Abstract
Dimers of conventional transforming growth factor–β (TGF-β) and bone morphogenetic protein (BMP) ligands are composed of two 100– to 140–amino acid peptides that are produced through the proteolytic processing of a proprotein precursor by proconvertases, such as furin. We report the identification of an evolutionarily conserved furin processing site in the amino terminus (NS) of the Glass bottom boat (Gbb; the Drosophila ortholog of vertebrate BMP5, 6, and 7) proprotein that generates a 328–amino acid, active BMP ligand distinct from the conventional 130–amino acid ligand. Gbb38, the large ligand form of Gbb, exhibited greater signaling activity and a longer range than the shorter form Gbb15. The abundance of Gbb15 and Gbb38 varied among different tissues, raising the possibility that differential processing could account for tissue-specific behaviors of BMPs. In human populations, mutations that abolished the NS cleavage site in BMP4, BMP15, or anti-Müllerian hormone were associated with cleft lip with or without cleft palate (BMP4), premature ovarian failure (BMP15), and persistent Müllerian duct syndrome (anti-Müllerian hormone), suggesting the importance of NS processing during development. The identification of this large BMP ligand form and the functional differences between large and small ligands exemplifies the potential for differential proprotein processing to substantially affect BMP and TGF-β signaling output in different tissue and cellular contexts.
INTRODUCTION
The family of transforming growth factor–β (TGF-β) and bone morphogenetic protein (BMP) signaling molecules affects various developmental processes, and a multitude of developmental abnormalities and disease states are associated with the misregulation of their signaling (1–3). Individual TGF-β superfamily members elicit different responses depending on tissue context. For example, patterning of the position of the longitudinal veins and the domains of intervein material in the developing Drosophila wing depends on a long-range signaling activity gradient established by two BMPs, Decapentaplegic (Dpp) and Glass bottom boat (Gbb) (4, 5). However, in the germline stem cell niche, these same BMPs instead act as short-range molecules to maintain stem cell identity (6). A mechanistic explanation for these differences in ligand function is still forthcoming, as is an understanding of how their activities are regulated.
The production of bioactive ligands responsible for the initiation of TGF-β and BMP signaling involves a series of poorly understood maturation events including proconvertase (furin) processing of an inactive dimeric proprotein at optimal (Arg-X-Lys/Arg-Arg) or minimal (Arg-X-X-Arg) sites to yield the C-terminally derived 100– to 140–amino acid growth factor–like peptides of ~15 to 20 kD (7). The high degree of sequence conservation observed between this C-terminal domain of more than 20 different gene products served to establish the now well-known TGF-β/BMP superfamily (8). Many of these family members have more than one furin cleavage consensus sequence just N-terminal to the first conserved cysteine of the ligand domain, and differential cleavage can influence the amount of BMP4 produced in mice and Xenopus and that of Dpp, the Drosophila ortholog of BMPs 2 and 4 (9–13). Such differences in ligand production, as well as in ligand turnover, can affect the functional range of active BMPs in developing tissues (10, 14–16). In addition, two different cleavage forms of Dpp differ in their ability to interact with the Drosophila heparin sulfate proteoglycan Dally, which may contribute to the difference in their in vivo stability (17).
We previously showed that the BMP signaling gradient in the Drosophila wing imaginal disc requires Gbb, the Drosophila ortholog of BMP5/6/7 (5, 18, 19). Dpp is present in a discrete stripe of cells along the anterior/posterior (A/P) boundary of the developing wing (20) where the source of morphogen responsible for the BMP activity gradient is thought to arise (21–23). We found that the activity of Gbb is critical for the breadth of the gradient (5), prompting us to investigate the basis of its long-range behavior to gain insight into how a morphogen gradient is established. As part of these investigations, we found that Gbb exists in two ligand forms. Gbb15 is the conventional ligand derived from the C-terminal domain of the proprotein and contains the conserved seven cysteine residues responsible for the cysteine knot fold characteristic of members of the TGF-β and BMP family (24, 25). Gbb38 is a previously unrecognized active ligand that is larger (with a predicted molecular mass of 38 kD) and also generated by proconvertase processing but at a furin consensus sequence that falls within the N-terminal portion of the prodomain (which we call the NS site). Gbb38 is secreted, binds to receptors, and stimulates signaling events distinct from that produced by Gbb15 alone. A survey of TGF-β family members revealed that the NS cleavage consensus sequence is conserved among many family members from cnidarians to humans. The association of NS site mutations in hBMP4, hBMP15, and AMH with developmental abnormalities [cleft lip with or without palate (CLP), premature ovarian failure (POF), and persistent Müllerian duct syndrome (PMDS), respectively (26–28)] suggests the functional importance of NS processing. Comparisons of the in vivo function of the large and small Gbb ligands uncovered distinct differences in their abilities to induce signaling and to do so over different cell distances. These findings underscore the impact that multiple ligand forms can have on signaling output and provide a new mechanistic explanation for how a single BMP family member can elicit different responses. Together, our data prompt a reevaluation of the identity of ligands that are responsible for activating TGF-β and BMP pathways in different tissue contexts, especially given the value of these molecules as therapeutics to treat and prevent various human diseases and developmental abnormalities.
