Abstract
Background
A genetic contribution to cannabis dependence (CaD) has been established, but susceptibility genes for CaD remain largely unknown.
Methods
We employed a multi-stage design to identify genetic variants underlying CaD. We first performed a genomewide linkage scan for CaD in 384 African-American (AA) and 354 European-American (EA) families ascertained for genetic studies of cocaine and opioid dependence. We then conducted association analysis under the linkage peak, first using data from a genomewide association study from the Study of Addiction: Genetics and Environment (SAGE), followed by replication studies of prioritized single nucleotide polymorphisms (SNPs) in independent samples.
Results
We identified the strongest linkage evidence with CaD (lod=2.9) on chromosome 8p21.1 in AAs. In the association analysis of the SAGE sample under the linkage peak, we identified one SNP (rs17664708) associated with CaD in both AAs (minor allele frequency (MAF) = 0.02, OR=2.93, 95% CI=1.47–5.85, P=0.0022) and EAs (MAF=0.096, OR=1.38, 95% CI=1.05–1.81, P=0.02). This SNP, located at NRG1, a susceptibility gene for schizophrenia, was prioritized for further study. We replicated the association of rs17664708 with CaD in an independent sample of AAs (MAF=0.013, OR=2.81, 95% CI=1.23–6.45, P=0.0068). The joint analysis of the two AA samples demonstrated highly significant association between rs17664708 and CaD with adjustment for either global (OR=2.34, 95% CI=1.42–3.85, P=0.00044) or local ancestry (OR=2.33, 95% CI=1.39–3.91, P=0.00075).
Conclusions
Our study shows that NRG1 is probably a susceptibility gene for CaD, based on convergent evidence of linkage and replicated associations in two independent AA samples.
Keywords: Cannabis dependence, linkage, association, candidate gene, SNP, NRG1
Introduction
Cannabis is the most commonly used illicit substance in the world with 143–190 million people in 2007 having used the drug at least once worldwide (1). An estimated 14.4 million Americans aged 12 or older reported cannabis use over the past month (2). Of these individuals, ~7% develop cannabis dependence (CaD) defined by DSM-IV criteria (3). With the expansion of legalization in the United States (generally for “medical” use), availability and use of cannabis is rising. Cannabis use is often accompanied by dependence on alcohol and other drugs (4), and is associated with serious consequences, including cognitive and psychomotor impairments (5, 6). The use of cannabis is associated with roughly twofold increased risk of schizophrenia; there is interindividual variability in susceptibility to cannabis-induced psychosis that could be, in part, genetic in origin (7, 8). Thus, it is important to identify factors that influence individual vulnerability to the development of CaD.
Family and twin studies have shown that CaD has an important genetic component. The heritability of cannabis abuse or dependence was estimated to be 45–78% (9). Genomewide linkage studies (GWLS) and candidate gene association studies have identified a list of possible chromosomal risk regions and candidate genes for cannabis use disorders (9–16). For example, linkage studies have identified genomic regions harboring candidate genes with biological relevance, such as the monoacylglycerol lipase gene (MGLL) on chromosome 3 and the gamma-aminobutyric acid receptor subunit alpha-2 gene (GABRA2) on chromosome 4 (14). The cannabinoid receptor gene (CNR1) and several other genes (CRN2, FAAH, and MGLL), which are specific to the endogenous cannabinoid system, have been selected for candidate gene association studies, although the results were largely inconclusive (14). Recently, a genomewide association study (GWAS) for CaD was conducted in 708 individuals with DSM-IV CaD cases and 2346 cannabis-exposed non-dependent controls, using a GWAS dataset from the Study of Addiction: Genetics and Environment (SAGE) (17). However, no results achieved genomewide significance in this study. Despite the effort that has been made in gene mapping for CaD, genetic factors underlying CaD susceptibility remain largely unknown.
With the growing evidence for the role of rare variants and copy number variation (CNV) in psychiatric disorders (18–20), linkage analysis remains a useful approach to gene discovery. An adequately powered linkage study can detect diverse kinds of genetic polymorphism that segregate in families, including common variants, multiple rare variants within one locus, and inherited CNVs. The apparent failure to identify association under linkage peaks could in part be attributable to the fact that often, only common variants are examined under the linkage peak, whereas the linkage signal could be caused by multiple rare variants with higher penetrance (21–23).
The current study employed a multi-stage design using a linkage scan, a GWAS dataset, and replication in independent samples to identify genetic variants associated with CaD. Specifically, the objectives of the current study were to: 1) conduct a genomewide linkage scan to detect genetic loci influencing CaD risk in African-Americans (AAs) and European-Americans (EAs); 2) assess the genetic association between CaD and single nucleotide polymorphisms (SNPs) under the strongest linkage peak using the GWAS dataset from SAGE; and 3) replicate prioritized SNPs in independent AAs and EAs from our samples.
Subjects and Methods
Study samples
The basic demographic information for the three samples involved in each stage of the study is summarized in Table 1. The following section provides details of the sample recruitment and characteristics for each sample set. Written informed consent was obtained from all subjects; the IRB at each recruitment site approved the study, and NIAAA and NIDA issued certificates of confidentiality for the work.
TABLE I.
