 1.92 | 0.14 | 0.43 | |||||
| FISH | 1.16 | 1.32 | 0.56 | <0.01 | |||||
| 18:1trans15 | CON | <0.01 | <0.01 | <0.01 | 0.20 | 2.07 | 0.42 | 0.44 | 0.47 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | <0.01 | <0.01 | 6.27 | <0.01 | |||||
| 18:1cis11 | CON | 0.53 | 0.66 | 1.03 | 0.64 | 0.16 | 0.55 | <0.01 | 0.36 |
| SFAT | 0.63 | 0.53 | 1.02 | 1.13 | |||||
| FISH | 0.666 | 0.45 | 0.97 | 1.06 | |||||
| 18:1cis12 | CON | 0.19 | 0.26 | 0.49 | 0.40 | 0.09 | 0.02 | <0.01 | 0.70 |
| SFAT | 0.26 | 0.27 | 0.49 | 0.54 | |||||
| FISH | 0.05 | <0.01 | 0.40 | 0.44 | |||||
| 18:1cis13 | CON | <0.01 | 0.02 | 0.12 | 0.14 | 0.04 | 0.17 | <0.01 | 0.98 |
| SFAT | <0.01 | 0.03 | 0.15 | 0.14 | |||||
| FISH | 0.05 | 0.09 | 0.18 | 0.15 | |||||
| 18:1trans16 | CON | <0.01 | 0.03 | 0.08 | 0.09 | 0.03 | 0.70 | <0.01 | 0.61 |
| SFAT | <0.01 | <0.01 | 0.12 | 0.09 | |||||
| FISH | 0.04 | <0.01 | 0.12 | 0.09 | |||||
| 18:1alltrans | CON | 2.07 | 2.43 | 2.09 | 1.92 | 2.02 | 0.31 | 0.36 | 0.25 |
| SFAT | 3.26 | 2.70 | 2.15 | 1.94 | |||||
| FISH | 1.91 | 2.79 | 9.23 | 2.21 | |||||
| 18:1allcis | CON | 15.00 | 14.77 | 22.53 | 21.90 | 2.70 | 0.96 | <0.01 | 0.23 |
| SFAT | 15.25 | 13.65 | 22.83 | 21.38 | |||||
| FISH | 20.80 | 14.55 | 16.47 | 23.34 | |||||
| 18:3n-6 | CON | 0.08α | 0.13a,α,β | 0.21b,β,γ | 0.35a,γ | 0.03 | <0.01 | <0.01 | 0.02 |
| SFAT | 0.11α | 0.10a,α | 0.12a,b,α | 0.37a,β | |||||
| FISH | 0.08α,β | 0.01b,α | 0.11a,α,β | 0.13b,β | |||||
| 18:2alltrans | CON | 0.05 | 0.06 | 0.09 | 0.11 | 0.02 | <0.01 | <0.01 | 0.08 |
| SFAT | 0.06 | 0.07 | 0.07 | 0.18 | |||||
| FISH | 0.03 | 0.02 | 0.06 | 0.04 | |||||
| 20:3n-3 | CON | 0.93 | 1.15 | 0.57 | 0.51 | 0.18 | 0.08 | 0.01 | 0.91 |
| SFAT | 0.66 | 0.89 | 0.50 | 0.53 | |||||
| FISH | 0.66 | 0.65 | 0.41 | 0.34 | |||||
| 22:1 | CON | <0.01 | <0.01 | <0.01 | 0.01 | 0.01 | 0.08 | 0.33 | 0.42 |
| SFAT | <0.01 | 0.03 | <0.01 | 0.02 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 22:2n-6 | CON | 0.36 | 0.18 | <0.01 | 0.02 | 0.10 | 0.14 | 0.22 | 0.46 |
| SFAT | 0.01 | <0.01 | <0.01 | 0.01 | |||||
| FISH | 0.08 | 0.13 | 0.04 | 0.02 | |||||
| 22:3n-3 | CON | <0.01 | <0.01b | <0.01 | 0.26 | 0.12 | 0.06 | 0.10 | 0.04 |
| SFAT | <0.01α | 0.61a,β | 0.05α | 0.14α | |||||
| FISH | <0.01 | <0.01b | <0.01 | 0.02 | |||||
| 24:1 | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.01 | 0.39 | 0.33 | 0.04 |
| SFAT | 0.04a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | 0.01 | 0.02 | |||||
| 22:4n-6 | CON | 0.51 | 0.48 | 0.22 | 0.19 | 0.21 | 0.31 | 0.01 | 0.15 |
| SFAT | 0.19 | 1.12 | 0.20 | 0.17 | |||||
| FISH | 0.33 | 0.34 | 0.12 | 0.13 | |||||
| 22:5n-6 | CON | 0.14 | 0.12 | 0.06 | 0.04 | 0.11 | 0.10 | 0.17 | 0.07 |
| SFAT | 0.08 | 0.49 | 0.06 | 0.04 | |||||
| FISH | 0.08 | 0.24 | 0.41 | 0.28 | |||||
| Unknown | CON | 9.53 | 6.28 | 6.44 | 5.63 | 1.70 | 0.08 | 0.86 | 0.30 |
| SFAT | 3.72 | 4.26 | 5.54 | 7.09 | |||||
| FISH | 8.44 | 8.61 | 690 | 6.38 | |||||
Notes:
Difference (P < 0.05) between different diet at same day.
