Abstract
Three-dimensional (3D) cell cultures are important tools in cell biology research and tissue engineering because they more closely resemble the architectural microenvironment of natural tissue, compared to standard two-dimensional cultures. Microscopy techniques that function well for thin, optically transparent cultures, however, are poorly suited for imaging 3D cell cultures. Three-dimensional cultures may be thick and highly scattering, preventing light from penetrating without significant distortion. Techniques that can image thicker biological specimens at high resolution include confocal microscopy, multiphoton microscopy, and optical coherence tomography. In this chapter, these three imaging modalities are described and demonstrated in the assessment of functional and structural features of 3D chitosin scaffolds, 3D micro-topographic substrates from poly-dimethyl siloxane molds, and 3D Matrigel cultures. Using these techniques, dynamic changes to cells in 3D microenvironments can be non-destructively assessed repeatedly over time.
Keywords: 3D culture, optical coherence tomography, multiphoton microscopy
1. Introduction
Culturing cells on two-dimensional (2D) substrates has been a standard technique in cell biology research for decades. However, the culture flasks and dishes typically used do not replicate the natural microenvironment of cells in tissue. Research has shown that the extracellular environment has a profound impact on cell biology, including differentiation of stem cells (1, 2). As a result, there has been an increased interest in developing culturing techniques that more closely resemble natural tissue. Many different types of three-dimensional (3D) cultures and 3D scaffold materials have been developed. A commonly used substance, Matrigel, is derived from the extra cellular matrix (ECM) of mouse tumor (sarcoma). Other techniques are based on seeding cells in porous 3D scaffolds constructed with synthetic materials. Three-dimensional cell cultures have become important for both research and clinical applications, with one goal being the development of advanced biocompatible engineered tissues (3, 4).
While 3D cell cultures more accurately model the natural environment of cells, their use also presents new challenges. Many of the tools used to visualize live cells in 2D cell cultures such as bright field and phase microscopy rely on light transmitted through the sample. These approaches are impractical for viewing cells in 3D cultures since the whole sample may be too thick for light to effectively pass through. For visualizing 3D cultures and thick tissue samples non-destructively, imaging techniques that rely on epi-illumination (with light collected in the backward direction) are more suitable. Different epi-illumination imaging techniques exist and can be based on the detection of fluorescence as well as backscattered light. Fluorescence-based techniques are most commonly used to visualize a fluorescent marker that has been targeted to a specific area or molecule of interest. Alternatively, the autofluorescent properties of cells and tissues can frequently provide sufficient contrast to identify structural and biochemical components. Imaging techniques based on the scattering of light are commonly used to observe the structural and dimensional characteristics of a sample. For example, by measuring the wavelength-dependent scattering properties of the sample, one can infer the sizes of the scatterers present, such as the size of individual cells or nuclei.
Confocal microscopy (CM) is an imaging technique that is capable of high resolution optical sectioning in relatively thick samples and can be used both in fluorescence as well as reflectance mode (see Note 1). The penetration depth of confocal microscopy is limited to roughly less than 100 µm (5). Deeper penetration depths for fluorescence imaging can be achieved by using multi-photon microscopy (MPM). This technique relies on non-linear optical effects and can perform high resolution optical sectioning in samples up to a millimeter thick, depending on tissue type (6). Scattering-based imaging with penetration depths of up to several millimeters can be achieved by using optical coherence tomography (OCT) (7). OCT constructs depth-resolved images by using interferometric techniques to measure the time-of-flight of scattered photons. Determining which imaging technique is appropriate for a specific 3D cell culture depends on the optical properties, type, and thickness of the culture, as well as the features of interest in the culture. Some of the imaging techniques described here can be used simultaneously, which provides a more comprehensive view of a sample based on differing contrast-enhancing mechanisms. Since these imaging techniques have little effect on the viability of living cells, they are well-suited for observing dynamic changes in 3D cell cultures.
This chapter will focus on imaging techniques for assessing cells grown in different 3D cultures. Optical coherence tomography, multiphoton microscopy, and confocal microscopy are used to visualize structural and functional properties of the samples. OCT and CM are used to observe the location and functional activity of cells over a period of several days in a chitosin scaffold. OCT and MPM are used to obtain high-resolution images of cells seeded on 3D micro-topographic substrates, as well as in 3D cultures using Matrigel as a scaffold material. The effect of mechanical stimuli on the cells in these cultures is also observed.
Understanding the principles and limitations of confocal microscopy, multiphoton microscopy, and optical coherence tomography is essential when choosing the appropriate technique for imaging a specific type of 3D culture, scaffold, or substrate. The most important parameters to consider for imaging 3D cultures are the optical contrast mechanism (fluorescence or scattering), the resolution, and the penetration depth. CM, MPM, and OCT instruments are all commercially available, but can also be constructed in a lab that is equipped with sufficient optical hardware components.
1.1. Confocal Microscopy
Confocal microscopy is a high resolution imaging technique that enables optical sectioning of non-transparent samples. There are several types of CM but the technique that has gained the most widespread use in the life sciences is confocal laser scanning microscopy. This technique is based on point illumination of the sample with a laser, and spatial filtering of the returning beam with a pinhole to block light from outside the focus. The beam is scanned across the sample and an image is constructed. CM can be used in both fluorescence mode as well as reflectance mode.
Spatial resolution in CM depends on the numerical aperture (NA) of the objective, the pinhole size, and the wavelength of the light. Sub-micrometer resolution can be achieved with commercial systems allowing even the smallest cells and sub-cellular features to be visualized. Penetration depth in thick samples is limited by the fact that scattering in the sample causes the illuminating beam to defocus. This will decrease the amount of light that passes through the pinhole as the imaging depth becomes deeper, effectively limiting the imaging depth. The penetration depth also depends on the optical properties of the sample, the NA of the objective, and wavelength of the light source. Generally, longer wavelength light penetrates deeper in a sample due to less absorption and scattering. Fluorescence CM typically uses shorter wavelength (visible) light to excite commercial fluorophores through absorption. Reflectance CM does not have stringent wavelength requirements and can readily be performed with longer wavelength light sources to improve penetration depth (8). Ongoing research is developing newer fluorescence-based probes or dyes that are excited by near-infrared wavelengths. The deepest penetration depth achievable with CM is typically limited to less than 100 µm (9, 10).
