Abstract
The human antibody repertoire is one of the most important defenses against infectious disease, and the development of vaccines has enabled the conferral of targeted protection to specific pathogens. However, there are many challenges to measuring and analyzing the immunoglobulin sequence repertoire, such as the fact that each B cell contains a distinct antibody sequence encoded in its genome, that the antibody repertoire is not constant but changes over time, and the high similarity between antibody sequences. We have addressed this challenge by using high-throughput long read sequencing to perform immunogenomic characterization of expressed human antibody repertoires in the context of influenza vaccination. Informatic analysis of 5 million antibody heavy chain sequences from healthy individuals allowed us to perform global characterizations of isotype distributions, determine the lineage structure of the repertoire and measure age and antigen related mutational activity. Our analysis of the clonal structure and mutational distribution of individuals’ repertoires shows that elderly subjects have a decreased number of lineages but an increased pre-vaccination mutation load in their repertoire and that some of these subjects have an oligoclonal character to their repertoire in which the diversity of the lineages is greatly reduced relative to younger subjects. We have thus shown that global analysis of the immune system’s clonal structure provides direct insight into the effects of vaccination and provides a detailed molecular portrait of age-related effects.
Introduction
The adaptive immune system produces a large and diverse set of antibodies, each with an individual evolutionary and clonal history. This so called “antibody repertoire” protects each individual against insults such as infection and cancer, and responds to vaccination with B cell proliferation in response to the antigenic stimulation. Hybridomas and antigen-specific FACS-based analysis have given us much insight on how the immune system generates the complex and diverse immune response required to protect the body from the wide variety of potential pathogens (1-3). However, these methods have not been sufficient to make global and unbiased characterizations of the clonal structure of the immune system of a particular individual, which could provide insights into how the diversity and clonal structures vary between individuals, with age or gender, and in response to specific antigen stimulation (4). With respect to antigen stimulation, although there is a great deal of data that has been obtained by sorting antigen specific B cells, there is less information on the effect of the antigen on the global response of the antibody repertoire (5-10).
We and others have begun applying high-throughput sequencing techniques to the immunogenetic characterization of antibody repertoire (11-17). Our previous work focused on zebrafish as a model organism, which enabled us to perform deep sequencing to exhaust the repertoire diversity in a manner that was independent of the physiology of the organism, i.e. independent of where the B cells were residing (11). This work revealed that the repertoire of individuals has a surprising high fraction of shared sequences, a universal structure and that the balance between determinism and stochasticity in the repertoire is tilted more towards determinism both in early development and in the primary repertoire of mature organisms than had previously been suggested (15). Others have used similar approaches to measure the amount of residual disease from lymphoma B cell clones (12), to study gene segment frequency after lymphocytic ablation (16), and to bypass cloning and directly synthesize antibody from mining the high-throughput sequencing data acquired by using bone marrow plasma cells from immunized mice (13). Attempts to use this approach to study vaccination have not been able to resolve lineage relationships and have not demonstrated a functional link between repertoire and immune response (18). Here, we address the question of how the human immune repertoire responds to specific antigen stimulation, in particular by influenza vaccination. We determine the lineage structure of the repertoire before and after vaccination and demonstrate that some sequences in the repertoire correspond to vaccine-specific immunoglobulins. We further observe age related changes in antibody isotype composition, lineage diversity and structure, as well as mutational load, thereby offering a molecular characterization of defects in humoral immune response resulting from aging.
Results
We analyzed antibody repertoires from peripheral blood drawn from 17 human volunteers who were immunized with 2009 or 2010 seasonal influenza vaccines (Supplementary Table 1). These volunteers were recruited from three age groups, children (8 to 17 years of age), young adults (18 to 30 years of age), and elderly (70 to100 years of age) and were randomly given either trivalent inactivated influenza vaccine (TIV) or live attenuated influenza vaccine (LAIV), except for subjects in the 70 to 100 years group who could only receive TIV (Supplementary Table 1 (9). TIV and LAIV contain antigenically equivalent virus strains, however LAIV is made of live attenuated viruses that are capable of limited proliferation after intranasal administration and are expected to induce a stronger mucosal immune response than TIV (19). The study included three pairs of identical twins in order to have repertoire control experiments with identical genetic background; these were in the age group of 8 to 17 and were randomly selected to receive either TIV or LAIV within a twin pair. Blood samples were collected from each participant at three time points: day 0 before vaccination (visit 1), day 7 or 8 (visit 2), and day 28 (± 4, visit 3) after vaccination. Peripheral blood mononuclear cells (PBMCs) were collected at both visit 1 and 3. Naïve B cells (NB) and plasmablasts (PB) were sorted from visit 2 blood samples by flow cytometry.