RESULTS
Proconvertase processing at an unconventional site is required for signaling
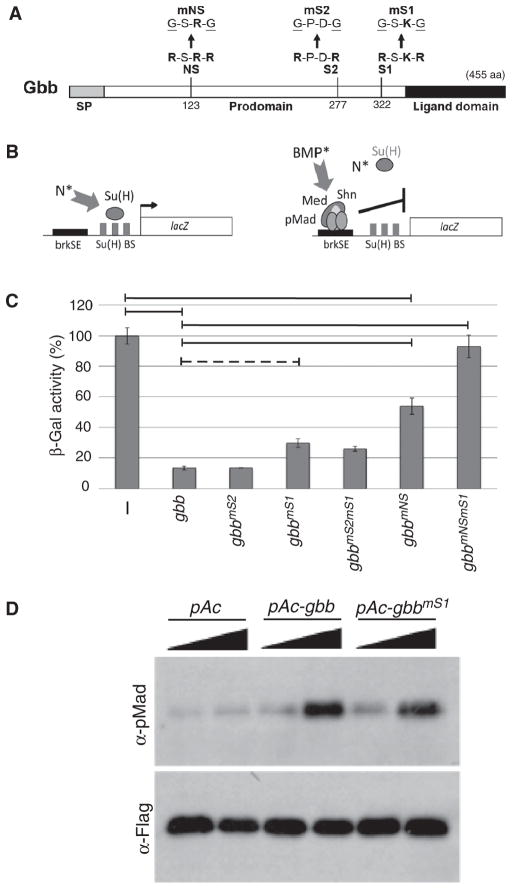
To assess the importance of proprotein processing at specific furin cleavage sites in producing an active Gbb ligand, we compared the signaling activity elicited by wild-type gbb and gbb cleavage mutant constructs. BMP signaling results in the phosphorylation of Mad (the Drosophila R-Smad ortholog) and activation of BMP signaling transcriptionally represses brinker through the direct binding of a phosphorylated Mad-Med-Shn complex to brinker silencer elements (brkSE) in the brinker cis-regulatory region (29). Thus, a brkSE-lacZ reporter transfected into Drosophila S2 cells quantitatively reports the direct transcriptional response to BMP signaling induced by cotransfected ligand or activated receptor constructs (29–31) (Fig. 1, A and B). Wild-type gbb reduced β-galactosidase activity, thus indicating robust BMP signaling, which repressed the brkSE-lacZ reporter (Fig. 1, B and C) (31). gbb cleavage mutant constructs containing mutations at either one or both of the two furin cleavage consensus sites located between the predicted pro- and ligand domains (S2 [Arg277-Pro-Asp-Arg] and S1 [Arg322-Ser-Lys-Arg]) were assayed for signaling activity (Fig. 1, A and C). Unexpectedly, mutations abolishing the S2 site (gbbmS2), previously shown to be essential for BMP4 and Dpp activity (11, 13, 32), had no effect on Gbb signaling activity (Fig. 1C). In addition, mutations at the S1 site (gbbmS1) had only a slight but consistent change in signaling compared to that elicited by wild-type gbb (Fig. 1C). Consistent with its ability to induce BMP signaling, the phosphorylation of Mad was increased in cells expressing gbbmS1 (Fig. 1D). We considered the possibility that processing at either the S2 or the S1 furin cleavage site was sufficient for full activity and that processing at the nonmutated site compensated for the lack of processing at the mutated site. However, we found that a construct lacking both S2 and S1 furin cleavage consensus sequences (gbbmS2mS1) did not show reduced Gbb signaling. This result suggested that either the Gbb proprotein has the ability to signal or an active Gbb ligand could be produced by some means other than the already characterized mode of proconvertase processing at sites adjacent to the conserved C-terminal domain.
Fig. 1.
Furin cleavage site mutations differentially affect Gbb signaling activity. (A) Schematic of Gbb pre-proprotein with amino acid (aa) position of furin cleavage consensus sequences. NS, S2, and S1 and disrupting mutations are noted. SP, signal peptide. (B) Schematic representation of cell-based BMP signaling assay. brkSE-lacZ reporter is activated in response to active Notch signaling (N*) through Su(H) binding sites (BS). Repression of brkSE-lacZ is a direct response to active BMP signaling (BMP*) resulting from binding of the Drosophila BMP signal transducer pMad-Med-Shn complex to brkSE. (C) Signaling activity induced by gbb cleavage mutants (gbb-CMs). In the absence of Gbb-induced signaling (−), β-galactosidase activity is set to 100%. Repression of brkSE-lacZ in response to BMP signaling is measured as reduction in β-gal activity. Horizontal lines indicate significance at P < 0.05 with the Tukey-Kramer HSD procedure for multiple comparisons. gbbmS1 is marginally different from gbb (dotted horizontal line) on the basis of its significant difference using Hsu’s MCB test but not at P < 0.05 with Tukey-Kramer. Each bar represents the average of three independent experiments; error bars indicate SE. (D) Induction of phosphorylated Mad (pMad) measured in S2 cells transfected with Mad-Flag and treated with 0.1× and 1× conditioned media from S2 cells expressing gbb, gbbmS1, or empty vector (pAc).
We identified two additional furin cleavage consensus sequences within the Gbb prodomain ([Arg120-Gly-His-Arg] and [Arg123-Ser-Arg-Arg]) at a position not previously recognized as a viable processing site (Fig. 1A). We found that signaling activity was reduced by 50% in forms of gbb with mutations in these N-terminal sites (NS) (gbbmNS: R123G and R126G), suggesting the importance of processing at the NS in the production of an active Gbb ligand (Fig. 1C). When both the NS and the conventional S1 site were mutated (gbbmNSmS1), Gbb signaling activity was attenuated (Fig. 1C), indicating that although the gbbmS1 mutant could induce signaling to nearly the same extent as the wild-type form, in the absence of processing at the NS site as well (namely, the gbbmNSmS1 double mutant) no signaling activity was retained. Thus, processing at both the S1 and the NS sites appears critical in generating wild-type amounts of Gbb signaling.
Cleavage at the N-terminal site produces a large BMP ligand
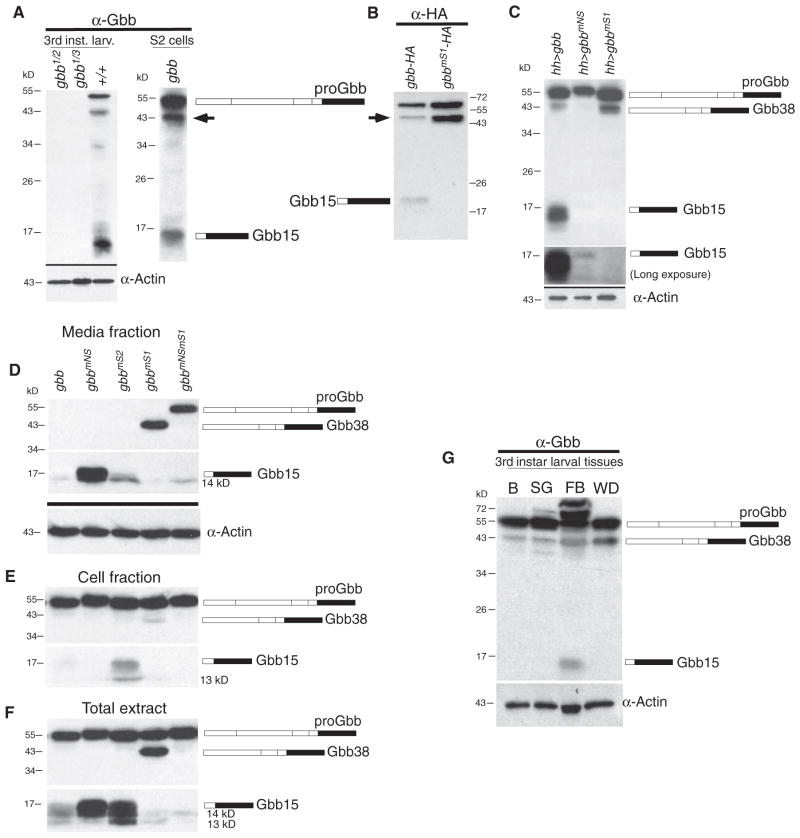
We produced a Gbb monoclonal antibody against a peptide in the C-terminal ligand domain. The specificity of the antibody for Gbb was indicated by the absence of cross-reactivity to proteins in extracts from gbb null larvae (gbb1/gbb2 and gbb1/gbb3) (Fig. 2A). Three prominent protein products at 54, 42, and 15 kD were identified by the Gbb antibody in both whole larval extracts and Drosophila S2 cells (Fig. 2A), and the same three bands were detected with a hemagglutinin (HA) antibody in S2 cell extracts expressing an HA-tagged construct (gbb-1xHA) (Fig. 2B). We had previously discovered that insertion of green fluorescent protein (GFP) into Gbb renders the resulting fusion protein inactive, so we sought to verify that the HA-tagged gbb construct retained signaling activity to enable us to conclude that protein products produced by this construct were functional signaling molecules (fig. S1). We found that Gbb with one HA epitope tag (Gbb-1xHA) retained considerable signaling activity, whereas the insertion of three HA tags (Gbb-3xHA) destroyed signaling activity.