1.2. Multiphoton Microscopy
Multiphoton microscopy is a high-resolution fluorescence imaging technique that has at least a twofold improvement in penetration depth over confocal microscopy (11). It is based on nonlinear optical processes that absorb two or more near-infrared photons and subsequently emit a single photon in the visible range. It is most commonly used to excite exogenous fluorescent dyes or probes through two-photon absorption. This process requires a high intensity of photons in both time and position (see Note 2). This is typically accomplished using a laser with ultra-short pulse duration (such as a titanium-sapphire laser) and a high numerical aperture objective. The high intensity requirement restricts the two-photon absorption to within the focal volume. This results in a high spatial resolution without the need for spatial filtering. In addition, MPM reduces photobleaching since fluorescence excitation is limited to a small volume (12). Because near-infrared wavelengths are used to excite the fluorescent molecules, imaging penetration is significantly better than CM, and there is little absorption or thermal effects. As a result, MPM is preferable to CM for imaging 3D cell cultures or tissue when they are thick or highly scattering.
1.3. Optical Coherence Tomography
Optical coherence tomography is a technique which is capable of obtaining high resolution scattering-based images of thick tissue samples (7, 13). OCT operates by focusing light from a broad bandwidth light source into a sample and determining the time it takes for the light to return to a detector. The echo time-of-flight information is used to determine the depth in the sample from where light was scattered. OCT is similar in principle to ultrasound imaging except that near-infrared light is used instead of acoustic waves. Compared to clinical ultrasound, OCT has a higher resolution but a shallower penetration depth. Because the speed of light is significantly faster than the speed of acoustic waves, the time information must be measured indirectly using interferometry. Light scattered from the sample is combined with a reference beam and the resulting interference pattern is measured. The interference pattern contains the time-of-flight information of the light in the sample arm. Light scattered from different depths can be distinguished because of the low coherence property of the laser or light source. Cross-sectional OCT images are constructed by scanning the laser beam across the sample and acquiring depth-dependent scattering profiles at each position.
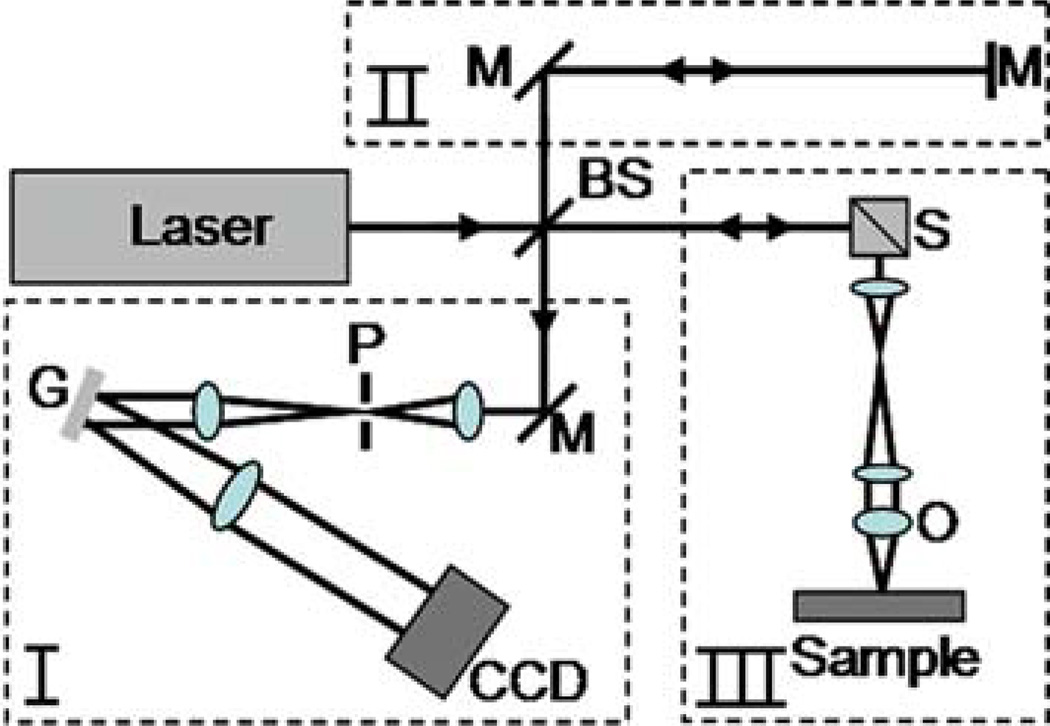
The setup of an OCT system can be divided into four parts: source, sample arm, reference arm, and detector arm. The source is commonly a near-infrared laser or superluminescent diode with a broad spectral bandwidth. The beam from the source is split by a beam splitter into the reference arm and the sample arm. Light reflected from the sample and the reference arm recombine at the beam splitter and is directed to the detector. The sample arm consists of focusing optics (which also function as collecting optics), the sample, and a mechanism for scanning the beam or the sample. The arrangement of the reference arm and the detection arm is dependent on the detection scheme being used. There are two basic schemes, time-domain OCT and frequency-domain OCT. Time-domain OCT consists of a single detector and mechanism for rapidly scanning the delay in the reference arm. At each delay of the reference arm, scattering from a specific depth in the sample can be determined. Frequency-domain OCT is performed by using a fixed reference arm path and measuring the spectral interference pattern in the detector arm. The entire depth-scattering profile can be obtained by taking one measurement of the spectrum and computing the inverse Fourier transform. Frequency-domain OCT does not require any mechanical scanning in the reference arm, which results in significantly improved speed and sensitivity (14). Frequency-domain OCT is further sub-divided into two categories: spectral (or Fourier) domain OCT and swept-source OCT (seeNote 3). In spectral-domain OCT the spectrum of the broad bandwidth light source is measured using a spectrometer (15). In swept-source OCT the wavelength of a tunable laser is rapidly swept over the bandwidth (16). The broad spectrum is thus encoded in time and a single detector is used instead of a spectrometer. A simplified schematic of a spectral-domain OCT system is shown in Fig. 13.1 to better demonstrate the principles of OCT.
Fig. 13.1.
Schematic of a spectral/Fourier-domain OCT system. The beam from the broad bandwidth light source is split into the reference arm (II) and the sample arm (III) using a beam splitter. The beam in the sample arm is focused onto a sample and scanned to acquire images. Light backscattered from the sample and the reference end mirror is recombined at the beam splitter and sent to the detector arm (I). The detector arm is a spectrometer that measures the spectral interference pattern. Scanning of the beam and acquisition from the charge-coupled device (CCD) camera is synchronized using a personal computer. Abbreviations: BS, beamsplitter; G, grating; M, mirror; O, objective; P, pinhole; S, beam scanners.