Denote significant interactions (P < 0.05) within a same diet at different days.
CON = control diet containing no supplemental lipid: SFAT = CON supplemented with Energy Booster: FISH = CON supplemented with fish oil;
standard error of the mean;
diet by time interaction.
References
- 1.Jump DB. N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr Opin Lipidol. 2008 Jun;19(3):242–7. doi: 10.1097/MOL.0b013e3282ffaf6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas GN, Rehage J, Beaulieu AD, Bahaa AO, Drackley JK. Prepartum nutrition alters fatty acid composition in plasma, adipose tissue, and liver lipids of periparturient dairy cows. J Dairy Sci. 2007 Jun;90(6):2941–59. doi: 10.3168/jds.2006-225. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JB, Ridder C, Larsen T. Priming the cow for mobilization in the periparturient period: effects of supplementing the dry cow with saturated fat or linseed. J Dairy Sci. 2008 Mar;91(3):1029–43. doi: 10.3168/jds.2007-0437. [DOI] [PubMed] [Google Scholar]
- 4.Carriquiry M, Weber WJ, Fahrenkrug SC, Crooker BA. Hepatic gene expression in multiparous Holstein cows treated with bovine somatotropin and fed n-3 fatty acids in early lactation. J Dairy Sci. 2009 Oct;92(10):4889–900. doi: 10.3168/jds.2008-1676. [DOI] [PubMed] [Google Scholar]
- 5.Loor JJ. Genomics of metabolic adaptations in the peripartal cow. Animal. 2010 Jul;4(7):1110–39. doi: 10.1017/S1751731110000960. [DOI] [PubMed] [Google Scholar]
- 6.Grum DE, Drackley JK, Younker RS, LaCount DW, Veenhuizen JJ. Nutrition during the dry period and hepatic lipid metabolism of periparturient dairy cows. J Dairy Sci. 1996 Oct;79(10):1850–64. doi: 10.3168/jds.S0022-0302(96)76553-0. [DOI] [PubMed] [Google Scholar]
- 7.Andersen JB, Ridder C, Larsen T. Priming the cow for mobilization in the periparturient period: effects of supplementing the dry cow with saturated fat or linseed. J Dairy Sci. 2008 Mar;91(3):1029–43. doi: 10.3168/jds.2007-0437. [DOI] [PubMed] [Google Scholar]
- 8.Pegorier JP, Le May C, Girard J. Control of gene expression by fatty acids. J Nutr. 2004 Sep;134(9):2444S–9S. doi: 10.1093/jn/134.9.2444S. [DOI] [PubMed] [Google Scholar]
- 9.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–92. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 10.Bionaz M, Thering BJ, Loor JJ. Fine metabolic regulation in ruminants via nutrient-gene interactions: saturated long-chain fatty acids increase expression of genes involved in lipid metabolism and immune response partly through PPAR-alpha activation. Br J Nutr. 2012 Jan;107(2):179–91. doi: 10.1017/S0007114511002777. [DOI] [PubMed] [Google Scholar]
- 11.Bionaz M, Thering BJ, Loor JJ. Fine metabolic regulation in ruminants via nutrient-gene interactions: saturated long-chain fatty acids increase expression of genes involved in lipid metabolism and immune response partly through PPAR-alpha activation. Br J Nutr. 2011 Jul;:1–13. doi: 10.1017/S0007114511002777. [DOI] [PubMed] [Google Scholar]
- 12.Loor JJ, Dann HM, Everts RE, et al. Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol Genomics. 2005 Oct 17;23(2):217–26. doi: 10.1152/physiolgenomics.00132.2005. [DOI] [PubMed] [Google Scholar]
- 13.Loor JJ, Dann HM, Guretzky NAJ, et al. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiol Genomics. 2006 Oct 3;27(1):29–41. doi: 10.1152/physiolgenomics.00036.2006. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt E, Ballou MA, Correa MN, DePeters EJ, Drackley JK, Loor JJ. Dietary lipid during the transition period to manipulate subcutaneous adipose tissue peroxisome proliferator-activated receptor-gamma co-regulator and target gene expression. J Dairy Sci. 2011 Dec;94(12):5913–25. doi: 10.3168/jds.2011-4230. [DOI] [PubMed] [Google Scholar]
- 15.Ballou MA, Gomes RC, Juchem SO, DePeters EJ. Effects of dietary supplemental fish oil during the peripartum period on blood metabolites and hepatic fatty acid compositions and total triacylglycerol concentrations of multiparous Holstein cows. J Dairy Sci. 2009 Feb;92(2):657–69. doi: 10.3168/jds.2008-1196. [DOI] [PubMed] [Google Scholar]