Fig. 2.
Large and small ligand forms are produced by differential processing of Gbb. (A) Extracts from gbb null (gbb1/gbb2 and gbb1/gbb3) and wild-type (+/+) third instar larvae (left) and S2 cells (right) probed for cross-reactivity to a Gbb monoclonal antibody (α-Gbb). In addition to Gbb proprotein (proGbb) and expected ligand (Gbb15), a 42-kD protein specific to Gbb was detected (arrow). Anti-actin (α-Actin) served as a loading control. (B) Extracts from gbb-1xHA– and gbb-1xHA–transfected S2 cells probed with anti-HA (α-HA). Gbb15 product is absent from gbbmS1-1xHA extracts. (C) Extracts from wing discs expressing gbb, gbbmNS, and gbbmS1 under the control of hhGal4 probed with α-Gbb and α-actin. The 42-kD product is lost when the NS cleavage site is mutated (gbbmNS). Predicted size of a product produced by processing at NS is 38 kD. Lower part of blot was exposed for 15 times longer to reveal Gbb15 in gbbmNS extracts and its absence in gbbmS1 extracts. (D to F) Extracts from S2 cells transfected with wild-type gbb and gbb-CM constructs analyzed for Gbb products. Media (D) and cell (E) fractions indicate secreted and cellular products, respectively. Total extracts (F). The abundance of endogenous Gbb38 and Gbb15 differs between different tissues from isolated third instar larval tissues, including brain (B), salivary glands (SG), fat body (FB), and wing discs (WD), which were probed with α-Gbb and α-actin (G). Actin is the loading control. The bottom panel of each Western blot was exposed twice as long as the top panel to visualize the lower–molecular weight products.
As expected, the S1 mutant (gbbmS1) resulted in the loss of the 15-kD band, henceforth referred to as Gbb15 (Fig. 2). This also occurred in wing discs from transgenic lines expressing gbbmS1 (lane 3, Fig. 2C). Extracts from wing discs expressing the NS mutant gbbmNS lacked the higher–molecular mass 42-kD band, indicating that the 42-kD product likely results from proconvertase processing at the NS site (lane 2, Fig. 2C). The predicted size of a Gbb product produced by cleavage at NS is 38 kD; thus, we refer to the NS cleavage product as Gbb38 despite its migration at a slightly higher molecular mass, which may be due to post-translational modifications. In wing discs expressing gbbmNS, Gbb15 is still present (as detected by a long exposure) albeit at a considerably lower amount (Fig. 2C), which may suggest that NS cleavage is important for Gbb15 production or stability. However, we observed the opposite phenomenon in S2 cells (see below; Fig. 2D).
The high signaling activity exhibited by gbbmS1 (Fig. 1C) prompted us to investigate whether Gbb38 was secreted as would be expected of a functional BMP ligand. Gbb38 was efficiently secreted from S2 cells expressing gbbmS1 (lane 4, Fig. 2D) and associated with the Sax type I receptor (fig. S2). As expected, no Gbb15 was apparent in the gbbmS1 media, although a small quantity of a 14-kD protein was detected in the media. The 14-kD protein appeared to be inactive because cells expressing the double mutant gbbmNSmS1, which contains the 14-kD band (lane 5, Fig. 2D), showed no signaling activity (Fig. 1C). Furthermore, the HA antibody did not detect a 14-kD product from the HA-tagged gbb construct, indicating that the 14-kD band was derived from a nonfunctional portion of the ligand domain because it must lack the first conserved cysteine residue. Gbb15 was secreted into the media by gbb-, gbbmNS-, and gbbmS2-expressing cells (lanes 1 to 3, Fig. 2D) and, as expected, bound to the Sax receptor (fig. S2) (33). The detection of Gbb38 in S2 cell extracts expressing wild-type gbb varied somewhat; however, it was reproducibly observed in tissue or whole-animal extracts (Fig. 2, A and C), suggesting that some factor(s) affecting its stability could be lacking in S2 cells. For example, we detected Gbb38 in several tissues dissected from third instar larvae, with more present in wing discs and fat body (Fig. 2G). The relative amounts of Gbb38 to Gbb15 varied between tissues, possibly reflecting site-specific processing in different tissues or differences in ligand stability. Together, our results indicate that Gbb38 is produced by processing at the NS site to yield a bona fide BMP ligand that elicits robust signaling.
Several other points of interest were also revealed by our biochemical analysis. First, the amount of ligand secreted into the media did not directly correlate with the amount of signaling induced by a given construct. For example, most of the large quantity of Gbb15 produced by gbbmNS was secreted into the media (lane 2, Fig. 2, D and F); however, gbbmNS exhibited significantly reduced signaling activity (Fig. 1C). Second, although gbbmS2 cells produced more total Gbb15 than wild-type gbb (lane 3, Fig. 2, D and F), the amount of secreted Gbb15 was comparable to that secreted from wild-type gbb–expressing cells (lane 1, Fig. 2, D and F). No aberrant Gbb products were detected in gbbmS2 cells consistent with their wild-type signaling activity (Fig. 1C). Thus, the Gbb15 secreted by gbbmS2-expressing cells appears to be more efficient at inducing signaling than that produced by gbbmNS-expressing cells. Finally, when both NS and S1 sites were mutated (gbbmNSmS1), the Gbb proprotein was secreted from cells (lane 5, Fig. 2D). Unexpectedly, we observed that proGbb could bind the Sax receptor (fig. S2) despite its inability to elicit signaling (Fig. 1C). Unlike the recent proposal that the structural “straitjacket” motif of the TGF-β1 prodomain blocks ligand-receptor binding (34), our data indicate that the presence of the prodomain in proGbb does not unequivocally disrupt receptor binding. Nevertheless, in wild-type cells and tissues, little to no proGbb is secreted, and therefore, we would not expect the unprocessed wild-type Gbb proprotein to occupy receptors.
