Abstract
Results of studies for the association of BRCA1 genotypes and haplotypes with sporadic breast cancer have been inconsistent. Therefore, a candidate SNP approach was used in a breast cancer case-control study to explore genotypes and haplotypes that have the potential to affect protein functions or levels. In a breast cancer case-control study, genotyping of BRCA1 polymorphisms Q356R, D693N, and E1038G was performed on 1005 cases and 1765 controls. Unconditional, polytomous logistic regression and chi square tests were used to examine the associations of breast cancer with genotypes and haplotypes. Additionally, interactions between genotype and smoking, benign breast disease, family history of breast cancer, BMI, alcohol consumption, and hormonal risk factors, hormone receptor status, and breast cancer pathology were calculated also using logistic regression and chi square. While, sporadic breast cancer was not associated with BRCA1 genotypes or haplotypes overall or by menopausal status, there was evidence of an interaction between the E1038G BRCA1 genotype, smoking, and BMI among premenopausal women (p for interaction=0.01 and 0.045, respectively) and between E1038G and D693N BRCA1 genotypes and hormone therapy use among postmenopausal women (p for interaction=0.01 and 0.02, respectively). There were no other associations found between BRCA1 genotypes and stage, histologic grade, or nuclear grade. However, the D693N SNP was associated with the risk of triple negative breast cancer (OR=2.31 95% CI: 1.08–4.93). The BRCA1 variants studied may play a role in the etiology of triple negative breast cancer and may interact with environmental factors such as hormone therapy or smoking and increase sporadic breast cancer risk.
Keywords: BRCA1, breast cancer, haplotypes, polymorphisms, gene × environment
INTRODUCTION
BRCA1 is a breast cancer tumor suppressor gene that plays a role in maintaining genomic stability. Specifically, BRCA1 plays a role in transcription, repair of double-stranded DNA breaks, and recombination (Bertwistle and Ashworth, 1998). Mutations in BRCA1 are responsible for approximately 40% of inherited breast cancers and more than 80% of families with both inherited breast and ovarian cancers (Easton et al., 1995;Thompson and Easton, 2002a;Thompson and Easton, 2002b;Liede et al., 2004;Easton et al., 2004). However, a role for BRCA1 in sporadic breast cancer, i.e., women without a strong family history of breast cancer, remains poorly understood. Many BRCA1 single nucleotide polymorphisms (SNPs) have been identified and approximately 20 SNPs result in amino acid changes. Yet these polymorphisms, with the exception of Q356R and D693N, are in significant linkage disequilibrium (LD) and are inherited as part of a shared haplotype (Freedman et al., 2005). Some studies have reported associations between BRCA1 SNPs and both familial and sporadic breast cancer risk; however there is a lack of consistency across studies (Durocher et al., 1996;Dunning et al., 1997;Cox et al., 2005;Freedman et al., 2005;Tommasi et al., 2005;Soucek et al., 2007;Dombernowsky et al., 2009;Sehl et al., 2009;The MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk, 2009).
Prior publications to date have not identified BRCA1 haplotypes as breast cancer risk factors (Cox et al., 2005;Soucek et al., 2007;The MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk, 2009). However, the haplotypes studied include coding and non-coding SNPs without regard to functional effects of the SNPs on protein function or levels. Therefore, we have chosen to study specific BRCA1 polymorphisms, Q356R, D693N, and E1038G, because they result in amino acid changes that could potentially influence the structure and function of the resulting protein. For example, Q356R is located in the ring finger domain that binds DNA (Supplementary Table II). In silico analysis of Q356R and E1038G revealed that these SNPs have regulatory effects of alternative splicing (Tommasi et al., 2008). In recent work by us, it was found that the E1038G and Q356R SNPs affected DNA repair because they were significantly associated with mean chromosomal breaks per cell in breast cancer patients from high-risk families using the mutagen sensitivity assay (Ricks-Santi et al., 2011). Additionally, these three SNPs tagging the majority of common variation in BRCA1 in women of European descent. For example, SNP E1038G is in high LD with P871L (rs799917), K1183R (rs16942), S1613G (rs1799966), and other synonymous and intronic SNPs in BRCA1 in women of European descent (Cox et al., 2005).
Breast cancer is a heterogeneous disease composed of several molecular subtypes that have been identified by gene expression analysis (Perou et al., 2000;Sorlie et al., 2001;Sorlie et al., 2003;Hu et al., 2006;Lehmann et al., 2011). The “aggressive” triple negative subtype is characterized by estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/neu) negativity and a high proliferative index (Bertucci et al., 2008;Carey et al., 2010;Foulkes et al., 2010;Lachapelle and Foulkes, 2011;Rios and Puhalla, 2011). This subtype is associated with poor prognosis, unresponsiveness to endocrine therapy, and shortened survival. Triple negative tumor is the most common subtype exhibited by BRCA1 mutation carriers (Lynch et al., 1998;Armes et al., 1999;Olopade and Grushko, 2001;Foulkes et al., 2001;Grushko et al., 2002;Lakhani et al., 2002;Sorlie et al., 2003;Palacios et al., 2003;Foulkes et al., 2003;Atchley et al., 2008).
In the present study, the associations between BRCA1 SNPs, haplotypes, sporadic breast cancer risk and molecular subtypes were investigated. Additionally, potential interactions were investigated for the genotypes with smoking, benign breast disease, family history of breast cancer, BMI, alcohol consumption, and hormonal risk factors such as age at menarche, parity, age at first birth, age at menarche, lactation, and hormone therapy (HT).