- 16.Graugnard DE, Piantoni P, Bionaz M, Berger LL, Faulkner DB, Loor JJ. Adipogenic and energy metabolism gene networks in longissimus lumborum during rapid post-weaning growth in Angus and Angus x Simmental cattle fed high-starch or low-starch diets. BMC Genomics. 2009;10:142. doi: 10.1186/1471-2164-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rukkwamsuk T, Geelen MJ, Kruip TA, Wensing T. Interrelation of fatty acid composition in adipose tissue, serum, and liver of dairy cows during the development of fatty liver postpartum. J Dairy Sci. 2000 Jan;83(1):52–9. doi: 10.3168/jds.S0022-0302(00)74854-5. [DOI] [PubMed] [Google Scholar]
- 18.Shimano H, Matsuzaka T, Yahagi N, et al. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPAR alpha. J Lipid Res. 2002 Jan;43(1):107–14. [PubMed] [Google Scholar]
- 19.Jenkins KJ. Factors affecting poor performance and scours in preruminant calves fed corn-oil. J Dairy Sci. 1988 Nov;71(11):3013–20. doi: 10.3168/jds.S0022-0302(88)79899-9. [DOI] [PubMed] [Google Scholar]
- 20.Laho T, Varadyova Z, Mihalikova K, et al. Effects of prefermented cereal-derived substrates (ground barley and rye bran) enriched with fungal gamma-linolenic acid on rumen fermentation parameters and lipid metabolism in vitro. J Appl Microbiol. 2011 Sep;111(3):537–46. doi: 10.1111/j.1365-2672.2011.05073.x. [DOI] [PubMed] [Google Scholar]
- 21.Mashek DG, Bertics SJ, Grummer RR. Metabolic fate of long-chain unsaturated fatty acids and their effects on palmitic acid metabolism and gluconeogenesis in bovine hepatocytes. J Dairy Sci. 2002 Sep;85(9):2283–9. doi: 10.3168/jds.S0022-0302(02)74308-7. [DOI] [PubMed] [Google Scholar]
- 22.Mashek DG, Grummer RR. Effects of long chain fatty acids on lipid and glucose metabolism in monolayer cultures of bovine hepatocytes. J Dairy Sci. 2003 Jul;86(7):2390–6. doi: 10.3168/jds.S0022-0302(03)73833-8. [DOI] [PubMed] [Google Scholar]
- 23.Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev. 2011 Oct 12;111(10):6359–86. doi: 10.1021/cr100404w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drackley JK. Biology of dairy cows during the transition period: The final frontier? J Dairy Sci. 1999 Nov;82(11):2259–73. doi: 10.3168/jds.s0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 25.Berger A, Roberts MA, Hoff B. How dietary arachidonic- and docosahexaenoic-acid rich oils differentially affect the murine hepatic transcriptome. Lipids Health Dis. 2006 Apr;20:5. doi: 10.1186/1476-511X-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawar A, Jump DB. Unsaturated fatty acid regulation of peroxisome proliferator-activated receptor alpha activity in rat primary hepatocytes. J Biol Chem. 2003 Sep 19;278(38):35931–9. doi: 10.1074/jbc.M306238200. [DOI] [PubMed] [Google Scholar]
- 27.Shearer GC, Savinova OV, Harris WS. Fish oil—How does it reduce plasma triglycerides? Biochim Biophys Acta. 2012 May;1821(5):843–51. doi: 10.1016/j.bbalip.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ide T. Interaction of fish oil and conjugated linoleic acid in affecting hepatic activity of lipogenic enzymes and gene expression in liver and adipose tissue. Diabetes. 2005 Feb;54(2):412–23. doi: 10.2337/diabetes.54.2.412. [DOI] [PubMed] [Google Scholar]
- 29.Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009 Jul;297(1):E28–37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofacer R, Magrisso IJ, Jandacek R, et al. Omega-3 fatty acid deficiency increases stearoyl-CoA desaturase expression and activity indices in rat liver: Positive association with non-fasting plasma triglyceride levels Prostag Leukotr Ess January–Feb2012861–271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velliquette RA, Gillies PJ, Kris-Etherton PM, Green JW, Zhao G, Vanden Heuvel JP. Regulation of human stearoyl-CoA desaturase by omega-3 and omega-6 fatty acids: implications for the dietary management of elevated serum triglycerides. J Clin Lipidol. 2009 Aug;3(4):281–8. doi: 10.1016/j.jacl.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Ntambi JM. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999 Sep;40(9):1549–58. [PubMed] [Google Scholar]
- 33.Sessler AM, Kaur N, Palta JP, Ntambi JM. Regulation of stearoyl-CoA desaturase 1 mRNA stability by polyunsaturated fatty acids in 3T3-L1 adipocytes. J Biol Chem. 1996 Nov 22;271(47):29854–8. doi: 10.1074/jbc.271.47.29854. [DOI] [PubMed] [Google Scholar]