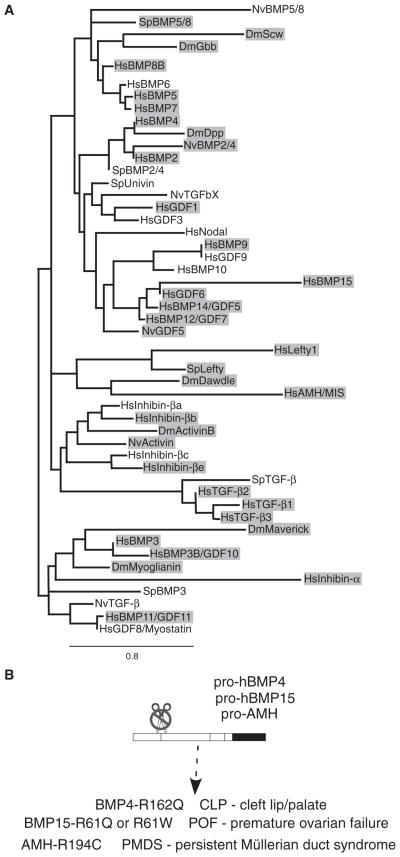
The NS processing site is evolutionarily conserved and appears to be required for human development
The presence of an NS proconvertase consensus sequence is not unique to Drosophila Gbb. We found that a proconvertase consensus sequence is present within the N terminus of the prodomains of many invertebrate and vertebrate TGF-β and BMP family members (Fig. 3A) from Nematostella to humans. (We have arbitrarily defined an NS site as one that would produce a protein larger than 200 amino acids including the highly conserved C-terminal domain.) In many cases, the NS site is conserved among orthologs (Fig. 3A, gray shading). The prevalence of NS sites suggests that alternative processing of TGF-β and BMP proproteins could be functionally conserved. Indeed, we found that point mutations in three human TGF-β superfamily members, AMH, hBMP15, and hBMP4, from patients with persistent Müllerian duct syndrome, premature ovarian failure, and CLP, respectively (26–28), specifically abolish an NS furin consensus sequence (Fig. 3B). The developmental abnormalities associated with these NS mutations suggest that the wild-type function of a particular TGF-β superfamily ligand for a specific developmental function, such as palate formation, could be compromised in the absence of NS cleavage. To test for a loss of signaling activity associated with an NS mutation in hBMP4, we assayed for the BMP signaling reporter BRE-luc in transiently transfected human embryonic kidney (HEK) 293 cells. Although we did not detect a significant reduction in signaling due to mutations in the hBMP4 NS site, either with hBMP4-R162Q (which is associated with CLP) or with hBMP4-R162G, R165G, we found that signaling activity was significantly reduced with the S1 site mutation (hBMP4-R289G, R292G) (fig. S3). Unexpectedly, we found that a mutant S2 site (hBMP4-R253G, R256G) did not show a loss in signaling. Given that processing at the S2 site of mouse BMP4 is critical for tissue-specific functions in vivo, in that both heterozygous and homozygous BMP4mS2 mutants show reduced numbers of primordial germ cells (11), it is conceivable that we cannot detect an effect on signaling activity due to mutations at the NS or S2 sites in HEK293 cells as a result of high autocrine BMP signaling (35).
Fig. 3.
NS cleavage sites are conserved in the TGF-β superfamily. (A) Phylogenetic tree of TGF-β family members from bilateria [Drosophila melanogaster (Dm, protostome), Strongylocentrotus purpuratus (Sp), and Homo sapiens (Hs; deuterostomes)] and radiata [Nematostella vectensis (Nv)]. Shaded proteins contain a putative NS cleavage site. (B) Specific mutations at NS sites in human BMP4, BMP15, and AMH proteins are associated with the indicated developmental abnormalities.
Cleavage mutants exhibit distinct signaling activities in vivo
The inability of the gbbmNSmS1 double mutant to provide functional signaling was borne out by the failure of a genomic gbb rescue construct with mutations in both the NS and the S1 sites (gbbR-gbbmNSmS1) to rescue the lethality associated with a gbb null, gbb1 (36). Whereas a wild-type gbb genomic construct (gbbR-gbb) restored viability with 99% of gbbR-gbb gbb1/gbbR-gbb gbb1 flies surviving (n = 447), 0% of the gbbR-gbbmNSmS1 gbb1/gbbR-gbbmNSmS1 gbb1 class survived (n = 363). Other cleavage mutants yielded survivors, with 60% of gbbR-gbbmNS gbb1, 7.4% of gbbR-gbbmS1 gbb1, and 0% of gbbR-gbb gbb1 homozygotes showing mild wing defects associated with abnormal BMP signaling (fig. S4). The higher percentage of gbbR-gbbmNS gbb1 flies exhibiting defective wings is consistent with the lower signaling activity of gbbmNS compared to that of gbbmS1 (Fig. 1C).
We next assessed the expression of gbb cleavage mutants in different tissue domains. Transgenic lines were generated such that each gbb cleavage mutant construct (like each gbb genomic rescue construct) was inserted into the same genomic position with the φC31 system (37) to eliminate variation in expression between lines due to positional effects. We first tested the impact of misexpressing each cleavage mutant with different Gal4 drivers on adult viability (hhGal4, ciGal4, apGal4, dppGal4; Table 1). Inappropriate BMP signaling during development is often associated with lethality, and viability resulting from gbb cleavage mutant misexpression (Table 1) inversely correlated with the different signaling activities attributed to each mutant as delineated in the S2 cell–based signaling assay (Fig. 1C). Those mutants exhibiting the highest signaling showed the lowest viability, such that wild-type gbb and gbbmS2 expressed under the control of apGal4 failed to produce adult flies, whereas misexpression of the signaling-inactive gbbmNSmS1 mutant did not adversely affect viability and produced 100% of the expected number of adult progeny (Table 1). Similar trends from most to least viable (gbb>gbbmS1>gbbmNS>gbbmNSmS1) were seen with the other Gal4 drivers. Misexpression of gbb cleavage mutants in the dpp expression domain was the exception (dppGal4>UAS-gbb-CM); for example, the expression of the double cleavage mutant (UAS-gbbmNSmS1) was completely lethal (Table 1). Like other TGF-β and BMP cleavage mutants (38–44), we found that the double mutant gbbmNSmS1 indeed affected BMP signaling in a dominant-negative fashion. Phosphorylation of Mad was reduced compared to that of the control (dppGAL4>UAS-GFP) when gbbmNSmS1 was specifically expressed in dpp-expressing cells (dppGal4>UAS-gbbmNSmS1) (fig. S5, A to C). Pharates (dppGal4>UAS-gbbmNSmS1) dissected out of their pupal cases showed macrochaetae and leg defects consistent with a reduction in BMP signaling (36, 45, 46) (table S1). We also found a cell-autonomous reduction in phosphorylated Mad when random clones overexpressing gbbmNSmS1 fell within the medial region of the wing disc (fig. S5, D and E). Defects associated with expression of uncleavable Gbb (gbbmNSmS1; proGbb) could reflect either its ability to form a nonproductive dimer with endogenous wild-type Dpp and Gbb, which would be expected to interfere with ligand processing or secretion, or its inability to elicit signaling if it is secreted and bound to receptors (see Figs. 1C and 2D).
Table 1.
