Abstract
Striated muscles that enable mouth opening and swallowing during feeding are essential for efficient energy acquisition, and are likely to have played a fundamental role in the success of early jawed vertebrates. The developmental origins and genetic requirements of these muscles are uncertain. Here, we determine by indelible lineage tracing in mouse that fibres of sternohyoid muscle (SHM), which is essential for mouth opening during feeding, and oesophageal striated muscle (OSM), which is crucial for voluntary swallowing, arise from Pax3-expressing somite cells. In vivo Kaede lineage tracing in zebrafish reveals the migratory route of cells from the anteriormost somites to OSM and SHM destinations. Expression of pax3b, a zebrafish duplicate of Pax3, is restricted to the hypaxial region of anterior somites that generate migratory muscle precursors (MMPs), suggesting that Pax3b plays a role in generating OSM and SHM. Indeed, loss of pax3b function led to defective MMP migration and OSM formation, disorganised SHM differentiation, and inefficient ingestion and swallowing of microspheres. Together, our data demonstrate Pax3-expressing somite cells as a source of OSM and SHM fibres, and highlight a conserved role of Pax3 genes in the genesis of these feeding muscles of vertebrates.
Keywords: Hypaxial muscle, Zebrafish, Mouse, Pax3, In vivo lineage tracing
INTRODUCTION
Energy acquisition in jawed vertebrates is accompanied by two processes: opening of the mouth and subsequent passage of food to the gastrointestinal tract. Two groups of striated muscles facilitate these processes. First, the hyoid musculature, including sternohyoid muscle (SHM), is essential for mouth opening. Second, oesophageal striated muscle (OSM) permits voluntary swallowing. The requirements for both SHM and OSM for efficient feeding are demonstrated by the inability to suckle after SHM fatigue (van Lunteren and Moyer, 2003) and by malnutrition in humans with oesophageal dysphagia (Perry, 2001). Given the physiological importance of OSM and SHM, it is surprising that their cellular origin and development are ill-defined.
Unlike the rest of the gut, the oesophagus of fish and mammals acquires a striated muscle phenotype correlating with voluntary feeding (Domeneghini et al., 1999; García Hernández et al., 2001; Wörl and Neuhuber, 2005; Carrassón et al., 2006; Elworthy et al., 2008; Katori et al., 2010). In mice, two layers of OSM appear sequentially during the foetal period among earlier-formed smooth muscle cells at the anterior end of the lengthening oesophagus (Fig. 1J). The source of OSM is controversial, with multiple hypotheses put forward, including a smooth-to-striated muscle transition (Patapoutian et al., 1995; Kablar et al., 2000; Stratton et al., 2000) or distinct smooth and striated muscle lineages (Rishniw et al., 2003; Wörl and Neuhuber, 2005). In fish and mammals, the SHM attaches to the pectoral girdle (clavicle in mammals, cleithrum in teleosts) and the hyoid bone, thus enabling complex movements associated with food acquisition, processing and swallowing (Cubbage and Mabee, 1996; Diogo et al., 2008; Konow et al., 2010). Although Noden (Noden, 1983) suggests that muscles in the SHM region derive from somites and gene expression in zebrafish supports this view (Schilling and Kimmel, 1997; Neyt et al., 2000; Lin et al., 2006), the embryological origin of SHM has not been proven.
Fig. 1.
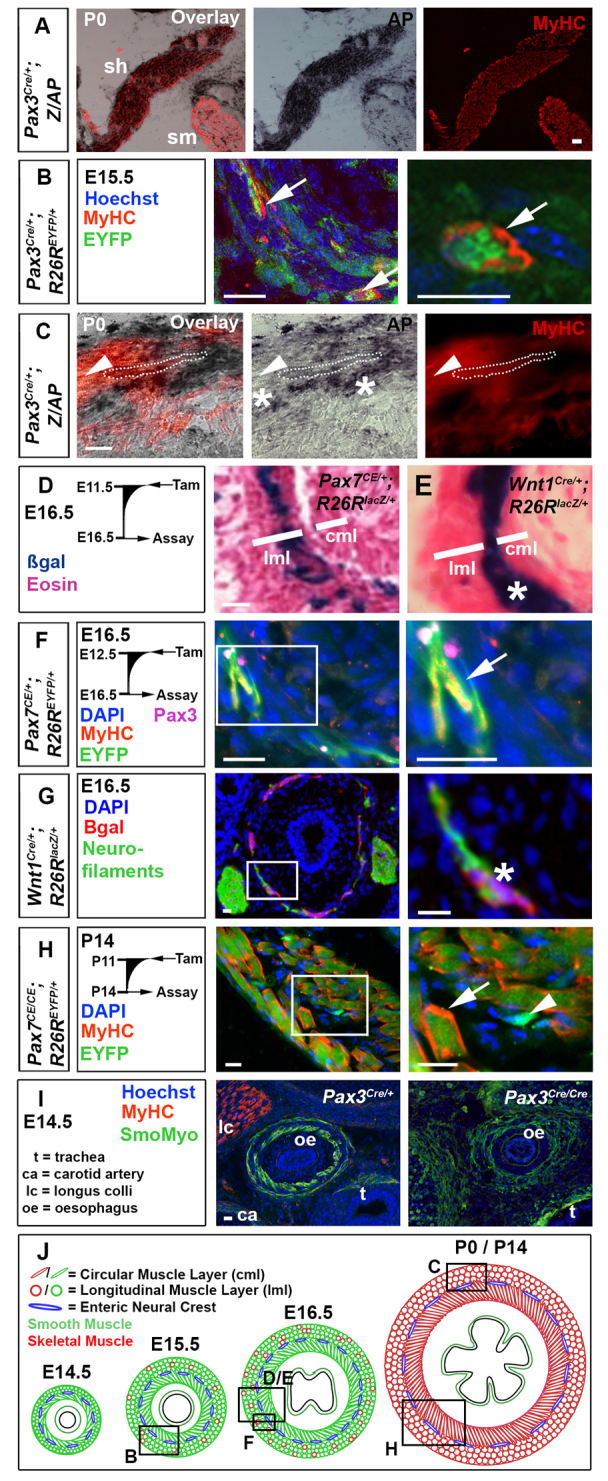
Pax3- and Pax7-expressing cells contribute to mouse OSM. (A-I) Immunodetection of MyHC (A-C,F,H,I), smooth muscle myosin (I), EYFP (B,F,H) and βgal (G); nuclei staining (DAPI/Hoechst; B,F-I); and histochemistry for alkaline phosphatase (AP; A,C) or βgal (D,E) on transverse cryosections of murine oesophagus. (A) The sternohyoid muscle (sh) was AP+, whereas the adjacent sternomastoid muscle (sm) was unlabelled. (B) Pax3Cre/+;R26REYFP mice show MyHC in some EYFP+ cells (arrows) at E15.5. (C) By P0, Pax3Cre/+;Z/AP mice have OSM cells marked with AP (dots outline an area of staining over a fibre). Some OSM is unmarked (arrowheads), and neural cells between the longitudinal and circumferential OSM layers are marked (asterisks). (D) Tamoxifen treatment of Pax7CE/+;R26RlacZ mice at E11.5 marked cells at E16.5 in outer longitudinal muscle layer (lml, where OSM is present) but only rare cells in inner circumferential muscle layer [cml, lacking OSM at this stage (see A)]. (E) Wnt1Cre;R26RlacZ (right panel) mice had neural crest cells between the muscle layers (asterisk). (F) Tamoxifen treatment of Pax7CE/+;R26REYFP mice at E12.5 marked OSM cells in lml at E16.5 with EYFP (arrow). Small rare Pax3+ cells are found adjacent to the OSM. (G) At E16.5, Wnt1Cre;R26RlacZ mice have βgal in a thin layer of neurofilament-containing cells between the muscle layers (asterisk). (H) Treatment of Pax7CE/CE;R26REYFP mice with tamoxifen at P11 led to abundant EYFP-marked OSM cells at P14 (arrow) and smaller EYFP-marked cells lacking MyHC (arrowhead). (I) At E14.5, Pax3Cre/Cre mice show slightly reduced and disorganised smooth muscle myosin (green), compared with Pax3Cre/+ littermates. There is a lack of longus colli striated muscle (lc) in mutant. ca, carotid artery; t, trachea; oe, oesophagus. (J) Mouse OSM development (red, sarcomeric myosin) among smooth muscle (green, smooth myosin), drawn approximately to scale. Boxed areas in the left panels are magnified on the right. Boxed areas in J are shown in panels above. Scale bars: 50 μm in A; 20 μm in B-I.
Undifferentiated, hypaxial somite-derived, migratory muscle precursors (MMPs) form all limb, tongue and diaphragm striated muscles (Noden, 1983; Dietrich et al., 1998; Dietrich, 1999). Pax3/7 transcription factors regulate MMP-derived muscle fate (Buckingham and Relaix, 2007). Klein-Waardenburg syndrome (OMIM #148820) is caused by PAX3 mutation and is characterised by reduced MMP-derived striated muscle (Goodman et al., 1982). Similarly, the Pax3 mouse mutant splotch (Sp), has greatly reduced MMP-derived striated muscle (Bober et al., 1994; Tajbakhsh et al., 1997; Tremblay et al., 1998; Borycki et al., 1999; Buckingham and Relaix, 2007; Zhou et al., 2008). pax3/7 genes are expressed in zebrafish somites (Groves et al., 2005; Devoto et al., 2006; Hammond et al., 2007; Minchin and Hughes, 2008) and MMP-derived myogenesis is morphologically and molecularly conserved with amniotes (Bladt et al., 1995; Brohmann et al., 2000; Gross et al., 2000; Neyt et al., 2000; Haines et al., 2004; Ochi and Westerfield, 2009). Although Pax7 regulates myoD expression in Xenopus somites (Maczkowiak et al., 2010), a role for Pax3 during myogenesis has not been elucidated beyond mammals, so the ancestral role of Pax3 in myogenesis is unclear. Here, we determine that Pax3-expressing somite cells give rise to OSM and SHM, a process that is Pax3b dependent in zebrafish and is required for efficient ingestion and swallowing.
