Abstract
Many tissue-engineering approaches for repair and regeneration involve transplants between species. Yet a challenge is distinguishing donor versus host effects on gene expression. This study provides a simple molecular strategy to quantify species-specific contributions in chimeras and xenografts. Species-specific primers for reverse transcription quantitative real-time PCR (RT-qPCR) were designed by identifying silent mutations in quail, duck, chicken, mouse and human ribosomal protein L19 (RPL19). cDNA from different pairs of species was mixed in a dilution series and species-specific RPL19 primers were used to generate standard curves. Then quail cells were transplanted into transgenic-GFP chick and resulting chimeras were analyzed with species-specific primers. Fluorescence-activated cell sorting (FACS) confirmed that donor- and host-specific levels of RPL19 expression represent actual proportions of cells. To apply the RPL19 strategy, we measured Runx2 expression in quail-duck chimeras. Elevated Runx2 levels correlated with higher percentages of donor cells. Finally, RPL19 primers also discriminated mouse from human and chick. Thus, this strategy enables chimeras and/or xenografts to be screened rapidly at the molecular level.
Keywords: Species-specific primers, RPL19, Quail-chick chimeras
INTRODUCTION
Strategies that combine cells in interspecific chimeras or in human-animal xenografts have revealed fundamental mechanisms in developmental biology and have provided a path forward for regenerative medicine. Given that a major clinical objective is to facilitate repair of tissues following disease or injury, more precise approaches are needed to track transplanted cells and to understand signaling interactions between donor- and host-derived tissues. Immunohistochemistry has long been the primary means for following transplanted cells and has been effective for interpreting data at the morphological level. However, crucial information is often needed at the molecular level, and the ability to distinguish donor versus host effects on gene expression has been limited by the lack of a simple technique for estimating species-specific contributions. Here, we provide a universal method based on the reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) (Bustin et al., 2009) that allows relative percentages of donor and host cells to be readily assessed in chimeras. This approach can be applied to xenografts, heterospecific cell pellets, and/or other strategies for repairing human tissues (Allon et al., 2012; Behringer, 2007; Cooke et al., 2011; Lin et al., 2012; Tisato and Cozzi, 2012).
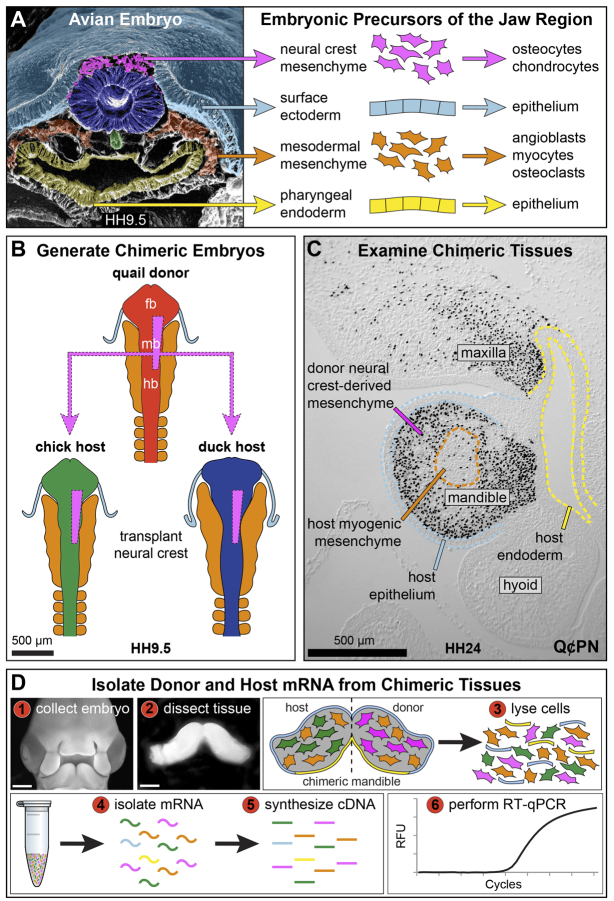
Chimeras have long been used to understand lineages, fates, and patterning abilities of cells (Le Douarin and McLaren, 1984; Lwigale and Schneider, 2008; Noden and Schneider, 2006). We employ quail-chick and quail-duck chimeras to understand patterning of neural crest mesenchyme (NCM), ectoderm, and mesoderm (Eames and Schneider, 2005; Eames and Schneider, 2008; Merrill et al., 2008; Schneider, 1999; Schneider, 2005; Solem et al., 2011; Tokita and Schneider, 2009). A goal is to define molecular programs that these embryonic precursors use to communicate with one another (Fig. 1A). To this end, we transplant NCM from quail embryos into either chick or duck embryos (Fig. 1B). Transplanted NCM then moves into the developing jaw and interacts with non-NCM-derived host tissues (Fig. 1C). Typically, we section and stain chimeras with an antibody (Q¢PN) that recognizes only quail cells. Although this approach is informative for extracting necessary spatial information, the number of genes that can be analyzed in any one sample is greatly dependent upon the number of available tissue sections. Other cell-labeling techniques used to distinguish donor from host such as dyes, transgenic reporters [e.g. green fluorescent protein (GFP) or lacZ], or probes to chromosomal markers (Cooke et al., 2011; Fontaine-Pérus et al., 1997; Matsuo et al., 2007) also have disadvantages. What is most needed for quantifying expression of numerous genes is a rapid molecular assay for screening samples by estimating numbers of donor cells.