- 34.Landschulz KT, Jump DB, MacDougald OA, Lane MD. Transcriptional control of the stearoyl-CoA desaturase-1 gene by polyunsaturated fatty acids. Biochem Biophys Res Commun. 1994 Apr 29;200(2):763–8. doi: 10.1006/bbrc.1994.1516. [DOI] [PubMed] [Google Scholar]
- 35.Man WC, Miyazaki M, Chu K, Ntambi J. Colocalization of SCD1 and DGAT2: implying preference for endogenous monounsaturated fatty acids in triglyceride synthesis. J Lipid Res. 2006 Sep;47(9):1928–39. doi: 10.1194/jlr.M600172-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Monetti M, Levin MC, Watt MJ, et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007 Jul;6(1):69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki T, Sasaki E, Kakinuma C, Yano T, Miura S, Ezaki O. Increased very low density lipoprotein secretion and gonadal fat mass in mice overexpressing liver DGAT1. J Biol Chem. 2005 Jun 3;280(22):21506–14. doi: 10.1074/jbc.M412989200. [DOI] [PubMed] [Google Scholar]
- 38.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008 Nov;49(11):2283–301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang BHJ, Chan L. Regulation of triglyceride metabolism. III. Emerging role of lipid droplet protein ADFP in health and disease. Am J Physiol-Gastr L. 2007 Jun;292(6):G1465–8. doi: 10.1152/ajpgi.00566.2006. [DOI] [PubMed] [Google Scholar]
- 40.Dalen KT, Ulven SM, Arntsen BM, Solaas K, Nebb HI. PPARalpha activators and fasting induce the expression of adipose differentiation-related protein in liver. J Lipid Res. 2006 May;47(5):931–43. doi: 10.1194/jlr.M500459-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Tobin KA, Harsem NK, Dalen KT, Staff AC, Nebb HI, Duttaroy AK. Regulation of ADRP expression by long-chain polyunsaturated fatty acids in BeWo cells, a human placental choriocarcinoma cell line. J Lipid Res. 2006 Apr;47(4):815–23. doi: 10.1194/jlr.M500527-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007 Dec;48(12):2547–59. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Ducharme NA, Bickel PE. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008 Mar;149(3):942–9. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- 44.Li LO, Ellis JM, Paich HA, et al. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J Biol Chem. 2009 Oct 9;284(41):27816–26. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frederiksen KS, Wulff EM, Sauerberg P, Mogensen JP, Jeppesen L, Fleckner J. Prediction of PPAR-alpha ligand-mediated physiological changes using gene expression profiles. J Lipid Res. 2004 Mar;45(3):592–601. doi: 10.1194/jlr.M300239-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Schoonjans K, Staels B, Grimaldi P, Auwerx J. Acyl-CoA synthetase mRNA expression is controlled by fibric-acid derivatives, feeding and liver proliferation. Eur J Biochem. 1993 Sep 1;216(2):615–22. doi: 10.1111/j.1432-1033.1993.tb18181.x. [DOI] [PubMed] [Google Scholar]
- 47.Thering BJ, Bionaz M, Loor JJ. Long-chain fatty acid effects on peroxisome proliferator-activated receptor-alpha-regulated genes in Madin-Darby bovine kidney cells: Optimization of culture conditions using palmitate. J Dairy Sci. 2009 May;92(5):2027–37. doi: 10.3168/jds.2008-1749. [DOI] [PubMed] [Google Scholar]
- 48.Bionaz M, Drackley JK, Dann HM, Loor JJ. Liver fatty acid binding protein (FABP) and acyl-CoA synthase (ACSL) isoform gene expression due to plane of dietary energy prepartum in dairy cows. J Dairy Sci. 2007;90:678–8. [Google Scholar]
- 49.Fan CY, Pan J, Chu R, et al. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J Biol Chem. 1996 Oct 4;271(40):24698–710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 50.Ide T, Kobayashi H, Ashakumary L, et al. Comparative effects of perilla and fish oils on the activity and gene expression of fatty acid oxidation enzymes in rat liver. Bba-Mol Cell Biol L. 2000 May 6;1485(1):23–35. doi: 10.1016/s1388-1981(00)00026-3. [DOI] [PubMed] [Google Scholar]