Resolution in the axial and transverse directions is decoupled in OCT. Axial resolution is governed by the coherence length of the light source which is inversely dependent on the spectral bandwidth. Axial resolution in a typical OCT system is less than 10 microns. Transverse resolution is dictated by the optics used to focus the beam onto the sample. Transverse resolution for a typical OCT system is around 10 |xm as well, so a large confocal parameter, or depth-of-focus, can be achieved. Just as with CM and MPM, imaging depth for OCT is highly dependent on the optical properties of the sample as well as the wavelength of the source. Penetration depth can be up to several millimeters, even in highly scattering samples, because OCT not only spatially filters out-of-focus light as in CM but also coherently rejects photons that are not singly backscattered from points in the tissue and therefore no longer remain coherent with the light reflected back in the reference arm (17).
A variation of OCT, called optical coherence microscopy (OCM), utilizes a high NA objective to achieve similar transverse spatial resolution as reflectance CM, at the expense of a short depth-of-focus (18). In this case, the axial resolution may then be determined by the focusing optics instead of the source bandwidth. Optical sectioning is achieved with both spatial and coherence gating, instead of spatial filtering alone. The advantage of OCM is that it has a greater penetration depth than CM. Because of the high NA and the narrow beam waist, the short focal region is effectively limited to a single en face plane in the sample. As a result, the beam in OCM is typically scanned in two lateral dimensions to acquire an en face image, as opposed to a cross-sectional (depth-resolved) image as in OCT. Because OCM uses a high NA objective and can use the same laser source (titanium:sapphire) as in MPM, it is possible to perform OCM at the same time as MPM to collect image data based on multiple contrast mechanisms (19–22). This enables simultaneous acquisition of high resolution scattering and fluorescence information from thick, highly scattering samples. OCT has also been used simultaneously with fluorescence CM to provide structural and functional information in engineered tissues (23).
Additional variations of OCT and OCM exist that enable imaging based of different contrast mechanisms. Spectroscopic OCT (24) and OCM (25) measure the depth-resolved spectrum of backscattered light, which allows wavelength-dependent properties of the sample to be assessed. Biomechanical properties can be spatially resolved using optical coherence elastography (26, 27). Doppler OCT is a technique which measures frequency shifts of backscattered light to measure dynamic events such as blood flow in vivo (28).
2. Materials
2.1. Chitosin Scaffold Culture
NIH 3T3 fibroblast cells (American Type Culture Collection, Manassas, VA).
4 µg of GFP-vinculin plasmid (provided by Dr. Benjamin Geiger, Weizmann Institute of Science, Israel) diluted with 50 µL of FreeStyle 293 expression (Invitrogen).
2 µL of lipofectamine 2000 reagent (Invitrogen) diluted with 50 µL of FreeStyle 293 expression.
2% (wt%) chitosan flakes (Sigma-Aldrich) dissolved in a 0.2 M acetic acid aqueous solvent.
Phosphate-buffered saline (PBS, 1×) solutions containing 70, 50, and 0% of ethanol.
Plastic 8 mm diameter and 3 mm depth cylindrical molds (cap of eppendorf tube).
Portable microincubator, LU-CPC (Harvard Apparatus, Holliston, MA).
2.2. Matrigel and Micro-topographic Culture
Matrigel solution (BD Bioscience, Bedford, MA).
Microtextured poly(dimethly-siloxane) (PDMS) substrates with 3D topographic features (provided by James Norman, Stanford University).
Hoechst 33342 nuclear dye (Invitrogen).
3D reconstruction software, Analyze 5.0 (Mayo Clinic, Rochester, MN).
Mechanical stimulation system Tissue Train and StageFlexer (Flexcell, NC).
2.3. Optical Coherence Tomography Instrument
The OCT setup used in this experiment is a custom built, fiber-based, time-domain OCT system. The axial and lateral resolutions of the system are 3 and 10 µm, respectively. The major components are as follows:
Titanium:sapphire laser (Kapteyn-Murnane Laboratories) pumped by a Nd:YVO4 laser (Coherent) that produces ~90-fs pulses with an 80-MHz repetition rate. The center wavelength is 800 nm with a bandwidth of 20 nm or greater.
Ultrahigh numerical aperture fiber UHNA4 (Thorlabs, Karslfeld, Germany) to broaden the spectrum from 20 to 100 nm.
Galvanometer-driven retroreflector delay line in reference arm operating at 30 Hz.
Galvanometer-controlled mirrors for scanning of beam across the sample.
Achromatic lens with 20 mm focal length and 12.5 mm diameter (Thorlabs).
Photodetector (Newport).
2.4. Integrated Optical Coherence and Multiphoton Microscope
The integrated OCM and MPM microscope is a custom-built system that uses free space optics (19). The OCM modality has a spectral-domain detection scheme. The axial and lateral MPM resolutions are 0.8 and 0.5 µm, respectively, while the axial and lateral OCM resolutions are 2.2 and 0.9 µm, respectively. The major components of the system are as follows:
Titanium:sapphire laser (Kapteyn-Murnane Laboratories) pumped by a Nd:YVO4 laser (Coherent) with center wavelength at 800 nm with a bandwidth of 120 nm.
Galvanometer-controlled mirrors for scanning of beam across the sample.
Microscope objective, 20 ×, 0.95 NA, water-immersion (Olympus).
Cold Mirror dichroic mirror (CVI Laser, Livermore, CA) and H7421-40 photo-multiplier tube (Hamamatsu, Inc.) for detection of multiphoton signal.
Emission filters to detect fluorescence from GFP (FF01-520/35-25, Semrock) and from Hoechst nuclear dye (BG39, CVILaser).
Custom-built spectrometer for detection of OCM signal. Consists of collimating optics, a blazed diffraction grating having 830.3 grooves/mm and a line-scan camera (L104k-2 k, Basler, Inc.) that contains a 2048-element CCD array maximum readout rate of 29 kHz.
3. Methods
3.1. Transfecting 3T3 Cell Line to Co-express GFP-Vinculin
Seed 7 × 105 3T3 fibroblast cells on each well of a six-well plate.
Mix 4 µg of diluted DNA and 2 µL of diluted Lipofectamine 2000 and add to each of the wells.
Incubate cells for 20 min then clean with PBS.
Resulting cells are a stable cell line of cells that express GFP-vinculin.
3.2. Chitosin Scaffold Cell Culture Preparation
Filter chitosan solution and transfer to cylindrical molds.
Freeze cylinders at –20°C and lyophilize for 4 days until solvent is completely removed (see Note 4).
Sterilize scaffold with ethanol/PBS solutions.
Hydrate scaffolds in cell culture media for 12 h before seeding cells.
Seed cells at a concentration of 5 × 106 cells/cm3.
Place scaffold into micro-incubator for imaging.
3.3. Matrigel and Micro-Topographic Culture Preparation
Stain GFP-transfected 3T3 fibroblast cells with Hoechst nuclear dye and incubate for 30 min.
Mix solution with 7 × 104 GFP-transfected 3T3 cells with thawed Matrigel solution at a 1:1 volume ratio.