Viable progeny resulting from misexpression of gbb and gbb cleavage mutants. Transgenes were integrated at chromosomal position 86Fb with the φC31 system (37). n = total number of GAL4/UAS progeny (including viable adult flies and/or dead pupae).
| GAL4 lines | Transgenic UAS lines
|
||||
|---|---|---|---|---|---|
| gbb | gbbmS2 | gbbmS1 | gbbmNS | gbbmNSmS1 | |
| ap-GAL4 | 0% (n = 57) | 0% (n = 52) | 14% (n = 45) | 64% (n = 58) | 100% (n = 31) |
| hh-GAL4 | 0% (n = 72) | 0% (n = 86) | 0% (n = 93) | 18% (n = 66) | 94% (n = 70) |
| ci-GAL4 | 0% (n = 40) | 0% (n = 42) | 0% (n = 52) | 0% (n = 65) | 3% (n = 67) |
| dpp-GAL4 | 39% (n = 107) | 0% (n = 68) | 97% (n = 117) | 99% (n = 100) | 0% (n = 172) |
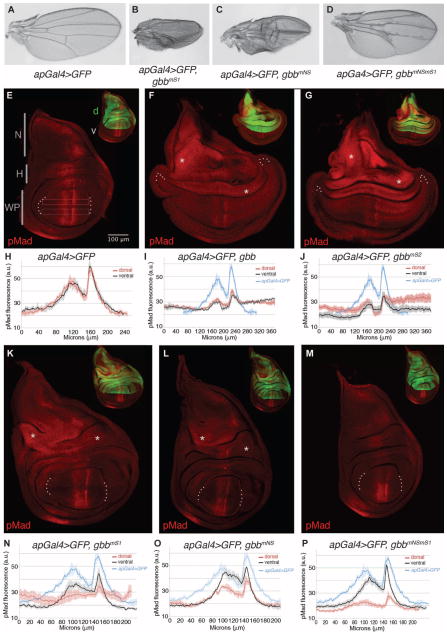
On the basis of the viability studies, we examined in greater detail the phenotypic consequences of expression of Gbb and the noncleavable mutant in the wings of apGal4>gbb-CM adult survivors and in the larval wing imaginal discs in which misexpression was directed to the dorsal compartment (apGal4; Fig. 4). Both apGal4>UAS-gbbmS1 and apGal4>UAS-gbbmNS were semilethal, with gbbmS1 being more lethal, consistent with its higher signaling activity in S2 cells (Fig. 1C and Table 1). Adult wings from wild-type flies showed a stereotyped pattern with five longitudinal veins and two cross veins (Fig. 4A). Wings from adult survivors expressing the cleavage mutants showed blistered and ectopic venation phenotypes commensurate with the magnitude of ectopic signaling elicited by each cleavage mutant (Fig. 1C), with apGal4>UAS-gbbmS1 (Fig. 4B) showing the most severe wing defects of the viable progeny, followed by apGal4>UAS-gbbmNS and apGal4>UASgbbmNSmS1 (Fig. 4, C and D). No adult survivors were found from apGal4>UAS-gbb or apGal4>UAS-gbbmS2 (Table 1).
Fig. 4.
Differential response of wing disc cells to dorsal expression of gbb cleavage mutants.(A to D)Wings from(A)apGal4>UAS-GFP, (B) apGal4>UAS-gbbmS1, (C) apGal4>UAS-gbbmNS, and (D) apGal4>UAS-gbbmNSmS1 adults. Ectopic venation, abnormal wing size, and the improper apposition of the dorsal and ventral wing surfaces are phenotypes that correlate with altered BMP signaling. (E to P) Distribution of pMad(red) in third instar larval wing discs expressing UAS-gbbCMs with inset of same disc at 0.4× shows UAS-GFP (green) under the control of apGal4 in the (E) dorsal compartment (d). v, ventral compartment; N, notum; H, hinge; WP, wing pouch. (H to J and N to P) pMad quantification [in arbitrary units (a.u.)] across the wing pouch (WP) in the dorsal (red) and ventral (black) compartments of multiple discs for each genotype as indicated by open boxes in (E). Tracing is the average intensity from multiple discs with SEs shown (vertical lines). n = number of discs. (F and I) apGal4> UAS-GFP, UAS-gbb (n=5); (G and J) apGal4>UAS-GFP, UAS-gbbmS2 (n = 5); (K and N) apGal4>UAS-GFP, UAS-gbbmNS(n=3); (L and O) apGal4>UAS-GFP, UAS-gbbmS1 (n = 4); (M and P) apGal4>UAS-GFP, UAS-gbbmNSmS1 (n= 4). Tracing of wild-type endogenous pMad gradient characterized by two peaks of higher pMad separated by a trough of lower accumulation is shown for comparison (blue, n = 3) in (I) and (J) and (N) to (P). Asterisks (*) highlight regions of ectopic pMad in the hinge (H) and notum (N) regions of the wing disc. All images obtained at the same magnification under identical confocal microscope settings.
In the wing imaginal disc, ectopic BMP signaling associated with gbb cleavage mutant misexpression was visible as an increase in the phosphorylation of Mad, especially in the lateral wing pouch, hinge, and notum regions of wing discs compared to the wild-type endogenous pattern of phosphorylated Mad (Fig. 4, E to P). The intensity of phosphorylated Mad, which reflects the BMP morphogen activity gradient in the wing pouch (47), was measured from the dorsal (the site of apGal4 expression) and the ventral compartment of multiple discs for each genotype (Fig. 4, H to J and N to P). Wing imaginal discs dissected from apGal4>UAS-gbb and apGal4>UAS-gbbmS2 larvae showed extensive phosphorylation of Mad throughout the entire disc (Fig. 4, F, G, I, and J), indicating that ligands produced by gbb and gbbmS2 in dorsal cells can act nonautonomously to activate BMP signaling in cells far from where they are expressed. In addition, apGal4>UAS-gbb and apGal4>UAS-gbbmS2 wing discs were overgrown, a phenotype consistent with increased BMP signaling (Fig. 4, F and G). The intensity of the phosphorylated Mad signal in the notum and hinge of apGal4>UAS-gbb and apGal4>UAS-gbbmS2 discs was higher than that observed in the wing pouch, indicating that different regions of the disc respond differently to increased ligand concentration. Although ectopic phosphorylated Mad was evident in the lateral wing pouch when gbb and gbbmS2 were overexpressed, the absolute amount of phosphorylated Mad in the medial wing pouch was reduced compared to that measured in control discs (Fig. 4, H to J), although the characteristic two peaks in the phosphorylated Mad gradient remained (47). Wing discs from apGal4>UAS-gbbmS1 larvae similarly exhibited a high degree of over-proliferation, but curiously, the wing pouch of this genotype was not expanded (Fig. 4K). The gradient of phosphorylated Mad in the dorsal compartment of larvae expressing apGal4>UAS-gbbmS1 extended into the lateral domains similar to larvae expressing apGal4>UAS-gbb and apGal4>UAS-gbbmS2. However, this ectopic phosphorylated Mad signal was not apparent in the ventral compartment (Fig. 4N). Both the amplitude and the breadth of the phosphorylated Mad gradient in the ventral compartment of apGal4>UAS-gbbmS1 discs were reduced, indicating that although Gbb38 produced by the gbbmS1 mutant can promote signaling in dorsal lateral cells, signaling is affected differently than in the ventral compartment. Similar to apGal4>UAS-gbb and apGal4>UAS-gbbmS2 wing imaginal discs, stronger ectopic phosphorylated Mad signals were detected in the hinge and notum compared to the wing pouch in apGal4>UAS-gbbmS1 discs (Fig. 4, K and N), again revealing a region-specific response. Consistent with its ability to induce signaling, albeit at a reduced amount, the notum and hinge regions of apGal4>UAS-gbbmNS wing discs showed a moderate increase in phosphorylated Mad, but within the wing pouch, the gradient of phosphorylated Mad was narrower, and the intensity of phosphorylated Mad was reduced in the dorsal compartment (Fig. 4, L and O). The double cleavage mutant apGal4>UAS-gbbmNSmS1 resulted in an overall reduction in the size of the wing disc, and as expected from the cell-based signaling assay (Fig. 1C), this construct did not induce ectopic phosphorylated Mad at other sites in the disc such as the notum or hinge domains (Fig. 4, M and P). The decrease in cell-autonomous phosphorylated Mad in the dorsal wing pouch of these discs was consistent with the expectation that an uncleavable form of Gbb would behave dominantly to disrupt the function of endogenous BMPs (Dpp and Gbb) or the Sax receptor, all of which are critical for signaling in the medialwing pouch (5, 18, 19). Strikingly, despite reduced phosphorylation of Mad in the dorsal wing pouch (Fig. 4, M and P), adult apGal4>UAS-gbbmNSmS1 wings (Fig. 4D) showed minimal patterning defects, suggesting that loss of BMP signaling in the dorsal cells of the larval wing disc could be compensated for by events during the pupal stage to generate a relatively normal wing pattern.