- 51.Leclercq S, Skrzypski J, Courvoisier A, et al. Effect of dietary polyun-saturated fatty acids on the expression of peroxisomal ABC transporters. Biochimie. 2008 Oct;90(10):1602–7. doi: 10.1016/j.biochi.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Vluggens A, Andreoletti P, Viswakarma N, et al. Reversal of mouse Acyl-CoA oxidase 1 (ACOX1) null phenotype by human ACOX1b isoform [corrected] Lab Invest. 2010 May;90(5):696–708. doi: 10.1038/labinvest.2010.46. [DOI] [PubMed] [Google Scholar]
- 53.Grum DE, Drackley JK, Younker RS, LaCount DW, Veenhuizen JJ. Nutrition during the dry period and hepatic lipid metabolism of periparturient dairy cows. J Dairy Sci. 1996 Oct;79(10):1850–64. doi: 10.3168/jds.S0022-0302(96)76553-0. [DOI] [PubMed] [Google Scholar]
- 54.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006 Jul;116(7):1784–92. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masterton GS, Plevris JN, Hayes PC. Review article: omega-3 fatty acids— a promising novel therapy for non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010 Apr;31(7):679–92. doi: 10.1111/j.1365-2036.2010.04230.x. [DOI] [PubMed] [Google Scholar]
- 56.Loor JJ, Everts RE, Bionaz M, et al. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics. 2007 Dec 19;32(1):105–16. doi: 10.1152/physiolgenomics.00188.2007. [DOI] [PubMed] [Google Scholar]
- 57.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007 Jun;5(6):426–37. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Overton TR, Drackley JK, Ottemann-Abbamonte CJ, Beaulieu AD, Emmert LS, Clark JH. Substrate utilization for hepatic gluconeogenesis is altered by increased glucose demand in ruminants. J Anim Sci. 1999 Jul;77(7):1940–51. doi: 10.2527/1999.7771940x. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds CK, Aikman PC, Lupoli B, Humphries DJ, Beever DE. Splanchnic metabolism of dairy cows during the transition from late gestation through early lactation. J Dairy Sci. 2003 Apr;86(4):1201–17. doi: 10.3168/jds.S0022-0302(03)73704-7. [DOI] [PubMed] [Google Scholar]
- 60.Greenfield RB, Cecava MJ, Donkin SS. Changes in mRNA expression for gluconeogenic enzymes in liver of dairy cattle during the transition to lactation. J Dairy Sci. 2000 Jun;83(6):1228–36. doi: 10.3168/jds.S0022-0302(00)74989-7. [DOI] [PubMed] [Google Scholar]
- 61.Velez JC, Donkin SS. Feed restriction induces pyruvate carboxylase but not phosphoenolpyruvate carboxykinase in dairy cows. J Dairy Sci. 2005 Aug;88(8):2938–48. doi: 10.3168/jds.S0022-0302(05)72974-X. [DOI] [PubMed] [Google Scholar]
- 62.Bradford BJ, Allen MS. Phlorizin administration increases hepatic gluconeogenic enzyme mRNA abundance but not feed intake in late-lactation dairy cows. J Nutr. 2005 Sep;135(9):2206–11. doi: 10.1093/jn/135.9.2206. [DOI] [PubMed] [Google Scholar]
- 63.Jump DB, Botolin D, Wang Y, Xu J, Christian B, Demeure O. Fatty acid regulation of hepatic gene transcription. J Nutr. 2005 Nov;135(11):2503–6. doi: 10.1093/jn/135.11.2503. [DOI] [PubMed] [Google Scholar]
- 64.White HM, Koser SL, Donkin SS. Differential regulation of bovine pyruvate carboxylase promoters by fatty acids and peroxisome proliferator-activated receptor-alpha agonist. J Dairy Sci. 2011 Jul;94(7):3428–36. doi: 10.3168/jds.2010-3960. [DOI] [PubMed] [Google Scholar]
- 65.Suh HN, Huong HT, Song CH, Lee JH, Han HJ. Linoleic acid stimulates gluconeogenesis via Ca(2+)/PLC, cPLA(2), and PPAR pathways through GPR40 in primary cultured chicken hepatocytes. Am J Physiol Cell Ph. 2008 Dec;295(6):C1518–27. doi: 10.1152/ajpcell.00368.2008. [DOI] [PubMed] [Google Scholar]
- 66.Armstrong MB, Towle HC. Polyunsaturated fatty acids stimulate hepatic UCP-2 expression via a PPARalpha-mediated pathway. Am J Physiol Endocrinol Metab. 2001 Dec;281(6):E1197–204. doi: 10.1152/ajpendo.2001.281.6.E1197. [DOI] [PubMed] [Google Scholar]
- 67.Ricquier D, Bouillaud F. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem J. 2000 Jan 15;345(Pt 2):161–79. [PMC free article] [PubMed] [Google Scholar]