Fig. 1.
Generation and examination of chimeric tissue. (A) Pseudocolored scanning electron micrograph showing precursors of the jaw complex. Image courtesy of K. Tosney. (B) NCM transplants from quail into either chick or duck. (C) Sagittal section through the jaw region showing quail cells stained with Qc/PN (black nuclei). Duck-host ectoderm (light blue), endoderm (yellow), and myogenic mesoderm (orange) remain unstained. (D) Work flow for quantifying gene expression.
Virology and hematology contain examples of using RT-qPCR to determine mtDNA mutant or viral load in an individual, mutations within a virus that confer drug resistance from individual to individual, genotypes of strains of virus within an infected population, and success versus failure of marrow engraftment based on ratios of individual recipient and donor genotypes (Bai and Wong, 2004; Gineikiene et al., 2009; Liu and Zhang, 2008; Waku-Kouomou et al., 2006). However, these approaches are limited in their applicability because they: rely on blood samples; are not especially useful for developmental biology given the few cell types and life-history stages that can be analyzed; require sequencing each donor and host every time; and necessitate prior knowledge of a nucleotide change that relates to a disease state in order to distinguish a mutant genotype from wild type within an individual or between individuals of the same species. Similarly, other work has employed Alu elements to identify human versus non-human tissues (Abellaneda et al., 2012; Walker et al., 2003), but this approach is fairly narrow because one sample must be human/primate, and there are often false positives because other species contain Alu templates at low levels.
What sets our strategy apart is its generic applicability to many animal species, cell types and contexts. The strategy involves collecting tissue from chimeras or site of xenograft integration, lysing cells, isolating mRNA, synthesizing cDNA, and performing RT-qPCR (Fig. 1D). The strategy uses ribosomal protein L19 (RPL19), which is normally expressed ubiquitously and equally in almost all cell types throughout the life of all individuals (Al-Bader and Al-Sarraf, 2005; Chari et al., 2010; Facci et al., 2011; Zhou et al., 2010). An exception is in tumors, where RPL19 expression is elevated (Bee et al., 2006; Henry et al., 1993; Kuroda et al., 2010; Nakamura et al., 1990; Roesch et al., 2003). Also, at the protein level RPL19 is highly conserved among major taxonomic groups (Brosius and Arfsten, 1978; Chan et al., 1987; Davies and Fried, 1995; Song et al., 1995; Van Dyck et al., 1994), but at the DNA level there are just enough nucleotide changes to enable different species to be distinguished from one another through primer design. Plotting levels of species-specific RPL19 expression against a standard curve then allows percentages of donor and host cells to be quantified. Percentages are validated using fluorescence-activated cell sorting (FACS). We also present species-specific primers for commonly used animals and show how others can be designed easily.
MATERIALS AND METHODS
Generating chimeras
Quail (Coturnix coturnix japonica), duck (Anas platyrhynchos domestica) (AA Labs, Westminster, CA), and transgenic GFP-chick (Gallus gallus) (Crystal Bioscience, Emeryville, CA) eggs were incubated at 37°C. Embryos were matched at stage 9.5 following Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1951). Embryos were handled according to NIH guidelines. NCM was excised from quail and transplanted into duck (Schneider and Helms, 2003) or GFP-chick. Tissues were processed as described (Schneider, 1999).
Preparing cell mixtures
Chick DF-1 (American Type Culture Collection, Manassus, VA), infected with RCAS virus (Morgan and Fekete, 1996) containing GFP, and mouse fibroblasts (NIH-3T3) were expanded in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin. Cells were trypsinized, re-suspended, and 50% GFP-chick were combined with 50% mouse for a total approximating six million. Approximately one million cells were isolated for FACS and five million for RNA isolation.
cDNA preparation
RNA was isolated using an RNeasy column purification kit (Qiagen, Valencia, CA). Concentration and purity of RNA were assessed using a Nanodrop ND-1000 (Thermo Scientific, Wilmington, DE). Approximately 250 ng of total RNA was converted to cDNA in a 20 μl reverse transcription reaction using 1 μl of iScript reverse transcriptase (Bio-Rad, Hercules, CA). The reaction involved: step 1, 25°C for 5 minutes; step 2, 42°C for 30 minutes; step 3, 85°C for 5 minutes; step 4, 4°C hold in a 2720 Thermal Cycler (Applied Biosystems, Carlsbad, CA). cDNA libraries were analyzed to ensure equal amplification efficiencies and then combined in known quantities to make dilution series. A given dilution series contained 0% to 100% cDNA of species ‘A’ mixed with 100% to 0% cDNA of species ‘B’ so that each well contained 2 μl (1.25 ng/μl) total cDNA of 100%. For example, ‘70% chick’ contained 70 μl (1.25 ng/μl) chick cDNA and 30 μl (1.25 ng/μl) quail cDNA.