Signaling activity of cleavage mutants is controlled in a domain-specific manner
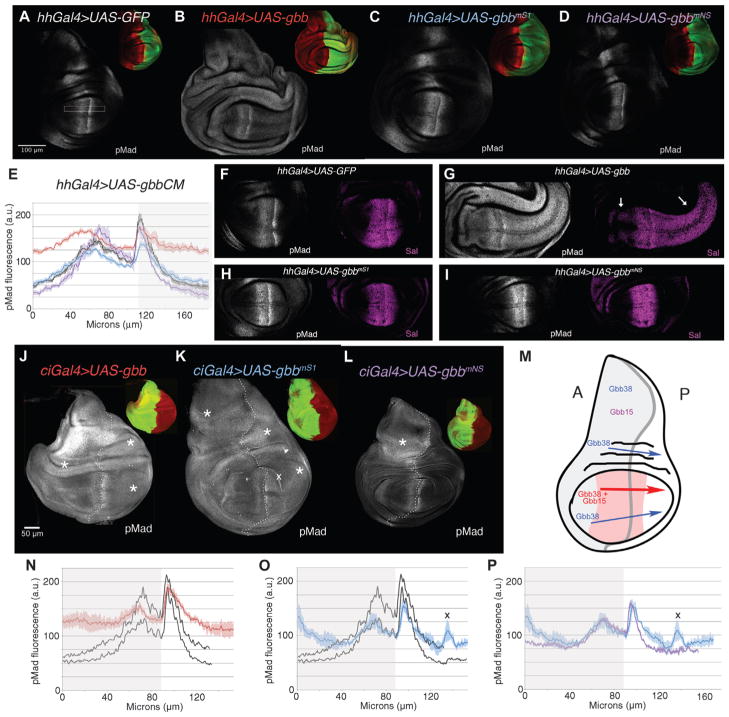
We next considered the effects of the Gbb cleavage mutants when expressed in the posterior (hhGal4) and anterior (ciGal4) compartments (Fig. 5, A to P) on BMP signaling by visualizing the distribution of phosphorylated Mad. We found that the expression of wild-type gbb led to ectopic phosphorylated Mad both within and outside gbb-expressing cells (Fig. 5, A, B, and J). Similar to our observations in apGal4>UAS-gbb discs (Fig. 4, F and I), when gbb was expressed throughout either the anterior (Fig. 5J) or the posterior (Fig. 5B) compartments, BMP signaling was induced in all cells of the wing disc. As predicted by our previous studies (5, 18, 19), the ectopic signaling induced in hhGal4>UAS-gbb, when Gbb was produced by posterior compartment cells where Dpp is not present, indicates that Gbb homodimers can act over a long range. Consistent with both the signaling activity and the long-range activity of Gbb homodimers, the expression domain of BMP target gene Spalt (Sal) was expanded in both the posterior and the anterior compartments of the wing pouch in hhGal4>UAS-gbb wing discs compared to its endogenous pattern (Fig. 5, F and G).
Fig. 5.
Long-range activity of Gbb38 is domain-dependent. BMP signaling [distribution of pMad (gray)] assessed in third instar larval wing discs expressing UAS-gbb and UAS-gbb-CMs in posterior (hhGal4) and anterior (ciGal4) compartments. Dorsal up, posterior (P), right. Insets of same disc at 0.4× show distribution of pMad (red) and GFP (green). (A) Control wild-type disc, hhGal4>UAS-GFP.(B)hhGal4>UASgbb, UAS-GFP. (C) hhGal4> UASgbbmS1 UAS-GFP (Gbb38). (D) hhGal4> UASgbbmNS UAS-GFP (Gbb15). All images collected at the same confocal settings. (E) pMad quantification in arbitrary units (a.u.) collected from dorsal wing pouch (open rectangle) from multiple discs. [hhGal4>UAS-GFP (black), n = 4; hhGal4> UASgbb (red), n = 6; hhGal4>UASgbbmS1 (blue), n = 4; hhGal4> UASgbbmNS (purple), n = 4]. SE indicated. (F to I) pMad and Sal distribution in the wing pouch of hhGal4>UAS-GFP (F), hhGal4>UASgbb (G), hhGal4>UASgbbmS1 (H), and hhGal4>UASgbbmNS (I) wing discs. Arrows in (G) denote expanded Sal expression. (J to M) ciGal4>UAS-gbb, UAS-GFP (J), ciGal4>UAS-gbbmS1, UAS-GFP (Gbb38) (K), ciGal4>UAS-gbbmNS, UAS-GFP (Gbb15) (L), schematic of sites of ectopic BMP signaling induced by gbbmS1 (Gbb38), gbbmNS (Gbb15), and wild-type gbb (Gbb38 + Gbb15) when expressed in the anterior compartment (gray) (M). Portion of wing pouch refractory to gbb overexpression, red. (N to P) pMad quantification in arbitrary units (a.u.) collected from dorsal wing pouch.(N)ciGal4>UAS-GFP(black), n=2; ciGal4>UAS-gbb (red), n = 3. (O) ciGal4>UAS-GFP (black), n = 2; ciGal4>UAS-gbbmS1 (blue), n = 3. (P) ciGal4> UAS-gbbmS1 (blue), n = 3; ciGal4>UAS-gbbmNS (purple), n = 3. All images collected at the same confocal settings. (O and P) X marks anomaly in pMad tracing due to fold at edge of wing pouch [see also (K)]. n = number of discs.