MATERIALS AND METHODS
Lines and maintenance
Pax3Cre [Pax3tm1(cre)Joe] (Engleka et al., 2005), inducible Pax7CE [Pax7tm2.1(cre/ERT2)Fan] (Lepper et al., 2009), Wnt1Cre [Tg(Wnt1-cre)11Rth] (Danielian et al., 1998), R26REYFP [Gt(ROSA)26Sortm1(EYFP)Cos] (Srinivas et al., 2001), R26RlacZ [Gt(ROSA)26Sortm1Sor] (Soriano, 1999) and Z/AP [Tg(CAG-Bgeo/ALPP)1Lbe] (Lobe et al., 1999) on a mixed genetic background were bred, genotyped by PCR and staged according to embryonic day. CreERT2 activation was achieved using tamoxifen (Sigma) injection prenatally (50 μl of 20 mg/ml tamoxifen intraperitoneally) (Lepper et al., 2009) or postnatally [20 μl of 2 mM 4OH-tamoxifen (Millipore) subcutaneously].
Wild-type zebrafish, mutant lines myf5hu2022 (Hinits et al., 2009) and myodfh261 (Hinits et al., 2011), and transgenic lines Tg(actc1b:egfp)zf13 (Higashijima et al., 1997), Tg(mylz2:egfp) (Ju et al., 2003) and Tg(kdrl:egfp)s843 (Jin et al., 2005) were kept on King’s College wild-type or Tupfel long fin backgrounds. Zebrafish maintenance, staging and husbandry were as previously described (Westerfield, 2000).
Morpholinos
Morpholinos (MOs, Gene-Tools) were injected into one-cell stage zebrafish embryos. Data showing MO1 and MO2 pairs behave similarly are given in supplementary material Table S1. MO sequences are included as supplementary material Table S2. pax3b MO knockdown efficacy was demonstrated by cloning pax3b 5′UTR and partial coding sequence in frame with GFP (supplementary material Fig. S9). pax3b MO1 induces a subtle small head phenotype that is rescued by co-injection with p53 MO (supplementary material Fig. S10) and was used unless otherwise stated.
In situ hybridisation and immunohistochemistry
Zebrafish in situ mRNA hybridisation was performed as previously described (Coutelle et al., 2001) with probes referenced in supplementary material Table S3. Immunodetection was performed as previously described (Hinits et al., 2009; Anderson et al., 2012). Alkaline phosphatase was revealed after antibody staining by washing into PBS and 2 mM MgCl2 at 65°C for 30 minutes, transferring into 100 mM NaCl, 100 mM Tris-Cl (pH 9.5), 50 mM MgCl2, 0.1% Tween and 2 mM levamisole for 20 minutes, and developing in NBT/BCIP (Roche). Antibodies used were against skeletal muscle myosin heavy chain (MyHC; MF20 or A4.1025, DSHB), smooth muscle myosin (Myh11, Biomedical Technologies) (Wallace et al., 2005), GFP (also binds EYFP; Torrey Pines), β-galactosidase (Cappel), Pax3 (DSHB), Pax7 (DSHB), paired-class homeodomain-containing proteins, including Pax3/7 (DP312) (Davis et al., 2005), phospho-Histone H3 (H3P, Millipore), β tubulin (βtub, Covance) and neurofilament (2H3, DSHB). H2O2 treatment during staining was used to diminish pigment in older larvae, but phenylthiourea was generally not used owing to potential toxicity (Bohnsack et al., 2011; Li et al., 2012). Phenotype quantification is included as supplementary material Table S1.
Muscle morphometry
OSM and gastrointestinal tract smooth muscle length were measured in projected z-stacks. Smooth muscle was defined according to Myh11 immunofluorescence and expressed as a fraction of total zebrafish length (standard length, SL).
Fluorescent microsphere swallowing and metabolic assays
Fluoresbrite plain YG 1.0 μm microspheres (Polysciences) were fed to 5 dpf larvae incubated in sterile GZM media (Pham et al., 2008) as previously described (Farber et al., 2001). Swimming activity was assessed over 5 minutes. Total swimming distance was measured using the Manual Tracking ImageJ plug-in. Time active was expressed as percentage of ‘movement frames’ (adjoining frames that contain movement), relative to total movie frames. Quantitative real-time PCR (qRT-PCR) was conducted as previously described (Kanther et al., 2011) with primers referenced in supplementary material Table S4.
Kaede lineage tracing
Kaede mRNA was transcribed using mMessage Machine (Ambion) and injected into one-cell stage embryos. Embryos with high levels of widespread Kaedegreen were selected and mounted as previously described (Minchin and Rawls, 2011). Somite cells were marked by photoconverting Kaede from green (Kaedegreen) to red (Kaedered) with 405 nm light (Hatta et al., 2006). Photoconversion from two directions, and at different stages of development, was undertaken to diminish the chance of non-somite photoconversion confounding interpretation. First, somite photoconversion from dorsal at 3-5 somite stage (ss) definitively excluded lateral tissue, including lateral mesoderm (Fig. 2A), as determined by expression of lateral mesoderm genes (Dooley et al., 2005). Second, photoconversion of a discrete region of the anteriormost somites from a lateral angle at 14-18 ss excluded labelling of both neural crest and endoderm (supplementary material Fig. S3). Both photoconversion strategies also labelled overlying epidermis and neural tube (supplementary material Fig. S3). After photoconversion, embryos were checked to ensure adjacent somites and surrounding tissue (excluding overlying ectoderm) remained green. Photoconversion and imaging was undertaken on a Zeiss LSM5 Exciter confocal with a Zeiss W Plan-Apochromat 20×/1.0 DIC (UV) VIS-IR objective. Images were processed using ImageJ.
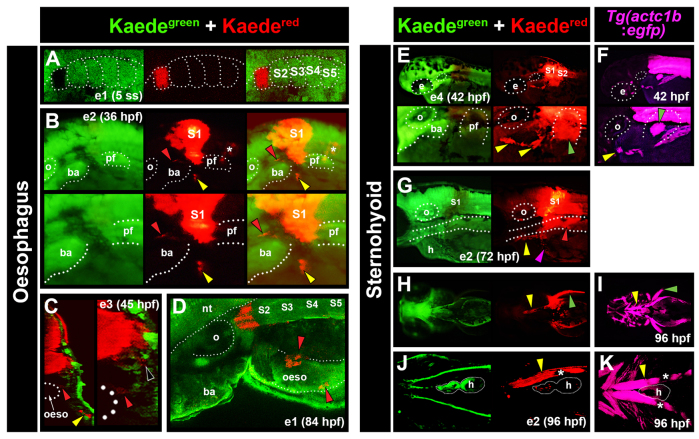
Fig. 2.
Lineage tracing of anterior somites in zebrafish marks OSM and SHM. Fluorescent images of primary green and photoconverted red Kaede (A-E,G,H,J). Images are from multiple embryos (e), photoconverted at either 3-5 ss (e1; A,D) or between 14 and 18 ss (e2-4; B,C,E,G,H,J). Tg(actc1b:egfp) labels muscle precursors (F,I,K). (A) Dorsal view confocal image showing photoconversion to Kaedered in S1 of e1 at 5 ss. (B) At 36 hpf, lateral view of e2 detected Kaedered in S1, and both anteriorly and medially migrating cells (yellow and red arrowheads, respectively). Marked cells were also observed associated with notochord (asterisks), and in neural tube as a result of photoconversion in overlying neural plate. (C) Cross-section confocal view of e3 at 45 hpf revealed medial cells localised to the oesophagus (oeso; red arrowheads; region magnified on right) and Kaedered in close proximity to the somite (black arrowhead). Anterior cells had intense label, reminiscent of SHM (yellow arrowhead). (D) e1 at 84 hpf. Dorso-lateral view confocal images showed Kaedered cells in close association with oesophagus. (E,F) Lateral view of e4 after photoconversion of S1+2 showing anteriorly located Kaedered (E, yellow arrowheads) in similar position to SHM primordium in Tg(actc1b:egfp) embryos (F, yellow arrowhead). (G) Lateral view of e2 reveals Kaedered in association with oesophagus (red arrowhead), SHM (yellow arrowhead) and common cardinal vein (magenta arrowhead) at 72 hpf. (H-K) Ventral confocal (J,K) and stereomicroscopic (H,I) views of e2 at 96 hpf showing Kaedered in left SHM (yellow arrowheads) and pectoral fin (green arrowheads), confirmed by Tg(actc1b:egfp). Melanophore pigment is located above SHM (J,K, asterisks). ba, branchial arches; e, eye; h, heart; nt, neural tube; oeso, oesophagus; o, otic vesicle; pf, pectoral fin.
Statistical analyses
Data were compared by one-way ANOVA followed by Tukey post hoc tests. In histograms, columns with distinct letters (A-C) differ significantly at the P-value shown in the legend. Columns with the same letter do not differ significantly (P>0.05). Error bars are s.e.m. Numbers of embryos are indicated on columns.
RESULTS
Pax3- and Pax7-expressing cells contribute to mouse SHM and OSM
To test the hypothesis that SHM and OSM derive from migratory somite cells, we genetically marked somite cells using Pax3/7 Cre-mediated lineage tracing in mouse. Pax3Cre labelled SHM, longus colli and many other muscles in the neck region (Fig. 1A; supplementary material Fig. S1). Cre driven from the Pax3 locus also labelled some, but not all, OSM cells (Fig. 1B,C). Similarly, treatment of Pax7CE mice with tamoxifen at E11.5-E12.5 marked OSM (Fig. 1D,F) and longus colli (supplementary material Fig. S2A-D). Thus, both Pax3- and Pax7-expressing cells contribute to OSM and SHM (supplementary material Table S5). The extent of labelling by Pax3Cre correlated with the level of origin of motor innervation (Fig. 1B,C; supplementary material Fig. S1). Muscles with innervation from C4-C6, such as the infraspinatus, deltoid and biceps were generally well labelled with alkaline phosphatase; like OSM, muscles innervated from C1-C3, such as longus colli, in the neck region of Pax3Cre;Z/AP mice, had mosaic labelling (supplementary material Fig. S1C-E; Fig. 1B,C). Nevertheless, longus colli was mostly absent in Pax3Cre/Cre mutants (Fig. 1I). Ventral to the oesophagus, SHM (innervated from C1-C3) were generally well labelled (Fig. 1A), whereas adjacent sternomastoid myofibres and also trapezius (innervated from the more anterior XIth cranial nerve) were entirely unlabelled (supplementary material Fig. S1B). By contrast, Wnt1Cre did not label OSM, SHM, longus colli or other neck muscles, indicating that neural crest did not contribute to OSM or other striated muscle (Fig. 1E,G; supplementary material Fig. S2A; data not shown). As Pax3 and Pax7 are only expressed in neural crest, non-migratory neuroepithelium and somites (Jostes et al., 1990; Daston et al., 1996; Borycki et al., 1999), these observations suggest that Pax3/7-expressing OSM, SHM and other neck muscle cells arise from the somite.