Cloning RPL19 and Runx2
Primers were designed to obtain full-length quail and duck RPL19 and partial-length Runx2 (RPL19 F: 5′-ATGAGTATGCTCCGGCTGCAG-3′; RPL19 R: 5′-TCACTTCTTGGTCTCTTCTTC-3′; Runx2 F: 5′-ACAGGACTTCCAGCCATCAC-3′; Runx2 R: 5′-TTGGGCAAGTTTGGGTTTAG-3′) based on RPL19 (Gene ID: 420003) and Runx2 (Gene ID: 373919) sequences published for chick. RPL19 was cloned using quail and duck cDNA libraries derived from HH27 mandibles, and Runx2 from HH34 whole heads. Pfu DNA polymerase (Stratagene, La Jolla, CA) was used for amplification. The protocol was: step 1, 95°C for 45 seconds; step 2, 95°C for 45 seconds; step 3, 50°C for 45 seconds; step 4, 72°C for 1 minute; steps 2, 3 and 4 were repeated 29 times; step 5, 72°C for 10 minutes. PCR products were examined on a 1% agarose gel to identify bands of interest, which were collected using QIAquick gel extraction (Qiagen) and then re-suspended in 30 μl of nuclease-free dH2O. PCR products were ligated using pGEM-T Easy Vector System I (Promega, Madison, WI), with 3 μl PCR product and ligase T4, and transformed using DH5α cells (Invitrogen, Grand Island, NY). Plasmid DNA was sequenced (McLab, South San Francisco, CA) using primers for promoter sites SP6 and T7 and analyzed using NCBI tools and Sequencher (Gene Codes Corporation, Ann Arbor, MI). PCR-based sequences were confirmed by sequencing several independent clones. Sequences were deposited in GenBank under accession numbers KF135654, KF135655, KF135656, KF135657.
Species-specific primers
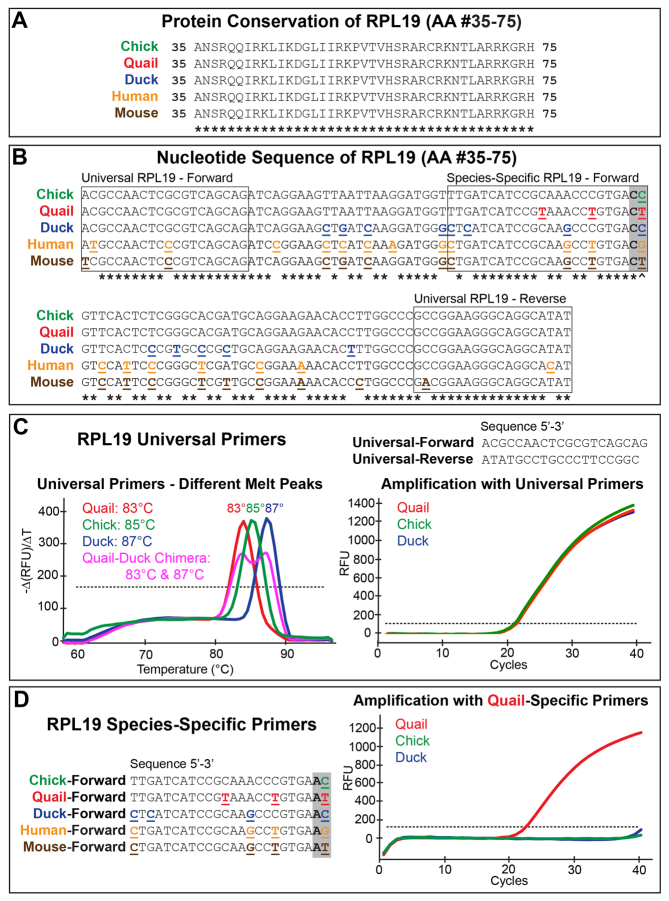
Species-specific primers were designed by identifying silent mutations in quail, duck, chick, mouse, and human RPL19 using Amplification Refractory Mutation System (ARMS) primer design (Bai and Wong, 2004; Newton et al., 1989). To increase species-specificity, one mismatch was introduced at the position immediately 5′ to species-specific silent mutations. Species-specific primers (Fig. 2) had an amplicon length of 83 bp.
Fig. 2.
Analysis of RPL19 and generation of species-specific primers. (A) RPL19 sequence between amino acids 35 through 75. (B) Corresponding nucleotide sequence contains between 3 and 19 mutations per species (underlined and emboldened) compared to chick. Binding locations of universal and species-specific primers are shown. (C) Universal primers amplify evenly, yet due to nucleotide differences among species, melt peaks vary when plotted as the rate of change of relative fluorescent units (RFU) with time (T). Quail-duck chimeras show two melt peaks. (D) Five RPL19 species-specific primers with the distinguishing region and one added mismatch (C to A) at the 3′ end (gray box). The quail-specific primer only amplifies quail cDNA and not duck or chick.
Reverse transcription quantitative real-time PCR
RT-qPCR was performed in a C1000 Thermal Cycler with a CFX96 Real-Time System (Bio-Rad). Forward and reverse primers, 2 μl of cDNA, RNase-free dH20, and iQ SYBR-Green Supermix (Bio-Rad), containing dNTPs, iTaq DNA polymerase, MgCl2, SYBR Green I, enhancers, stabilizers, and fluorescein, were manually mixed in a 25 μl reaction to amplify each cDNA of interest. Samples were run in triplicate on hard-shell PCR white 96-well plates (Bio-Rad, catalog #HSP9601). Results were normalized to universal RPL19, which was checked to make sure amplification efficiencies were equal among samples. The protocol was: step 1, 95°C for 3 minutes; step 2, 95°C for 10 seconds; step 3, 60°C for 30 seconds and a plate read; steps 2 and 3 were repeated 39 times; step 4, 95°C for 10 seconds; step 5, melt curve of 60-90°C for 5 seconds at each 0.5°C with a plate read. Melt curves were checked for specificity. PCR products amplified after 35 cycles were considered to be false positives. For avian Runx2 primers, we used F: 5′-TGGACCTTTCCAGACCAGCAGCA-3′ and R: 5′-GGCAAGTTTGGGTTTAGCAGCGT-3′ with an amplicon length of 162 bp. Universal RPL19 primers (Fig. 2) produced an amplicon length of 127 bp.