- 68.Arsenijevic D, Onuma H, Pecqueur C, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000 Dec;26(4):435–9. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 69.Yonezawa T, Sanosaka M, Haga S, Kobayashi Y, Katoh K, Obara Y. Regulation of uncoupling protein 2 expression by long-chain fatty acids and hormones in bovine mammary epithelial cells. Biochem Biophys Res Commun. 2008 Oct 17;375(2):280–5. doi: 10.1016/j.bbrc.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 70.Bossaert P, Leroy JLMR, De Vliegher S, Opsomer G. Interrelations between glucose-induced insulin response, metabolic indicators, and time of first ovulation in high-yielding dairy cows. J Dairy Sci. 2008 Sep;91(9):3363–71. doi: 10.3168/jds.2008-0994. [DOI] [PubMed] [Google Scholar]
- 71.Krones-Herzig A, Mesaros A, Metzger D, et al. Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. J Biol Chem. 2006 Feb 10;281(6):3025–9. doi: 10.1074/jbc.M509770200. [DOI] [PubMed] [Google Scholar]
- 72.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003 Apr;284(4):E671–8. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- 73.Bai L, Jia Y, Viswakarma N, et al. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011 Apr;53(4):1164–74. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herbein JH, Aiello RJ, Eckler LI, Pearson RE, Akers RM. Glucagon, insulin, growth hormone, and glucose concentrations in blood plasma of lactating dairy cows. J Dairy Sci. 1985 Feb;68(2):320–5. doi: 10.3168/jds.S0022-0302(85)80828-6. [DOI] [PubMed] [Google Scholar]
- 75.Jepsen K, Hermanson O, Onami TM, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000 Sep 15;102(6):753–63. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 76.Fritah A, Christian M, Parker MG. The metabolic coregulator RIP140: an update. Am J Physiol Endocrinol Metab. 2010 Sep;299(3):E335–40. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- 77.Leonardsson G, Steel JH, Christian M, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8437–42. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006 Mar 15;119(Pt 6):973–6. doi: 10.1242/jcs.02866. [DOI] [PubMed] [Google Scholar]
- 79.Horton JD, Shah NA, Warrington JA, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li JZ, Lei Y, Wang Y, et al. Control of cholesterol biosynthesis, uptake and storage in hepatocytes by Cideb. Biochim Biophys Acta. 2010 May;1801(5):577–86. doi: 10.1016/j.bbalip.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 81.Bommer GT, MacDougald OA. Regulation of lipid homeostasis by the bifunctional SREBF2-miR33a locus. Cell Metab. 2011 Mar 2;13(3):241–7. doi: 10.1016/j.cmet.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lessard M, Gagnon N, Petit HV. Immune response of postpartum dairy cows fed flaxseed. J Dairy Sci. 2003 Aug;86(8):2647–57. doi: 10.3168/jds.S0022-0302(03)73860-0. [DOI] [PubMed] [Google Scholar]
- 83.Miles EA, Calder PC. Modulation of immune function by dietary fatty acids. P Nutr Soc. 1998 May;57(2):277–92. doi: 10.1079/pns19980042. [DOI] [PubMed] [Google Scholar]
- 84.Fitzgerald KA, McWhirter SM, Faia KL, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003 May;4(5):491–6. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 85.Calder PC. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am J Clin Nutr. 2006 Jun;83(Suppl 6):1505S–19S. doi: 10.1093/ajcn/83.6.1505S. [DOI] [PubMed] [Google Scholar]
- 86.Curtis CL, Rees SG, Little CB, et al. Pathologic indicators of degradation and inflammation in human osteoarthritic cartilage are abrogated by exposure to n-3 fatty acids (Retraction of vol 46, pg 1544, 2002) Arthritis Rheum-Us. 2006 Sep;54(9):2933–2933. doi: 10.1002/art.10305. [DOI] [PubMed] [Google Scholar]
- 87.Curtis JL, Huffnagle GB, Chen GH, et al. Experimental murine pulmonary cryptococcosis. Differences in pulmonary inflammation and lymphocyte recruitment induced by 2 encapsulated strains of Cryptococcus-neoformans. Lab Invest. 1994 Jul;71(1):113–26. [PubMed] [Google Scholar]
- 88.Graugnard DE, Moyes KM, Trevisi E, et al. Liver lipid content and inflammometabolic indices in peripartal dairy cows are altered in response to pre-partal energy intake and postpartal intramammary inflammatory challenge. J Dairy Sci. 2013 Feb;96(2):918–35. doi: 10.3168/jds.2012-5676. [DOI] [PubMed] [Google Scholar]
- 89.Li JZ, Ye J, Xue B, et al. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 2007 Oct;56(10):2523–32. doi: 10.2337/db07-0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2.
Concentration of fatty acids in hepatic triacylglycerol during the peripartal period.