Pax7 expression in OSM precursors increases during foetal life
Temporal analysis of Pax7CE embryos suggested that Pax7 expression accumulates gradually in OSM precursors. Exposure to tamoxifen at successively later embryonic stages marked increasing numbers of OSM fibres at E16.5, particularly in the outer longitudinal layer, where OSM first arises (supplementary material Fig. S2). Tamoxifen administration at E9.5 or E10.5, marked only a limited number of cells in the outer muscle layer at E16.5, suggesting that, around E10.5, most OSM precursors do not express Pax7 (supplementary material Fig. S2A,B). Nevertheless, tamoxifen administration at E11.5 and onwards marked increasing numbers of OSM fibres in the outer muscle layer at E16.5, indicating that a significant fraction of OSM precursors express Pax7 at E12.5, prior to OSM differentiation (Fig. 1D,F; supplementary material Fig. S2C,D; Table S6). Neonatally, most OSM fibres were marked 3 days after tamoxifen administration, even in Pax7CE/CE mutants (Fig. 1H; supplementary material Fig. S2E). These findings suggest that Pax7 expression in OSM precursors increases during foetal life, becoming more uniform around birth, as occurs in myogenic cell populations in somite and limb.
Anterior somites contribute to zebrafish OSM
To ascertain which somite(s) contribute to OSM, we used Kaede lineage tracing in zebrafish. Single anterior somites were photoconverted to Kaedered at 3-5 ss (somite stage), or groups of somites were converted at 14-18 ss (Fig. 2A; supplementary material Fig. S3; Table 1). Consistent with Wood and Thorogood (Wood and Thorogood, 1994), early conversion of somite 1 (S1) marked, at 84 hpf, the anteriormost epaxial muscle fibres that extend from the posterior end of the otic vesicle to the cleithrum, having lost a morphological anterior somite border (Fig. 2D). S2 conversion marked the second somite, immediately behind the cleithrum (Table 1). Strikingly, both conversion strategies labelled Kaedered cells in two stereotypical clusters of migratory cells rostral to the marked somite between 36 and 45 hpf (Fig. 2B; Table 1). By 45 hpf, a medial subpopulation of rostrally migrating cells was associated with the oesophagus (Fig. 2C). Subsequently, robust Kaedered labelling was observed in the oesophagus at 84 hpf, suggesting somite cells migrate rostrally then medially to the oesophagus (Fig. 2D; Table 1). We conclude that anterior somite-derived cells migrate to the oesophagus and most likely correspond to OSM.
Table 1.
Kaede lineage tracing of anterior somites
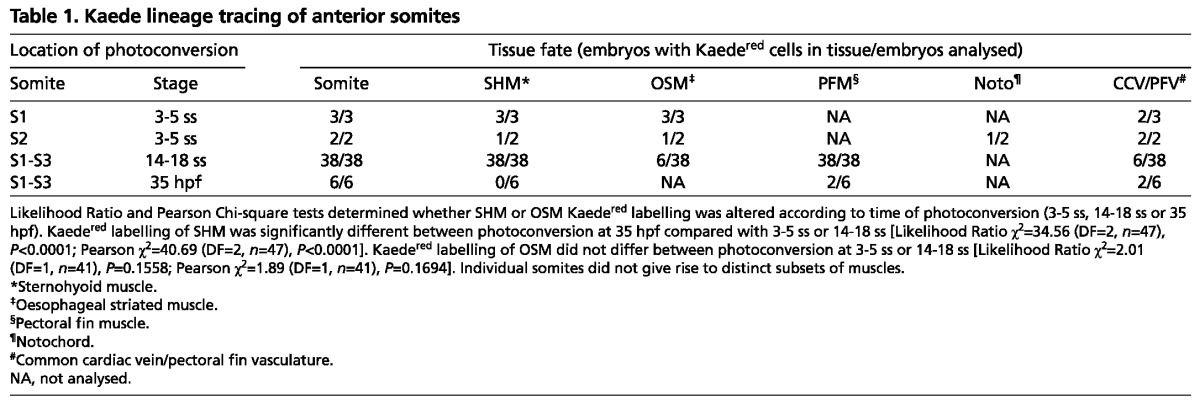
Zebrafish SHM derives from anterior somites
Kaedegreen photoconversion within S1 or S2 at 3-5 ss, or S1-S3 at 14-18 ss, yielded migrating mesenchymal Kaedered cells at 42 hpf that resemble SHM marked by an actin:GFP transgene (Fig. 2E,F; Table 1). Moreover, analysis of Kaedered cells at 72-96 hpf, revealed strong labelling of the terminally differentiated SHM fibres, demonstrating that SHM derives from S1-S3 (Fig. 2G,H,J; Table 1). Interestingly, S1 marked more anterior regions of SHM compared with S2, suggesting anterior-posterior positioning is retained in the migrating SHM primordium (data not shown). Later photoconversion of anterior somites failed to label SHM, indicating that emigration of SHM precursors occurs before 35 hpf (P<0.0001; Table 1). Pectoral fin was routinely labelled, consistent with a S2-S3 origin for pectoral fin muscle (PFM; Fig. 2E,H) (Neyt et al., 2000; Haines et al., 2004). Somite photoconversion resulted in Kaedered within blood vessels of the common cardinal vein (CCV) and pectoral fin vasculature (PFV; Fig. 2G; supplementary material Fig. S4B,C; Table 1). Finally, S1-derived marked cells were also associated with the surface of the notochord, but were always located posterior to the marked somite (Fig. 2B; Table 1), presumably reflecting sclerotome fate and posteriorward movement of the notochord (Wood and Thorogood, 1994). Taken together, our lineage tracing analyses identified both presumptive OSM and SHM as novel derivatives of somites in zebrafish.
MRF expression analysis reveals migration of somite-derived OSM precursors
Lineage tracing suggested that OSM precursors migrate rostrally from S1-S3 prior to 35 hpf before cascading medially onto the oesophagus (Fig. 2B; Table 1). To investigate the temporal and spatial development of zebrafish OSM, we analysed expression dynamics of genes important for striated myogenesis. At 30 hpf, myf5 was expressed in bilateral cell clusters immediately rostral to the anterior-most somites (Fig. 3A). These myf5+ striated muscle myoblasts were located within mesenchymal tissue, did not express other MRFs (data not shown) and appeared to move medially at 38 hpf (Fig. 3B,C). By 48 hpf, myf5+ myoblasts converged at the midline, at the level of S1-S3, forming a tube-like structure in the region of the developing oesophagus (Fig. 3D,G). Some myf5+ cells remained lateral at 48 hpf, thus forming a distinctive seagull shape immediately rostral to S1, and suggesting that migration towards the oesophagus is ongoing in an ‘inverse fountain’ (Fig. 3D,Q). Based on anatomical location during 30-48 hpf, we suggest these myf5+ cells are OSM precursors.
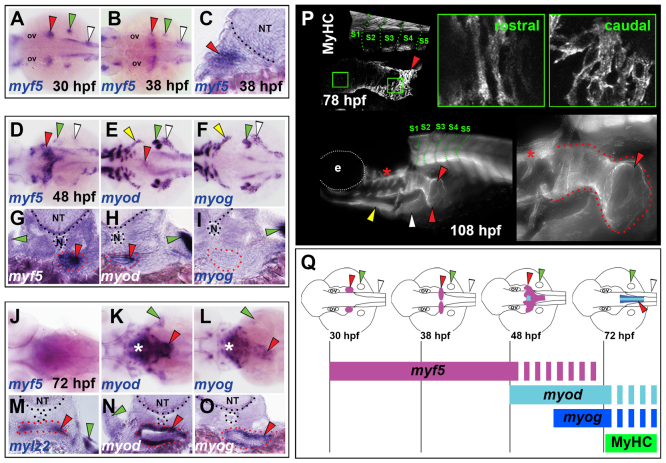
Fig. 3.
Developmental dynamics of OSM development in zebrafish. In situ hybridisation for myf5 (A-D,G,J), myod (E,H,K,N), myog (F,I,L,O) and mylz2 (M), or MyHC immunodetection (P). Images are either dorsal view wholemounts with anterior towards the left (A,B,D,F,H,J,K,M), transverse cryosections with dorsal towards the top (C,E,G,I,L,N,O) or lateral view wholemounts with anterior towards left and dorsal towards the top (P). (A-C) At 30-38 hpf, myf5 is expressed in somites (white arrowheads), pectoral fin (green arrowheads) and bilateral patches anterior to the pectoral fins (red arrowheads). (D-I) By 48 hpf, myf5 expression had coalesced at the midline, forming a distinctive chevron structure (red arrowheads) that had begun expression of myod but not myog, and was associated with oesophageal tissue (red dots). The anterior hypaxial muscle anlagen expressed both myod and myog (yellow arrowheads). (J-O) At 72 hpf, expression in OSM region of myf5 was greatly reduced, myod increased and myog appeared strongly. Asterisks indicate hypobranchial muscle in pharynx. At 72 hpf, weak oesophageal mylz2 expression commenced (red arrowhead). (P) MyHC was detected in OSM (red arrowheads) at 78 hpf and extended posteriorly to somite 4 (green dotted lines). By 108 hpf, OSM (red dotted lines) had a distinctive funnel shape still extending from above SHM (yellow arrowhead) and PHM (white arrowhead) only to somite 4. Asterisks indicate hypobranchial muscles in pharynx. (Q) OSM morphogenesis and temporal onset of MRF gene expression from a dorsal view. Arrowheads indicate OSM (red), SHM (yellow), pectoral fin muscle (green) and PHM (white). e, eye; N, notochord; NT, neural tube; ov, otic vesicle.