Calculating species-specific percentages
Threshold cycle (Ct) values were obtained using universal and species-specific RPL19 primer sets for each sample. Fold changes in species-specific expression were calculated relative to the 100% sample for each species using the delta-delta Ct method (Livak and Schmittgen, 2001). Fold changes were multiplied by 100 to represent values as percentages. The dilution series was then used to determine the linear regression for values of 0% to 100% by intervals of 10%. For each dilution series, linear regressions were analyzed for a best fit by calculating R2 values. To determine contributions of each species, the delta-delta Ct * 100 value was plotted on the linear regression. Confidence intervals were calculated at one standard deviation.
Validation using FACS
Mandibles were isolated as described (Merrill et al., 2008) and digested for 30 minutes at 4°C in 0.25% trypsin with EDTA in Saline A (UCSF Cell Culture Facility). Digestion was stopped in RNase-free 1× PBS. Tissues were pipetted, vortexed, and passed through a 70 μm filter. One million cells were separated for FACS and analyzed within 24 hours. Remaining cells were stored in liquid nitrogen at -80°C for RNA isolation (as described above). Cells from each sample (e.g. GFP-chick and quail, and GFP-chick and mouse) were sorted on a FACSAria Flow Cytometer (BD Biosciences, San Jose, CA). For all sorts, doublets were excluded via gating discrimination using FSC-A and FSC-W. The FSC-SSC gate was used to eliminate debris. At least 10,000 cells were analyzed in each run.
RESULTS AND DISCUSSION
RT-qPCR primers detect species-specific transcripts of RPL19
To analyze tissues from chimeras or xenografts quantitatively, we developed a strategy involving species-specific primers that amplify RPL19. Protein sequence of RPL19 from amino acid #35-75 is 100% conserved among chick, quail, duck, mouse and human (Fig. 2A). Yet the corresponding nucleotide sequence shows multiple silent mutations (Fig. 2B). Our ‘universal’ primers equally amplified transcripts for RPL19 (Fig. 2C) but also produced unique melt peaks for each species due to nucleotide differences (Fig. 2B). The quail melt peak was 83°C, chick 85°C and duck 87°C. Interestingly, we observed two melt peaks when we analyzed chimeras using universal RPL19 primers. One corresponded to the donor and the other to the host (Fig. 2C).
Realizing that different melt peaks arose due to nucleotide changes across species, we used ARMS (Newton et al., 1989) to amplify RPL19 in a manner that was specific to each species (Fig. 2B, gray box). For example, the quail-specific primer was effective because quail have a ‘T’ in the same position occupied by a ‘C’ in chick. Yet, this same primer sequence did not discriminate chick from duck, as both contain a ‘C’ in the same position. To increase primer specificity, a mismatch was deliberately introduced near the 3′ end, adjacent to the silent mutation (Fig. 2B,D). These forward primers allowed for exclusive amplification of the species-specific target when combined with universal reverse primers. For example, the quail-specific primer amplified RPL19 from quail but not from chick or duck (Fig. 2D). Equivalent amplification plots were generated for each species (data not shown).
Species-specific RPL19 can estimate the number of cells from donor versus host
To use RPL19 expression as a means to estimate cell number, we generated dilution series containing known quantities of cDNA from each species. Universal, quail-specific and chick-specific primers were utilized to quantify levels of RPL19 expression. After obtaining Ct values for each sample, fold changes in RPL19 expression were calculated relative to pure 100% samples. For example, percentages calculated for the 70% chick/30% quail sample were 72% by the chick-specific primer and 29% by the quail-specific primer. R2 values for both quail- and chick-specific linear regressions were greater than 0.98 and P-values were less than 0.0001 (Fig. 3A). Thus, species-specific RPL19 primers accurately quantify the proportions of each species in samples containing known quantities of cDNA from different species.
Fig. 3.