Basic demographic information for the three samples included in the current study
| African-Americans | European-Americans | |
|---|---|---|
| Linkage sample | ||
| No. of families | 384 | 355 |
| No. of genotyped individuals | 1022 | 874 |
| No. of pedigrees with 1 CaD | 134 | 136 |
| No. of pedigrees with ≥ 2 CaD | 40 | 54 |
| Age (years ±SD) (CaD) | 38.6±6.5 | 33.5±9.6 |
| Males (%) (CaD) | 57.8 | 60.4 |
| SAGE sample | ||
| CaD | 275 | 422 |
| Males (%) | 66.5 | 67.3 |
| Age (years ±SD) | 38.7±7.8 | 34.8±8.5 |
| Control | 401 | 1049 |
| Males (%) | 38.2 | 33.1 |
| Age (years ±SD) | 39.8±7.5 | 39.1±9.7 |
| Replication Sample | ||
| CaD | 758 | 568 |
| Males (%) | 67.7 | 70.4 |
| Age (years ±SD) | 40.1±8.7 | 34.7 ±10.8 |
| Control | 280 | 318 |
| Males (%) | 26.4 | 40.3 |
| Age (years± SD) | 37.3±13.2 | 38.7±13.9 |
CaD, cannabis dependence
Linkage scan sample
Subjects were originally ascertained for genetic studies of cocaine dependence (CD) and opioid dependence (OD) using the affected sibling pair (ASP) linkage approach (24, 25). The recruitment procedure has been previously described in detail (26–28). Briefly, there were four recruitment sites: University of Connecticut Health Center, Yale University School of Medicine, Medical University of South Carolina, and McLean Hospital. Families were selected on the basis of having at least two siblings affected with either cocaine and/or opioid dependence. The distribution of family numbers recruited at each site is presented in Supplementary Table 1. Cannabis use played no role in proband selection or pedigree extension. We evaluated these subjects with the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (29, 30), a polydiagnostic instrument that assesses a range of psychiatric diagnoses, including DSM-IV CaD. Probands with an axis I clinical diagnosis of a major psychotic disorder such as schizophrenia or schizoaffective disorder were excluded from participation. Subjects were classified as AA or EA on the basis of a Bayesian model-based clustering method with ancestry informative genetic markers using STRUCTURE (31, 32), as described previously (28).
Study of Addiction: Genetics and Environment (SAGE)
We obtained the individual level SAGE GWAS dataset from the database of Genotypes and Phenotypes (dbGaP). SAGE aims to identify genetic risk factors and the interplay of genes and environmental factors for addiction. Cases and controls were selected from three large, complementary cohorts: the Collaborative Study on the Genetics of Alcoholism (COGA), the Family Study of Cocaine Dependence (FSCD), and the Collaborative Genetic Study of Nicotine Dependence (COGEND), all of which have been previously described (33–36). The current study included 4036 unrelated self-reported AA (1297 in total including 275 CaD and 422 healthy controls) or EA (2740 in total including 401 CaD and 1049 healthy controls) subjects. Lifetime CaD was defined in accordance with the DSM-IV diagnosis. Controls used for association analysis in the current study were defined as subjects without dependence on any substances, including cannabis, alcohol, cocaine, opioid, nicotine, and other substances.
Replication sample
Subjects were recruited for participation in studies of the genetics of CD, OD, and alcohol dependence (AD) from the communities around four sites listed above for linkage scan sample plus the University of Pennsylvania. The number of samples recruited at each site for the current study is presented in Supplementary Table 1. Part of the set of samples genotyped at the replication stage overlapped with the samples used for linkage analysis. To obtain an independent sample set for replication study, the overlapping samples (72 CaD cases) were excluded from analysis in the replication stage. Subjects were interviewed with the SSADDA and the diagnosis of CaD was derived based on DSM-IV diagnostic criteria. Most controls were recruited at the same recruitment sites (excluding McLean) and were screened to exclude those with a diagnosis of DSM-IV substance abuse or dependence, and a major Axis I psychiatric disorders. All subjects included in the current study are self-reported AA or EA. Subjects were re-classified as AA or EA on the basis of 41 ancestry informative genetic markers (AIMs) using STRUCTURE, as described previously (20, 37). Among the subjects included in current study, 3% of the subjects reporting to be of AA descent clustered in the EA group, and 2% of subjects reporting to be EA clustered in the AA group.
Genotyping and Quality Control
Linkage analysis
Genotyping and quality control (QC) for linkage analysis have been previously described in detail (28). Briefly, 1630 subjects were genotyped at the Center for Inherited Disease Research (CIDR) for the 6,008 SNPs Illumina Human Linkage IVb Marker Panel. An additional 266 subjects were genotyped at the Yale Keck Center with the 6,090 SNP Illumina Infinium-12 Human Linkage Marker Panel. We limited our analyses to 4,518 autosomal SNPs available in both panels. After QC (genotyping rate ≥ 95%, minor allele frequency (MAF) ≥0.1, and HWE P ≥0.01), 4,133 and 4,395 autosomal SNPs were retained for analysis in AAs and EAs, respectively. Mendelian inconsistencies and potential genotyping errors were identified and set as missing data using PedCheck (38) and Merlin (39) programs. We used the Pedigree RElationship Statistical Test (PREST) (40) to verify family relationships, which showed pedigree errors in two AA families and five EA families. Of these, the relationships in one AA family and five EA families were corrected based on the shared identical by decent (IBD) patterns and the re-assigned family relationships were verified by PREST. One AA family relationship could not be resolved and the family was excluded from further analysis.