Reduction of relative IgM abundance after vaccination decreases with age
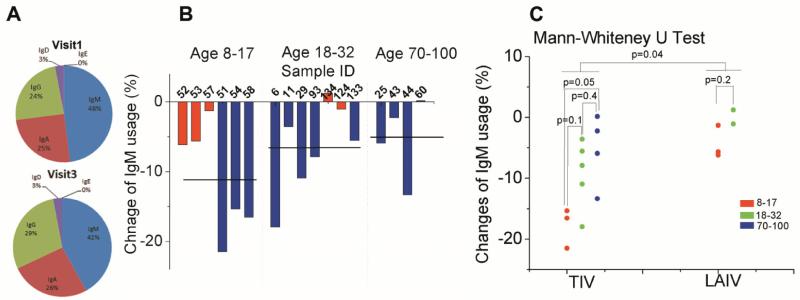
All five isotypes were detected in all the samples processed, however, with different relative amounts. IgM, IgA and IgG are more abundant than IgD and IgE, which together account for less than 4% of all sequencing reads (Fig. 1A). Most naïve B cells express IgM on their membrane, and then upon antigen stimulation undergo an activation process that changes their constant region from IgM to other isotypes and increases the antibody transcript copy numbers in each cell (20-22). We tracked the changes of isotype distribution between visit 1 and visit 3 and noticed a decrease of relative IgM abundance for all volunteers except one (Fig. 1B). On average, relative IgM usage decreased 11.1% ± 3.2 (SEM) (age 8-17), 6.5% ± 2.4 (SEM) (age 18-32) and 6.0% ± 2.9 (SEM) (age 70-100) at visit 3 (Fig. 1B, black lines). An independent measurement by digital PCR (23, 24) was used to verify the relative isotype abundance in visit 1 and 3 samples and the reduction of relative IgM abundance from visit 1 to visit 3 (fig. S3). This decrease coincided with an increase of relative IgA and IgG abundance (fig. S3), suggesting that a portion of the naïve B cells may have undergone isotype switching and the antibody transcript copy numbers of these isotypes may also have increased as a result of the vaccine stimulation. This interpretation is supported by flow cytometry data which did not show changes in the relative abundances of IgM expressing cells between visits 1 and 3. Therefore, the large difference in relative IgM usage is due to antibody transcript copy number changes as a result of antigen stimulation. We also directly observed isotype switched lineages in a small fraction of the sequence data. These sequences contained common CDR3 sequences and had extensive mutations throughout the variable regions (fig. S5), which suggests that they are not template-switched PCR artifacts. The number of lineages containing isotype-switches decreased with age, (fig. S4) which is consistent with our observation that reduction of relative IgM transcript abundance from visit 1 to visit 3 decreases with age regardless of vaccine types (Fig. 1C). While LAIV receivers have less change in relative IgM usage than individuals who received TIV, there is a strong age dependence of isotype relative abundance change in TIV receivers – children who received TIV were more likely to have an increased relative IgA usage compared to young adults (p=0.03, Mann-Whitney U test) or elderly (p=0.05, Mann-Whitney U test) (fig. S3).
Fig. 1. Antibody isotype distribution changes after vaccination.
(A), antibody isotype composition in PBMCs at visit 1 (before vaccination, top) and visit 3 (28 ± 4 days after vaccination, bottom) averaged for all subjects. (B), percent change of individual’s relative IgM usage in PBMCs from visit 1 to visit 3. The subject IDs were labeled on horizontal axis. (C), comparison of relative change in IgM in different age group and vaccine types. p value was calculated by Mann-Whitney U test (3 samples for TIV receivers of age 8-17, 5 samples for TIV receivers of age 18-32, 4 samples for TIV receivers of age 70-100, 3 samples for LAIV receivers of age 8-17 and 2 samples for LAIV receivers of age 18-32). Red, LAIV receivers; blue, TIV receirvers. Percent change = (IgM reads in visit 1/total reads in visit 1) – (IgM reads in visit 3/total reads in visit 3)
Single linkage clustering enables informatic definition of lineages
One crucial aspect of clonal expansion after antigen stimulation is that activated B cells undergo a somatic hypermutation process during which random mutations are introduced to the antibody genes. Clonal expansion is therefore not truly clonal as a key aspect is also the introduction of new diversity, followed by selection for the mutants with higher antibody affinity. Distinguishing mutations, grouping sequences that differ by somatic hypermutation to the same clonal lineage, and following the sequence evolution within a lineage are critical steps in understanding B cell biology and the relationship between B cell repertoire and vaccine stimulation. High throughput sequenching enables one to directly measure these relationships at a scale that was not possible with earlier approaches.