METHODS
Case-Control Study of Sporadic Cancer Risk
Subjects were recruited for the Western New York Exposures and Breast Cancer (WEB) study, a case-control study, between 1996 and 2001. This study has been described in detail elsewhere (McCann et al., 2004). Briefly, cases (n=1170) were women with primary, incident, pathologically confirmed breast cancer between the ages of 35 and 79 years, and all were residents of Erie and Niagara counties in western New York State. Population-based controls (n=2115) were randomly selected from the same counties using lists of driver’s license enrollees provided by the New York State Department of Motor Vehicles for those less than 65 years of age, and the Health Care Finance Administration for those 65 years of age and older. Controls were frequency matched by age and race to cases. The protocol was approved by the Institutional Review Boards at the University at Buffalo and Georgetown University, as well as by participating hospitals. A detailed interviewer-administered questionnaire was used to assess breast cancer risk factors. We report here only on Caucasian Americans (n= 1005 cases, n= 1765 controls) because of differences in genetic variant frequencies by racial groups and because the number of women from other racial/ethnic groups was too small for meaningful statistical analyses.
SNP Prioritization Algorithm
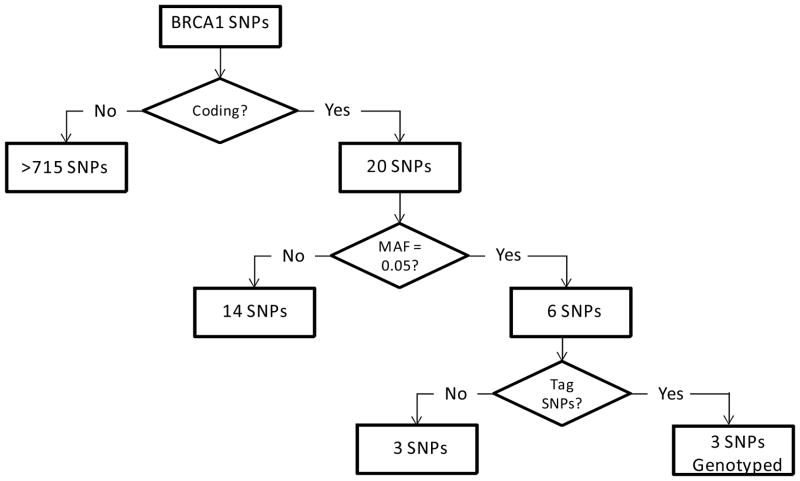
HapMap genotype data for Caucasians (www.HapMap.org) and Haploview© (Haploview 3.32, Broad Institute of MIT and Harvard, Boston, MA; www.broad.mit.edu/mpg/haploview/) (Barrett et al., 2005) were used to identify SNPs in the coding region of BRCA1 (Figure 1). Haploview was used to select haplotype tagging SNPs that capture the common variation in the BRCA1 gene (Figure 2). Only tagging SNPs with a ≥0.05 minor allele frequency were genotyped.
Figure 1.
BRCA1 SNP Prioritization Algorithm
Figure 2.
Genotyping and Haplotyping
We genotyped three tagging SNPs in BRCA1: D693N (rs4986850), Q356R (rs1799950), and E1038G (rs16941). Genotyping was carried by allelic discrimination real time PCR with TaqMan probes using primers and probes from Applied Biosystems (Applied Biosystems, Foster City, CA) as previously described (Kadouri et al., 2004). The three SNPs resulted in six total haplotypes.
Statistical Analysis
SNP allele frequencies in cases and controls were tested for Hardy-Weinberg equilibrium. Haplotypes were constructed using the software program PHASE version 2.1 (Stephens et al., 2001;Stephens and Donnelly, 2003). The most frequent haplotype in controls was considered the reference haplotype. The association of disease status with each SNP and haplotype was analyzed using logistic regression, stratifying by menopausal status (SAS/STAT® software version 9.1, SAS Institute Inc., Cary, NC). Because of the small number of variant homozygotes, dominant genetic modeling was used for D693N (rs4986850) and Q356R (rs1799950); the codominant model was used for E1038G (rs16941). Odds ratios (OR) and 95% confidence intervals (CI) were only adjusted for age because no additional covariate altered OR by 5% or more. Two-sided p-values ≤0.05 were considered as statistically significant. The interaction between SNPs, smoking, benign breast disease, family history of breast cancer, BMI, alcohol consumption, and hormonal risk factors such as age at menarche, parity, age at first birth, age at menarche, lactation, and hormone therapy (HT) were also examined in cases versus controls using logistic regression. For most variables, the cutoffs for the categories were defined by the median in all controls. For parity, the two categories were women with children vs. women without children. For BMI, the cutoff used was 30 kg/m2.
The interaction between HT and SNPs in cases and controls was only analyzed in post-menopausal women. Along with exploring the association with risk of breast cancer, the association between SNPs and breast cancer molecular subtype (Perou et al., 2000;Sorlie et al., 2001;Sorlie et al., 2003;Hu et al., 2006), hormone receptor status, histologic grade, nuclear grade, and stage were also analyzed using polytomous regression. Power calculations were performed using PS Power and Sample Size Calculations© (Version 3.0) (http://biostat.mc.vanderbilt.edu/PowerSampleSize) for main effects and menopausal subset analysis (Dupont and Plummer, Jr., 1990;Dupont and Plummer, Jr., 1998).
RESULTS
The characteristics of the WEB study participants have been reported elsewhere (Brasky et al., 2010) and a summary of subjects reported in this study are shown in Table I. Results from analyses of BRCA1 genotypes, haplotypes and breast cancer risk in pre- and post-menopausal breast cancer are presented in Tables II and III. We examined BRCA1 polymorphisms Q356R (rs1799950), D693N (rs4986850), and E1038G (rs16941). All three SNPs were in Hardy-Weinberg equilibrium (p=0.92, 0.65, and 0.59, respectively). There was no evidence of an association between the genotypes and breast cancer risk in either pre- or post- menopausal women (Table II). The use of phasing software in this study resulted in determination of six different BRCA1 haplotypes. Haplotype frequencies did not differ between cases and controls (Table III).
Table I.