Expression of myod (myod1 - Zebrafish Information Network) first became evident in OSM precursors at the midline towards the end of the myf5+ fountain at 48 hpf, indicating maturation of OSM precursors in this region (Fig. 3E,H). myog mRNA was not detected in OSM at 48 hpf (Fig. 3F,I). By 72 hpf, levels of myf5 mRNA were significantly reduced (Fig. 3J). However, OSM now strongly expressed myod (Fig. 3K,N), myog (Fig. 3L,O) and mef2ca (data not shown), suggesting that differentiation of OSM was occurring. Indeed, weak expression of both slow (smyhc1) and fast (mylz2) myosin mRNA at 72 hpf suggested differentiated fibres were present in OSM (Fig. 3M; data not shown) (Elworthy et al., 2008). By 78 hpf, robust MyHC immunoreactivity was observed in the oesophagus, which extended caudally to the level of somite 4 (Fig. 3P). Fibres in rostral OSM had elongated morphology, whereas those in caudal OSM appeared less mature (Fig. 3P). To conclude, MRF expression analysis revealed a progression of OSM precursors, initially located in bilateral myf5+ clusters immediately rostral to anterior somites, which then appear to converge, express other MRFs and move posteriorly to localise at the oesophagus, where they undergo further differentiation (Fig. 3Q).
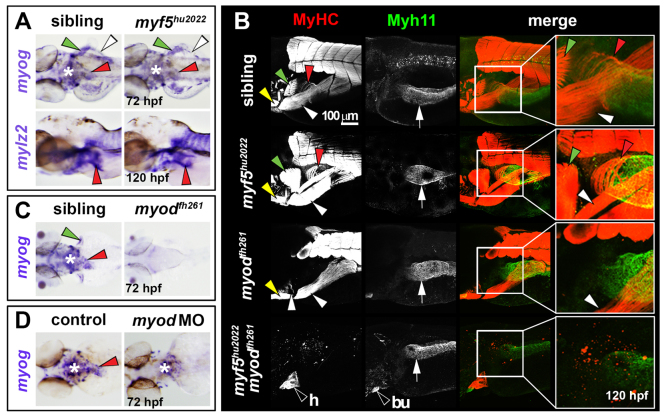
Zebrafish OSM development is dependent on Myod, but not on Myf5
MRFs, including myf5, myod and myog, are essential for striated myogenesis in both mouse and zebrafish (Rudnicki et al., 1993; Hinits et al., 2009; Hinits et al., 2011). In mouse, OSM development is Myf5/Mrf4 (Myf6) dependent, but Myod1 independent (Kablar et al., 2000). The zebrafish myf5hu2022 and myodfh261 mutants are predicted nulls (Hinits et al., 2009; Hinits et al., 2011). At 72 and 120 hpf, myf5hu2022 revealed no obvious defect in OSM myod, myog or myosin mRNA or MyHC protein accumulation (Fig. 4A,B; data not shown). However, in myodfh261 mutants and myod morphants at 72 hpf, myog mRNA within OSM precursors was dramatically reduced, and substantially reduced in other cranial and somite-derived migratory muscles (Fig. 4C,D). Therefore, unlike mouse, Myod is required for myog expression within OSM in zebrafish. At 5 dpf, MyHC accumulation is strongly reduced in OSM of both myodfh261 mutants and myod morphants, demonstrating that Myod is required for OSM differentiation (Fig. 4B; data not shown). Double myf5hu2022;myodfh261 mutants entirely lack OSM and all other skeletal muscle, although both striated cardiac and gut smooth muscle remain intact (Fig. 4B). Thus, in contrast to mouse, development of the OSM in zebrafish appears primarily Myod dependent, with some compensatory role of Myf5.
Fig. 4.
OSM is Myod dependent in zebrafish. In situ hybridisation for myog (A,C,D) or mylz2 (A), and immunodetection of MyHC (B) and Myh11 (B). Dorsal view wholemounts are oriented anterior towards the left (A, upper panels; C,D). Lateral view wholemounts are oriented anterior towards the left and dorsal towards the top (A, lower panels; B). (A) At 72 hpf, myog mRNA expression was indistinguishable from that in wild-type siblings in OSM of myf5hu2022 mutants (red arrowheads). PHM, PFM and pharynx muscle were also unchanged (white arrowheads, green arrowheads and asterisk, respectively). Myosin mRNA (mylz2) at 120 hpf was unaltered in myf5hu2022 mutants, suggesting OSM terminal differentiation occurs normally after reduction of Myf5 (red arrowheads). (B) OSM and smooth muscle differentiation was unaffected in myf5hu2022 mutants (red arrowheads and white arrows, respectively). In myodfh261 mutants, MyHC within OSM was absent, but SHM and PHM differentiated well (yellow and white arrowheads, respectively), as did oesophageal smooth muscle (white arrows). Double myf5hu2022;myodfh261 mutants had no striated skeletal muscle, but cardiac striated muscle (h) was readily observed, as was smooth muscle in the gastrointestinal tract and bulbus arteriosus (bu). (C,D) myog mRNA reduction in the oesophageal region of myodfh261 mutants (C) and myod morphants (D). Asterisk indicates pharyngeal muscle. Arrowheads indicate OSM (red), SHM (yellow), pectoral fin muscle (green) and PHM (white). White arrow indicates oesophageal smooth muscle.
Pax3 mouse mutants die prior to OSM formation
Little is known regarding genetic factors involved in OSM development prior to MRF expression. We hypothesised that development of somite-derived OSM is dependent on Pax3/7 genes. Mutation of Pax7 in mouse does not affect early OSM formation (Fig. 1H) (Wörl et al., 2009). At E14.5, Pax3Cre/Cre mutants fail to form many neck muscles (Fig. 1I). Oesophageal smooth muscle was present but disorganised in Pax3Cre/Cre mutants and no OSM was observed (Fig. 1I). However, as such mutants appear developmentally delayed prior to foetal death from heart defects, it was not possible to determine whether OSM was formed at the equivalent of E15.5, as observed in wild type.
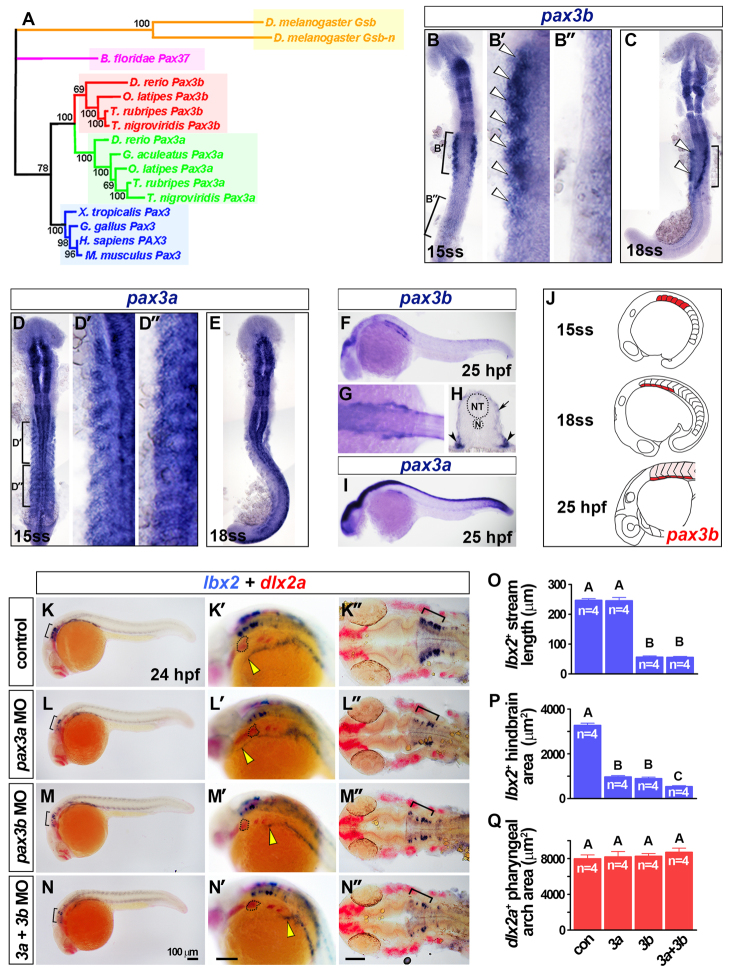
Somitic pax3b is restricted to the anteriormost somites in zebrafish
We turned to zebrafish to assess the role of pax3/7 in OSM. Duplicated teleost pax3 genes form discrete pax3a and pax3b clades (Fig. 5A; supplementary material Figs S5, S6). Strikingly, following early neural expression (supplementary material Fig. S7A-G), somitic pax3b mRNA was restricted to anterior somites at 15 ss, in contrast to more widespread pax3a somite expression (Fig. 5B,D). Furthermore, by 18 ss, pax3b mRNA became restricted to the ventrolateral region of anteriormost somites, the putative site of MMPs (Fig. 5C), and persisted in this region until 55 hpf (Fig. 5F-H; supplementary material Fig. S7H). pax3a mRNA was detected widely in somite, but persisted in the ventrolateral region containing MMPs at 25 hpf (Fig. 5D,E,I). However, neither pax7 mRNA was detected in MMPs (Minchin and Hughes, 2008). In conclusion, pax3b mRNA is restricted to regions containing MMPs, suggesting a function for pax3b in the development of MMPs.
Fig. 5.