SAGE dataset
SAGE samples were genotyped on the ILLUMINA Human 1 M platform at CIDR. We included 4036 unrelated self-reported AA (1297) or EA (2740) subjects (60 duplicate genotype samples were excluded from analysis). We used PLINK software to perform basic data cleaning steps before analysis (41). After QC(sample call rate ≥ 97%; SNP call rate ≥ 95%; MAF ≥ 0.005 in controls; HWE P ≥ 0.00001 in controls), a total of 1297 (2740) unrelated subjects and 953,258 (888,092) autosomal SNPs for AAs (EAs) were available for further analysis. To obtain a more genetically homogeneous sample and correct for population stratification in the association analysis, we computed principal components (PC) using the EIGENSOFT package (42). Specifically, 172,891 pruned SNPs common to AA and EA samples and in low linkage disequilibrium (LD) (genotypic correlation < 0.5) with one another, were fed into EIGENSOFT. The top two PCs of AA and EA samples and with the Phase II Hapmap CEU and YRI samples are shown in Supplementary Figure 1. Outliers were defined as subjects whose ancestry was at least 3 standard deviations from the mean of the two largest PCs. This step removed 33 AAs and 127 EAs, retaining 1264 AAs and 2613 EAs in the final cleaned dataset.
Replication study
The prioritized SNP, rs17664708, was genotyped for a replication study in 2543 AAs and 2042 EAs (ethnicity verified or re-classified by STRUCTURE using 41 mostly short tandem repeat (STR) AIMs (20, 37). Genotyping was performed with a fluorogenic 5′ nuclease assay method (TaqMan technique), using the ABI 7900HT real time PCR system (ABI, Foster City, CA). The missing rate for genotyping was 0.03 in the replication sample. For genotyping quality control, 8% of samples were re-genotyped with 100% concordance.
A subset of the replication sample (n=931, including 672 cases and 259 controls) of AAs were also genotyped by the Illumina Omni1-Quad platform at CIDR (690 subjects) or the Yale Keck center (241 subjects) for our ongoing GWAS study of alcohol, cocaine, opioid, and nicotine dependence. Genotype information from these subjects were used to control for the potential population stratification in association test in AAs (The prioritized SNP rs17664708 was not included on the Illumina Omni1-Quad chip). After QC (SNP call rate ≥ 95%, sample call rate ≥ 97%, MAF ≥ 0.005 in controls, HWE P ≥ value 0.00001 in controls), 2745 AIMs across the whole genome and 188 SNPs located at NRG1 were extracted to estimate the global and local ancestry in this subset of AA samples, respectively.
Statistical analysis
Linkage analysis
We used Merlin (39) to perform the linkage scan using a nonparametric allele-sharing model. Allele frequencies were estimated by counting all genotyped individuals. The Kong and Cox linear allele-sharing model (43) was used to estimate the lod score. To minimize the inflation of linkage signals caused by marker-marker LD, we grouped SNPs by LD into clusters using the Merlin “--rsq” option (44). Analyses were repeated with r2 thresholds of 0.05, 0.2, and 0.3 to evaluate the robustness of the linkage results. We assessed the thresholds for autosomal genomewide suggestive and significant linkages, and the autosomal genomewide empirical significance of an observed lod score, by 1000 computer simulations, as described previously (28).
Global ancestry estimation in AAs
Spurious association between a marker and a phenotype can arise from population stratification, especially in admixed populations such as AAs. To account for the effect of population stratification in AAs, we estimated the individual global ancestry using STRUCTURE program and included it as a covariate in the association analysis. To obtain a more consistent estimate of individual global ancestry for SAGE and our replication AA samples, we selected 2475 AIMs that were genotyped in both the SAGE AA sample and a subset of our replication sample (931 subjects). These AIMs were common to a reported AIMs panel for AAs based on a subset of SNPs on the ILLUMINA Human 1 M platform (45). The log likelihood of each analysis at varying numbers of assumed population groups (k) was estimated from the average of 3 independent runs (5,000 burn in and 5,000 iterations). As expected, the results favored a two-ancestry population model. The average proportion of European ancestry was 0.186 in the SAGE AA sample and 0.166 in our replication AA sample.
Local ancestry estimation in AAs
Because the global ancestry information obtained across the whole genome may not reflect the variation of ancestry at the tested genomic locus, methods that adjust the global ancestry to control population stratification may be insufficient. However, methods that conditioned on local ancestry at the tested locus more fully account for the confounding effect of hidden population structure (46). Therefore, we estimated the local ancestry at the NRG1 locus using the HAPMIX program (47), and the overall ancestry across SNPs at the NRG1 locus was included as a covariate in the association analysis to control for the local population stratification, (i.e., the estimated ancestry of the specific genomic region under consideration). Briefly, we downloaded the phased YRI and CEU data from Hapmap Phase II as the parental reference haplotype input for HAPMIX. After QC and filtering the monomorphic SNPs in the phased YRI and CEU data, 188 SNPs that were located at the NRG1 gene and were genotyped in both the SAGE AA sample and in the subset of the replication AA sample (931), were used to estimate local ancestry at the NRG1 locus. The estimated average European ancestries at the NRG1 locus were 0.194 and 0.171 for the SAGE and the replication AA samples, respectively, which approximated the values of the global European ancestry proportion in each sample.