Characteristics of Study Sample by Case-Control and Menopausal Status, among Whites, WEB Study
| Characteristics* | Pre-menopausal (n=811)
|
Post-menopausal (n=1959)
|
||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| N | 279 | 532 | 726 | 1233 |
| Age (years) | 45.0 (4.6)a | 44.2 (4.6)a | 63.4 (8.4) | 62.9 (8.9) |
| Education (years) | 13.9 (2.3)b | 14.3 (2.2)b | 13.3 (2.6)c | 13.1 (2.3)c |
| Body Mass Index | 27.2 (6.9) | 27.2 (6.6) | 28.7 (5.9) | 28.2 (6.0) |
| Age at menarche (years) | 12.5 (1.5) | 12.6 (1.6) | 12.6 (1.6)d | 12.7 (1.7)d |
| Age at menopause | / | / | 48.4 (5.4)e | 47.6 (6.0)e |
| Number of births | 1.9 (1.3) | 2.0 (1.3) | 2.5 (1.7)f | 3.0 (1.9)f |
| Percentage of women without child (%) | 16.8 | 15.4 | 17.4g | 9.8g |
| First-degree relative with breast cancer (% of Yes) | 19.4h | 10.5h | 19.3i | 13.8i |
| Previous benign breast disease (% of Yes) | 37.3j | 22.2j | 33.5k | 23.4k |
Values for continuous variables are mean (SD); for categorical variables (race, first-degree relative with breast cancer, history of benign breast disease) values are for percentages among either the cases or controls. Two-sided comparisons of means between the cases and controls were computed by T-test for continuous and by χ square test for categorical variables.
The letters indicate statistical significant differences between comparison groups (P<0.05).
Table II.
Risk of Breast Cancer by Smoking Status and BRCA1 Genotypes, WEB Study Participants
| Cases | Controls | Crude OR (95% CI) | Adjusted OR1 (95% CI) | |
|---|---|---|---|---|
| E1038G (rs16941) | ||||
| Pre-menopausal | ||||
| nonsmoker + TT | 47 | 151 | 1.00 | 1.00 |
| nonsmoker + CT | 59 | 113 | 1.68 (1.07–2.64) | 1.63 (1.03–2.57) |
| nonsmoker + CC | 14 | 23 | 1.96 (0.93–4.10) | 1.93 (0.92–4.06) |
| smoker + TT | 74 | 104 | 2.29 (1.47–3.56) | 2.25 (1.45–3.51) |
| smoker + CT | 65 | 114 | 1.83 (1.17–2.87) | 1.87 (1.19–2.93) |
| smoker + CC | 8 | 20 | 1.29 (0.53–3.11) | 1.34 (0.55–3.25) |
| P for interaction | 0.01 | 0.01 | ||
| Post-menopausal | ||||
| nonsmoker + TT | 147 | 257 | 1.00 | 1.00 |
| nonsmoker + CT | 130 | 258 | 0.88 (0.66–1.18) | 0.88 (0.66–1.18) |
| nonsmoker + CC | 34 | 54 | 1.10 (0.69–1.77) | 1.11 (0.69–1.79) |
| smoker + TT | 172 | 282 | 1.07 (0.81–1.41) | 1.09 (0.82–1.44) |
| smoker + CT | 173 | 280 | 1.08 (0.82–1.43) | 1.10 (0.83–1.45) |
| smoker + CC | 49 | 77 | 1.11 (0.74–1.68) | 1.12 (0.74–1.70) |
| P for interaction | 0.81 | 0.85 | ||
Odds ratios and 95% confidence intervals adjusted for age.
N=972 and 1733, for cases and controls, respectively
Table III.
Risk of Breast Cancer by HT & BRCA1 Genotypes among White WEB Study Participants
| Cases | Controls | Crude OR (95% CI) | Adjusted OR1 (95% CI) | |
|---|---|---|---|---|
| Q356R (rs1799950) | ||||
| Post-menopausal2 | ||||
| Never HRT + TT | 279 | 490 | 1.00 | 1.00 |
| Never HRT + CT&CC | 33 | 77 | 0.75 (0.49–1.16) | 0.75 (0.49–1.16) |
| HRT + TT | 345 | 533 | 1.14 (0.93–1.39) | 1.18 (0.97–1.45) |
| HRT + CT&CC | 47 | 85 | 0.97 (0.66–1.43) | 1.01 (0.68–1.49) |
| P for interaction | 0.67 | 0.68 | ||
|
| ||||
| D693N (rs4986850) | ||||
| Post-menopausal3 | ||||
| Never HRT + CC | 285 | 474 | 1.00 | 1.00 |
| Never HRT + CT&TT | 32 | 93 | 0.57 (0.37–0.88) | 0.57 (0.37–0.87) |
| HRT + CC | 335 | 535 | 1.04 (0.85–1.27) | 1.09 (0.89–1.34) |
| HRT + CT&TT | 58 | 80 | 1.21 (0.83–1.74) | 1.27 (0.87–1.84) |
| P for interaction | 0.01 | 0.01 | ||
|
| ||||
| E1038G (rs16941) | ||||
| Post-menopausal4 | ||||
| Never HRT + TT | 145 | 244 | 1.00 | 1.00 |
| Never HRT + CT | 137 | 243 | 0.95 (0.71–1.27) | 0.95 (0.71–1.27) |
| Never HRT + CC | 29 | 77 | 0.63 (0.40–1.02) | 0.63 (0.39–1.02) |
| HRT + TT | 171 | 280 | 1.03 (0.78–1.36) | 1.07 (0.81–1.42) |
| HRT + CT | 166 | 279 | 1.00 (0.76–1.33) | 1.04 (0.78–1.38) |
| HRT + CC | 53 | 52 | 1.72 (1.11–2.65) | 1.79 (1.16–2.78) |
| P for interaction | 0.02 | 0.02 | ||
Odds ratios and 95% confidence intervals adjusted for age.