Solidify the mixture in a 37°C incubator.
Place Matrigel culture into micro-incubator for imaging.
Seed GFP-transfected 3T3 cells at concentration of 5 × 10 cell/ml on a micro-topographic PDMS substrate with 10 |xm height and diameter cylindrical pegs.
3.4. OCT and CM Imaging of Cell Dynamics in Chitosin Scaffold Culture
OCT and CM images of the chitosin scaffold cell culture were taken at several time points after seeding to observe dynamic changes to the distribution of the cell population (29, 30). OCT is used to visualize changes to the cell population that occur throughout the entire volume of the 3D chitosin scaffold culture. CM, in comparison, provides cellular-level resolution and functional information near the surface of the cell culture.
The micro-incubator is placed in the sample arm of the OCT system allowing the chitosin scaffold to be imaged while maintaining the sterile environment. Three-dimensional OCT images of the same chitosin scaffold are taken at 1, 3, 5, 7, and 9 days after seeding (see Note 5).
To verify the dynamic changes observed in the OCT images taken on different days, different identical samples are histo-logically prepared after being incubated for the same durations (see Note 6). The histological samples are viewed with a standard light microscope and compared to cross-sectional OCT images (see Fig. 13.2).
To more effectively visualize the whole cell population, 3D OCT data images are reconstructed and viewed from different angles (see Fig. 13.3).
To visualize the culture at cellular resolution and to observe functional characteristics (see Note 7), CM images of the superficial layers of the chitosin scaffold are taken in fluorescence mode using a commercial system, DM-IRE (Leica Microsystems, Bensheim, Germany). CM images are taken at 1, 3, 5, and 7 days to observe dynamic changes to the culture (see Fig. 13.4).
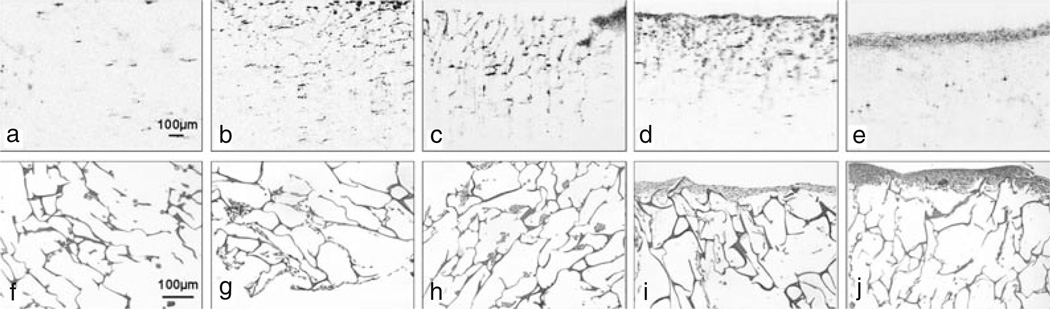
Fig. 13.2.
Cross-sectional OCT (a–e) and corresponding hematoxylin and eosin-stained histology (f–j) images of chitosin scaffold cultures at day 1, 3, 5, 7, and 9, respectively. Cells are relatively evenly distributed throughout the scaffold after seeding. After several days, it is evident that the cells have migrated to the surface of the culture and have formed a dense, highly scattering layer. This demonstrates the ability of OCT to observe morphological features in a 3D cell culture at different times during the development of the 3D culture. Note the difference in image scale between OCT and histology images. Modified figure used with permission (20).
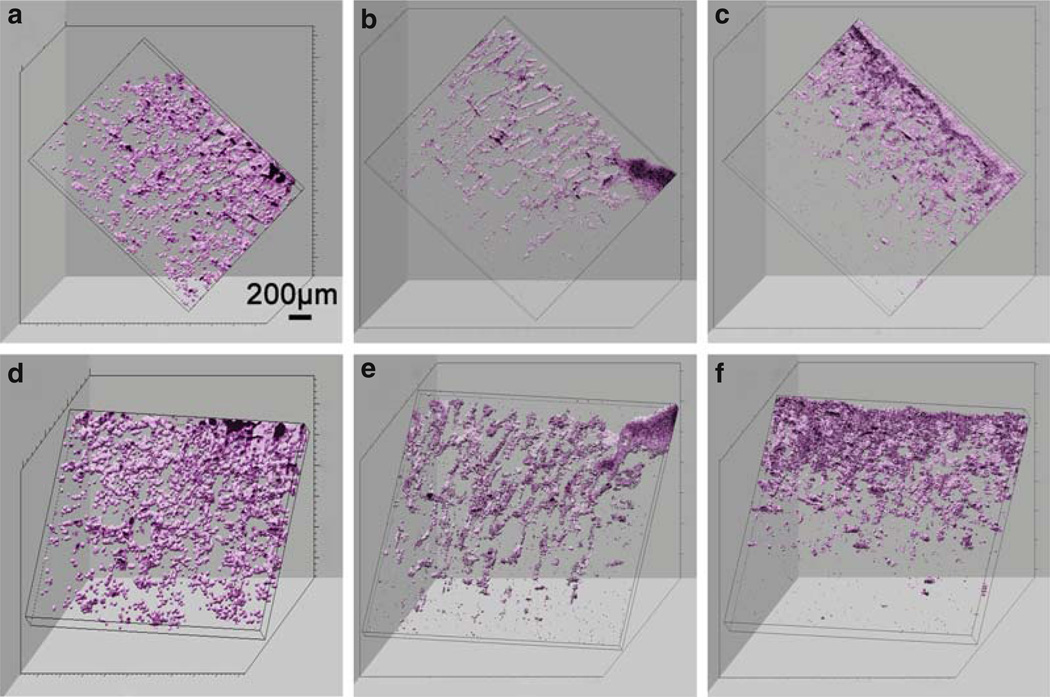
Fig. 13.3.
Three-dimensional OCT reconstructions of chitosin scaffolds from two different viewing angles taken at 3 (a, d), 5 (b, e), and 7 (c, f) days. Rotational angles of image a–c are 40° (y), 20° (z), 70° (x) while d–f are 0° (y), 10° (z), 50° (x). Viewing a 3D reconstruction from many angles, and computationally sectioning out planes from arbitrary angles, enables a more complete view and assessment of the 3D characteristics of a sample, compared to single cross-sectional images. Modified figure used with permission (20).