Somitic pax3b is localised to the ventrolateral somite and required for MMP migration. (A) Phylogeny of Pax3 proteins using the Bayesian Monte Carlo Markov chain method with 10,000 bootstrap replicates (MrBayes v3.1.2) (Ronquist and Huelsenbeck, 2003). Tetrapod Pax3 genes are blue; teleost Pax3a and Pax3b clades are green and red, respectively; a cephalochordate Pax3/7 homolog is magenta; and orange indicates insect homologs. (B-N) In situ hybridisation for pax3b (B,C,F-H), pax3a (D,E,I), lbx2 and dlx2a (K-N′). Dorsal view flatmounts have anterior towards the top (B-E) or anterior towards the left (K′-N′). Lateral view wholemounts have anterior towards the left and dorsal towards the top (F,I,K-N′), dorsal view, wholemount has anterior towards the left (G). Transverse cryosection has dorsal towards the top (H). (B,C) Somitic pax3b mRNA was restricted to the anterior somites at 15 and 18 ss (white arrowheads). (D,E) Somitic pax3a was expressed in all somites at 15 and 18 ss. (F-I) At 25 hpf, somitic pax3b was localised to the ventrolateral region of anterior somites (arrowheads, H) and dermomyotome (arrow, H), as was pax3a (I). (J) Somitic pax3b expression dynamics (pink indicates dermomyotome at 25 hpf). (K-N′) Rostral extension of lbx2+ cells (yellow arrowheads) had reached the hyoid arch (dotted outline) at 24 hpf in controls, but was severely reduced in pax3b morphants and pax3a+pax3b double morphants. Scale bars: 100 μm. (O-Q) Rostral migration of lbx2+ cells at 24 hpf was significantly reduced (O; group A versus B; P<0.0001) as was the area of hindbrain lbx2+ expression (brackets in K-N and K′-N′) (P; P<0.0001; groups A-C differ significantly P<0.05). By contrast, the area of dlx2a expression in pharyngeal arches was unaltered after pax3 manipulation (Q; P=0.7282). Data are mean+s.e.m.
Pax3b is required for OSM formation in zebrafish
To investigate the role of the pax3 genes during OSM formation, previously verified MOs targeting pax3a were used to reduce specifically Pax3a protein (supplementary material Fig. S8) (Minchin and Hughes, 2008). Injection of a chimaeric 5′UTRpax3b:GFP RNA revealed strong efficacy of translation blocking by either of two non-overlapping pax3b MOs (supplementary material Fig. S9). In pax3a morphants, residual Pax3/7 immunoreactivity remained in the location of pax3b mRNA in rhombomere 4, and could be ablated by either pax3b MO (supplementary material Fig. S8H,H′; data not shown). Thus, we conclude that pax3b MOs reduce Pax3b protein.
In mouse, Pax3 is required for Lbx1 expression in MMPs, and both genes are essential for normal MMP migration into the limb (Mennerich et al., 1998; Gross et al., 2000). In zebrafish, lbx2 marks MMPs from 22 ss (Neyt et al., 2000; Ochi and Westerfield, 2009). At 24 hpf, a stream of lbx2+ cells projected rostrally from S1, and presumably corresponds to OSM and SHM progenitors (Fig. 5K). Comparison with dlx2a expression within cranial neural crest showed that the anteriormost lbx2+ cells had migrated to the hyoid arch in controls (Fig. 5K), but were substantially reduced and had progressed less far anteriorly in pax3b morphants (Fig. 5M,O-Q). Injection of pax3a MO alone had little effect on early migrating lbx2+ myoblasts and double pax3a+pax3b morphants had a similar phenotype to pax3b single morphants (Fig. 5L-Q). Therefore, Pax3b is required for the rostral projection of lbx2+ presumptive MMPs.
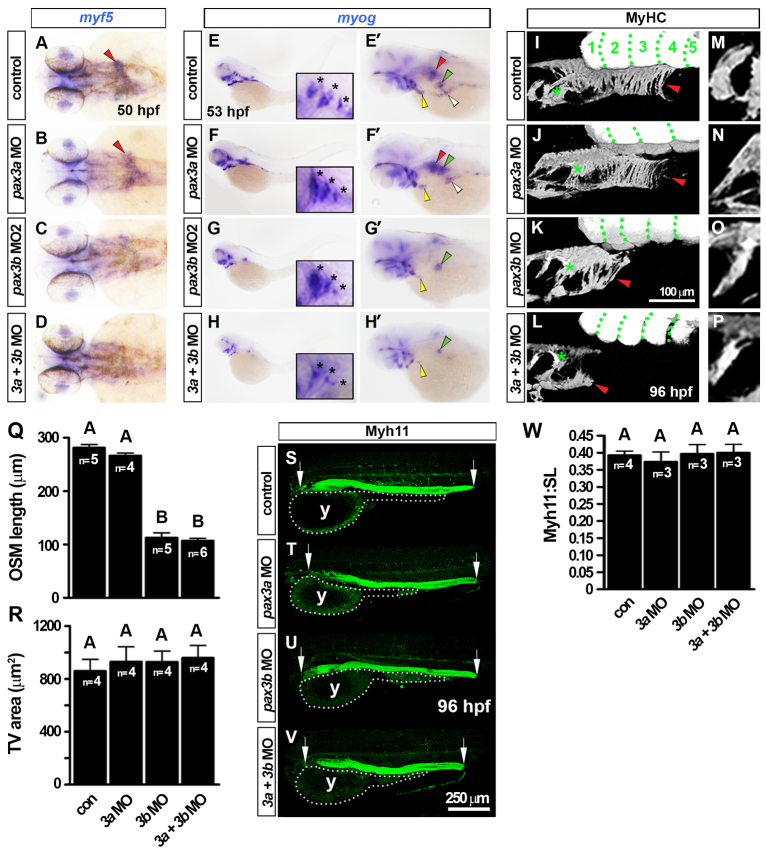
Knockdown of Pax3b also resulted in a severe reduction in MRF mRNAs in OSM myoblasts around 50 hpf (Fig. 6A-H′; supplementary material Fig. S8J) and significantly reduced OSM at 96 hpf (Fig. 6I-L,R). Depletion of Pax3a alone did not affect OSM specification or differentiation, whereas pax3a+pax3b double morphants showed disruption of OSM differentiation similar to single pax3b morphants (Fig. 6). This suggests that reduced MMP migration in pax3b morphants leads to defective OSM differentiation. Other myogenic processes in pax3b morphants appeared unperturbed. These included early somitic myogenesis (supplementary material Fig. S11A-F), presence of Pax3/7+ cells within the myotome at 120 hpf (supplementary material Fig. S11G-W) and proliferation of Pax3/7+ dermomyotome cells at 53 hpf (supplementary material Fig. S11X,Y). Thus, knockdown of Pax3b preferentially affects myogenesis in muscle near to, but outside of, rostral somites.
Fig. 6.
pax3b morphants have reduced OSM but unaffected development of gastrointestinal smooth muscle. In situ hybridisation for myf5 (A-D) and myog (E-H′) and immunodetection of MyHC (I-P) and Myh11 (S-V). Wholemounts have anterior towards the left in dorsal (A-D), lateral (E-H, I-P,S-V; dorsal to top) or dorsal oblique views (E′-H′). (A-D) Loss of myf5 mRNA from the ‘inverse fountain’ OSM region in pax3b morphants (red arrowheads). (E-H′) Reduction in myog mRNA in pax3b morphant OSM (red arrowheads), SHM (yellow arrowheads), PHM (white arrowheads) and PFM (green arrowheads). In pax3a morphants, myog expression in all muscles was indistinguishable from controls. Insets and asterisks show comparable opercular muscle developmental in pax3b morphants and controls. (I-L) At 96 hpf, MyHC had accumulated in control and pax3a morphant OSM (red arrowheads), extending caudally to somite 3-4. Green dots delineate somites. pax3b and pax3b+pax3a morphants had severely reduced OSM (red arrowheads). (M-Q) Shape, orientation and area of tranversus ventralis (TV) muscles were unaffected after pax3 manipulation. (R) OSM length was measured from the caudal-most hypobranchial muscle in the pharynx (asterisks in I-L) to the caudal edge of OSM (red arrowheads in I-L). A and B groups differ significantly (P<0.0001). (S-V) Myh11 was unchanged after pax3 manipulation. Arrows indicate anterior and posterior extent for Myh11 length measurements. (W) Fraction of larvae standard length (SL) occupied by Myh11+ was not altered after pax3 manipulation. y, yolk. Data are mean+s.e.m.
pax3b morphants have unaltered smooth muscle differentiation
It has been suggested that OSM arises from smooth muscle (Patapoutian et al., 1995). We asked whether reduction of OSM in pax3b morphants led to increased oesophageal smooth muscle. Oesophageal smooth muscle, as assayed by either transgelin mRNA at 72 hpf (supplementary material Fig. S8I) or Myh11 accumulation in the oesophagus at 96 hpf (Fig. 6S-V), was not overtly affected by pax3 knockdown. Furthermore, the length of the gastrointestinal tract smooth muscle was not increased, showing that smooth muscle is not augmented in the absence of OSM (Fig. 6W). Thus, Pax3b depletion does not influence oesophageal smooth muscle formation.