Association analysis
The association between each SNP and the binary trait was estimated in a multivariate logistic regression framework under a log-additive genetic model. We used PLINK for the SNP-trait association test in the region of the linkage peak in the SAGE sample, with sex, age and the top 10 PCs as covariates. For the replication analysis and joint data analysis stages for rs17664708, we included sex, age, and global or local ancestry estimates as covariates where appropriate, and analysis was performed using the R package “SNPassoc” (48).
Results
Genomewide linkage analysis for CaD
Empirical genomewide thresholds for suggestive and significant linkage for non-parametric linkage analyses in our family dataset were determined based on 1000 simulations. The thresholds for genomewide suggestive and significant linkage in AAs (EAs) were 1.79 (1.76) and 3.23 (3.22), respectively.
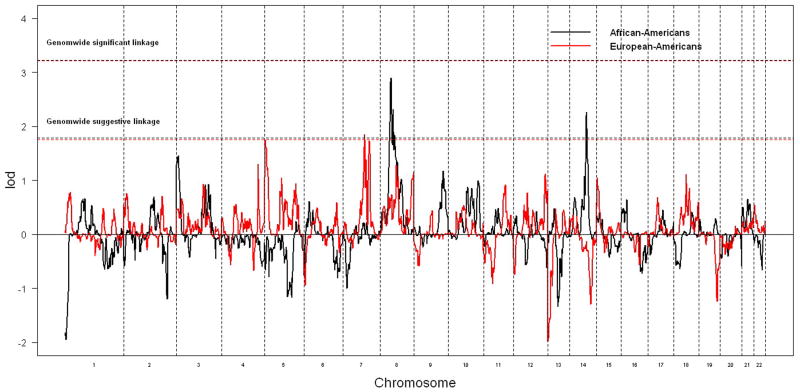
The genomewide non-parametric linkage results for AAs and EAs are presented in Figure 1. The strongest linkage signal was identified on chromosome 8p21.1 at 54.9 cM (lod=2.9, pointwise P = 0.00013, empirical genomewide P = 0.097) in AAs, and weak linkage evidence was detected at the same location in EAs (lod=0.62, pointwise P = 0.05). We identified a second genomewide suggestive linkage peak in AAs on chromosome 14 with a peak lod of 2.26 at 89.9 cM, though no linkage evidence was detected at this peak region in EAs. In EAs, only one region showed genomewide suggestive linkage on chromosome 7 at 107 cM (lod=1.85), where weak linkage signal was observed in AAs (lod=0.19). The locations and values of these suggestive linkage peaks did not change when analyses were repeated using different r2 values to group SNPs into clusters.
Figure 1.
Genomewide non-parametric linkage results in AAs and EAs. The black and red virtue lines represent the empirical genomewide suggestive (significant) linkage thresholds for AAs and EAs, respectively.
Association analysis under the linkage peak using SAGE GWAS dataset
Utilizing the SAGE GWAS dataset, we examined the genetic association between CaD and 4853 SNPs under the strongest linkage peak from our family sample (lod > 2, from 48.9 cM to 65.2 cM) on chromosome 8. All of these SNPs passed QC and were tested for genetic association with adjustment for sex, age and the top 10 PCs in AAs and EAs separately. Eleven SNPs were nominally significantly associated with CaD in both AAs and EAs (Table 2). One SNP (rs17664708), which is relatively rare in AA controls (MAF = 0.02) but common in EA controls (MAF=0.096), showed consistent evidence for association in both AAs (OR=2.93, 95% CI=1.47–5.85, P=0.0022) and EAs (OR=1.38, 95% CI=1.05–1.81, P=0.02). This SNP is located at NRG1, which has been previously implicated in the risk for schizophrenia. Considering the high degree of commorbidity between cannabis use and schizophrenia (7, 8, 49–53), this SNP (rs17664708) was prioritized for further study.
TABLE 2.