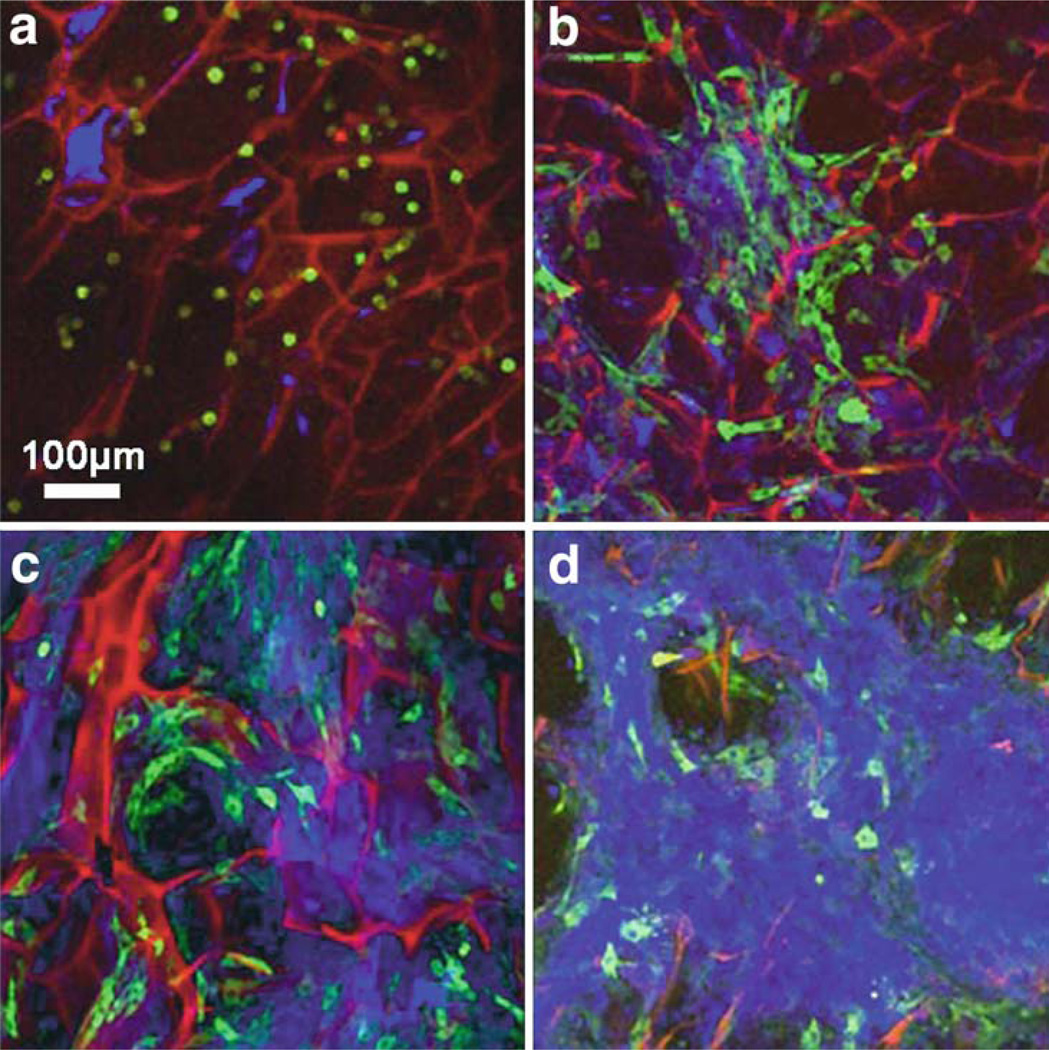
Fig. 13.4.
Fluorescence CM images of GFP-transfected fibroblasts in a 3D chitosin scaffold culture taken at day 1, 3, 5, and 7 (a–d, respectively). The red channel is the autofluorescence signal from the scaffold, the green channel is the GFP fluorescence from the cells, and the blue channel is the backscattering signal predominantly from the deposited extracellular matrix and over time, the higher scattering cell density near the surface of the scaffold. One day after seeding, the fibroblasts are spherical in shape and have a low GFP signal. As the cells grow and attach to the substrate, there is an increase in expression of the GFP-vinculin gene. The increase in the backscattering signal by day 7 verifies that the cells have formed a dense, highly scattering layer near the surface over a period of several days. Modified figure used with permission (20).
3.5. Imaging Effects of Mechanical Stimuli on 3D Cultures with OCM and MPM
Mechanical forces are known to affect morphology and genetic expression in cells. To observe the effects of mechanical forces on cells in 3D cultures both micro-topographic substrates and Matrigel cultures (see Note 8) were imaged before and after being subjected to uniaxial stretching (31). Integrated OCM and MPM allows simultaneous acquisition of fluorescence and scattering-based images. For these samples, this enables visualization of individual cells (via MPM) and the local structural microenvironment of the substrates (via OCM).
En face OCM and MPM images of the 3D micro-topographic substrate are taken 3 days after seeding (see Fig. 13.5a). Two MPM channels are obtained by acquiring the same image twice using different emission filters for fluorescence detection (see Note 9).
The culture is mechanically stimulated with the FlexCell apparatus with 5% cyclic, 1 Hz equibiaxial sinusoidal stretching for a period of 18 h.
En face OCM and MPM images are again taken to observe the changes to cell morphology caused by the mechanical stimulation (see Fig. 13.5b).
3D OCM and MPM images of the Matrigel culture are taken prior to mechanical stimulation. Three-dimensional data sets are obtained by acquiring a series of en face planes at different depths in the sample. Using the Analyze software, en face planes are computationally extracted from the 3D data to observe the structure (see Fig. 13.6a) and functional (see Fig. 13.6b) properties of cells in the culture.
The Matrigel culture is subjected to the same mechanical strain as in step 2.
3D OCM and MPM images are taken after stimulation for comparison. Different projections of the overlaid 3D data set are viewed using the Analyze software (see Fig. 13.7).
Analysis of the before and after OCM and MPM images allowed the effects of mechanical stimulation on the function and morphology of cells in these two types of cultures to be observed (see Note 10).