To dissect differences between lineages and analyze the detailed mutations within a lineage, we developed a clustering scheme that focused on the complementarity determining region 3 (CDR3) of the antibody sequence, which covers the region between the end of V and beginning of J gene segments. We first converted the nucleotide sequences into amino acid sequences for each read. Translation rescue was performed for out-of-frame sequences that were mostly due to insertions and/or deletions in the V, D, or J segments (fig. S1). In order to set the clustering threshold, we analyzed the amino acid distance between reads in the CDR3 region. The resulting distribution showed two distinct peaks; the first is at 1 amino acid and the other covers 4 to 10 amino acids (fig. S6). This suggests that the first peak contained sequencing reads that were mostly mutated and the second peak contained sequencing reads that had distinct CDR3 sequences that were generated during the VDJ recombination process. The amplitude of these two peaks changed between different samples that were collected at different time points and had varying NB and PB cell fractions. Sorted NBs had the lowest first peak and highest second peak, while sorted PBs has the highest first peak and the lowest second peak. PBMC samples from visit 1 and 3 fell in between NBs and PBs while visit 2 PBMCs (available only for selected subjects) were similar to visit 2 PBs. These trends are consistent with dynamics of antibody mediated immune response and distribution of NBs and PBs in peripheral blood. These trends further demonstrated that the first peak is due to mutation and second peak is due to junctional diversity.
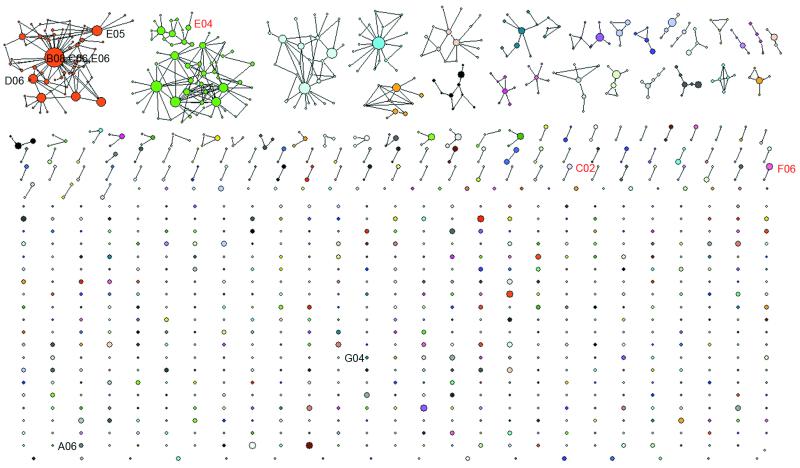
This distribution provides a natural threshold when clustering and was used to group sequences according to their lineage identity. We also performed clustering directly on nucleotide sequences with varying thresholds (fig. S15). Here “lineage” refers to antibody sequences that originated from the same VDJ recombination event and have the same junctional sequence, but may be further diversified because of antigen stimulation and somatic hypermutation. We clustered all sequencing reads from sorted PBs at visit 2 using one amino acid difference at CDR3 as a threshold. This means that two sequences will be grouped into the same lineage if they are in the same V and J family and their protein sequence in the CDR3 region differ by no more than one amino acid. Using this lineage data, one can construct a graphical representation of the clonal structure of the immune repertoire (Fig. 2 and fig. S7). The central functional question regarding these informatically defined sequence lineages is to what extent they include influenza specific antibodies. To examine this, we amplified influenza-specific antibody sequences from single sorted PBs for two of the subjects in the 70-100 years old group, 017-043 and 017-044. We then expressed monoclonal antibodies according to these sequences and verified their binding to each of the three virus strains in the vaccine. 11 out of 16 heavy chain sequences from single cell cloned PBs were found within the lineages we measured, especially for the anti-vaccine high affinity antibodies (Fig. 2). For subjects 017-044, the single cell cloned sequences overlap with lineages containing smaller number of reads compared to sequences cloned from 017-043, where many single cell cloned sequences are in the top lineage containing most of the reads (fig. S11). This may reflect structural differences in repertoire between these two subjects as one has a dominant lineage and the other one has a more even distribution (fig. S11). Taken together, these data confirm that the influenza specific antibody responses are contained within the globally measured immune repertoire sequences as well as the informatically defined lineages we derived from them.
Fig. 2. Informatically defined lineages with influenza specificity.
The intra- and inter-lineage structure of all IgG lineages visualized by sequencing the PBs sorted from blood sample collected at visit 2 (7 days after vaccination) from a volunteer in the 70-100 year-old group received TIV (subject 017-043). In this network representation, each cluster of dots connected by lines represents a lineage. Different colors were used to distinguish different lineages. Each dot represents a unique CDR3 protein sequence. Two dots are linked if they differ by one amino acid in the CDR3 region. This is the threshold used when performing the single linkage clustering. The area of a dot is proportional to the number of reads with identical CDR3 protein sequences. Single cell cloned antibodies are labeled with text. Red text indicates antibodies having a high affinity towards one of the virus strains used in the flu vaccine. Black text indicates antibodies with a low affinity towards one of the virus strains used in the flu vaccine or background level of binding towards all three virus strains. 8 of 10 single cell cloned antibodies were found in the 454 sequences, except G04 and A06. All reads from 454 sequencing were used for this plot.