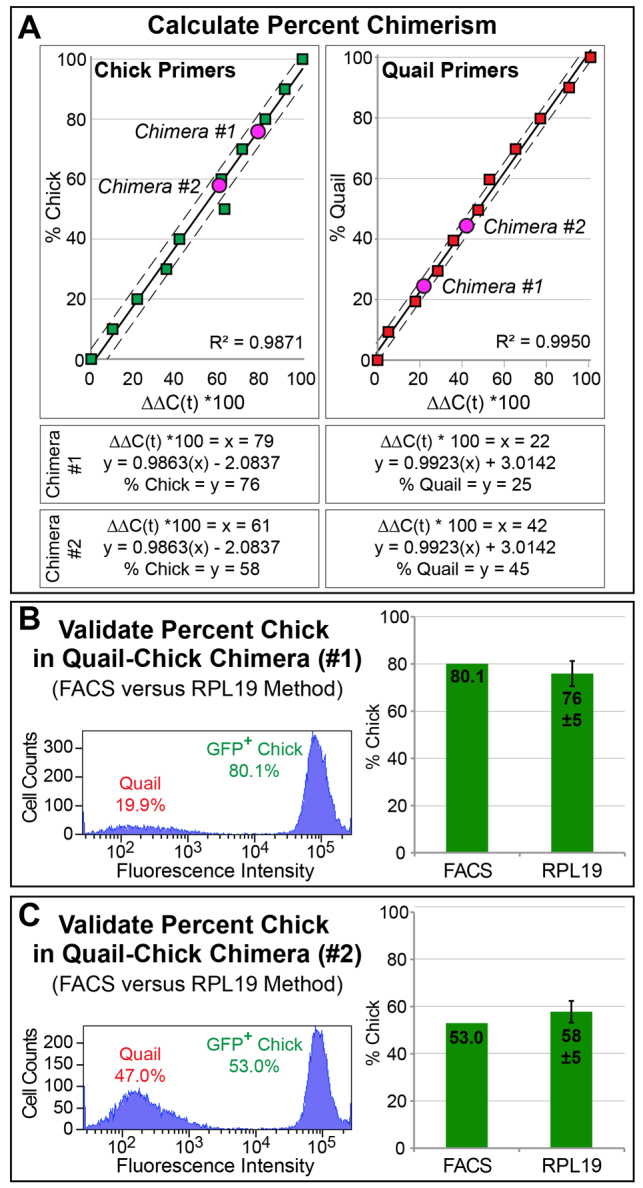
Determining species-specific percentages of cells. (A) Chick- and quail-specific primers were used with a dilution series of cDNA from both chick (green squares) and quail (red squares). Linear regressions were calculated (solid line), as were confidence intervals (dashed lines). Values for Chimera #1 and Chimera #2 (pink circles) were calculated from the regression lines. (B,C) Percentages calculated for Chimera #1 and Chimera #2 with RPL19 were validated using FACS to count GFP-positive chick cells.
To test if RPL19 could also be used to estimate percent contributions of each species within a chimera, we used universal, quail-specific, and chick-specific primers with cDNA from two quail-chick chimeras. In Chimera #1 we found that the fold-change value multiplied by 100 was 79% when using the chick-specific primer and 22% with the quail-specific primer. Using the formula for the species-specific linear regression (Fig. 3A) we could estimate that Chimera #1 contained 76% chick cells and 25% quail cells. Importantly, in Chimera #1 and Chimera #2 the percentage of quail plus percentage of chick approximates 100%.
To validate percentages measured in each quail-chick chimera, we performed FACS. Chimeras were created using transgenic-GFP chick. FACS demonstrated that the percentage of chick in each chimera was within one standard deviation of the value calculated from the linear regression. For example, in Chimera #1 (Fig. 3B), FACS revealed that 80.1% of cells in the sample were GFP-positive chick, whereas the RPL19 method had predicted that 76±5% of cells would be derived from chick. Thus, FACS confirms that species-specific RPL19 primers can accurately measure cellular contributions from different species.
We expect that this RPL19 strategy would work for any highly conserved and ubiquitously expressed reference gene (e.g. GAPDH or β-Actin). By contrast, most other genes would probably not be suitable for discriminating between and/or making predictions about numbers of donor and host cells. Whereas species-specific primers could probably be designed for any gene of interest and allow transcripts from the donor to be distinguished from those of the host, this would be most applicable in situations where only one or a few donor- or host-derived cell types needed to be followed using lineage-specific genes (such as osteocyte or myocyte markers) with the caveat that expression might not remain constant over time as cells progress from their progenitor to terminally differentiated states, and thus not be an accurate predictor of cell number. Nonetheless, designing species-specific primers would be informative for many types of studies, and our strategy provides a blueprint for how to accomplish this task.
RPL19 strategy can distinguish donor- versus host-effects on gene expression
Previously we have shown that Runx2 is elevated on the donor-side of chimeric mandibles owing to accelerated activation of molecular programs in faster-developing quail cells (Fig. 4A) (Merrill et al., 2008). However, when we collect multiple samples of chimeras, we often see a range of Runx2 expression phenotypes, probably as a consequence of varying amounts of donor cells. To test if we could use the RPL19 strategy to screen chimeras and make predications about the effects of donor cell number on gene expression, we transplanted different-sized populations of quail NCM into duck hosts. We collected eight chimeric mandibles and used species-specific RPL19 primers to estimate the ratios of quail and duck cells. For each chimeric sample we also measured the fold change for total Runx2 expression (i.e. from both quail and duck). When we ranked samples based on number of quail donor cells, we observed a significant fold increase in Runx2 expression when comparing the average of the group containing more donor and all donor cells, with the average of the group containing fewer donor and no donor cells (Fig. 4A). We did find some variation in Runx2 expression levels within each group, probably due to the fact that donor cells not only differ in number but also in spatial distribution, which may or may not fall within normal domains of Runx2 expression. Thus, this RPL19 strategy allows chimeric tissues to be screened rapidly, enables trends in gene expression to be measured against percentage of chimerism, and provides an objective criterion for removing potential outliers from datasets, such as those that arise from unsuccessful transplants or poor engraftment of donor cells.