| Fatty acid | Diet† | Day | SEM‡ | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| −21 | −10 | 1 | 11 | Diet | Time | D × T¶ | |||
| 14:0 | CON | 3.94 | 5.34 | 5.06 | 3.98 | 0.53 | 0.06 | 0.08 | 0.62 |
| SFAT | 5.24 | 5.30 | 5.17 | 5.15 | |||||
| FISH | 4.00 | 5.39 | 4.17 | 4.16 | |||||
| 14:1trans | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.07 | 0.11 | 0.08 | 0.04 |
| SFAT | 0.33a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | <0.01 | <0.01 | |||||
| 14:1cis | CON | 0.06a,α | 0.14α | 0.43β | 0.35β | 0.06 | 0.16 | 0.01 | 0.03 |
| SFAT | 0.05a,α | 0.08α | 0.34β | 0.34β | |||||
| FISH | 0.32b,α | <0.01β | 0.39α | 0.43α | |||||
| 15:0 | CON | 1.50 | 1.58 | 1.22 | 1.24 | 0.24 | 0.37 | 0.01 | 0.72 |
| SFAT | 2.00 | 1.50 | 1.11 | 1.19 | |||||
| FISH | 1.35 | 1.53 | 2.00 | 1.06 | |||||
| 15:1trans | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.64 | 0.11 | 0.08 | 0.04 |
| SFAT | 0.31a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | <0.01 | <0.01 | |||||
| 16:1trans | CON | 0.65a,b | 0.68 | 0.64a | 0.66 | 0.11 | 0.02 | 0.36 | 0.02 |
| SFAT | 0.90a | 0.58 | 0.74a | 0.60 | |||||
| FISH | 0.58b,α | 0.85α,β | 1.12b,β | 0.88β | |||||
| 16:1cis | CON | 1.70 | 2.01 | 4.03 | 3.30 | 0.42 | 0.05 | <0.01 | 0.11 |
| SFAT | 1.87 | 1.74 | 2.64 | 3.80 | |||||
| FISH | 2.44 | 3.03 | 3.90 | 3.41 | |||||
| 17:0 | CON | 1.87 | 1.77 | 0.84 | 1.04 | 0.25 | 0.83 | <0.01 | 0.98 |
| SFAT | 1.73 | 1.79 | 0.98 | 0.84 | |||||
| FISH | 1.59 | 1.70 | 0.91 | 0.91 | |||||
| 17:1trans | CON | <0.01 | <0.01 | 0.05 | 0.05 | 0.02 | 0.40 | <0.01 | 0.71 |
| SFAT | 0.04 | <0.01 | 0.07 | 0.09 | |||||
| FISH | 0.03 | <0.01 | 0.06 | 0.06 | |||||
| 17:1cis | CON | <0.01 | <0.01 | 0.12 | <0.01 | 0.08 | 0.58 | 0.32 | 0.55 |
| SFAT | 0.07 | <0.01 | <0.01 | 0.10 | |||||
| FISH | <0.01 | <0.01 | 0.13 | 0.20 | |||||
| 18:1trans5 | CON | <0.01 | <0.01 | <0.01 | <0.01 | 0.10 | 0.37 | 0.63 | 0.48 |
| SFAT | 0.28 | <0.01 | 0.07 | 0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | 0.09 | |||||
| 18:1trans7 | CON | <0.01 | <0.01 | <0.01 | 0.01 | 0.11 | 0.40 | 0.45 | 0.48 |
| SFAT | 0.32 | <0.01 | 0.01 | 0.01 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 18:1trans68 | CON | 0.04 | <0.01 | <0.01 | 0.11 | 0.34 | 0.55 | <0.01 | 0.71 |
| SFAT | <0.01 | 0.04 | 0.09 | 0.11 | |||||
| FISH | <0.01 | <0.01 | 0.04 | 0.06 | |||||
| 18:1trans9 | CON | <0.01 | 0.07 | 0.12 | 0.02 | 0.05 | 0.67 | 0.03 | 0.52 |
| SFAT | <0.01 | 0.04 | 0.12 | 0.15 | |||||
| FISH | 0.03 | 0.04 | 0.09 | 0.09 | |||||
| 18:1trans10 | CON | <0.01 | <0.01 | 0.06 | 0.20 | 0.07 | 0.61 | 0.66 | 0.53 |
| SFAT | <0.01 | 0.07 | <0.01 | <0.01 | |||||
| FISH | 0.05 | 0.04 | 0.04 | 0.04 | |||||
| 18:1trans12 | CON | <0.01 | 0.04 | 0.27 | 0.21 | 0.12 | 0.36 | <0.01 | 0.89 |
| SFAT | 0.14 | <0.01 | 0.39 | 0.28 | |||||
| FISH | 0.20 | <0.01 | 0.34 | 0.40 | |||||
| 18:1t13-14 | CON | 1.21 | 1.30 | 0.49 | 0.35 | 0.38 | 0.51 | <0.01 | 0.71 |
| SFAT | 1.68 | 1.92 | 0.14 | 0.43 | |||||
| FISH | 1.16 | 1.32 | 0.56 | <0.01 | |||||
| 18:1trans15 | CON | <0.01 | <0.01 | <0.01 | 0.20 | 2.07 | 0.42 | 0.44 | 0.47 |