N=704 and 1185 for cases and controls, respectively
N=710 and 1182 for cases and controls, respectively
N=701 and 1175 for cases and controls, respectively
We estimated our power to detect genetic associations based on the following parameters: our available sample size (1005 cases and 1765 controls), SNP minor allele frequencies ranging from 0.05–0.35, reported haplotype frequencies (14), alpha of 0.05, and a detectable odds ratio (OR) of 1.5; our statistical power was estimated at 70% for a minor allele frequency of 0.05 and 99% for a minor allele frequency of 0.35. The power was also calculated for the pre- and post-menopausal status with and available sample size for the pre-menopausal cases and controls (n=279 cases and 532 controls, respectively) and for the post-menopausal cases and controls (n=726 cases and 1233 controls); our statistical power was estimated at 23.6–75% in the pre-menopausal strata and 52%–99% for the post-menopausal strata.
While no significant results were found for BRCA1 SNPs or haplotypes and risk of breast cancer (Supplemental table 1 and 2), significant genotype interactions were observed for smoking, HT, and BMI. For rs16941 (E1038G), there was an interaction between genotype and smoking status with breast cancer risk among the pre- (p for interaction = 0.01), but not post-menopausal women (p for interaction = 0.85). Specifically, smokers with the TT genotype were at a higher risk (smoking + TT: OR=2.25, 95% CI: 1.45–3.51) (Table II).
Significant interactions with HT were found in postmenopausal women for the rs4986850 (D693N) and rs16941 (E1038G) SNPs (p for interactions = 0.01 and 0.02, respectively) (Table III). For the E1038G SNP, there was an increased risk for carriers of the CC genotype among the postmenopausal HRT users (HRT + CC: OR=1.79, 95% CI: 1.16–2.78; p for interaction = 0.02). An association between BMI and BRCA1 genotypes was also found. In premenopausal women with a BMI over 30 kg/m2, there was a trend toward increased risk of breast cancer with the C allele (>30 BMI + CC: OR=3.319, 95% CI: 1.12–9.09; p for interaction = 0.045; p for trend < 0.01) (Table IV).
Table IV.
Risk of Breast Cancer by BMI & BRCA1 Genotypes, among Whites, WEB Study
| Cases | Controls | Crude OR (CI) | Adjusted OR1 (CI) | |
|---|---|---|---|---|
| E1038G (rs16941) | ||||
| Pre-menopausal2 | ||||
| ≤30 BMI + TT | 89 | 179 | 1.00 | 1.00 |
| ≤30 BMI + CT | 93 | 169 | 1.11 (0.77–1.58) | 1.12 (0.78–1.60) |
| ≤30 BMI + CC | 12 | 37 | 0.65 (0.32–1.31) | 0.67 (0.33–1.36) |
| >30 BMI + TT | 32 | 76 | 0.85 (0.52–1.38) | 0.84 (0.51–1.36) |
| >30 BMI + CT | 32 | 59 | 1.09 (0.66–1.80) | 1.06 (0.64–1.75) |
| >30 BMI + CC | 10 | 6 | 3.35 (1.18–9.52) | 3.19 (1.12–9.09) |
| P for interaction | 0.03 | 0.045 | ||
| Post-menopausal3 | ||||
| ≤30 BMI + TT | 208 | 374 | 1.00 | 1.00 |
| ≤30 BMI + CT | 200 | 381 | 0.94 (0.74–1.20) | 0.94 (0.74–1.20) |
| ≤30 BMI + CC | 45 | 86 | 0.94 (0.63–1.40) | 0.95 (0.63–1.41) |
| >30 BMI + TT | 112 | 167 | 1.21 (0.90–1.62) | 1.21 (0.90–1.62) |
| >30 BMI + CT | 103 | 158 | 1.17 (0.87–1.58) | 1.17 (0.87–1.59) |
| >30 BMI + CC | 38 | 45 | 1.52 (0.96–2.42) | 1.51 (0.95–2.40) |
| P for interaction | 0.43 | 0.44 | ||
Odds ratios and 95% confidence intervals adjusted for age (the matching variable).
N=268 and 526 for cases and controls, respectively
N=706 and 1211 for cases and controls, respectively
There was no evidence of an interaction for any of the three BRCA1 SNPs with benign breast disease, family history of breast cancer, alcohol consumption, age at menarche, parity, age at first birth, age at menarche, or lactation either in pre-menopausal or post-menopausal women.
Furthermore, no significant associations emerged between individual SNPs and estrogen receptor status (Additional File III), progesterone receptor status (Additional file IV) or HER2/neu status (Additional file V). However, when examining the association of SNPs and molecular subtypes in case-only analyses, the D693N CT and TT genotypes were associated with the triple negative subtype when compared to the other subtypes combined (OR=2.31, 95% CI: 1.08–4.93; p=0.03) (Table V). There were no associations found between BRCA1 genotypes and stage (Additional file VI), histologic grade (Additional file VII), or nuclear grade (Additional file VIII).
Table V.
Risk of Molecular Breast Cancer Subtype by BRCA1 Genotypes among White WEB Study Participants (Triple negative vs. other subtypes)
| BRCA1 genotypes | Other Subtypes1 | Triple Negative2 | Crude OR (95% CI) | Adjusted OR3 (95% CI) |
|---|---|---|---|---|
| Q356R (rs1799950)4 | ||||
| 1: TT | 469 | 95 | 1.00 | 1.00 |
| 2: CT+CC | 68 | 12 | 1.15 (0.60–2.20) | 1.15 (0.60–2.22) |
|
| ||||
| D693N (rs4986850)5 | ||||
| 1: CC | 456 | 98 | 1.00 | 1.00 |
| 2: CT+TT | 84 | 8 | 2.26 (1.06–4.81) | 2.31 (1.08–4.93) |
|
| ||||
| E1038G (rs16941)6 | ||||
| 1: TT | 236 | 53 | 1.00 | 1.00 |
| 2: CT | 229 | 43 | 1.20 (0.77–1.86) | 1.20 (0.77–1.87) |
| 3: CC | 65 | 8 | 1.83 (0.83–4.03) | 1.79 (0.81–3.97) |
Other Subtypes includes: Luminal A: ER+ or PR+, HER2-; Luminal B: ER+ or PR+, HER2+; HER2 Expressing: ER−/PR−, HER2+
Triple-negative: ER−/PR−/HER2−
Odds ratios and 95% confidence intervals adjusted for age (the matching variable).