Pax3b promotes zebrafish sternohyoid myogenesis
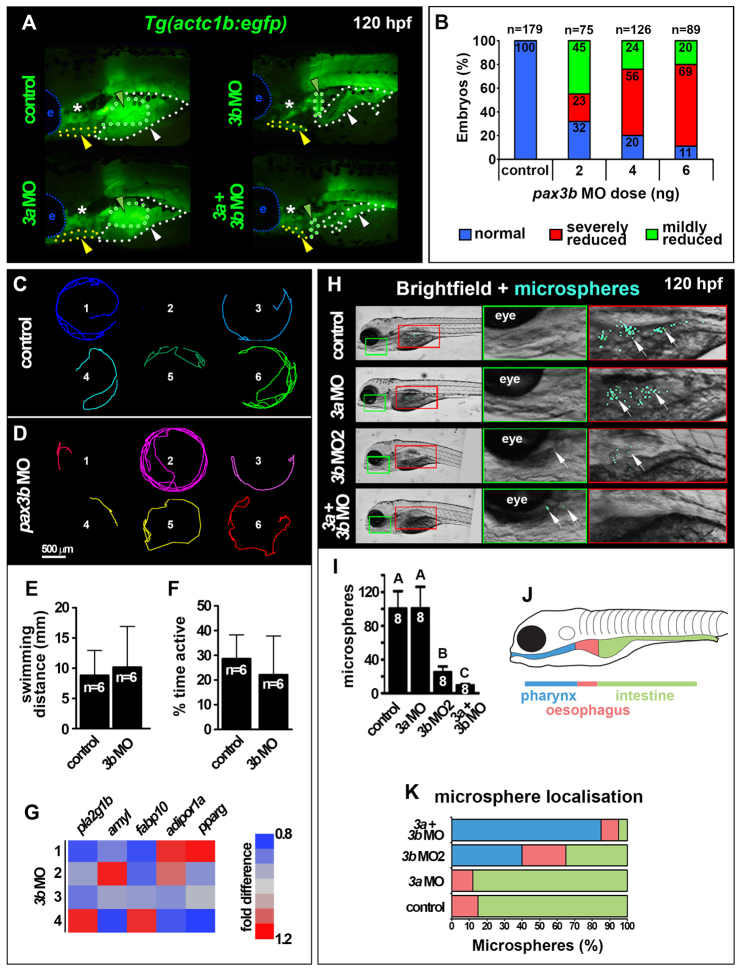
Our lineage analysis identified SHM as an anterior somite derivative (Fig. 2E,G,H,J). Knockdown of Pax3b, but not of Pax3a, caused a significant reduction of myog mRNA in all migratory hypaxial muscles at 53 hpf, including SHM, PFM and posterior hypaxial muscle (PHM, Fig. 6G). We next used the Tg(actc1b:egfp) transgenic line to examine the consequence of Pax3b knockdown in hypaxial myogenesis. At 120 hpf, terminally differentiated hypaxial muscle was dose-dependently reduced and disorganised upon injection of pax3b MO (Fig. 7A,B). By contrast, pax3a morphants did not exhibit a severe hypaxial muscle phenotype and pax3a+pax3b double morphants were similar to pax3b single morphants (Fig. 7A). Thus, pax3b appears the primary Pax3 required for efficient hypaxial muscle differentiation in zebrafish. Curiously, formation of the common cardinal vein, which also received cells from anterior somites, was delayed in pax3b, but not in pax3a, morphants at 30 hpf (supplementary material Fig. S12). Knockdown of both Pax7a and Pax7b proteins by MO injection, which ablated Pax7 immunoreactivity (supplementary material Fig. S13A), had no discernible effect either upon MRF expression in MMPs or the later differentiation of OSM or SHM (supplementary material Fig. S13B-D). Together, these data demonstrate that Pax3b is required for migration of normal hypaxial muscle precursors.
Fig. 7.
pax3b morphants are metabolically normal but exhibit reduced swallowing capability. All images are lateral view wholemounts with anterior towards the left and dorsal towards the top. (A) Somite-derived hypaxial muscles of 120 hpf Tg(actc1b:egfp) larvae showing PFM (green dots and arrowheads), PHM (white dots and arrowheads) and SHM (yellow dots and arrowheads) muscle reduction and disorganization in pax3b and double morphants. e, eye; asterisk, cranial muscles. (B) Dose dependence of Tg(actc1b:egfp) hypaxial muscle defect in pax3b morphants. (C,D) Migration tracks over 5 minutes of six control (C) and pax3b MO 120 hpf larvae (D). (E,F) Distance swum (E, P=0.87) and time active (F, P=0.73) in a 5-minute period were not significantly different between control and pax3b morphants (unpaired t-tests). Data are mean+s.e.m. (G) qRT-PCR of metabolic marker mRNAs showed no significant difference (pairwise t-tests) between control and pax3b morphants at 120 hpf. Values from four biological replicates (rows 1-4, 10 larvae/sample normalized to 18S rRNA). (H) Fluorescent microspheres fed to 120 hpf larvae were swallowed and detected in the GI tract (arrows). (I) Number of swallowed microspheres in controls and pax morphants. Groups A, B and C differ significantly (P<0.0001). Data are mean+s.e.m. (J,K) Colour-coded regions of GI tract (J) in which microspheres were distributed in control and morphants (K).
pax3b morphants are metabolically normal but have reduced ability to internalise and swallow microspheres
To test whether Pax3b-dependent migratory muscles are required for effective feeding, we assessed internalisation of fluorescent microspheres (Farber et al., 2001). At 5 dpf, pax3b morphants exhibited equivalent levels of physical activity in comparison with controls (Fig. 7C-F), and unchanged accumulation of mRNAs associated with lipid and carbohydrate metabolism (Fig. 7G). Thus, pax3b morphants appear physiologically normal at 5 dpf. Strikingly, however, pax3b single and pax3a+pax3b double morphants showed significant reduction in microsphere internalisation, indicating that ingestion is impaired after Pax3b knockdown (Fig. 7H,I). In control larvae and pax3a morphants, microspheres were mainly in the intestine, showing that internalised microspheres quickly pass through the oesophagus (Fig. 7J,K). In pax3b or double morphants a large increase in pharynx-localised microspheres was evident, suggesting that reduced OSM leads to defective oesophageal swallowing (Fig. 7J,K). pax3b morphants did not consistently show defects in neural crest (supplementary material Fig. S14), head or pharyngeal arch development (Fig. 5K′-N′,Q; Fig. 6), suggesting that defects in feeding behaviour of pax3b morphants were not related to neural crest problems. Clearly, zebrafish Pax3b is required for effective microsphere internalisation and swallowing.
DISCUSSION
The findings in this study demonstrate four major points. First, cells contributing to OSM and SHM come from somites in both zebrafish and mouse. Second, OSM formation is Myod dependent in zebrafish. Third, normal formation of OSM and SHM require Pax3b function. Fourth, loss of Pax3 function leads to defective ingestion and swallowing. As human PAX3 is abundantly expressed in oesophagus (Tsukamoto et al., 1994), and Waardenburg Syndrome, which is caused by mutations in PAX3 (Goodman et al., 1982), can be associated with oesophageal abnormalities (Nutman et al., 1986), our findings raise the possibility that PAX3 may regulate oesophageal myogenesis in humans.
The ontogeny of OSM: from anterior somites to oesophagus
Our work addresses the long-standing issue of the origin of OSM. In mouse, Pax3 and Pax7-expressing cells contribute to OSM, suggesting a somitic origin. Kaede lineage tracing in zebrafish allowed us to gain a more precise view of this event: bilateral streams of lbx2+ OSM and SHM precursors emanate from pax3a/b+ hypaxial regions of the most anterior somites (S1-3) and migrate rostrally along a shared path. These streams bifurcate as myf5 expression commences and OSM precursors then move medially towards the oesophagus in an ‘inverse fountain’ where they undergo striated muscle differentiation (supplementary material Fig. S15). As myosin is not detected in OSM until after 72 hpf, cell migration appears to occur via MMP intermediates. In mouse, Pax3 is expressed in presomitic mesoderm, dermomyotome and during MMP migration (Relaix et al., 2004; Relaix et al., 2005; Horst et al., 2006). By contrast, Pax7 only becomes evident in limb MMPs at ∼E12, after migration (Relaix et al., 2004; Relaix et al., 2005). We propose that mouse OSM progenitors undergo canonical MMP development, expressing Pax3 within the somite and during migration, with some expressing Pax7 at the site of differentiation. Zebrafish in situ hybridisation did not detect mesodermal pax3b mRNA outside the somite. Furthermore, mouse Pax3 regulates MMP emanation from somites (Epstein et al., 1996; Dietrich, 1999). Therefore, we speculate that Pax3 functions within somites at early stages of OSM development.
It remains possible that OSM has multiple developmental origins. Our lineage analyses marked small numbers of cells in zebrafish oesophagus, suggesting either additional, non-somitic, origins for OSM or limited tracer detection. Several factors conspire to limit detectability of the Kaede lineage tracer: unilateral photoconversion, MMP proliferation, rapid increase in cell volume after muscle terminal differentiation, fusion with unmarked cells, and the flattened monolayer topology and deep location of OSM. Despite these limitations, genetic lineage tracing in mice produced a similar picture: only some OSM fibres were marked by Pax3-driven Cre. Therefore, our data suggest multiple developmental origins for OSM. The residual presence of OSM in Pax3Sp mice (Kablar et al., 2000) and after pax3b knockdown in zebrafish supports this view. Other potential non-somitic sources of OSM include lateral cranial mesoderm, as loss of tbx1 function also reduces larval oesophageal muscle (Piotrowski and Nüsslein-Volhard, 2000). Perhaps OSM forms from multiple cell populations, as has been suggested for limb muscle (van Swearingen and Lance-Jones, 1993). We find no evidence for smooth muscle transdifferentiation into OSM. First, Pax3Cre did not mark oesophageal smooth muscle. Second, reduced OSM in Pax3b-deficient zebrafish was not accompanied by noticeably increased smooth muscle, although our methods may not detect small increases in smooth muscle. Thus, our data support existing evidence against smooth muscle transdifferentiation into OSM (Zhao and Dhoot, 2000; Rishniw et al., 2003; Rishniw et al., 2009).
Genetic regulation of OSM
Our study demonstrates a role for pax3b, uniquely among pax3/7 genes, during initial development of zebrafish MMPs, SHM and OSM. Pax3Cre/Cre and Pax3Sp mutants die prior to OSM formation with severe neck muscle defects (Tajbakhsh et al., 1997). Pax3Sp mutants on a non-C57BL background survive longer, however, and have some OSM (Kablar et al., 2000). Interestingly, an 80% reduction of Pax3 in Pax3neo/neo mice leads to defective tongue but functional diaphragm muscle, suggesting that loss of Pax3 has stronger effects on specific MMP derivatives (Zhou et al., 2008). Pax3a appears to have little role in OSM development, in contrast to its essential role in neural crest (Minchin and Hughes, 2008). However, pax3a+pax3b double morphants had a more severe reduction of hindbrain lbx2 mRNA, increased H3P+ cells within the neural tube, developmental retardation and more severely reduced microsphere ingestion than pax3b single morphants. Thus, knockdown of both Pax3a and Pax3b elicits unique phenotypes that warrant further investigation.
In zebrafish, some somite-derived muscles are myod dependent (Hinits et al., 2009; Hinits et al., 2011). In accordance, zebrafish OSM is also myod dependent. In mice, OSM is Myf5;Mrf4-dependent but Myod independent (Kablar et al., 2000). It is currently unclear whether double myf5;mrf4 mutant zebrafish have OSM defects. However, the diminished role for Myod in murine OSM development probably reflects shifting MRF use during vertebrate evolution, as discussed elsewhere (Hinits et al., 2009).