| SFAT | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| FISH | <0.01 | <0.01 | 6.27 | <0.01 | |||||
| 18:1cis11 | CON | 0.53 | 0.66 | 1.03 | 0.64 | 0.16 | 0.55 | <0.01 | 0.36 |
| SFAT | 0.63 | 0.53 | 1.02 | 1.13 | |||||
| FISH | 0.666 | 0.45 | 0.97 | 1.06 | |||||
| 18:1cis12 | CON | 0.19 | 0.26 | 0.49 | 0.40 | 0.09 | 0.02 | <0.01 | 0.70 |
| SFAT | 0.26 | 0.27 | 0.49 | 0.54 | |||||
| FISH | 0.05 | <0.01 | 0.40 | 0.44 | |||||
| 18:1cis13 | CON | <0.01 | 0.02 | 0.12 | 0.14 | 0.04 | 0.17 | <0.01 | 0.98 |
| SFAT | <0.01 | 0.03 | 0.15 | 0.14 | |||||
| FISH | 0.05 | 0.09 | 0.18 | 0.15 | |||||
| 18:1trans16 | CON | <0.01 | 0.03 | 0.08 | 0.09 | 0.03 | 0.70 | <0.01 | 0.61 |
| SFAT | <0.01 | <0.01 | 0.12 | 0.09 | |||||
| FISH | 0.04 | <0.01 | 0.12 | 0.09 | |||||
| 18:1alltrans | CON | 2.07 | 2.43 | 2.09 | 1.92 | 2.02 | 0.31 | 0.36 | 0.25 |
| SFAT | 3.26 | 2.70 | 2.15 | 1.94 | |||||
| FISH | 1.91 | 2.79 | 9.23 | 2.21 | |||||
| 18:1allcis | CON | 15.00 | 14.77 | 22.53 | 21.90 | 2.70 | 0.96 | <0.01 | 0.23 |
| SFAT | 15.25 | 13.65 | 22.83 | 21.38 | |||||
| FISH | 20.80 | 14.55 | 16.47 | 23.34 | |||||
| 18:3n-6 | CON | 0.08α | 0.13a,α,β | 0.21b,β,γ | 0.35a,γ | 0.03 | <0.01 | <0.01 | 0.02 |
| SFAT | 0.11α | 0.10a,α | 0.12a,b,α | 0.37a,β | |||||
| FISH | 0.08α,β | 0.01b,α | 0.11a,α,β | 0.13b,β | |||||
| 18:2alltrans | CON | 0.05 | 0.06 | 0.09 | 0.11 | 0.02 | <0.01 | <0.01 | 0.08 |
| SFAT | 0.06 | 0.07 | 0.07 | 0.18 | |||||
| FISH | 0.03 | 0.02 | 0.06 | 0.04 | |||||
| 20:3n-3 | CON | 0.93 | 1.15 | 0.57 | 0.51 | 0.18 | 0.08 | 0.01 | 0.91 |
| SFAT | 0.66 | 0.89 | 0.50 | 0.53 | |||||
| FISH | 0.66 | 0.65 | 0.41 | 0.34 | |||||
| 22:1 | CON | <0.01 | <0.01 | <0.01 | 0.01 | 0.01 | 0.08 | 0.33 | 0.42 |
| SFAT | <0.01 | 0.03 | <0.01 | 0.02 | |||||
| FISH | <0.01 | <0.01 | <0.01 | <0.01 | |||||
| 22:2n-6 | CON | 0.36 | 0.18 | <0.01 | 0.02 | 0.10 | 0.14 | 0.22 | 0.46 |
| SFAT | 0.01 | <0.01 | <0.01 | 0.01 | |||||
| FISH | 0.08 | 0.13 | 0.04 | 0.02 | |||||
| 22:3n-3 | CON | <0.01 | <0.01b | <0.01 | 0.26 | 0.12 | 0.06 | 0.10 | 0.04 |
| SFAT | <0.01α | 0.61a,β | 0.05α | 0.14α | |||||
| FISH | <0.01 | <0.01b | <0.01 | 0.02 | |||||
| 24:1 | CON | <0.01b | <0.01 | <0.01 | <0.01 | 0.01 | 0.39 | 0.33 | 0.04 |
| SFAT | 0.04a,α | <0.01β | <0.01β | <0.01β | |||||
| FISH | <0.01b | <0.01 | 0.01 | 0.02 | |||||
| 22:4n-6 | CON | 0.51 | 0.48 | 0.22 | 0.19 | 0.21 | 0.31 | 0.01 | 0.15 |
| SFAT | 0.19 | 1.12 | 0.20 | 0.17 | |||||
| FISH | 0.33 | 0.34 | 0.12 | 0.13 | |||||
| 22:5n-6 | CON | 0.14 | 0.12 | 0.06 | 0.04 | 0.11 | 0.10 | 0.17 | 0.07 |
| SFAT | 0.08 | 0.49 | 0.06 | 0.04 | |||||
| FISH | 0.08 | 0.24 | 0.41 | 0.28 | |||||
| Unknown | CON | 9.53 | 6.28 | 6.44 | 5.63 | 1.70 | 0.08 | 0.86 | 0.30 |
| SFAT | 3.72 | 4.26 | 5.54 | 7.09 | |||||
| FISH | 8.44 | 8.61 | 690 | 6.38 | |||||
Notes:
Difference (P < 0.05) between different diet at same day.
Denote significant interactions (P < 0.05) within a same diet at different days.
CON = control diet containing no supplemental lipid: SFAT = CON supplemented with Energy Booster: FISH = CON supplemented with fish oil;
standard error of the mean;
diet by time interaction.