N=537 and 107 for cases and controls, respectively
N=540 and 106 for cases and controls, respectively
N=530 and 104 for cases and controls, respectively
DISCUSSION
In this case-control study of breast cancer there were detectable interactions for overall breast cancer risk in women carrying BRCA1 SNPs related to smoking, BMI, and hormone therapy use. An increased risk of triple negative breast cancer was also identified in women carrying the BRCA1 D693N SNP. Furthermore, No BRCA1 haplotype associations were found with breast cancer risk.
There has been limited study of BRCA1 genotypes and haplotypes in relation to sporadic breast cancer risk. However, mostly null results have been reported (Menzel et al., 2004;Freedman et al., 2005;Dombernowsky et al., 2009), which has also been the case for familial breast cancer and low penetrant SNPs (Durocher et al., 1996;Dunning et al., 1997;Tommasi et al., 2005;Seymour et al., 2008;Dombernowsky et al., 2009),potentially due to sample size issues and limited statistical power, or the choice of SNPs without regard to functional effects on the protein. Herein, the BRCA1 SNPs were chosen for investigation because in silico analyses indicated that they could affect protein functionIn a previous study, we determined that the E1038G SNP, Q356R SNP, and a three SNP haplotype (Q356R-D693N-E1038G) were associated with DNA repair capacity in women with hereditary breast cancer (Ricks-Santi et al., 2011). Therefore, we sought to determine if the same SNPs and haplotypes would be associated with risk of sporadic breast cancer.
The E1038G SNP is highly linked (r2>0.80) in samples of European descent to other SNPs such as P871L, K1183R, and S1613G, and they are represented in the same haplotype block (Figure 2). Consequently, the polymorphism has not been examined independently, but only as part of a haplotype. Notably, the E1038G SNP has been found to be associated with altered BRCA1 protein expression in breast cancer tissues of those who carried the polymorphism (Mangia et al., 2009;Cox et al., 2011). Notably, bioinformatic functional analysis of the E1038G SNP reveals that the SNP is deleterious and may change the secondary structure of the protein using Sorting Intolerant From Tolerant (SIFT) (Ng and Henikoff, 2001;Ng and Henikoff, 2003;Ng and Henikoff, 2006;Sim et al., 2012). Additionally, the variant allele (1038G) is associated with the disappearance of a putative splice enhancer sequence using Putative Exonic Splicing Enhancers/Silencers (PESX), a computational resource that searches for exonic splicing signals (Zhang and Chasin, 2004;Zhang et al., 2005). Nonetheless, like other studies (Durocher et al., 1996;Dunning et al., 1997;Menzel et al., 2004;Freedman et al., 2005;Tommasi et al., 2005;Dombernowsky et al., 2009), the E1038G SNP was not independently associated with breast cancer status in this study, even after stratifying by menopausal status. However, in this study, a genotype interaction with smoking was found with pre-menopausal breast cancer for E1038G SNP carriers. An interaction between BRCA1 mutations and smoking has been previously reported in BRCA1 mutation carriers (Bissonauth et al., 2009;Lecarpentier et al., 2011). Still, the relationship between smoking and breast cancer risk remains somewhat controversial (Ginsburg et al., 2009), although other gene-smoking interactions have been reported (Ambrosone et al., 1996;Ambrosone et al., 2008). Moreover, the California Environmental Protection Agency (Cal/EPA) Office of Environmental Health Hazard Assessment (OEHHA) has concluded that environmental tobacco smoke (ETS) exposure is causally associated with breast cancer in younger, primarily premenopausal women (Miller et al., 2007). With regard to biological plausibility, a number of investigators have shown that breast tissue from smokers have a higher level of DNA adducts resulting from ETS and that mammary epithelia is capable of metabolic activation of carcinogens (Li et al., 1996;Firozi et al., 2002;Faraglia et al., 2003). Since the ability of BRCA1 to repair DNA damage may be hampered by this effect of the E1038G polymorphism, it remains plausible that the gene-smoking interaction reported here could be real. It is worth mentioning that although there a trend for the increased risk of breast cancer in nonsmokers with the addition of a C allele, the opposite is true in smokers. Potentially, the expression of the genotype is strictly dependent on the environment in which it acts. Here, the same allele that increased risk in one environment (non-smokers) may decrease it in a different one (smokers). It is also possible that the interaction is spurious due to the small sample size in those with the CC genotype. Nevertheless, the interaction, while still controversial, warrants additional investigations.
Another interaction identified here was for the E1038G SNP and obesity in premenopausal women. This is the first report where obesity was found to increase the risk for breast cancer in E1038G SNP carriers. Notably, in studies of BRCA1 mutation carriers, a high body weight and BMI increased the risk of postmenopausal breast cancer (Brunhuber et al., 2008;Bissonauth et al., 2009;Manders et al., 2011). However, less consistent is the data for premenopausal women (Manders et al., 2011). The mechanisms that link BMI and cancer risk are also not fully understood. It is hypothesized that BMI influences hormonal systems linked through insulin pathways and it may be that chronic hyperinsulinemia decreases concentrations of IGF binding proteins resulting in increased free IGF-I and that these conditions favor tumor formation (Renehan et al., 2006a;Renehan et al., 2006b). Further, a direct interaction between BRCA1 and acetyl-CoA carboxylase (ACC) which is involved in lipogenesis has also been shown (Magnard et al., 2002;Sinilnikova et al., 2004); there is also evidence to suggest that BRCA1 helps to maintain fatty acid biosynthesis and lipogenesis under control in normal cells, which is an important requirement for cancer cell growth (Ray et al., 2009). Although there was a small sample size for those that were obese with the CC genotype, our data provide further evidence that supports the interaction between obesity and breast cancer risk, especially in post-menopausal BRCA1 mutation carriers.