Summary of association analysis results under the linkage peak for SNPs nominally significantly associated with CaD in both AAs and EAs using SAGE GWAS dataset
| SNP | Position1 | Function | Gene | African-Americans (275 cases/401 controls) | European-Americans (422 cases/1049 controls) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MA | MAF (Cases) | MAF (Conts) | OR | 95% CI | P | MA | MAF (Cases) | MAF (Conts) | OR | 95% CI | P | ||||
| rs931874 | 25301315 | Intronic | DOCK5 | T | 0.19 | 0.24 | 0.74 | 0.56–0.99 | 0.043 | T | 0.38 | 0.36 | 1.20 | 1.00–1.43 | 0.05 |
| rs6558049 | 28148960 | Intergenic | - | A | 0.23 | 0.18 | 1.41 | 1.05–1.89 | 0.021 | A | 0.026 | 0.045 | 0.54 | 0.33–0.90 | 0.017 |
| rs7015363 | 28159947 | Intergenic | - | G | 0.23 | 0.18 | 1.36 | 1.02–1.81 | 0.037 | G | 0.026 | 0.046 | 0.53 | 0.32–0.88 | 0.013 |
| rs7011660 | 30405716 | Intronic | RBPMS | A | 0.19 | 0.23 | 0.74 | 0.56–0.98 | 0.038 | A | 0.19 | 0.22 | 0.77 | 0.62–0.95 | 0.016 |
| rs7833229 | 30542430 | Intronic | RBPMS | A | 0.024 | 0.042 | 0.45 | 0.22–0.90 | 0.025 | A | 0.11 | 0.14 | 0.74 | 0.57–0.97 | 0.031 |
| rs17664708 | 32556559 | Intronic | NRG1 | T | 0.049 | 0.020 | 2.93 | 1.47–5.85 | 0.0022 | T | 0.13 | 0.096 | 1.38 | 1.05–1.81 | 0.020 |
| rs12155970 | 34064572 | Intergenic | - | C | 0.065 | 0.034 | 2.20 | 1.26–3.86 | 0.0057 | C | 0.093 | 0.062 | 1.51 | 1.09–2.09 | 0.012 |
| rs2593095 | 38587762 | Intergenic | - | T | 0.54 | 0.48 | 1.27 | 1.00–1.61 | 0.049 | T | 0.33 | 0.39 | 0.77 | 0.64–0.93 | 0.0056 |
| rs6989042 | 40301460 | Intergenic | - | G | 0.30 | 0.23 | 1.32 | 1.01–1.73 | 0.041 | G | 0.23 | 0.20 | 1.25 | 1.01–1.55 | 0.037 |
| rs7826645 | 40924370 | Intergenic | - | A | 0.13 | 0.16 | 0.68 | 0.49–0.96 | 0.026 | A | 0.16 | 0.11 | 1.50 | 1.17–1.93 | 0.0016 |
| rs12547609 | 41091390 | Intergenic | - | C | 0.15 | 0.21 | 0.68 | 0.50–0.93 | 0.015 | C | 0.21 | 0.17 | 1.31 | 1.05–1.63 | 0.018 |
CaD, cannabis dependence; SNP, single nucleotide polymorphism; MA, minor allele; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; Conts, Controls;
Coordinate based on Genome Build 36.3. Association tests are adjusted by sex, age and the top 10 principal components. Prioritized SNP (rs17664708) for follow up study is shown in bold.
Replication of the prioritized SNP in independent AAs and EAs
We tested the association between rs17664708 and CaD in independent AAs (758 CaD and 280 healthy controls) and EAs (568 CaD and 318 healthy controls) from our own sample. The genotype distribution of this SNP was in HWE in both cases and controls for AAs and EAs (In AAs, P=1 for both cases and controls; In EAs, P=0.17 and P=0.80 for cases and controls, respectively). We replicated the association of rs17664708 with CaD in AAs after adjusting for sex and age (OR=2.81, 95% CI=1.23–6.45, P=0.0068), but not in EAs (OR=1.04, 95% CI=0.77–1.40, P=0.82). The MAF of rs17664708 in the replication AA sample was 0.013 and 0.038 in controls and cases, respectively, which approximated the values from the SAGE AA sample.
Joint analysis in AAs
Considering that the strongest linkage signal on chromosome 8 was observed only in AAs, we performed a joint analysis for the association of rs17664708 to CaD in AAs including sex, age, and global or local ancestry as covariates (Table 3). To permit comparison, analysis results with adjustment for global or local ancestry in each AA sample are also listed in Table 3. The association results remained significant after adjusting for either global (P=0.00044) or local ancestry (P=0.00075), which argues against the possibility that the significant association was caused by population stratification in AAs.
TABLE 3.
Association analysis for rs17664708 with CaD in the joint samples of AAs
| Datasets (Cases/Controls) | MAF (case) | MAF (cont) | Association test adjusted by | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex and age | Sex, age and global ancestry | Sex, age and Local ancestry | |||||||||
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |||
| SAGE (275/401) | 0.049 | 0.020 | 2.60 | 1.37±4.92 | 0.0028 | 2.71 | 1.38±5.32 | 0.0031 | 2.73 | 1.35±5.51 | 0.0043 |
| Replication (672/259)1 | 0.038 | 0.013 | 2.70 | 1.17±6.24 | 0.010 | 2.15 | 0.92±5.03 | 0.059 | 2.17 | 0.91±5.16 | 0.063 |
| Joint (947/660) | 0.042 | 0.018 | 2.45 | 1.49±4.02 | 0.00017 | 2.34 | 1.42±3.85 | 0.00044 | 2.33 | 1.39–3.91 | 0.00075 |
CaD, cannabis dependence;
To estimate the local ancestry at NRG1 locus and the global ancestry in the replication AA sample, only a subset of individuals (931) who were genotyped by Illumina Omni1-Quad platform were included.
Discussion
The current study demonstrated how linkage analysis could inform genetic association studies and lead to discovery of a rare variant in NRG1 associated with CaD in AAs. We first conducted a dense GWLS for CaD in small nuclear families recruited on the basis of ASPs with either CD and/or OD. The strongest evidence for linkage in our family sample was observed on 8p21.1 (lod =2.9) in AAs. To our knowledge, this region has not been previously reported by linkage studies of any substance dependence disorder. However, chromosome 8p22-p21, which overlaps our linkage signal, has been repeatedly implicated in several neuropsychiatric disorders including schizophrenia, bipolar affective disorder, and major depression (54). Using the SAGE GWAS dataset, we found that rs17664708 located at NRG1 under the linkage peak was associated with CaD in both AAs and EAs. The association was further replicated in an independent AA sample. Our rigorous QC and statistical analysis adjusting for both global and local ancestry argues against the possibility that the significant association between the rs17664708 and CaD is from the effect of population stratification in AAs.