We observed minor but reproducible differences in the ability of Gbb38 (gbbmS1) and Gbb15 (gbbmNS) to induce signaling in posterior cells when expressed in those cells (hhGal4>UAS-CM), as well as in the cells of the anterior compartment (Fig. 5, C to E). When gbbmNS was expressed in posterior cells, the width of the phosphorylated Mad gradient and the expression of Sal were slightly reduced (Fig. 5, D, E, and I). Although the narrowing in the phosphorylated Mad gradient could reflect a reduction in the size of the wing pouch given that the gradient of phosphorylated Mad scales with disc size (48, 49) and the average width of the hhGal>UAS-gbbmNS wing pouch was 0.77 times that of the hhGal4>UAS-GFP wing pouch (n = 7 discs, P < 0.05), the significant increase in the amplitude of the anterior peak of phosphorylated Mad suggested that Gbb15 is active and may alter the dynamics of signaling in this medial-most portion of the disc. The reduction in phosphorylated Mad in the lateral regions may indicate a dominant-negative effect. As we have observed previously, dpp and gbb genetically interact, and the balance of Dpp to Gbb is likely critical to the generation of the phosphorylated Mad gradient (5, 18, 19). When gbbmNS is expressed in the posterior compartment, it is possible that its mild negative impact on signaling could result from a shift in the stoichiometry of Gbb15 to Gbb38, although further studies will be required to test this hypothesis.
When expressed in the anterior compartment, gbbmS1 induced ectopic signaling in the lateral cells of the anterior compartment as well as throughout the posterior compartment (Fig. 5, K and O). However, gbbmNS induced increased signaling only in the lateral anterior cells (Fig. 5, L and P). As summarized in Fig. 5M, wild-type Gbb (Gbb38 and Gbb15) induced phosphorylated Mad in cells far from where it was produced, whereas Gbb38 induced ectopic phosphorylated Mad at a reduced amount and in a more selective pattern. In contrast, Gbb15 did not exhibit the ability to induce signaling outside of the anterior compartment where it was produced (ciGal4>UAS-gbbmNS); furthermore, it induced minimal ectopic phosphorylated Mad outside of the notum region. The narrow signaling range of Gbb15 and the long-range signaling ability of both Gbb38 and wild-type Gbb are further supported by the induction of ectopic phosphorylated Mad outside random clones expressing gbb (wild-type Gbb) and gbbmS1 (Gbb38) compared to gbbmNS (Gbb15)–expressing clones (fig. S6, A to F). Together, all of our results from the domain-specific expression of gbb cleavage mutants indicate that (i) the response of cells varies with the region within the disc; (ii) Gbb38 can influence cells distant from where it is produced, but Gbb15 in general does not; (iii) the expression of wild-type gbb elicits a more pronounced, long-range influence on cells; and (iv) the phosphorylated Mad gradient within the medial wing pouch is resilient to increases in ligand abundance.
DISCUSSION
To date, active TGF-β and BMP family dimeric ligands are known to consist of two peptides processed from the C-terminal 100 to 110 amino acids of two larger proproteins. All ligands adopt a similar “inverted clasped hands” or “butterfly” fold because of the high conservation in this domain, especially of the hallmark cysteine residues. Here, we report the identification of a BMP ligand form containing the highly conserved C-terminal domain contiguous with a portion of the less conserved prodomain. This large ligand (Gbb38), one of two forms produced by gbb, arises from proconvertase processing at a previously unrecognized furin consensus sequence (NS) at residue 123 of the 455–amino acid Gbb proprotein. A second ligand, Gbb15, is produced by cleavage at the canonical S1 site located at residue 322, which is N-terminal to the conventional ligand domain. Both Gbb38 and Gbb15 are active, with Gbb38 exhibiting a higher level of signaling activity both in cell culture (Fig. 1C) and in vivo (Figs. 4 and 5 and fig. S6). The ability of Gbb38 and Gbb15 to signal is influenced by the tissue domain in which they are expressed. When expressed in either the dorsal or the anterior compartment of the wing imaginal disc, Gbb38 and Gbb15 induce ectopic signaling commensurate with their relative signaling abilities as measured in cell culture (Figs. 1C, 4, and 5). However, when expressed in the posterior compartment, their ability to elicit ectopic signaling is impaired (Fig. 5). This finding is particularly interesting in light of our previous observations that Gbb produced in the anterior compartment behaves differently from that generated in the posterior compartment (5). On the basis of the fact that Dpp is produced solely in the anterior compartment, one could envision that the compartmental difference in the behavior of Gbb may be explained by the action of Dpp-Gbb heterodimers formed only in the anterior compartment. However, we found that Gbb38 induced signaling from clones lacking Dpp (fig. S6, C and D) and that Gbb15 induced a significant increase in the amplitude of the anterior phosphorylated Mad peak when expressed in the posterior compartment (Fig. 5, D and E). Furthermore, the expression of wild-type gbb (hhGal4>UAS-gbb) in the posterior compartment, which produces both Gbb38 and Gbb15 (Fig. 2C), induced considerable ectopic phosphorylated Mad in all cells of the disc in both posterior and anterior compartments (Fig. 5, B and E). Thus, the differential response to expression of gbb and gbb cleavage mutants in various wing disc domains cannot be explained solely by the presence of Dpp. Given that both Gbb38 and Gbb15 are produced by posterior cells (Fig. 2C), the factors responsible for this compartment-specific behavior are likely to be involved in the reception or integration of their independent signals.