Lineage structure analysis reveals distortion in some elderly subjects
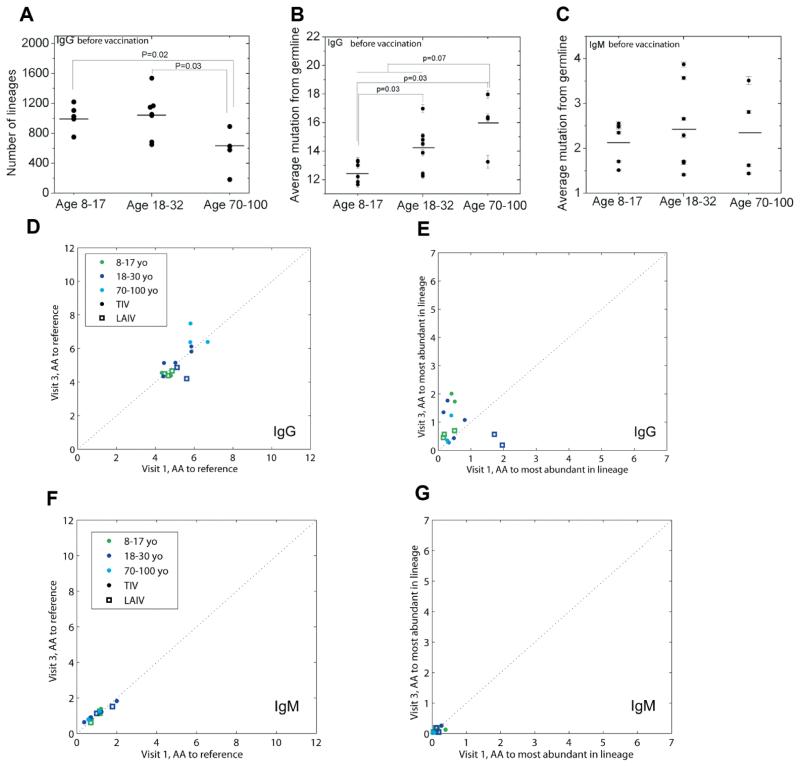
Lineages belonging to plasmablasts exhibit an apparent power law distribution with a few lineages that dominate the repertoire, whereas those belonging to naïve cells do not (fig. S13). This is consistent with long-tailed distributions observed previously (11) and are the direct consequence of clonal expansion. The elderly have fewer lineages than other age groups both before (Fig. 3A) and after (fig. S12) vaccination, indicating an altered repertoire structure and potentially a smaller pool of diversity for the immune repertoire to draw upon in vaccine response.
Fig. 3. Age related repertoire diversity and mutation changes.
(A), repertoire diversity changes with age as measured by number of lineages in IgG from visit 1 PBMCs. (B and C), before vaccination mutation load as measured by averaging mutations at nucleotide level for IgG (B) and IgM (C) in visit 1 PBMCs respectively. Mutations for each read were defined as the number of mismatches to germline reference in V, D and J regions. (D-G), lineage analysis, performed with 80% nucleotide-sequence identity at the VDJ junctional region, gives measurements of amino acid mutations-per-read at V and J gene segments measured either to the germline reference (D and F) or from the most abundant sequence of the lineage to which each belongs (E and G) for IgG (D and E) and IgM (F and G). X-axes denote the measurement at visit 1, and the Y-axes denote the measurement at visit 3. Elderly patients show a higher number of IgG mutations from the germline (comparing 8-30 year-olds to 70-100 year-olds gives p<0.075 before vaccination and p<0.0044 after; restricting this analysis to TIV-patients alone gives p<0.18 and p<0.017, respectively). 3000-read of subsampling was applied to all panels. All error bars are the standard error. p-values were calculated by Mann-Whitney U test.
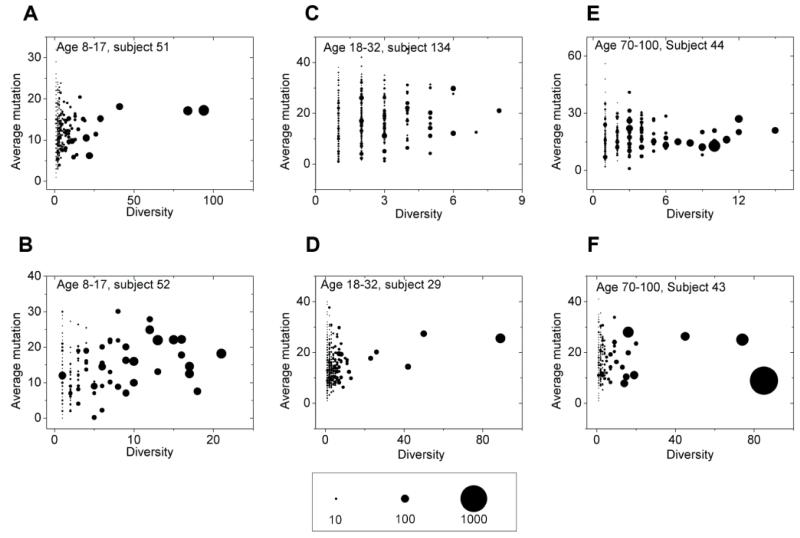
Using the three parameters of diversity (unique protein sequences), average mutation, and number of reads in each lineage, one can visualize and compare the antibody repertoire in a quantitative manner (Fig. 4A-F). In each individual, the majority of the lineages contain less than 10 reads and less than two unique amino acid sequences. The elderly vaccine recipients can be separated into two groups: one group had a distribution of lineages similar to the children (Fig. 4A and B) and young adults (Fig. 4C and D); the other group had a very different distribution of lineages compared to other age groups (Fig. 4E and F). Elderly subjects in the second group had a few lineages that encompassed more than 80% of the reads. This is exemplified by subject 017-43 (Fig. 4F, and fig. S8 and S11). Detailed sequence analysis revealed that 58% (subject 017-043) and 90% (subject 017-060) of the reads within the biggest lineage for these elderly were identical. This is consistent with the overall observation that influenza vaccination resulted in expansion of far fewer B cell lineages in the elderly compared to the other age groups (fig. S12A). This reduced clonal diversity when weighted with abundance may be related to a reduced antibody response to influenza vaccine in the elderly. Lineage analysis on IgG from visit 1 and 3 PBMCs also suggested that in general the elderly have a reduced B cell clonal diversity compared to the younger age groups (Fig. 3A and fig. S12B), which might explain the reduced clonal diversity in vaccine-activated B cells in the elderly.