None of the SNPs that were nominally significant (P ≤ 0.05) in both AAs and EAs can survive multiple testing correction in the discovery stage of association analysis under the linkage peak using the SAGE dataset. However, our prioritization strategy, designed to order SNPs for follow up studies, relies on both statistical evidence and prior knowledge. Accordingly, the SNP rs17664708 located at NRG1 was prioritized for further studies. NRG1 is a well established susceptibility gene for schizophrenia in many populations supported by genetic linkage, association studies, and meta-analysis (55–57). There are at least three lines of prior knowledge which prompt us to consider NRG1 as a candidate gene for CaD as well. First, elevated rates of cannabis use have repeatedly been reported among individuals with schizophrenia (49, 50), and epidemiological studies suggest that frequent cannabis use is associated with about 2-fold increased risk for developing schizophrenia and related disorders (7,8,51–53). The significant commorbidity between cannabis use and schizophrenia may be attributable in part to a shared underlying genetic component for CaD and schizophrenia. Second, the neurobiology of CaD and schizophrenia overlap; for example, the mesolimbic pathway is heavily implicated in the neurobiology of CaD (and other substance dependence) and schizophrenia (58, 59). Third, animal studies have provided evidence supporting the role of NRG1 in CaD. NRG1 heterozygous mice have increased sensitivity to the acute neurobehavioral effects of cannabinoids (60, 61); NRG1 modulates the development of tolerance to cannabinoids in mice (62).
The gene NRG1 encodes neuregulin 1, a pleiotropic growth factor that is important in nervous system development and function. It has been implicated in the modulation of many processes of neural development, including neuronal migration, synapse formation, synaptic plasticity and neuronal survival (63). The SNP rs17664708 we reported here is located at the intron region of NRG1. We did not find any evidence supporting the functional role of this SNP involved in CaD. As it is the case for most genetic association studies, the associated SNP may not be the causal SNP but could represent a tag SNP which is in linkage disequilibrium with the surrounding causal variants. Further studies using a higher density SNP panel and deep sequencing technology are needed to fully characterize the genetic architecture of NRG1 and pinpoint the functional variants that could be involved in CaD.
Cannabinoids have been shown to produce greater behavioral effects in female than in male rats (64, 65). There is also evidence showing that male Nrg1 HET mice being more sensitive to the acute effects of the psychotropic cannabis constitute Δ9-tetrahydrocannabinol (THC) which is not observed in females (66). We investigated whether there is a sex-specific effect for the rs17664708 in CaD susceptibility by sex-stratified analysis. We observed a stronger effect for rs17664708 in females (OR=3.08, 95% CI=1.56–6.07, P=0.0007) than in males (OR=1.88, 95% CI=0.95–3.74, P=0.055). The stronger effect of rs17664708 in females provides further evidence for the interactions between NRG1 and sex in CaD susceptibility.
Samples included in the current study were ascertained for DSM-IV cocaine or opioid dependence (linkage sample), and AD (SAGE), and CD, OD or AD (in the replication sample). Therefore, the linkage and association signals detected for CaD in AAs could be attributable to other substance dependence (SD). We examined this alternate explanation for the findings in several ways. First, we examined the linkage signals across other SD traits (AD, CD, OD, and nicotine dependence (ND)) in the same family dataset (Supplementary Figure 2). The only linkage signal that we found with a lod greater than 1 in the chromosome 8 region reported above for CaD was a modest signal (lod = 1.4) for ND at 41 cM (i.e., ~14 cM away from the CaD linkage peak). Second, we tested the association of rs17664708 with other SD in each AA sample and the joint datasets (Supplementary Table 2). In the joint analysis, we observed highly significant associations between rs17664708 and CD (P=0.0023), AD (P=0.0076), ND (P=0.0026) and OD (P=0.00018). However, significant associations disappeared when subjects affected with CaD were excluded from AD (P=0.3), ND (P=0.069) and CD (P=0.053). Nonetheless, the association remained highly significant for OD when the CaD cases were excluded (P=0.0013), which suggests that rs17664708 could also be a risk variant for OD in AAs independent of CaD. Finally, to remove the confounding effect of OD, we tested the association between rs17664708 and CaD by either controlling for OD status in the regress model or excluding participants who are OD. The results remain significant in the joint datasets by OD status adjusted analysis (OR=2.03, 95% CI=1.17–3.55, P=0.011) or after excluding OD participants (OR=2.47, 95% CI=1.48–4.11, P=0.0003) arguing against the possibility that the significant associations between rs17664708 and CaD arise solely from OD. The association results from other SD might indicate that the association is not specific for CaD and could reflect a shared liability between different substances. The role of NRG1 in other SD remains to be further investigated.
In summary, our study shows that NRG1 is probably a susceptibility gene for CaD, based on convergent evidence of linkage and replicated associations in two independent samples of AAs. Further studies using a high density SNP panel or deep sequencing are necessary to confirm the role of NRG1 in CaD.