Null results for the BRCA1 D693N (rs4986850) SNP have also been reported elsewhere (Durocher et al., 1996). However, the D693N SNP, specifically the T allele, was found to be a risk factor for triple negative breast cancers, which is a particularly aggressive form of breast cancer (Bertucci et al., 2008;Carey et al., 2010;Foulkes et al., 2010;Lachapelle and Foulkes, 2011;Rios and Puhalla, 2011). These tumors are particularly common among BRCA1 mutation carriers (Lynch et al., 1998;Armes et al., 1999;Olopade and Grushko, 2001;Foulkes et al., 2001;Grushko et al., 2002;Lakhani et al., 2002;Sorlie et al., 2003;Palacios et al., 2003;Foulkes et al., 2003;Atchley et al., 2008).
The results of the study herein indicate that HT may increase risk of developing breast cancer in the presence of the BRCA1 D693N and E1038G SNPs, but not the Q356R SNP. This interaction has not been studied previously for breast cancer risk. It has previously been demonstrated that HT increases the risk of breast cancer (Rossouw, 2002;Beral, 2003) and that estrogens and estrogen metabolites have been shown to damage DNA (Tsutsui et al., 2000;Schnyder et al., 2009).
The Q356R SNP is of particular interest because of its location in the ring finger domain and its interaction with several genes such as p53, RAD51, and pRB which are all related to DNA repair (Wang et al., 2000;Rebbeck et al., 2011). Also, in silico analyses also reveal this SNP to be damaging and deleterious using bioinformatics analyses (Ng and Henikoff, 2001;Ramensky et al., 2002;Ng and Henikoff, 2003;Ng and Henikoff, 2006;Tommasi et al., 2008;Sim et al., 2012). While positive associations of the Q356R SNP with sporadic (Soucek et al., 2007) and familial (Dunning et al., 1997) breast cancer risk have been reported, agreeing with the results reported herein, null results have also been reported in sporadic (Menzel et al., 2004;Freedman et al., 2005) (Menzel et al., 2004;Freedman et al., 2005;Soucek et al., 2007) and familial (Seymour et al., 2008) breast cancer risk studies. But this could be a reflection of sample size and population stratification.
For BRCA1 haplotype analyses, both null (Freedman et al., 2005;Soucek et al., 2007;The MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk, 2009) and positive (Cox et al., 2005) haplotype results have been reported for sporadic breast cancer risk. Although previous studies in our laboratories identified a haplotype associated with DNA repair capacity in hereditary breast cancer patients (Ricks-Santi et al., 2011), there was no association with sporadic breast cancer examined here. This is the first report of a haplotype study for the Q356R, D693N, and E1038G SNPs, and the rationale for studying these was because of their potential functional impact.
In assessing these study results, it is important to consider the strengths and limitations of the study. The population-based study design, large sample size, and the detailed epidemiologic information on known risk factors for breast cancer and disease characteristics are strengths. However, several limitations should be considered when evaluating our results. Although the sample size and power of this study was sufficient to study main effects by menopausal status, subset analysis by some of the less common risk factors, e.g., HT, smoking, and family history, and analysis for gene interactions, reduced the sample size, increasing the likelihood of null findings. Also, chance must be considered as an explanation for positive associations found herein, e.g., HT, smoking, obesity and triple negative breast cancer. As in all epidemiologic studies, additional studies in other populations are needed to replicate our finding and to elucidate the underlying biological mechanisms. Although we chose to examine three SNPs in one gene associated with familial breast cancer risk, other genes that interact with BRCA1 (Rebbeck et al., 2011) may be more informative for examination, specifically as it relates to the effect of DNA repair deficiencies associated with breast cancer predisposition.
We conclude that although genetic variation in BRCA1 may be associated with the etiology of hereditary breast cancer, BRCA1 genetic variation by itself may also play a role in the etiology of sporadic breast cancer in the context of some breast cancer risk factors and for triple negative breast cancers.
Supplementary Material
Acknowledgments
The authors would also like to thank Shiva Krishnan, David Goerlitz, and Leoni Leondaridis, for their technical assistance.
GRANT SUPPORT
This project was supported in part or in whole by the Department of Defense grants (LSR pre-doctoral training grant BC030134, and BC02234, and DAMD179616292), the National Cancer Institute (CA092040 and AA09802), National Center for Research Resources (NCRR, National Institutes of Health, through the Clinical and Translational Science Awards Program (CTSA), a trademark of DHHS, part of the Roadmap Initiative, “Re-Engineering the Clinical Research Enterprise,” (UL1RR031975), the RCMI Program at Howard University, Division of Research Infrastructure, National Center for Research Resources (G12 RR003048), and the Howard University Cancer Center/ Johns Hopkins Cancer Center Partnership, National Cancer Institute, NIH (U54 CA091431).
Footnotes
COMPETING INTERESTS: The authors declare that there are no competing interests.
AUTHORS’ CONTRIBUTIONS
LJR-S conceived the study, carried out the molecular genetic studies, and drafted the manuscript. JN and HMO-B carried out the final statistical analysis of the WEB study. CM carried out the haplotype analysis. MT, SBE and JLF participated in the design of the study and coordinated the WEB study. PGS conceived the study, participated in the design of the study and edited the manuscript. All authors read and approved the manuscript.