Fig. 4. Inter-lineage structure of IgGs in visit 2 PBMCs.
Inter-lineage structure of IgGs in visit 2 PBMCs is presented for six randomly selected subjects (A-B, age 8-17; C-D, age 18-32; E-F, age 70-100). Each dot represents a lineage of antibody sequences defined by single linkage clustering with 1 amino acid difference at CDR3 as the threshold. The area of the dot is proportional to the number of reads belonging to this lineage, as indicated in the scale bar. X-axis is the diversity of the lineage which measures number of unique protein sequences (full protein sequence, not just the CDR3 region) within the lineage. Y-axis is the number of mutations at nucleotide level of the lineage averaged over reads. 3000 reads of subsampling was applied.
Age affects somatic hypermutation and lineage diversity
One interesting question about reduced immune response to influenza vaccination in the elderly is whether the B cells that respond to the current vaccine had been primed by prior infections or vaccinations. If so, those B cells from the elderly will mostly likely be memory B cells that have a higher baseline mutation than responding B cells from younger volunteers where most of these cells should be of naïve phenotype or relatively less antigen experienced, therefore, have fewer mutations. Another important question is whether those responding memory B cells in the elderly have the same ability to introduce new mutation upon antigen stimulation compared to responding B cells from young volunteers.
In order to answer these questions, we performed a detailed analysis of mutation statistics. Although 454 sequencing has a high error rate, around 1%, most of them are insertions and deletions (indels) (11, 15) and can be repaired (fig. S1). The substitution error rate (from sequencing and/or PCR) is estimated to be 0.065% per nucleotide (fig. S16 and Control Library section in the Supplementary Materials). This is lower than the estimated somatic hypermutation rate, which is approximately 0.1% measured in nucleotides per cell division (25). Also, any B cells undergoing somatic hypermutation are likely to have several rounds of division, which increases their overall mutations per sequence. To analyze mutation statistics, we performed single linkage clustering by comparing the peptide sequences of CDR3 regions, using 1 amino acid as a threshold. We compared the average mutations-per-read from visit 1 PBMC across different age groups. This number consistently increased with age in the IgG fraction (Fig. 3B) while remaining at the background level and with no difference between age groups in the IgM fraction (Fig. 3C), which is consistent with the fact that most of the IgM expressing B cells are in a naïve state. We also applied the antibody-lineage clustering performed previously using junctional nucleotide sequences, thresholded at 80% identity (15).
We found that mutations in general are far higher in IgG than IgM, both when these mutations are measured relative to the germline reference sequences (Fig. 3D and 3F) as well as to the most abundant sequence in each lineage. These observations point to mutational excursions of abundant class-switched sequences, as well as to diversification within the most abundant IgG lineages, respectively. In addition, there is a far greater parity between mutation-loads measured at visits 1 and 3 among IgM antibodies (R2=0.92) than among IgG (R2=0.54). In other words, even accounting for the variability amongst individuals, the IgM repertoire is more similar between before and after vaccine samples than the IgG repertoire. This demonstrates that IgG antibodies undergo a far greater change in composition between the two time points compared to IgM. Furthermore, the elderly had the highest number of amino acid mutations in both visit 1 and visit 3 IgG fraction (Fig. 3D) while remaining low and similar to the IgM fraction in both visits of other age groups (Fig. 3F). This trend is consistent with our mutation analysis using clustering performed on amino acid sequences. IgG sequences had a higher number of mutations at visit 3 than visit 1 when these were tallied in reference to the most abundant sequence in each lineage (off diagonal line towards visit 3) suggesting that somatically hypermutated sequences persisted within the bloodstream 28 days after vaccination. At the same time, the elderly were not necessarily the group possessing the greatest number of mutations anymore relative to these most abundant sequences (Fig. 3E). Therefore, because they lack any indications of greater intraclonal mutation compared to other age-groups, these data suggest that the higher numbers of somatic mutations observed earlier in elderly individuals arise from clonal expansions that draw upon a pool of B-cells having more somatic mutations to begin with.