Supplementary Material
Acknowledgments
The authors are grateful to the volunteer families and individuals who participated in this research study. We gratefully acknowledge the assistance in recruitment and assessment provided at McLean Hospital by Roger Weiss, M.D., at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D. Genotyping services of linkage analysis and our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University (Keck Center). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Michelle Cucinelli, Christa Robinson, and Greg Kay for their excellent technical assistance, to the SSADDA interviewers who devoted substantial time and effort to phenotype the study sample and to John Farrell for database management assistance. This study was supported by National Institutes of Health grants, R01 DA12690, R01 DA12849, RC2 DA028909, R01 DA18432, R01 AA11330, R01 AA017535, K01 DA24758, and the VA Connecticut REAP center, a VA MERIT grant, VA National Center for PTSD Research, and the VA Connecticut and VISN4 MIRECC Centers. It was also partially supported by the Alcoholic Beverage Medical Research Foundation (ABMRF) grant (S Han).
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Footnotes
Financial Disclosures
Dr. Kranzler has been a paid consultant for Alkermes, GlaxoSmithKline, Gilead, Lilly, Lundbeck, and Roche. Dr. Anton has received honoraria or grant support from Eli Lilly, Alkermes, GlaxoSmithKline, Merck, Lundbeck and Roche and is a shareholder in Alcomed. Drs. Kranzler and Anton also report associations with Eli Lilly, Janssen, Schering Plough, Lundbeck, Alkermes, GlaxoSmithKline, Abbott, and Johnson & Johnson, as these companies provide support to the American College of Neuropsychopharmacology Alcohol Clinical Trials Initiative and both receive support from the Alcohol Clinical Trials Initiative. The other authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.United Nations Office of Drug Control; Publications, UN, editor. World Drug Report 2009. 15. UNODC; 2009. [Google Scholar]
- 2.Substance Abuse and Mental Health Services Administration. Results from the 2007 National Survey on Drug Use and Health. National Findings, Office of Applied Studies; Rockville, MD: 2007. [Google Scholar]
- 3.Agrawal A, Lynskey MT. Does gender contribute to heterogeneity in criteria for cannabis abuse and dependence? Results from the national epidemiological survey on alcohol and related conditions. Drug Alcohol Depend. 2007;88:300–307. doi: 10.1016/j.drugalcdep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stinson FS, Ruan WJ, Pickering R, Grant BF. Cannabis use disorders in the USA: prevalence, correlates and co-morbidity. Psychol Med. 2006;36:1447–1460. doi: 10.1017/S0033291706008361. [DOI] [PubMed] [Google Scholar]
- 5.Ramaekers JG, Moeller MR, van Ruitenbeek P, Theunissen EL, Schneider E, Kauert G. Cognition and motor control as a function of Delta9-THC concentration in serum and oral fluid: limits of impairment. Drug Alcohol Depend. 2006;85:114–122. doi: 10.1016/j.drugalcdep.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Wadsworth EJ, Moss SC, Simpson SA, Smith AP. Cannabis use, cognitive performance and mood in a sample of workers. J Psychopharmacol. 2006;20:14–23. doi: 10.1177/0269881105056644. [DOI] [PubMed] [Google Scholar]
- 7.Casadio P, Fernandes C, Murray RM, Di Forti M. Cannabis use in young people: The risk for schizophrenia. Neurosci Biobehav Rev. 2011;35:1779–1787. doi: 10.1016/j.neubiorev.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Malone DT, Hill MN, Rubino T. Adolescent cannabis use and psychosis: epidemiology and neurodevelopmental models. Br J Pharmacol. 2010;160:511–522. doi: 10.1111/j.1476-5381.2010.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal A, Lynskey MT. The genetic epidemiology of cannabis use, abuse and dependence. Addiction. 2006;101:801–812. doi: 10.1111/j.1360-0443.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- 10.Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, et al. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89:34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal A, Morley KI, Hansell NK, Pergadia ML, Montgomery GW, Statham DJ, et al. Autosomal linkage analysis for cannabis use behaviors in Australian adults. Drug Alcohol Depend. 2008;98:185–190. doi: 10.1016/j.drugalcdep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal A, Pergadia ML, Saccone SF, Lynskey MT, Wang JC, Martin NG, et al. An autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project. Arch Gen Psychiatry. 2008;65:713–721. doi: 10.1001/archpsyc.65.6.713. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction. 2009;104:518–532. doi: 10.1111/j.1360-0443.2009.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addict Biol. 2009;14:338–348. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehlers CL, Gizer IR, Vieten C, Wilhelmsen KC. Linkage analyses of cannabis dependence, craving, and withdrawal in the San Francisco family study. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:802–811. doi: 10.1002/ajmg.b.31050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, et al. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011;16:514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie P, Kranzler HR, Krauthammer M, Cosgrove KP, Oslin D, Anton RF, et al. Rare Nonsynonymous Variants in Alpha-4 Nicotinic Acetylcholine Receptor Gene Protect Against Nicotine Dependence. Biol Psychiatry. 2011;70:528–536. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss LA, Arking DE. Gene Discovery Project of Johns Hopkins & the Autism Consortium, Daly MJ, Chakravarti A. (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature. 461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho MK, Goldman D, Heinz A, Kaprio J, Kreek MJ, Li MD, et al. Breaking barriers in the genomics and pharmacogenetics of drug addiction. Clin Pharmacol Ther. 2010;88:779–791. doi: 10.1038/clpt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Gene. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 24.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 25.Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J, et al. Confirmation and generalization of an alcohol-dependence locus on chromosome 10q. Neuropsychopharmacology. 2010;35:1325–1332. doi: 10.1038/npp.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang BZ, Han S, Kranzler HR, Farrer L, Gelernter J. A Genomewide Linkage Scan of Cocaine Dependence and Major Depressive Episode in Two Populations. Neuropsychopharmacology. 2011;36:2422–2430. doi: 10.1038/npp.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, et al. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- 34.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, et al. Genomewide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 35.Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Z, Alvarado GF, Hatsukami DK, Johnson EO, Bierut LJ, et al. Race differences in nicotine dependence in the Collaborative Genetic study of Nicotine Dependence (COGEND) Nicotine Tob Res. 2008;10:1223–1230. doi: 10.1080/14622200802163266. [DOI] [PubMed] [Google Scholar]
- 37.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Farrer LA, et al. Interaction of FKBP5 with childhood adversity on risk for post-traumatic stress disorder. Neuropsychopharmacology. 2010;35:1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 40.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Kong A, Cox NJ. Allele-sharing models: lod scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tandon A, Patterson N, Reich D. Ancestry informative marker panels for African Americans based on subsets of commercially available SNP arrays. Genet Epidemiol. 2011;35:80–83. doi: 10.1002/gepi.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Zhu X, Qin H, Cooper RS, Ewens WJ, Li C, et al. Adjustment for local ancestry in genetic association analysis of admixed populations. Bioinformatics. 2011;27:670–677. doi: 10.1093/bioinformatics/btq709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5(6):e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González JR, Armengol L, Solé X, Guinó E, Mercader JM, Estivill X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 49.Green B, Young R, Kavanagh D. Cannabis use and misuse prevalence among people with psychosis. Br J Psychiatry. 2005;187:306–313. doi: 10.1192/bjp.187.4.306. [DOI] [PubMed] [Google Scholar]
- 50.Compton WM, Grant BF, Colliver JD. Prevalence of marijuana use disorders in the United States: 1991–1992 and 2001–2002. JAMA. 2004;291:2114–2121. doi: 10.1001/jama.291.17.2114. [DOI] [PubMed] [Google Scholar]
- 51.Smit F, Bolier L, Cuijpers P. Cannabis use and the risk of later schizophrenia: a review. Addiction. 2004;99:425–430. doi: 10.1111/j.1360-0443.2004.00683.x. [DOI] [PubMed] [Google Scholar]
- 52.Moore TH, Zammit S, Lingford-Hughes A, Barnes TR, Jones PB, Burke M, et al. Cannabis use and risk of psychotic or affective mental health outcomes: a systematic review. Lancet. 2007;370:319–332. doi: 10.1016/S0140-6736(07)61162-3. [DOI] [PubMed] [Google Scholar]
- 53.Hall W, Degenhardt L. Cannabis use and the risk of developing a psychotic disorder. World Psychiatry. 2008;7:68–71. doi: 10.1002/j.2051-5545.2008.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabarés-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 55.Munafò MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 56.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 57.Ng MY, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volkow ND, Wang GJ, Fowler JS, Tomasi D, Telang F. Addiction: beyond dopamine reward circuitry. Proc Natl Acad Sci U S A. 2011;108:15037–15042. doi: 10.1073/pnas.1010654108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laviolette SR. Dopamine modulation of emotional processing in cortical and subcortical neural circuits: evidence for a final common pathway in schizophrenia? Schizoprenia Bulletin. 2007;33:971–981. doi: 10.1093/schbul/sbm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boucher AA, Arnold JC, Duffy L, Schofield PR, Micheau J, Karl T. Heterozygous neuregulin 1 mice are more sensitive to the behavioural effects of Delta9-tetrahydrocannabinol. Psychopharmacology (Berlin) 2007;192:325–336. doi: 10.1007/s00213-007-0721-3. [DOI] [PubMed] [Google Scholar]
- 61.Boucher AA, Hunt GE, Karl T, Micheau J, McGregor IS, Arnold JC. Heterozygous neuregulin 1 mice display greater baseline and Delta(9)-tetrahydrocannabinol-induced c-Fos expression. Neuroscience. 2007;149:861–870. doi: 10.1016/j.neuroscience.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Boucher AA, Hunt GE, Micheau J, Huang X, McGregor IS, Karl T, et al. The schizophrenia susceptibility gene neuregulin 1 modulates tolerance to the effects of cannabinoids. Int J Neuropsychopharmacol. 2011;14:631–643. doi: 10.1017/S146114571000091X. [DOI] [PubMed] [Google Scholar]
- 63.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tseng AH, Craft RM. Sex differences in antinociceptive and motoric effects of cannabinoids. Eur J Pharmacol. 2001;430:41–47. doi: 10.1016/s0014-2999(01)01267-5. [DOI] [PubMed] [Google Scholar]
- 65.Tseng AH, Harding JW, Craft RM. Pharmacokinetic factors in sex differences in [Delta]9-tetrahydrocannabinol-induced behavioral effects in rats. Behav Brain Res. 2004;154:77–83. doi: 10.1016/j.bbr.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 66.Long LE, Chesworth R, Arnold JC, Karl T. A follow-up study: acute behavioural effects of Delta(9)-THC in female heterozygous neuregulin 1 transmembrane domain mutant mice. Psychopharmacology (Berl) 2010;211:277–289. doi: 10.1007/s00213-010-1896-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.