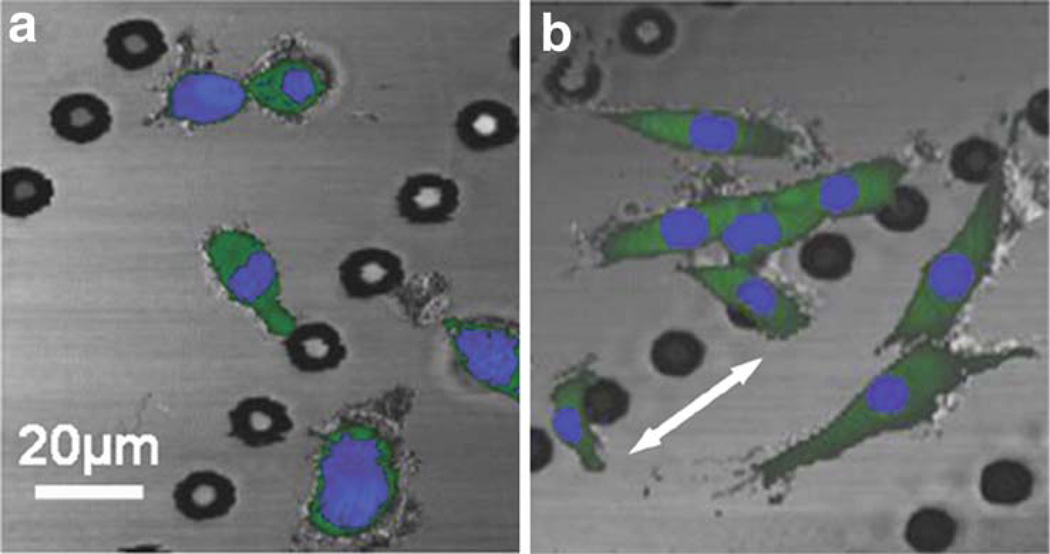
Fig. 13.5.
OCM/MPM images of GFP-vinculin fibroblast cells on a micro-topographic substrate (linear arrays of micropegs) before (a) and after (b) mechanical stimulation. OCM shows the features of the substrate (grey) while MPM images show the GFP fluorescence (green channel) and the nuclear stain (blue channel). Visualizing the nucleus enables individual cells to be clearly identified while the GFP signal shows the extent of the cell body, the level of adhesion of the cell to the substrate, and interactions with adjacent cells. The flexible polymer (PDMS) substrate was repeatedly stretched in the direction of the white arrow in (b). Mechanical stimulation results in an elongation of the fibroblast cells, roughly along the direction of stretch. The stretching causes an elongation of the cells and an increase in the distribution of the GFP signal indicating that there is an increase in expression of vinculin. Modified figure used with permission (31).
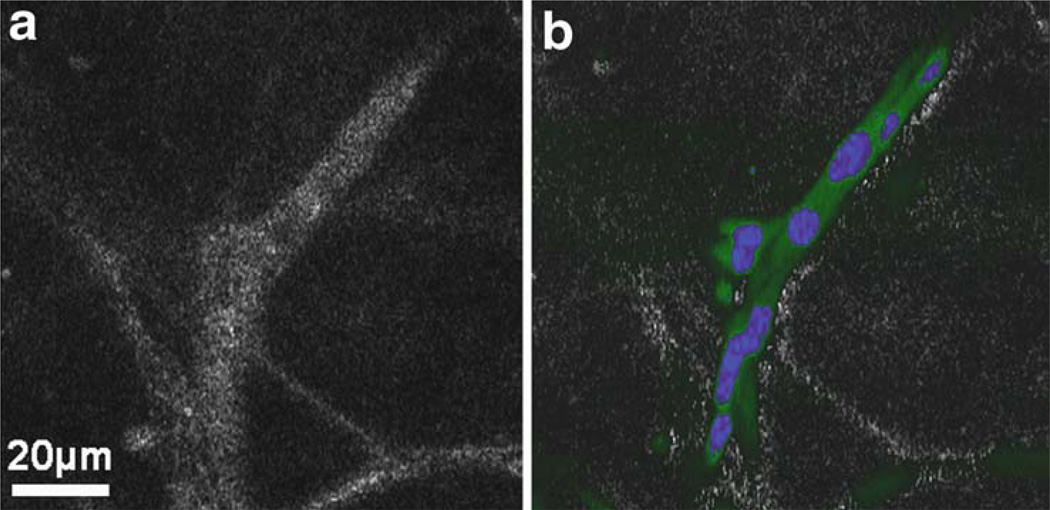
Fig. 13.6.
OCM (a) and MPM overlaid on OCM (b) images from a 3D Matrigel scaffold seeded with GFP-vinculin fibroblast cells stained with a nuclear dye prior to mechanical stimulation. The green signal in the MPM image is from GFP and indicates the relative degree of cell adhesion. The blue signal is the nuclear stain which identifies individual cells. Scattering in this culture, as seen in the OCM image, is caused by both the cells and the extra-cellular environment (collagen secreted by the cells). It is observed that the cells have organized into an interconnected architecture along the fibrous collagen structure of the Matrigel. Modified figure used with permission (31).
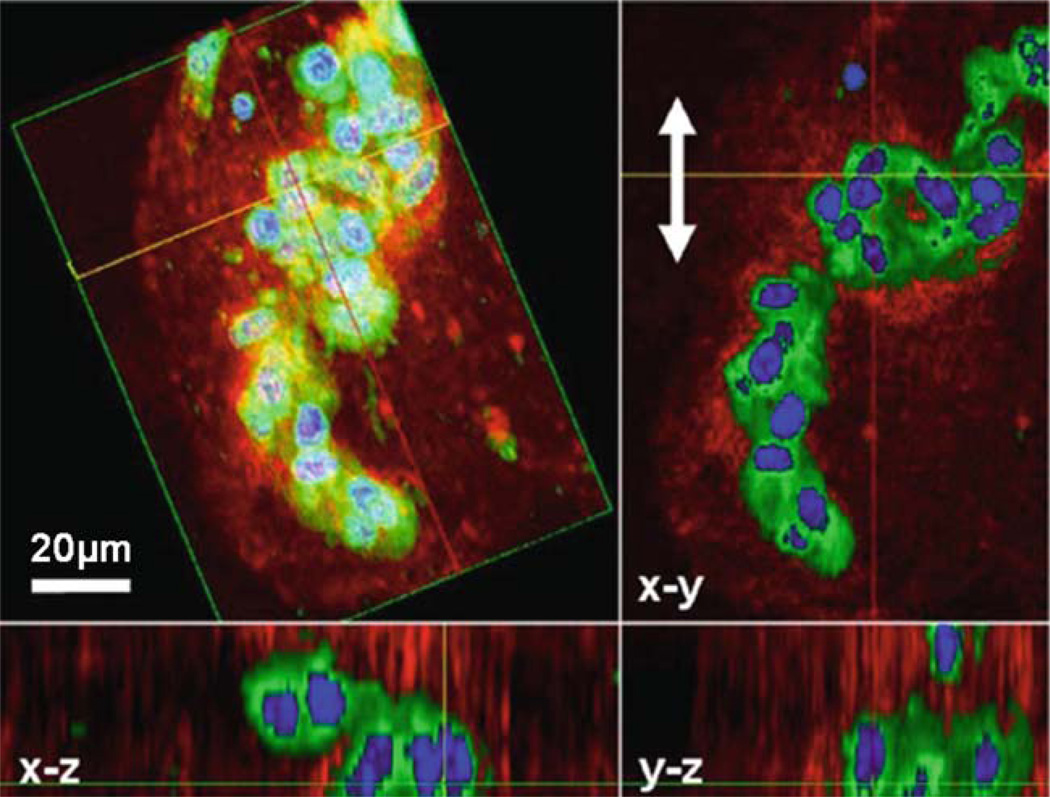
Fig. 13.7.