References Cited
- Ambrosone CB, et al. Cigarette smoking, N-acetyltransferase 2 genetic polymorphisms, and breast cancer risk. JAMA. 1996;276(18):1494–1501. [PubMed] [Google Scholar]
- Ambrosone CB, Kropp S, Yang J, Yao S, Shields PG, Chang-Claude J. Cigarette smoking, N-acetyltransferase 2 genotypes, and breast cancer risk: pooled analysis and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17(1):15–26. doi: 10.1158/1055-9965.EPI-07-0598. [DOI] [PubMed] [Google Scholar]
- Armes JE, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59(8):2011–2017. [PubMed] [Google Scholar]
- Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, Hortobagyi GN, Arun BK. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26(26):4282–4288. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362(9382):419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- Bertwistle D, Ashworth A. Functions of the BRCA1 and BRCA2 genes. Curr Opin Genet Dev. 1998;8(1):14–20. doi: 10.1016/s0959-437x(98)80056-7. [DOI] [PubMed] [Google Scholar]
- Bissonauth V, Shatenstein B, Fafard E, Maugard C, Robidoux A, Narod S, Ghadirian P. Weight History, Smoking, Physical Activity and Breast Cancer Risk among French-Canadian Women Non-Carriers of More Frequent BRCA1/2 Mutations. J Cancer Epidemiol. 2009;2009:748367. doi: 10.1155/2009/748367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasky TM, et al. Non-steroidal anti-inflammatory drug (NSAID) use and breast cancer risk in the Western New York Exposures and Breast Cancer (WEB) Study. Cancer Causes Control. 2010;21(9):1503–1512. doi: 10.1007/s10552-010-9579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunhuber T, Haybaeck J, Schafer G, Mikuz G, Langhoff E, Saeland S, Lebecque S, Romani N, Obrist P. Immunohistochemical tracking of an immune response in mammary Paget’s disease. Cancer Lett. 2008;272(2):206–220. doi: 10.1016/j.canlet.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7(12):683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- Cox DG, Kraft P, Hankinson SE, Hunter DJ. Haplotype analysis of common variants in the BRCA1 gene and risk of sporadic breast cancer. Breast Cancer Res. 2005;7(2):R171–R175. doi: 10.1186/bcr973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DG, et al. Common variants of the BRCA1 wild-type allele modify the risk of breast cancer in BRCA1 mutation carriers. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombernowsky SL, Weischer M, Freiberg JJ, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Missense polymorphisms in BRCA1 and BRCA2 and risk of breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(8):2339–2342. doi: 10.1158/1055-9965.EPI-09-0447. [DOI] [PubMed] [Google Scholar]
- Dunning AM, et al. Common BRCA1 variants and susceptibility to breast and ovarian cancer in the general population. Hum Mol Genet. 1997;6(2):285–289. doi: 10.1093/hmg/6.2.285. [DOI] [PubMed] [Google Scholar]
- Dupont WD, Plummer WD., Jr Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11(2):116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- Dupont WD, Plummer WD., Jr Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19(6):589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- Durocher F, Shattuck-Eidens D, McClure M, Labrie F, Skolnick MH, Goldgar DE, Simard J. Comparison of BRCA1 polymorphisms, rare sequence variants and/or missense mutations in unaffected and breast/ovarian cancer populations. Hum Mol Genet. 1996;5(6):835–842. doi: 10.1093/hmg/5.6.835. [DOI] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56(1):265–271. [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Hopper JL, Thomas DC, Antoniou A, Pharoah PD, Whittemore AS, Haile RW. Breast cancer risks for BRCA1/2 carriers. Science. 2004;306(5705):2187–2191. doi: 10.1126/science.306.5705.2187c. [DOI] [PubMed] [Google Scholar]
- Fan S, et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene. 2001;20(1):77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- Fan S, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284(5418):1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- Faraglia B, et al. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24(4):719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- Firozi PF, Bondy ML, Sahin AA, Chang P, Lukmanji F, Singletary ES, Hassan MM, Li D. Aromatic DNA adducts and polymorphisms of CYP1A1, NAT2, and GSTM1 in breast cancer. Carcinogenesis. 2002;23(2):301–306. doi: 10.1093/carcin/23.2.301. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Rosenblatt J, Chappuis PO. The contribution of inherited factors to the clinicopathological features and behavior of breast cancer. J Mammary Gland Biol Neoplasia. 2001;6(4):453–465. doi: 10.1023/a:1014791115760. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, I, Smith E, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, I, Stefansson M, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Penney KL, Stram DO, Riley S, Kean-Cowdin R, Le ML, Altshuler D, Haiman CA. A haplotype-based case-control study of BRCA1 and sporadic breast cancer risk. Cancer Res. 2005;65(16):7516–7522. doi: 10.1158/0008-5472.CAN-05-0132. [DOI] [PubMed] [Google Scholar]
- Ginsburg O, et al. Smoking and the risk of breast cancer in BRCA1 and BRCA2 carriers: an update. Breast Cancer Res Treat. 2009;114(1):127–135. doi: 10.1007/s10549-008-9977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushko TA, Blackwood MA, Schumm PL, Hagos FG, Adeyanju MO, Feldman MD, Sanders MO, Weber BL, Olopade OI. Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res. 2002;62(5):1481–1488. [PubMed] [Google Scholar]
- Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri L, Kote-Jarai Z, Hubert A, Durocher F, Abeliovich D, Glaser B, Hamburger T, Eeles RA, Peretz T. A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br J Cancer. 2004;90(10):2002–2005. doi: 10.1038/sj.bjc.6601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachapelle J, Foulkes WD. Triple-negative and basal-like breast cancer: implications for oncologists. Curr Oncol. 2011;18(4):161–164. doi: 10.3747/co.v18i4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani SR, Van d, Jacquemier VJ, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- Lecarpentier J, et al. Variation in breast cancer risk with mutation position, smoking, alcohol, and chest X-ray history, in the French National BRCA1/2 carrier cohort (GENEPSO) Breast Cancer Res Treat. 2011;130(3):927–938. doi: 10.1007/s10549-011-1655-3. [DOI] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang M, Dhingra K, Hittelman WN. Aromatic DNA adducts in adjacent tissues of breast cancer patients: clues to breast cancer etiology. Cancer Res. 1996;56(2):287–293. [PubMed] [Google Scholar]
- Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J Clin Oncol. 2004;22(4):735–742. doi: 10.1200/JCO.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Lynch BJ, Holden JA, Buys SS, Neuhausen SL, Gaffney DK. Pathobiologic characteristics of hereditary breast cancer. Hum Pathol. 1998;29(10):1140–1144. doi: 10.1016/s0046-8177(98)90427-0. [DOI] [PubMed] [Google Scholar]
- Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, Venezia ND. BRCA1 interacts with acetyl-CoA carboxylase through its tandem of BRCT domains. Oncogene. 2002;21(44):6729–6739. doi: 10.1038/sj.onc.1205915. [DOI] [PubMed] [Google Scholar]
- Manders P, et al. Body weight and risk of breast cancer in BRCA1/2 mutation carriers. Breast Cancer Res Treat. 2011;126(1):193–202. doi: 10.1007/s10549-010-1120-8. [DOI] [PubMed] [Google Scholar]
- Mangia A, Chiriatti A, Tommasi S, Menolascina F, Petroni S, Zito FA, Simone G, Schittulli F, Paradiso A. BRCA1 expression and molecular alterations in familial breast cancer. Histol Histopathol. 2009;24(1):69–76. doi: 10.14670/HH-24.69. [DOI] [PubMed] [Google Scholar]
- McCann SE, Ip C, Ip MM, McGuire MK, Muti P, Edge SB, Trevisan M, Freudenheim JL. Dietary intake of conjugated linoleic acids and risk of premenopausal and postmenopausal breast cancer, Western New York Exposures and Breast Cancer Study (WEB Study) Cancer Epidemiol Biomarkers Prev. 2004;13(9):1480–1484. [PubMed] [Google Scholar]
- Menzel HJ, Sarmanova J, Soucek P, Berberich R, Grunewald K, Haun M, Kraft HG. Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer. 2004;90(10):1989–1994. doi: 10.1038/sj.bjc.6601779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Marty MA, Broadwin R, Johnson KC, Salmon AG, Winder B, Steinmaus C. The association between exposure to environmental tobacco smoke and breast cancer: a review by the California Environmental Protection Agency. Prev Med. 2007;44(2):93–106. doi: 10.1016/j.ypmed.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Olopade OI, Grushko T. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344(26):2028–2029. doi: 10.1056/NEJM200106283442613. [DOI] [PubMed] [Google Scholar]
- Palacios J, et al. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin Cancer Res. 2003;9(10 Pt 1):3606–3614. [PubMed] [Google Scholar]
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H, Suau F, Vincent A, Dalla VN. Cell cycle regulation of the BRCA1/acetyl-CoA-carboxylase complex. Biochem Biophys Res Commun. 2009;378(3):615–619. doi: 10.1016/j.bbrc.2008.11.090. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, et al. Modification of BRCA1-Associated Breast and Ovarian Cancer Risk by BRCA1-Interacting Genes. Cancer Res. 2011;71(17):5792–5805. doi: 10.1158/0008-5472.CAN-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renehan AG, Frystyk J, Flyvbjerg A. Obesity and cancer risk: the role of the insulin-IGF axis. Trends Endocrinol Metab. 2006a;17(8):328–336. doi: 10.1016/j.tem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Renehan AG, Harvie M, Howell A. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and breast cancer risk: eight years on. Endocr Relat Cancer. 2006b;13(2):273–278. doi: 10.1677/erc.1.01219. [DOI] [PubMed] [Google Scholar]
- Ricks-Santi LJ, et al. Association of Rad51 polymorphism with DNA repair in BRCA1 mutation carriers and sporadic breast cancer risk. BMC Cancer. 2011;11:278. doi: 10.1186/1471-2407-11-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology (Williston Park) 2011;25(11):1014–1025. [PubMed] [Google Scholar]
- Rossouw JE. Hormones, genetic factors, and gender differences in cardiovascular disease. Cardiovasc Res. 2002;53(3):550–557. doi: 10.1016/s0008-6363(01)00478-3. [DOI] [PubMed] [Google Scholar]
- Schnyder S, Du NT, Le HB, Singh S, Loredo GA, Vaughan AT. Estrogen treatment induces MLL aberrations in human lymphoblastoid cells. Leuk Res. 2009;33(10):1400–1404. doi: 10.1016/j.leukres.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehl ME, et al. Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clin Cancer Res. 2009;15(6):2192–2203. doi: 10.1158/1078-0432.CCR-08-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour IJ, et al. Disease family history and modification of breast cancer risk in common BRCA2 variants. Oncol Rep. 2008;19(3):783–786. [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(W1):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinilnikova OM, et al. Acetyl-CoA carboxylase alpha gene and breast cancer susceptibility. Carcinogenesis. 2004;25(12):2417–2424. doi: 10.1093/carcin/bgh273. [DOI] [PubMed] [Google Scholar]
- Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek P, Borovanova T, Pohlreich P, Kleibl Z, Novotny J. Role of single nucleotide polymorphisms and haplotypes in BRCA1 in breast cancer: Czech case-control study. Breast Cancer Res Treat. 2007;103(2):219–224. doi: 10.1007/s10549-006-9367-9. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. Polymorphisms in the BRCA1 and ABCB1 genes modulate menopausal hormone therapy associated breast cancer risk in postmenopausal women. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0489-8. [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton D. Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomarkers Prev. 2002a;11(4):329–336. [PubMed] [Google Scholar]
- Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002b;94(18):1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- Tommasi S, et al. BRCA1 mutations and polymorphisms in a hospital-based consecutive series of breast cancer patients from Apulia, Italy. Mutat Res. 2005;578(1–2):395–405. doi: 10.1016/j.mrfmmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Tommasi S, et al. Molecular and in silico analysis of BRCA1 and BRCA2 variants. Mutat Res. 2008;644(1–2):64–70. doi: 10.1016/j.mrfmmm.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Tamura Y, Yagi E, Barrett JC. Involvement of genotoxic effects in the initiation of estrogen-induced cellular transformation: studies using Syrian hamster embryo cells treated with 17beta-estradiol and eight of its metabolites. Int J Cancer. 2000;86(1):8–14. doi: 10.1002/(sici)1097-0215(20000401)86:1<8::aid-ijc2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang H, Fishel R, Greene MI. BRCA1 and cell signaling. Oncogene. 2000;19(53):6152–6158. doi: 10.1038/sj.onc.1203974. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18(11):1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25(16):7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.