Discussion
Although the antibody repertoire is encoded by gene segments that are common to each individual human being, the various processes of immunoglobulin diversity generation create a repertoire where the number of distinct immunoglobulin sequences in an individual exceeds the number of distinct genes in their consensus genome. The antibody repertoire is constantly evolving; it records the pathogenic exposure that one has experienced in the past and retains information on what it can protect us from. Therefore, it is of great interest to quantify and measure this dynamic system to understand how the repertoire responds to infection and vaccination and provide potential metrics for immune monitoring.
In this study, we used seasonal influenza vaccine as a means of stimulation, and measured and quantified the changes in the antibody repertoire. First, we observed that the relative percentage of IgM sequences dropped after vaccination across all volunteers except for one. This reduction in IgM usage decreased with age, which is consistent with the hypothesis that elderly are more likely to use memory B cells than naïve B cells to respond to influenza vaccination (26). We noted that children appear likely to increase relative IgA percentage in PBMCs compared to IgG.
A challenge of analyzing and quantifying the antibody repertoire is clonal expansion after antigen stimulation is not truly clonal as random mutations are introduced to the antibody genes at a rate of approximately 10−3 mutations per base pair per cell division (25). Using high-throughput sequencing in combination with informatics analysis, we were able to distinguish mutations, group sequences that differ by somatic hypermutation to the same clonal lineage, and follow the sequence evolution within a lineage. This approach enabled us an unbiased measurement of the relative size among different lineages within one individual and the sequence diversities within each lineage.
A network representation of lineages allowed visual comparison of the intricate intra- and inter-lineage structure. Many of the top lineages were composed of extensively connected CDR3 sequences, each with varying number of sequencing reads. Sequence data from our single cell cloning also confirmed that many of the top lineages are influenza specific (Fig. 2 and fig. S11). Some single-cell-generated sequences did not have a high affinity towards any one of the virus strains used in the vaccine; it is possible that they may not be representative or the recombinant antibodies may have been specific to internal viral proteins rather than the whole virus used in the ELISA tests. The detailed topology of each lineage may contain information about how antigen selection and antibody affinity maturation work in concert in shaping the antibody repertoire. Studying the function of those informatically defined lineages may provide insight into this process.
Having several twin pairs among our subjects provides an interesting genetic control for the data. As one might expect, for the IgM repertoire the twins have closely related mutational loads but these values diverge substantially for the IgG repertoire (fig. S19). We attribute this to the notion that the naïve repertoire is probably more strongly influenced by the background genetics of the individual while the secondary repertoire incorporates a larger degree of stochasticity and randomness (15, 16). The mutational load versus diversity graphs for twins show little correlation (fig. S8, Age 8-17 group); this is also to be expected as these data represent strong environmental and stochastic contributions to the immune system.
In conclusion, we have shown that it is possible to make personalized individual-specific measurements of immune repertoire with high throughput DNA sequencing technology. These global repertoires contain a wealth of information and can be used to study individual-specific vaccine responses, and we have shown that analysis of the clonal structure provides direct insight into the effects of vaccination and provides a detailed molecular portrait of age-related effects. This approach to immune system characterization may be generally applicable to the development of new vaccines and may also help identify which individuals respond to a given vaccine.
Materials and Methods
Human participants, vaccination protocol, blood sample collection and cell sorting
Human participants, vaccination protocol, blood sample collection and cell sorting were described by Sasaki et al. (9). Samples from a subgroup of volunteers were used in this study and the demographical information of human participants was listed in Supplementary Table 1. The study protocols were approved by the institutional review boards at Stanford University. Informed consent was obtained from participants and the parents of pediatric participants. In addition, assent was obtained from the child participants. Participants were immunized with one dose of either the 2009 or 2010 seasonal TIV (Fluzone, from Sanofi Pasteur) or LAIV (FluMist, from MedImmune). The 2009 vaccine contained an A/Brisbane/59/2007 (H1N1)-like virus, an A/Brisbane/10/2007 (H3N2)-like virus, and a B/Brisbane/60/2008-like virus. The 2010 vaccine contained an A/California/7/2009 (H1N1)-like virus, an A/Perth/16/2009 (H3N2)-like virus and a B/Brisbane/60/2008-like virus. Blood samples were collected from each participant at three time points: day 0 before vaccination, day 7 or 8, and day 28 (± 4) after vaccination. PBMCs were isolated from the day 0 and day 28 blood samples using Ficoll-Paque Plus (GE Healthcare) following the manufacturer’s instruction. Sorting of plasmablasts was performed as previously described (9). In brief, B- cells were isolated by negative selection using the RosetteSep Human B-cell Enrichment Cocktail (Stemcell Technologies) following the manufacturer’s instructions from the day 7-8 whole blood samples. Plasmablasts were then sorted based on the phenotype of CD3−CD19+CD20−CD27+CD38+ and naïve B cells were sorted based on the phenotype of CD3− CD19+CD20+CD27−CD38−. Both populations reached a purity of 95%. Cells were lysed in RLT buffer (Qiagen) supplemented with 1% β-mercaptoethanol (Sigma) and stored at −80°C.