Multimodality 3D OCM/MPM image of GFP-vinculin fibroblasts in a 3D Matrigel scaffold acquired after mechanical stimulation. The 3D reconstruction is shown (top left) as well as projections along different axes giving a comprehensive view of this cluster of cells. The red channel corresponds to the scattering from the sample (OCM) while the green and blue channels are fluorescence images (MPM). Three-dimensional reconstruction, rotation, and computational sectioning at arbitrary plane angles allow for a more comprehensive view of a 3D cell culture. Mechanical stimulation in the direction of the arrows in the x-y plane has induced the cells to become more spherical and form clusters, potentially indicating that adhesion sites with the scaffold were broken during mechanical stimulation. Modified figure used with permission (31).
Footnotes
Traditional microscopy requires that a specimen be fixed and cut into thin slices and placed on a glass slide for imaging. Optical sectioning refers to imaging thin sections of the intact specimen. In addition to enabling visualization of samples in 3D, optical sectioning has the advantage of eliminating the need for the harsh processing steps that physically destroy the specimen and may alter some of its structural properties. The ability to obtain optical sections in live specimens allows one to observe dynamic events in biology. Instead of viewing different specimens at fixed points in time, single specimens can be observed longitudinally over time with minimal disturbance. The optical sectioning techniques discussed in this chapter are particularly well-suited for imaging thick, highly scattering samples that do not allow light to be transmitted.
MPM is typically less harmful to biological specimen than fluorescence mode CM because it minimizes photobleaching and photodamage (32). However, MPM requires focusing ultrashort laser pulses into a sub-femtoliter volume of a sample. Although power levels needed for MPM are moderate (2–20 mW), the high instantaneous intensity of light could possibly damage a specimen through different mechanisms. Exposure to ultrashort pulses of light has been shown to cause cell death through oxidative photodamage (33, 34). In practice, damage can be minimized by limiting the power levels and reducing the exposure time of the sample to the laser beam. The later can be achieved by rapidly scanning the beam or blocking the beam path to the sample when the beam is not scanning.
A key advantage of swept-source OCT over spectral-domain OCT is the imaging speed. Imaging speed for spectral-domain OCT is limited by the read-out speed of the line camera in the spectrometer. Currently, maximum readout rates are ~100 kHz, or ~100,000 columns in OCT or points in OCM, per second). Swept-source OCT using Fourier-domain mode-locked lasers can achieve speeds of several hundred kHz (16). However, these lasers currently operate in the 1.3 µm wavelength range. Spectral-domain OCT can be performed with a broad bandwidth laser source centered at any wavelength; however, line cameras sensitive to wavelengths greater than 1000 nm use indium-gallium-arsenide detection elements and can be expensive. Very high imaging speeds are needed when imaging in vivo or when observing fast dynamic events.
Resulting porous scaffold should have an interconnected porosity of >80% with an average pore size of 100 µm.
With this OCT system, the resolution is sufficient so that the location and structure of the cell population can be visualized, even though individual cellular features cannot be identified. The chitosin scaffold itself is not visible due to the low scattering properties of the material. OCT was capable of imaging throughout the depth of the sample (~1 mm).
Histological preparation included fixing samples with 3.7% formaldehyde, embedding in paraffin, cutting into 5 µm slices with a microtome, and staining with hematoxylin and eosin.
These fibroblast cells are transfected to co-express GFP with vinculin. Vinculin is a surface adhesion protein and thus the presence of a GFP signal correlates to cell–cell and cell–substrate adhesion.
Although cells in 3D micro-topographic cultures are located on a surface of the polymer substrate, these substrates have defined 3D features and characteristics, compared to flat planar cultures, and have been shown to affect cellular organization (35). Cells in the Matrigel culture are in a true 3D microenvironment.
All fluorescent dyes used with single photon excitation can be used for MPM. The emission spectrum of a dye excited with either one photon or with two simultaneous photons is similar and the same filters can often be used. However, the two-photon excitation spectrum of a fluorescent molecule is not easily predicted from its one-photon spectrum and must therefore be determined experimentally in many cases. The two-photon emission spectra for many commonly used dyes and biomolecules have been characterized (36, 37).
The response of the cells in the Marigel culture to stretching is opposite to what was observed with the cells on the micro-topographic substrates, where the fibroblasts were initially more spherical, and became elongated following mechanical stimulation. The different responses could potentially be explained by differences in adhesion strength to the PDMS and to the Matrigel, or to the differences in strain experienced at the cellular level in each type of 3D culture. These images demonstrate that OCM and MPM can enable the observation of the complex cell-scaffold interactions in 3D cultures. With this integrated imaging technique, 3D multi-modality information from cultures can be acquired, providing a comprehensive view of the structural and functional characteristics of the sample.