Anterior somite origin for SHM
Our lineage analysis demonstrates an anterior somite origin for zebrafish SHM, thus confirming suppositions based on gene expression (Schilling and Kimmel, 1997; Lin et al., 2006). Zebrafish SHM structure retains vestiges of its multi-segmental origin: two separate compartments have aligned fibre ends reminiscent of myotome segment borders (Hinits et al., 2011). No other cranial muscles in zebrafish had a somitic contribution or depended on Pax3. By contrast, Pax3Cre lineage tracing in mouse indicates a somitic contribution to many neck muscles, including SHM and longus colli muscles which have similar innervation, flank the oesophagus and are mostly absent in Pax3 mutants. Other neck muscles, such as sternomastoid and trapezius, are not marked by Pax3Cre. Although fish do not have necks, SHM and OSM are the only larval muscles that can be homologised with the more complex neck region of mammals (Diogo et al., 2008). We speculate that the evolutionary descendant muscles of SHM in primitive fish are defective after Pax3 knockout in mouse.
Like MRFs, Pax3/7 function was differentially partitioned in early amniotes (Franz et al., 1993; Monsoro-Burq et al., 2005; Sato et al., 2005; Basch et al., 2006). As OSM and SHM occur in chondrichthyes, but not agnathans (Kusakabe et al., 2004; Chatchavalvanich et al., 2006), it seems that in early bony fish the Pax3/7 and MRF gene families retained substantial evolutionary flexibility, which may have allowed migratory hypaxial muscle to co-evolve with the vertebrate jaw. We speculate that evolution of the jaw occurred concomitant with somite production of OSM/SHM, facilitating voluntary control of swallowing.
Supplementary Material
Acknowledgments
We thank D. Stemple and C. Moens for mutant fish, J. Epstein and M. Logan for mouse lines, and F. Conlon for useful discussions.
Footnotes
Funding
J.E.N.M. was supported by an Medical Research Council (MRC) PhD studentship and an American Heart Association Postdoctoral Fellowship [11POST7360004]. C.-M.F. and S.L. were supported by a National Institutes of Health grant [AR060042] and the Carnegie Endowment. J.F.R. was supported by a Pew Scholars in the Biomedical Sciences Award. S.M.H. is a member of MRC External Scientific Staff with Programme Grant [G1001029] support. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
J.E.N.M. and S.M.H. designed the study. J.E.N.M. performed the fish experiments with help from Y.H., S.M.H. and P.T. V.C.W. performed most mouse experiments. S.L. and C.-M.F. did the tamixofen and Wnt1Cre experiments. S.M.H., C.-M.F. and J.F.R. obtained the funding. J.E.N.M. and S.M.H. wrote the manuscript with input from all authors.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.090050/-/DC1
References
- Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C., Williams V. C., Moyon B., Daubas P., Tajbakhsh S., Buckingham M. E., Shiroishi T., Hughes S. M., Borycki A. G. (2012). Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 26, 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch M. L., Bronner-Fraser M., García-Castro M. I. (2006). Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218–222 [DOI] [PubMed] [Google Scholar]
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. (1995). Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768–771 [DOI] [PubMed] [Google Scholar]
- Bober E., Franz T., Arnold H. H., Gruss P., Tremblay P. (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120, 603–612 [DOI] [PubMed] [Google Scholar]
- Bohnsack B. L., Gallina D., Kahana A. (2011). Phenothiourea sensitizes zebrafish cranial neural crest and extraocular muscle development to changes in retinoic acid and IGF signaling. PLoS ONE 6, e22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki A. G., Li J., Jin F., Emerson C. P., Epstein J. A. (1999). Pax3 functions in cell survival and in pax7 regulation. Development 126, 1665–1674 [DOI] [PubMed] [Google Scholar]
- Brohmann H., Jagla K., Birchmeier C. (2000). The role of Lbx1 in migration of muscle precursor cells. Development 127, 437–445 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Relaix F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 23, 645–673 [DOI] [PubMed] [Google Scholar]
- Carrassón M., Grau A., Dopazo L. R., Crespo S. (2006). A histological, histochemical and ultrastructural study of the digestive tract of Dentex dentex (Pisces, Sparidae). Histol. Histopathol. 21, 579–593 [DOI] [PubMed] [Google Scholar]
- Chatchavalvanich K., Marcos R., Poonpirom J., Thongpan A., Rocha E. (2006). Histology of the digestive tract of the freshwater stingray Himantura signifer Compagno and Roberts, 1982 (Elasmobranchii, Dasyatidae). Anat. Embryol. (Berl.) 211, 507–518 [DOI] [PubMed] [Google Scholar]
- Coutelle O., Blagden C. S., Hampson R., Halai C., Rigby P. W., Hughes S. M. (2001). Hedgehog signalling is required for maintenance of myf5 and myoD expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev. Biol. 236, 136–150 [DOI] [PubMed] [Google Scholar]
- Cubbage C. C., Mabee P. M. (1996). Development of the cranium and paired fins in the zebrafish, Danio rerio (Ostariophysi, Cyprinidae). J. Morphol. 229, 121–160 [DOI] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 [DOI] [PubMed] [Google Scholar]
- Daston G., Lamar E., Olivier M., Goulding M. (1996). Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122, 1017–1027 [DOI] [PubMed] [Google Scholar]
- Davis G. K., D’Alessio J. A., Patel N. H. (2005). Pax3/7 genes reveal conservation and divergence in the arthropod segmentation hierarchy. Dev. Biol. 285, 169–184 [DOI] [PubMed] [Google Scholar]
- Devoto S. H., Stoiber W., Hammond C. L., Steinbacher P., Haslett J. R., Barresi M. J., Patterson S. E., Adiarte E. G., Hughes S. M. (2006). Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol. Dev. 8, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich S. (1999). Regulation of hypaxial muscle development. Cell Tissue Res. 296, 175–182 [DOI] [PubMed] [Google Scholar]
- Dietrich S., Schubert F. R., Healy C., Sharpe P. T., Lumsden A. (1998). Specification of the hypaxial musculature. Development 125, 2235–2249 [DOI] [PubMed] [Google Scholar]
- Diogo R., Hinits Y., Hughes S. M. (2008). Development of mandibular, hyoid and hypobranchial muscles in the zebrafish: homologies and evolution of these muscles within bony fishes and tetrapods. BMC Dev. Biol. 8, 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeneghini C., Arrighi S., Radaelli G., Bosi G., Mascarello F. (1999). Morphological and histochemical peculiarities of the gut in the white sturgeon, Acipenser transmontanus. Eur. J. Histochem. 43, 135–145 [PubMed] [Google Scholar]
- Dooley K. A., Davidson A. J., Zon L. I. (2005). Zebrafish scl functions independently in hematopoietic and endothelial development. Dev. Biol. 277, 522–536 [DOI] [PubMed] [Google Scholar]
- Elworthy S., Hargrave M., Knight R., Mebus K., Ingham P. W. (2008). Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115–2126 [DOI] [PubMed] [Google Scholar]
- Engleka K. A., Gitler A. D., Zhang M., Zhou D. D., High F. A., Epstein J. A. (2005). Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396–406 [DOI] [PubMed] [Google Scholar]
- Epstein J. A., Shapiro D. N., Cheng J., Lam P. Y., Maas R. L. (1996). Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. USA 93, 4213–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S. A., Pack M., Ho S. Y., Johnson I. D., Wagner D. S., Dosch R., Mullins M. C., Hendrickson H. S., Hendrickson E. K., Halpern M. E. (2001). Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science 292, 1385–1388 [DOI] [PubMed] [Google Scholar]
- Franz T., Kothary R., Surani M. A., Halata Z., Grim M. (1993). The Splotch mutation interferes with muscle development in the limbs. Anat. Embryol. (Berl.) 187, 153–160 [DOI] [PubMed] [Google Scholar]
- García Hernández M. P., Lozano M. T., Elbal M. T., Agulleiro B. (2001). Development of the digestive tract of sea bass (Dicentrarchus labrax L). Light and electron microscopic studies. Anat. Embryol. (Berl.) 204, 39–57 [DOI] [PubMed] [Google Scholar]
- Goodman R. M., Lewithal I., Solomon A., Klein D. (1982). Upper limb involvement in the Klein-Waardenburg syndrome. Am. J. Med. Genet. 11, 425–433 [DOI] [PubMed] [Google Scholar]
- Gross M. K., Moran-Rivard L., Velasquez T., Nakatsu M. N., Jagla K., Goulding M. (2000). Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127, 413–424 [DOI] [PubMed] [Google Scholar]
- Groves J. A., Hammond C. L., Hughes S. M. (2005). Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development 132, 4211–4222 [DOI] [PubMed] [Google Scholar]
- Haines L., Neyt C., Gautier P., Keenan D. G., Bryson-Richardson R. J., Hollway G. E., Cole N. J., Currie P. D. (2004). Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development 131, 4857–4869 [DOI] [PubMed] [Google Scholar]
- Hama K., Provost E., Baranowski T. C., Rubinstein A. L., Anderson J. L., Leach S. D., Farber S. A. (2009). In vivo imaging of zebrafish digestive organ function using multiple quenched fluorescent reporters. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G445–G453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C. L., Hinits Y., Osborn D. P., Minchin J. E., Tettamanti G., Hughes S. M. (2007). Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 302, 504–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K., Tsujii H., Omura T. (2006). Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 1, 960–967 [DOI] [PubMed] [Google Scholar]
- Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G. (1997). High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 192, 289–299 [DOI] [PubMed] [Google Scholar]
- Hinits Y., Osborn D. P., Hughes S. M. (2009). Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development 136, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y., Williams V. C., Sweetman D., Donn T. M., Ma T. P., Moens C. B., Hughes S. M. (2011). Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish. Dev. Biol. 358, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst D., Ustanina S., Sergi C., Mikuz G., Juergens H., Braun T., Vorobyov E. (2006). Comparative expression analysis of Pax3 and Pax7 during mouse myogenesis. Int. J. Dev. Biol. 50, 47–54 [DOI] [PubMed] [Google Scholar]