Primer design, RNA preparation, cDNA synthesis and PCR
244 human heavy-chain variable gene segment sequences were downloaded from ImMunoGeneTics (IMGT) (27), excluding pseudogenes. The leader regions of these sequences were used to design the 11 forward primers. The first 100bp of the IgA, IgD, IgE, IgG and IgM constant domain were used to design the reverse primers. Gene specific primers were also designed for the reverse transcription step; these were located about 50bp downstream from the PCR reverse primers. All primer sequences are listed in the Supplementary Table 2.
10 million PBMCs or sorted cells with varying numbers lysed in RLT buffer was used as input material for RNA purification. This was done by using the All prep DNA/RNA purification kit (Qiagen) following manufacture’s instruction. The concentration of the RNA was determined using a Nanodrop spectrophotometer.
cDNA was synthesized using SuperScript™ III reverse transcriptase (Invitrogen). One fifth of the RNA purified from each sample was used for cDNA synthesis reactions with a total volume of 60ul. All five constant region reverse transcription primers were added to the same reaction together with SUPERase·In™ (Ambion). RNase H (Invitrogen) was added to each reaction to remove RNA at the end of the cDNA synthesis step. All enzyme concentrations, reaction volumes and the incubation temperature were based on the manufacturer’s protocol for synthesis of cDNA using gene specific primers.
For each sample, 11 PCR reactions were set up corresponding to 11 forward primers with a mixture of 5 reverse primers in each reaction. 2ul of reverse transcription mixture was used in each PCR reaction of 50ul. Final concentration of 200nM was used for each primer. The PCR program began with an initial denaturation at 94°C for 2 minutes, followed by 35 cycles of denaturation at 94°C for 30 s, annealing of primer to DNA at 60°C for 30 s, and extension by Platinum® Taq DNA Polymerase High Fidelity (Invitrogen) at 68°C for 2 minutes. PCR products were first cleaned using QIAquick PCR Purification Kit (Qiagen) and then purified using the QIAquick gel extraction kit (Qiagen). Concentration was measured using the nanodrop spectrophotometer.
454 library preparation and sequencing
About 0.5μg of QIAquick cleaned PCR product for each sample was used to start the 454 library preparation process. 454 Titanium shotgun library construction protocol was followed for all samples. Briefly, double stranded DNA was end polished and ligated to sequencing adaptors which contained a molecular identifier (MID, a nucleotide based barcode system). The rest of the Roche 454 protocol was followed which includes library immobilization, fill-in reaction and single stranded template DNA (sstDNA) library isolation. The sstDNA was quantified using a digital-PCR method (28). Up to 16 libraries were pooled for one sequencing run and Roche 454 emulsion PCR and sequencing protocols were followed for the rest of the sequencing procedure.
Data analysis
Detailed information on data analysis is included in the Supplementary Material. In summary, reads from Roche 454 first entered into the primary analysis which includes matching MID, filtering for minimum length of 250bp and then truncating to 220bp. After V, D, and J assignment, protein translation rescue was formed on each reads. Sequencing reads that could not be rescued were discarded, which was about 13% of the sequencing reads. Single linkage cluster was performed at both nucleic acid and amino acid level with varying thresholds. Subsampling was applied in all analysis related to sequence and lineage diversity, mutation and lineage structure except when looking for overlapping lineages between single cell cloned sequences and high throughput sequencing data.
Supplementary Material
Acknowledgements
We thank Norma Neff, Ben Passarelli, Jennifer Okamoto, Nicolas Gobet and Gary Mantalas at the Stanford Stem Cell Genome Center for assistance with sequencing. We would also like to thank Sally Mackey for clinical project management, regulatory and data management, Sue Swope and Cynthia Walsh for research nurse support, Garry Swam and members at the Stanford Research Institute for help with twin volunteer recruitment, Holden Maecker, Jackie Bierre, and Ben Varasteh at the Stanford Human Immune Monitoring Core for sample banking.
Funding: This research was supported by the National Institutes of Health grant U19 AI057229 (M.M.D., X.H., H.B.G. and S.R.Q.), a National Institutes of Health Pathway to Independence Award K99 AG040149 (N.J.) and a National Science Foundation graduate fellowship (J.A.W.).