Fig. 4.
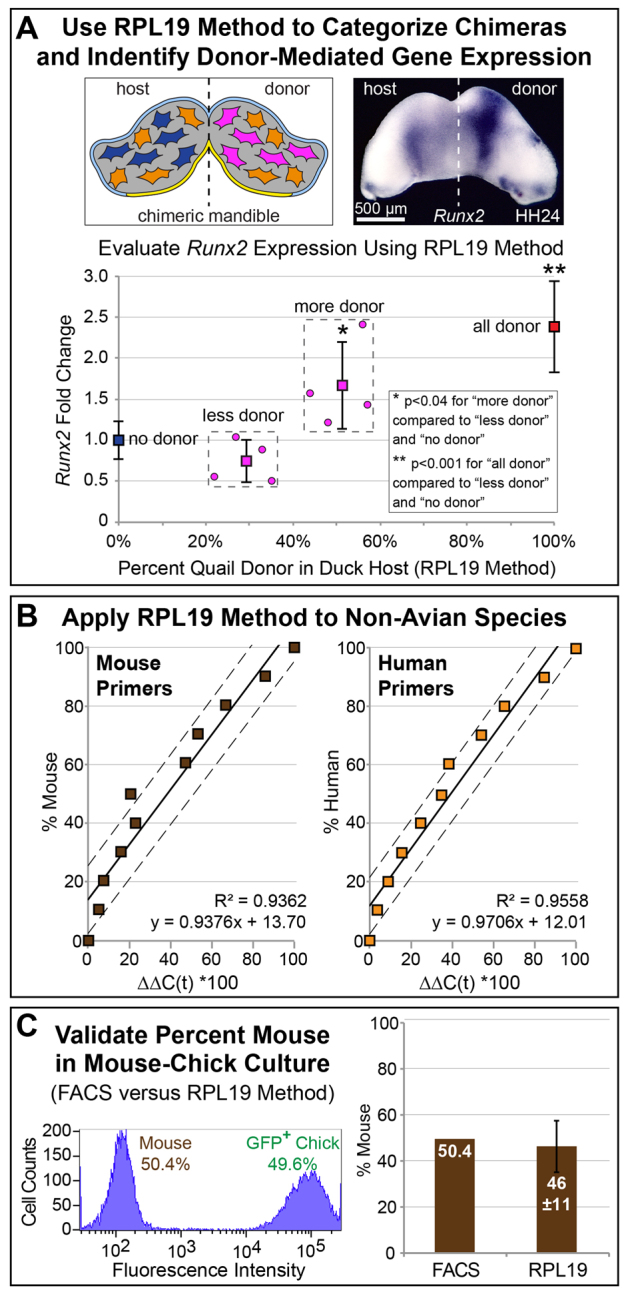
RPL19 strategy for gene expression and other animal models. (A) Quail-duck mandible shown schematically and after performing whole-mount in situ hybridization for Runx2. Expression is elevated on the donor side although the number of quail cells remains unknown (Merrill et al., 2008). Runx2 fold change is measured relative to calculated percentages of quail donor cells in duck hosts. Chimeras with high percent quail NCM show elevated Runx2. (B) RPL19 strategy in mouse and human. (C) Percentage of mouse versus GFP-chick is validated using FACS.
Species-specific RPL19 primers can be used with other chimeras and xenografts
To demonstrate applicability of the RPL19 strategy for non-avian chimeras and xenografts, we developed species-specific primers for human and mouse RPL19. By amplifying RPL19 in a dilution series made from known quantities of human and mouse cDNA, we found that we could easily determine contributions from each species (Fig. 4B). As with avian chimeras, human-mouse samples had an R2 greater than 0.90 and a P-value less than 0.0001. We also successfully extended the RPL19 strategy to the context of mouse-chick chimeras. First, we combined 50% GFP-positive chick fibroblasts with 50% mouse fibroblasts, and then used mouse and chick RPL19 primers to estimate cell number, and FACS to validate (Fig. 4C). Our results show that we can measure accurately percentage of mouse within one standard deviation of the value calculated from the linear regression. Thus, this RPL19 strategy can be used reliably with non-avian species.
Importantly, the RPL19 protein sequence (#35-75) that we analyzed is also 100% conserved in chimp (Pan troglodytes) (XP_51150.1), pig (Sus scrofa) (kXP_00313557.1), cow (Bos taurus) (NP_001035606.1), rat (Rattus norvegicus) (NP_112365.1), zebrafish (Danio rerio) (NP_998373.1), and frog (Xenopus laevis) (NP_001080267.1). Therefore, this strategy can include almost any combination of animal models for various tissue-engineering approaches and future clinical therapies. For example, putting human cells into animals can provide an in vivo system for studying differentiation without experimenting on human individuals (Behringer, 2007; Goldstein, 2010). Ultimately, RPL19 could be used in the clinical setting to evaluate success for many kinds of xenografts where small biopsies could be taken from patients and quickly analyzed for the contributions of donor versus host. Thus, this RT-qPCR-based method addresses a crucial need to quantify cells that each species contributes in chimeras or xenografts when tissues must be processed for gene and protein expression. Moreover, the ability to measure donor- versus host-mediated changes in gene expression on a larger scale will undoubtedly facilitate discovery of more key mechanisms in developmental biology and allow for advances in tissue engineering and regenerative medicine.