- Imrie D., Sadler K. C. (2010). White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev. Dyn. 239, 3013–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. W., Beis D., Mitchell T., Chen J. N., Stainier D. Y. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 [DOI] [PubMed] [Google Scholar]
- Jostes B., Walther C., Gruss P. (1990). The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech. Dev. 33, 27–37 [DOI] [PubMed] [Google Scholar]
- Ju B., Chong S.-W., He J., Wang X., Xu Y., Wan H., Tong Y., Yan T., Korzh V., Gong Z. (2003). Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter. Dev. Dyn. 227, 14–26 [DOI] [PubMed] [Google Scholar]
- Kablar B., Tajbakhsh S., Rudnicki M. A. (2000). Transdifferentiation of esophageal smooth to skeletal muscle is myogenic bHLH factor-dependent. Development 127, 1627–1639 [DOI] [PubMed] [Google Scholar]
- Kanther M., Sun X., Mühlbauer M., Mackey L. C., Flynn E. J., 3rd, Bagnat M., Jobin C., Rawls J. F. (2011). Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology 141, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori Y., Cho B. H., Song C. H., Fujimiya M., Murakami G., Kawase T. (2010). Smooth-to-striated muscle transition in human esophagus: an immunohistochemical study using fetal and adult materials. Ann. Anat. 192, 33–41 [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Schmid B., Eisen J. S. (2000). Genetic analysis of melanophore development in zebrafish embryos. Dev. Biol. 225, 277–293 [DOI] [PubMed] [Google Scholar]
- Kioussi C., Mamalaki A., Jessen K., Mirsky R., Hersh L. B., Matsas R. (1995). Expression of endopeptidase-24.11 (common acute lymphoblastic leukaemia antigen CD10) in the sciatic nerve of the adult rat after lesion and during regeneration. Eur. J. Neurosci. 7, 951–961 [DOI] [PubMed] [Google Scholar]
- Konow N., Thexton A., Crompton A. W., German R. Z. (2010). Regional differences in length change and electromyographic heterogeneity in sternohyoid muscle during infant mammalian swallowing. J. Appl. Physiol. 109, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe R., Takechi M., Tochinai S., Kuratani S. (2004). Lamprey contractile protein genes mark different populations of skeletal muscles during development. J. Exp. Zool. B 302, 121–133 [DOI] [PubMed] [Google Scholar]
- Lepper C., Conway S. J., Fan C. M. (2009). Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ptak D., Zhang L., Walls E. K., Zhong W., Leung Y. F. (2012). Phenylthiourea specifically reduces zebrafish eye size. PLoS ONE 7, e40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Y., Yung R. F., Lee H. C., Chen W. T., Chen Y. H., Tsai H. J. (2006). Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev. Biol. 299, 594–608 [DOI] [PubMed] [Google Scholar]
- Lobe C. G., Koop K. E., Kreppner W., Lomeli H., Gertsenstein M., Nagy A. (1999). Z/AP, a double reporter for cre-mediated recombination. Dev. Biol. 208, 281–292 [DOI] [PubMed] [Google Scholar]
- Maczkowiak F., Matéos S., Wang E., Roche D., Harland R., Monsoro-Burq A. H. (2010). The Pax3 and Pax7 paralogs cooperate in neural and neural crest patterning using distinct molecular mechanisms, in Xenopus laevis embryos. Dev. Biol. 340, 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennerich D., Schäfer K., Braun T. (1998). Pax-3 is necessary but not sufficient for lbx1 expression in myogenic precursor cells of the limb. Mech. Dev. 73, 147–158 [DOI] [PubMed] [Google Scholar]
- Minchin J. E., Hughes S. M. (2008). Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 317, 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin J. E., Rawls J. F. (2011). In vivo analysis of white adipose tissue in zebrafish. Methods Cell Biol. 105, 63–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Wang E., Harland R. (2005). Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell 8, 167–178 [DOI] [PubMed] [Google Scholar]
- Neyt C., Jagla K., Thisse C., Thisse B., Haines L., Currie P. D. (2000). Evolutionary origins of vertebrate appendicular muscle. Nature 408, 82–86 [DOI] [PubMed] [Google Scholar]
- Nishio S., Gibert Y., Bernard L., Brunet F., Triqueneaux G., Laudet V. (2008). Adiponectin and adiponectin receptor genes are coexpressed during zebrafish embryogenesis and regulated by food deprivation. Dev. Dyn. 237, 1682–1690 [DOI] [PubMed] [Google Scholar]
- Noden D. M. (1983). The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am. J. Anat. 168, 257–276 [DOI] [PubMed] [Google Scholar]
- Nutman J., Steinherz R., Sivan Y., Goodman R. M. (1986). Possible Waardenburg syndrome with gastrointestinal anomalies. J. Med. Genet. 23, 175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi H., Westerfield M. (2009). Lbx2 regulates formation of myofibrils. BMC Dev. Biol. 9, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patapoutian A., Wold B. J., Wagner R. A. (1995). Evidence for developmentally programmed transdifferentiation in mouse esophageal muscle. Science 270, 1818–1821 [DOI] [PubMed] [Google Scholar]
- Perry L. (2001). Dysphagia: the management and detection of a disabling problem. Br. J. Nurs. 10, 837–844 [DOI] [PubMed] [Google Scholar]
- Pham L. N., Kanther M., Semova I., Rawls J. F. (2008). Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3, 1862–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T., Nüsslein-Volhard C. (2000). The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio). Dev. Biol. 225, 339–356 [DOI] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2004). Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 18, 1088–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953 [DOI] [PubMed] [Google Scholar]
- Rishniw M., Xin H. B., Deng K. Y., Kotlikoff M. I. (2003). Skeletal myogenesis in the mouse esophagus does not occur through transdifferentiation. Genesis 36, 81–82 [DOI] [PubMed] [Google Scholar]
- Rishniw M., Fisher P. J., Doran R. M., Bliss S. P., Kotlikoff M. I. (2009). Striated myogenesis and peristalsis in the fetal murine esophagus occur without cell migration or interstitial cells of Cajal. Cells Tissues Organs 189, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007). p53 activation by knockdown technologies. PLoS Genet. 3, e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993). MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- Sato T., Sasai N., Sasai Y. (2005). Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 132, 2355–2363 [DOI] [PubMed] [Google Scholar]
- Schilling T. F., Kimmel C. B. (1997). Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 124, 2945–2960 [DOI] [PubMed] [Google Scholar]
- Seger C., Hargrave M., Wang X., Chai R. J., Elworthy S., Ingham P. W. (2011). Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Dev. Dyn. 240, 2440–2451 [DOI] [PubMed] [Google Scholar]
- Seo H. C., Saetre B. O., Håvik B., Ellingsen S., Fjose A. (1998). The zebrafish Pax3 and Pax7 homologues are highly conserved, encode multiple isoforms and show dynamic segment-like expression in the developing brain. Mech. Dev. 70, 49–63 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton C. J., Bayguinov Y., Sanders K. M., Ward S. M. (2000). Ultrastructural analysis of the transdifferentiation of smooth muscle to skeletal muscle in the murine esophagus. Cell Tissue Res. 301, 283–298 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. (1997). Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127–138 [DOI] [PubMed] [Google Scholar]
- Tremblay P., Dietrich S., Mericskay M., Schubert F. R., Li Z., Paulin D. (1998). A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev. Biol. 203, 49–61 [DOI] [PubMed] [Google Scholar]
- Tsukamoto K., Nakamura Y., Niikawa N. (1994). Isolation of two isoforms of the PAX3 gene transcripts and their tissue-specific alternative expression in human adult tissues. Hum. Genet. 93, 270–274 [DOI] [PubMed] [Google Scholar]
- van Lunteren E., Moyer M. (2003). Sternohyoid muscle fatigue properties of dy/dy dystrophic mice, an animal model of merosin-deficient congenital muscular dystrophy. Pediatr. Res. 54, 547–553 [DOI] [PubMed] [Google Scholar]
- van Swearingen J. M., Lance-Jones C. (1993). Spatial and temporal patterns of muscle formation in the limb of the avian embryo. Prog. Clin. Biol. Res. 383B, 553–562 [PubMed] [Google Scholar]
- Wallace K. N., Dolan A. C., Seiler C., Smith E. M., Yusuff S., Chaille-Arnold L., Judson B., Sierk R., Yengo C., Sweeney H. L., et al. (2005). Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev. Cell 8, 717–726 [DOI] [PubMed] [Google Scholar]
- Weinberg E. S., Allende M. L., Kelly C. S., Abdelhamid A., Murakami T., Andermann P., Doerre O. G., Grunwald D. J., Riggleman B. (1996). Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271–280 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide For The Laboratory Use Of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Wood A., Thorogood P. (1994). Patterns of cell behaviour underlying somitogenesis and notochord formation in intact vertebrate embryos. Dev. Dyn. 201, 151–167 [DOI] [PubMed] [Google Scholar]
- Wörl J., Neuhuber W. L. (2005). Ultrastructural analysis of the smooth-to-striated transition zone in the developing mouse esophagus: emphasis on apoptosis of smooth and origin and differentiation of striated muscle cells. Dev. Dyn. 233, 964–982 [DOI] [PubMed] [Google Scholar]
- Wörl J., Breuer C., Neuhuber W. L. (2009). Deletion of Pax7 changes the tunica muscularis of the mouse esophagus from an entirely striated into a mixed phenotype. Dev. Dyn. 238, 864–874 [DOI] [PubMed] [Google Scholar]
- Xu Y., He J., Wang X., Lim T. M., Gong Z. (2000). Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev. Dyn. 219, 201–215 [DOI] [PubMed] [Google Scholar]
- Zhao W., Dhoot G. K. (2000). Both smooth and skeletal muscle precursors are present in foetal mouse oesophagus and they follow different differentiation pathways. Dev. Dyn. 218, 587–602 [DOI] [PubMed] [Google Scholar]
- Zhou H. M., Wang J., Rogers R., Conway S. J. (2008). Lineage-specific responses to reduced embryonic Pax3 expression levels. Dev. Biol. 315, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.