References
- 1.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 3.Friedrich MJ. Studying cancer in 3 dimensions: 3D models foster new insights into tumorigenesis. J. Am. Med. Assoc. 2003;290:1977–1979. doi: 10.1001/jama.290.15.1977. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng. Part B. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 5.Pawley J. Handbook of Biological Confocal Microscopy. New York, NY: Springer; 1995. [Google Scholar]
- 6.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt JM. Optical coherence tomography (OCT): a review. IEEE J. Sel. Top. Quantum Electron. 1999;5:1205–1215. doi: 10.1109/2944.796347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajadhyaksha M, Grossman M, Esterowitz D, Webb RH, Anderson RR. In vivo confocal scanning laser microscopy of human skin: melanin provides strong contrast. J. Invest. Dermatol. 1995;104:946–952. doi: 10.1111/1523-1747.ep12606215. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt JM, Knüttel A, Yadlowsky M. Confocal microscopy in turbid media. J. Opt. Soc. Am. A. 1994;11:2226–2235. doi: 10.1364/josaa.11.002226. [DOI] [PubMed] [Google Scholar]
- 10.Smithpeter CL, Dunn AK, Welch AJ, Richards-Kortum R. Penetration depth limits of in vivo confocal reflectance imaging. Appl. Opt. 1998;37:2749–2754. doi: 10.1364/ao.37.002749. [DOI] [PubMed] [Google Scholar]
- 11.Centonze VE, White JG. Multiphoton excitation provides optical sectionsfrom deeper within scattering specimens than confocal imaging. Biophys. J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubart M. Two-photon microscopy of cells and tissue. Circ. Res. 2004;95:1154–1166. doi: 10.1161/01.RES.0000150593.30324.42. [DOI] [PubMed] [Google Scholar]
- 13.Huang D, Swanson EA, Lin CP, Schu-man JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitgeb R, Hitzenberger CK, Fercher AF. Performance of Fourier-domain vs. time-domain optical coherence tomography. Opt. Express. 2003;11:889–894. doi: 10.1364/oe.11.000889. [DOI] [PubMed] [Google Scholar]
- 15.Nassif N, Cense B, Park BH, Yun SH, Chen TC, Bouma BE, Tearney GJ, de Boer JF. In vivo human retinal imaging by ultrahigh-speed spectral domain optical coherence tomography. Opt. Lett. 2004;29:480–482. doi: 10.1364/ol.29.000480. [DOI] [PubMed] [Google Scholar]
- 16.Huang SW, Aguirre AD, Huber RA, Adler DC, Fujimoto JG. Swept source optical coherence microscopy using a Fourier domain mode-locked laser. Opt. Express. 2007;15:6210–6217. doi: 10.1364/oe.15.006210. [DOI] [PubMed] [Google Scholar]
- 17.Welzel J. Optical coherence tomography in dermatology: a review. Skin Res. Tech. 2008;7:1–9. doi: 10.1034/j.1600-0846.2001.007001001.x. [DOI] [PubMed] [Google Scholar]
- 18.Aguirre AD, Hsiung P, Ko TH, Hartl I, Fujimoto JG. High-resolution optical coherence microscopy for high-speed, in vivo cellular imaging. Opt. Lett. 2003;28:2064–2066. doi: 10.1364/ol.28.002064. [DOI] [PubMed] [Google Scholar]
- 19.Vinegoni C, Ralston T, Tan W, Luo W, Marks DL, Boppart SA. Integrated structural and functional optical imaging combining spectral-domain optical coherence and multiphoton microscopy. Appl. Phys. Lett. 2006;88:053901. [Google Scholar]
- 20.Tang S, Sun CH, Krasieva TB, Chen Z, Tromberg BJ. Imaging sub-cellular scattering contrast by using combined optical coherence and multiphoton microscopy. Opt. Lett. 2007;32:503–505. doi: 10.1364/ol.32.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang S, Krasieva TB, Chen Z, Tromberg BJ. Combined multi-photon microscopy and optical coherence tomography using a 12-fs broadband source. J. Biomed. Opt. 2006;11:020502. doi: 10.1117/1.2193428. [DOI] [PubMed] [Google Scholar]
- 22.Beaurepaire E, Moreaux L, Amblard F, Mertz J. Combined scanning optical coherence and two-photon excited fluorescence microscopy. Opt. Lett. 1999;24:969–971. doi: 10.1364/ol.24.000969. [DOI] [PubMed] [Google Scholar]
- 23.Dunkers J, Cicerone M, Washburn N. Collinear optical coherence and confocal fluorescence microscopies for tissue engineering. Opt. Express. 2003;11:3074–3079. doi: 10.1364/oe.11.003074. [DOI] [PubMed] [Google Scholar]
- 24.Morgner U, Drexler W, Kärtner FX, Li XD, Pitris C, Ippen EP, Fujimoto JG. Spectroscopic optical coherence tomography. Opt. Lett. 2000;25:111–113. doi: 10.1364/ol.25.000111. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Vinegoni C, Ralston T, Luo W, Tan W, Boppart S. Spectroscopic spectral-domain optical coherence microscopy. Opt. Lett. 2006;31:1079–1081. doi: 10.1364/ol.31.001079. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt J. OCT elastography: imaging microscopic deformation and strain of tissue. Opt. Express. 1998;3:199–211. doi: 10.1364/oe.3.000199. [DOI] [PubMed] [Google Scholar]
- 27.Liang X, Oldenburg AL, Crecea V, Chaney EJ, Boppart SA. Optical micro-scale mapping of dynamic biomechanical tissue properties. Opt. Express. 2008;16:11052–11065. doi: 10.1364/oe.16.011052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milner TE, Srinivas S, Wang X, Malekafzali A, Van Gemort MJ, Nelson JS, Chen Z. Noninvasive imaging of in vivo blood flow velocity using optical Doppler tomography. Opt. Lett. 1997;22:1119–1121. doi: 10.1364/ol.22.001119. [DOI] [PubMed] [Google Scholar]
- 29.Tan W, Sendemir-Urkmez A, Fahrner LJ, Jamison R, Leckband D, Boppart SA. Structural and functional optical imaging of three-dimensional engineered tissue development. Tissue Eng. 2004;10:1747–1756. doi: 10.1089/ten.2004.10.1747. [DOI] [PubMed] [Google Scholar]
- 30.Tan W, Oldenburg AL, Norman JJ, Desai TA, Boppart SA. Optical coherence tomography of cell dynamics in three-dimensional tissue models. Opt. Express. 2006;14:7159–7171. doi: 10.1364/oe.14.007159. [DOI] [PubMed] [Google Scholar]
- 31.Tan W, Vinegoni C, Norman JJ, Desai TA, Boppart SA. Imaging cellular responses to mechanical stimuli within three-dimensional tissue constructs. Microsc. Res. Tech. 2007;70:361–371. doi: 10.1002/jemt.20420. [DOI] [PubMed] [Google Scholar]
- 32.Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat. Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirlapur UK, König K, Peuckert C, Krieg R, Halbhuber KJ. Femtosecond near-infrared laser pulses elicit generation of reactive oxygen species in mammalian cells leading to apoptosis-like death. Exp. Cell. Res. 2001;263:88–97. doi: 10.1006/excr.2000.5082. [DOI] [PubMed] [Google Scholar]
- 34.Sun C, Chen C, Chu S, Tsai T, Chen Y, Lin B. Multiharmonic-generation biopsy of skin. Opt. Lett. 2003;28:2488–2490. doi: 10.1364/ol.28.002488. [DOI] [PubMed] [Google Scholar]
- 35.Norman JJ, Desai TA. Control of cellular organization in three dimensions using a microfabri-cated polydimethylsiloxane-collagen composite tissue scaffold. Tissue Eng. 2005;11:378–386. doi: 10.1089/ten.2005.11.378. [DOI] [PubMed] [Google Scholar]
- 36.Bestvater F, Spiess E, Stobrawa G, Hacker M, Feurer T, Porwol T, Berchner-Pfannschmidt U, Wotzlaw C, Acker H. Two-photon fluorescence absorption and emission spectra of dyes relevant for cell imaging. J. Microsc. 2002;208:108–115. doi: 10.1046/j.1365-2818.2002.01074.x. [DOI] [PubMed] [Google Scholar]
- 37.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. PNAS. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]