Acknowledgments
We thank K. Butcher, J. Fish, J. Yu, E. Berthet, J. Allen, T. Rambaldo and K. Hemati for technical assistance; T. Alliston for helpful discussions; T. Dam at AA Lab Eggs; and M. van de Lavoir at Crystal Bioscience.
Footnotes
Funding
Funded in part, by National Institute of Dental and Craniofacial Research (NIDCR) [T32 DE007306 and K08 DE021705 to E.L.E.]; and by NIDCR [R01 DE016402 to R.A.S.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
E.L.E. conceived and coordinated the project, established the method, performed the experiments and compiled the data. E.L.E. and R.A.S. designed the experiments, analyzed the data and co-wrote the manuscript.
References
- Abellaneda J. M., Ramis G., Martínez-Alarcón L., Majado M. J., Quereda J. J., Herrero-Medrano J. M., Mendonça L., García-Nicolás O., Reus M., Insausti C., et al. (2012). Generation of human-to-pig chimerism to induce tolerance through transcutaneous in utero injection of cord blood-derived mononuclear cells or human bone marrow mesenchymals cells in a preclinical program of liver xenotransplantation: preliminary results. Transplant. Proc. 44, 1574–1578 [DOI] [PubMed] [Google Scholar]
- Al-Bader M. D., Al-Sarraf H. A. (2005). Housekeeping gene expression during fetal brain development in the rat-validation by semi-quantitative RT-PCR. Brain Res. Dev. Brain Res. 156, 38–45 [DOI] [PubMed] [Google Scholar]
- Allon A. A., Butcher K., Schneider R. A., Lotz J. C. (2012). Structured coculture of mesenchymal stem cells and disc cells enhances differentiation and proliferation. Cells Tissues Organs 196, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R. K., Wong L. J. (2004). Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: a single-step approach. Clin. Chem. 50, 996–1001 [DOI] [PubMed] [Google Scholar]
- Bee A., Ke Y., Forootan S., Lin K., Beesley C., Forrest S. E., Foster C. S. (2006). Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin. Cancer Res. 12, 2061–2065 [DOI] [PubMed] [Google Scholar]
- Behringer R. R. (2007). Human-animal chimeras in biomedical research. Cell Stem Cell 1, 259–262 [DOI] [PubMed] [Google Scholar]
- Brosius J., Arfsten U. (1978). Primary structure of protein L19 from the large subunit of Escherichia coli ribosomes. Biochemistry 17, 508–516 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Lin A., McNally J., Peleg D., Meyuhas O., Wool I. G. (1987). The primary structure of rat ribosomal protein L19. A determination from the sequence of nucleotides in a cDNA and from the sequence of amino acids in the protein. J. Biol. Chem. 262, 1111–1115 [PubMed] [Google Scholar]
- Chari R., Lonergan K. M., Pikor L. A., Coe B. P., Zhu C. Q., Chan T. H., MacAulay C. E., Tsao M. S., Lam S., Ng R. T., et al. (2010). A sequence-based approach to identify reference genes for gene expression analysis. BMC Med. Genomics 3, 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M. E., Allon A. A., Cheng T., Kuo A. C., Kim H. T., Vail T. P., Marcucio R. S., Schneider R. A., Lotz J. C., Alliston T. (2011). Structured three-dimensional co-culture of mesenchymal stem cells with chondrocytes promotes chondrogenic differentiation without hypertrophy. Osteoarthritis Cartilage 19, 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B., Fried M. (1995). The L19 ribosomal protein gene (RPL19): gene organization, chromosomal mapping, and novel promoter region. Genomics 25, 372–380 [DOI] [PubMed] [Google Scholar]
- Eames B. F., Schneider R. A. (2005). Quail-duck chimeras reveal spatiotemporal plasticity in molecular and histogenic programs of cranial feather development. Development 132, 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames B. F., Schneider R. A. (2008). The genesis of cartilage size and shape during development and evolution. Development 135, 3947–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facci M. R., Auray G., Meurens F., Buchanan R., van Kessel J., Gerdts V. (2011). Stability of expression of reference genes in porcine peripheral blood mononuclear and dendritic cells. Vet. Immunol. Immunopathol. 141, 11–15 [DOI] [PubMed] [Google Scholar]
- Fontaine-Pérus J., Halgand P., Chéraud Y., Rouaud T., Velasco M. E., Cifuentes Diaz C., Rieger F. (1997). Mouse-chick chimera: a developmental model of murine neurogenic cells. Development 124, 3025–3036 [DOI] [PubMed] [Google Scholar]
- Gineikiene E., Stoskus M., Griskevicius L. (2009). Single nucleotide polymorphism-based system improves the applicability of quantitative PCR for chimerism monitoring. J. Mol. Diagn. 11, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. S. (2010). Transplantation of mammalian embryonic stem cells and their derivatives to avian embryos. Stem Cell Rev. 6, 473–483 [DOI] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 [PubMed] [Google Scholar]
- Henry J. L., Coggin D. L., King C. R. (1993). High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 53, 1403–1408 [PubMed] [Google Scholar]