Footnotes
Author contributions: N.J., J.A.W., M.M.D., and S.R.Q. conceived initial idea; N.J., J.A.W., X.H., C.L.D., H.B.G., M.M.D. and S.R.Q. designed research; C.L.D. was responsible for regulatory and clinical aspects of the study protocols; S.S. and X.H. performed cell sorting; N.Z., M.H., M.S. and P.C.W. generated recombinant monoclonal antibodies and performed antibody functional assay; N.J., J.H., J.A.W., and L.P., performed high-throughput sequencing related research; N.J., J.H., J.A.W., D.S.F., and S.R.Q. analyzed data; and N.J., J.H., J.A.W., D.S.F., and S.R.Q. wrote the paper.
Competing interests: A patent application entitled “Measurement and Comparison of Immune Diversity by High-Throughput Sequencing” (Application number: PCT/US2011/035507. Inventors: S.R.Q., J.A.W., N.J., and D.S.F.) was filed by Stanford University based on this work. It has been licensed to ImmuMetrix, LLC, of which S.R.Q. is a founder and advisor, and M.M.D., D.S.F., and N.J. are advisors. The other authors declare that they have no competing interests.
Data and materials: The sequence data sets published in this paper can be found in the Sequence Read Archive, accession no. SRA058972.
List of Supplementary Material
Fig. S1. Flowchart of bioinformatics pipeline.
Fig. S2. The composition of five antibody isotypes from PBMCs for each subject at visits 1 and 3.
Fig. S3. Isotype changes from visit 1 to visit 3.
Fig. S4. Young subjects have more lineages that are isotype switched.
Fig. S5. Nucleotide sequence alignment for VDJ region exemplified for one isotype switched lineage.
Fig. S6. Reads distribution based on relative sequence distance.
Fig. S7. The inter- and intra-lineage structure of all IgG lineages revealed by sequencing plasmablasts sorted from the visit 2 blood samples for selected subjects.
Fig. S8. The inter-lineage structure of IgG from plasmablasts sorted for all subjects at visit 2.
Fig. S9. The inter-lineage structure of IgG from PBMCs purified from subject 017-060 at visit 1.
Fig. S10. The inter-lineage structure of IgM and IgG for naïve B cells and plasmablasts from one subject at visit 2.
Fig. S11. Overlapping of single-cell cloned antibody sequences with lineages.
Fig. S12. Repertoire diversity changes with age.
Fig. S13. Distribution of lineage size observes the power-law distribution.
Fig. S14. Mutation pattern of IgG for three age groups in visit 1 PBMCs.
Fig. S15. The mutation patterns for different age groups at threshold of 90% of nucleotide similarity in the CDR3 region.
Fig. S16. Zebrafish control data.
Fig. S17. Diversity and reads of IgG lineages of human plasmablasts at visit 2.
Fig. S18. Synthetic sequence control data.
Fig. S19. Figures 3B and 3C, respectively, from the main text with twin status indicated by arrows.
Table S1. Demographical information of human participants.
Table S2. Primer sequences.
Table S3. Summary of cell numbers and filtered reads for all samples.
Table S4. Raw reads for five isotypes in each sample.
Table S5. Summary of identifiable VJ reads for IgG in visit 2 plasmablasts.
Table S6. Summary of single cell cloned sequences.
Table S7. Control data information.
References and Notes
- 1.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 2.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed R, Oldstone MB, Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8:1188. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrammert J, et al. [Google Scholar]
- 6.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki S, et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest. 2011;121:3109. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187:3704. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 14.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang N, et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci U S A. 2011;108:5348. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glanville J, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A. 2011;108:20066. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaout R, et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YC, Kipling D, Dunn-Walters DK. Age-Related Changes in Human Peripheral Blood IGH Repertoire Following Vaccination. Front Immunol. 2012;3:193. doi: 10.3389/fimmu.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox RJ, Brokstad KA, Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol. 2004;59:1. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 20.Schibler U, Marcu KB, Perry RP. The synthesis and processing of the messenger RNAs specifying heavy and light chain immunoglobulins in MPC-11 cells. Cell. 1978;15:1495. doi: 10.1016/0092-8674(78)90072-7. [DOI] [PubMed] [Google Scholar]
- 21.Jack HM, Wabl M. Immunoglobulin mRNA stability varies during B lymphocyte differentiation. Embo J. 1988;7:1041. doi: 10.1002/j.1460-2075.1988.tb02911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 23.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan HC, Quake SR. Detection of aneuploidy with digital polymerase chain reaction. Analytical chemistry. 2007;79:7576. doi: 10.1021/ac0709394. [DOI] [PubMed] [Google Scholar]
- 25.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 26.Dormitzer PR, et al. Influenza vaccine immunology. Immunol Rev. 2011;239:167. doi: 10.1111/j.1600-065X.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 27.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. ed. 5 Vol. 1. Public Health Service, National Institutes of Health; Washington, DC: 1991. [Google Scholar]
- 30.Kleinstein SH, Louzoun Y, Shlomchik MJ. Estimating hypermutation rates from clonal tree data. J Immunol. 2003;171:4639. doi: 10.4049/jimmunol.171.9.4639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.