- Kuroda K., Takenoyama M., Baba T., Shigematsu Y., Shiota H., Ichiki Y., Yasuda M., Uramoto H., Hanagiri T., Yasumoto K. (2010). Identification of ribosomal protein L19 as a novel tumor antigen recognized by autologous cytotoxic T lymphocytes in lung adenocarcinoma. Cancer Sci. 101, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N., McLaren A. (1984). Chimeras in Developmental Biology. London/Orlando: Academic Press; [Google Scholar]
- Lin C. S., Lin G., Lue T. F. (2012). Allogeneic and xenogeneic transplantation of adipose-derived stem cells in immunocompetent recipients without immunosuppressants. Stem Cells Dev. 21, 2770–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. B., Zhang Z. (2008). Development of SYBR Green I-based real-time PCR assay for detection of drug resistance mutations in cytomegalovirus. J. Virol. Methods 149, 129–135 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lwigale P. Y., Schneider R. A. (2008). Other chimeras: quail-duck and mouse-chick. Methods Cell Biol. 87, 59–74 [DOI] [PubMed] [Google Scholar]
- Matsuo S., Kurisaki A., Sugino H., Hashimoto I., Nakanishi H. (2007). Analysis of skin graft survival using green fluorescent protein transgenic mice. J. Med. Invest. 54, 267–275 [DOI] [PubMed] [Google Scholar]
- Merrill A. E., Eames B. F., Weston S. J., Heath T., Schneider R. A. (2008). Mesenchyme-dependent BMP signaling directs the timing of mandibular osteogenesis. Development 135, 1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B. A., Fekete D. M. (1996). Manipulating gene expression with replication-competent retroviruses. In Methods in Avian Embryology (ed. Bronner-Fraser M.), pp. 186–217 San Diego, CA: Academic Press; [DOI] [PubMed] [Google Scholar]
- Nakamura T., Onno M., Mariage-Samson R., Hillova J., Hill M. (1990). Nucleotide sequence of mouse L19 ribosomal protein cDNA isolated in screening with tre oncogene probes. DNA Cell Biol. 9, 697–703 [DOI] [PubMed] [Google Scholar]
- Newton C. R., Graham A., Heptinstall L. E., Powell S. J., Summers C., Kalsheker N., Smith J. C., Markham A. F. (1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D. M., Schneider R. A. (2006). Neural crest cells and the community of plan for craniofacial development: historical debates and current perspectives. Adv. Exp. Med. Biol. 589, 1–23 [DOI] [PubMed] [Google Scholar]
- Roesch A., Vogt T., Stolz W., Dugas M., Landthaler M., Becker B. (2003). Discrimination between gene expression patterns in the invasive margin and the tumour core of malignant melanomas. Melanoma Res. 13, 503–509 [DOI] [PubMed] [Google Scholar]
- Schneider R. A. (1999). Neural crest can form cartilages normally derived from mesoderm during development of the avian head skeleton. Dev. Biol. 208, 441–455 [DOI] [PubMed] [Google Scholar]
- Schneider R. A. (2005). Developmental mechanisms facilitating the evolution of bills and quills. J. Anat. 207, 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R. A., Helms J. A. (2003). The cellular and molecular origins of beak morphology. Science 299, 565–568 [DOI] [PubMed] [Google Scholar]
- Solem R. C., Eames B. F., Tokita M., Schneider R. A. (2011). Mesenchymal and mechanical mechanisms of secondary cartilage induction. Dev. Biol. 356, 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M., Cheung E., Rabinowitz J. C. (1995). Nucleotide sequence and characterization of the Saccharomyces cerevisiae RPL19A gene encoding a homolog of the mammalian ribosomal protein L19. Yeast 11, 383–389 [DOI] [PubMed] [Google Scholar]
- Tisato V., Cozzi E. (2012). Xenotransplantation: an overview of the field. Methods Mol. Biol. 885, 1–16 [DOI] [PubMed] [Google Scholar]
- Tokita M., Schneider R. A. (2009). Developmental origins of species-specific muscle pattern. Dev. Biol. 331, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosney K. W. (1982). The segregation and early migration of cranial neural crest cells in the avian embryo. Dev. Biol. 89, 13–24 [DOI] [PubMed] [Google Scholar]
- Van Dyck L., Jonniaux J. L., de Melo Barreiros T., Kleine K., Goffeau A. (1994). Analysis of a 17.4 kb DNA segment of yeast chromosome II encompassing the ribosomal protein L19 as well as proteins with homologies to components of the hnRNP and snRNP complexes and to the human proliferation-associated p120 antigen. Yeast 10, 1663–1673 [DOI] [PubMed] [Google Scholar]
- Waku-Kouomou D., Alla A., Blanquier B., Jeantet D., Caidi H., Rguig A., Freymuth F., Wild F. T. (2006). Genotyping measles virus by real-time amplification refractory mutation system PCR represents a rapid approach for measles outbreak investigations. J. Clin. Microbiol. 44, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. A., Kilroy G. E., Xing J., Shewale J., Sinha S. K., Batzer M. A. (2003). Human DNA quantitation using Alu element-based polymerase chain reaction. Anal. Biochem. 315, 122–128 [DOI] [PubMed] [Google Scholar]
- Zhou L., Lim Q. E., Wan G., Too H. P. (2010). Normalization with genes encoding ribosomal proteins but not GAPDH provides an accurate quantification of gene expressions in neuronal differentiation of PC12 cells. BMC Genomics 11, 75 [DOI] [PMC free article] [PubMed] [Google Scholar]