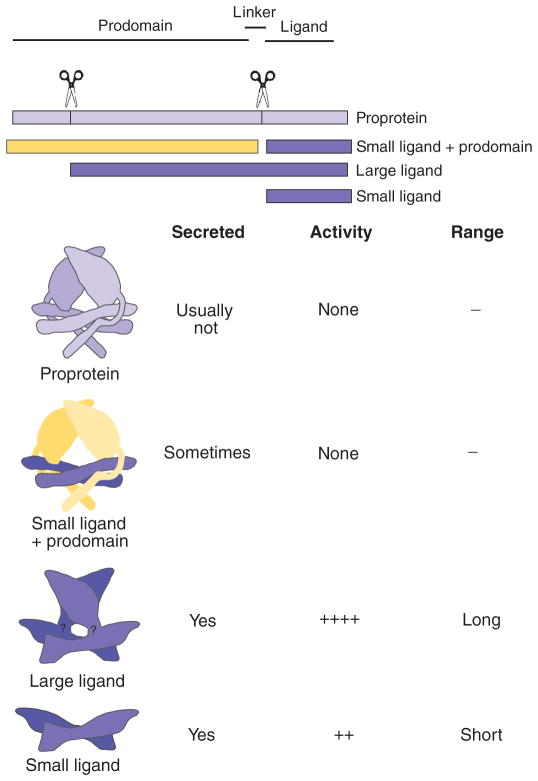
In addition to compartment-specific behavior, we also found that Gbb38 promoted signaling in cells far from where it is produced (Fig. 5, K and O, and fig. S6, C and D), as did wild-type Gbb (Figs. 4, F and I, and 5, B, E, J, and N, and fig. S6, A and B). However, Gbb15 in general did not because it induced high signaling in the notum and hinge of the anterior compartment only when it was expressed in the anterior compartment (Fig. 5, L and P). Tissue-specific processing at the conventional linker domain cleavage sites can regulate the amount of BMP4 and Dpp ligand produced and thus influence signaling strength and ligand function in Xenopus and Drosophila (11, 13). In the case of the larger Gbb38 ligand, the presence of considerable additional “pro-domain” sequences not found in the conventional ligands adds a new set of parameters to be considered in understanding the mechanisms that regulate TGF-β signaling. The additional protein domains Gbb38 could allow for or block interactions with extracellular factors that regulate ligand movement, such as the heparan sulfate proteoglycans Dally and Dally-like protein (Dlp), Pentagone (Pent) and Larval Translucida (Ltl), and different combinations of receptors (50, 51). In this vein, the existence of an active large ligand form is of particular interest in light of the structure of the TGF-β1 prodomain (34). Cleaved prodomains are thought to assist in intracellular ligand folding and secretion, but for some family members, the ligand remains noncovalently associated with its two prodomains after secretion. In most cases, this association confers latency (inactivity) as a result of structural changes, with ligand activation requiring proteolytic cleavage of the prodomain in the latent complex or a force-dependent mechanism through integrin interactions (such as is the case for TGF-β, BMP2, and GDF8) (34, 52–55). The prodomain-BMP7 complex is not latent, and the prodomains are displaced upon ligand binding to the BMP type II receptor (56). In this case, ligand activation does not require extracellular matrix molecules or proteases, an observation that may hold true for AMH as well (56, 57). Despite the apparent differences in TGF-β and BMP family members and their extracellular associations with their prodomains, structural studies indicate that prodomains in TGF-β and BMP family ligands are expected to share the same cysteine knot fold (58). In the case of TGF-β1, the prodomain dimer envelops the C-terminally derived ligand dimer to prevent ligand activation, with a straitjacket motif created by the overlapping N-terminal 45 residues of the prodomain (Fig. 6). If processing occurs at the NS site in Gbb, the N-terminal 90 amino acids would be removed, eliminating the straitjacket domain and the “latency lasso” that encircles the residues critical for receptor binding. As our analyses have shown, the removal of this N-terminal–most domain must allow for receptor binding of the Gbb ligand without requiring any further processing and subsequent signaling. Indeed, the 73 amino acids at the extreme N terminus of the GDF8 prodomain confer its inhibitory effect over GDF8 (53). Although the structural conformation of the large ligand is not yet known, it is conceivable that the butterfly structure of the C-terminally derived ligand domain will fold on the basis of the conserved cysteine knot structure, with the receptor interaction faces unimpeded by the linker domain (Fig. 6). Given the more robust signaling elicited by Gbb38 than Gbb15 and the therapeutic uses of BMPs, it will be interesting to resolve the structure of a large ligand form.
Fig. 6.
Model summarizing the signaling capacity and range of action for products produced by TGF-β and BMP family members. (Top) Linear schematic of TGF-β/BMP protein domains (prodomain, linker, and ligand domains), NS and linker cleavage sites (vertical lines, scissors), and cleavage products. (Bottom) Model schematic of structure of products based on Shi et al. (34) (color-coded to linear representations) with secretion, signaling activity, and range of action properties noted. No structure is known for the linker region, which is designated in the large ligand model.
Although we found that the signaling ability of cleavage mutants that exclusively produced Gbb38 (gbbmS1) or Gbb15 (gbbmNS) depended on the domain in which they were expressed, the expression of wild-type gbb led to increased BMP signaling in all cells of disc with the exception of the medial wing pouch, which appears to be under tight regulation. Although the phosphorylated Mad gradient in the medial wing pouch is sensitive to the loss of gbb and other signaling components, it is resistant to their increases most likely in part due to a negative feedback mechanism involving Dad, an inhibitory Smad (5, 59). In addition, the phosphorylated Mad gradient scales with disc size in the medial wing pouch (48, 49), an effect that we did not observe in other regions of the disc upon overexpression of gbb or the gbb cleavage mutants. Furthermore, we found that scaling was disrupted by wild-type gbb expression, which produced an overgrown disc with relatively minor changes to the phosphorylated Mad gradient (Figs. 4 and 5 and fig. S6). Future studies will be important to resolve the molecular basis of these domain-specific signaling responses and the inability of either Gbb38 or Gbb15 to recapitulate the effects of wild-type gbb, but it is clear that the different Gbb ligands exhibit distinct effects.
The inability of either Gbb38 or Gbb15 to recapitulate wild-type gbb when expressed under endogenous gbb control (fig. S4) indicates that to attain full Gbb signaling, processing must be possible at both the S1 and the NS sites. The simplest explanation for this observation is that wild-type signaling requires the presence of both Gbb38 and Gbb15 ligands either as homodimers or as Gbb38:Gbb15 heterodimers. An alternative explanation is that to produce a canonical active Gbb ligand (presumably Gbb15), processing must occur at both sites either sequentially or simultaneously. Our data do not favor the latter alternative because Gbb38 alone exhibits robust signaling and both forms are present endogenously with differential enrichment in various tissues, suggesting that the cell type or tissue environment may influence proGbb processing (Fig. 2G). Instead, we envision that the production, maturation, and stability of the array of active ligands under normal conditions depends on the presence of both NS and S1 cleavage sites and that the reception and integration of the various combinations of ligands occurs in a domain-specific manner.
The differential requirement for different sets of ligands could explain the apparent specificity of the NS mutation in hBMP4 (hBMP4-R162Q) associated with CLP (28). The associated failure in the fusion of the medial nasal and maxillary processes during orofacial development is a fairly specific temporal and spatial disruption of hBMP4 signaling. BMP4 is required for mouse lip and palate fusion (60) as well as many other fundamental roles in early development including axis determination and the specification of different germ layers (3, 61, 62). Given that BMP4 is essential for embryonic development, this relatively minor defect suggests that either processing at the NS site of hBMP4 occurs only in a specific set of cells, at a specific time in development, and individuals carrying the hBMP4R162Q allele fail to produce the proper hBMP4 ligand type(s) necessary to elicit the proper magnitude of BMP signaling required for lip and palate fusions, or compensatory mechanisms, such as BMP redundancy and feedback regulation, restore BMP signaling activity to its normal magnitude in other tissues of patients with CLP. Bmp4S2G/S2G mice also survive to adulthood with severe defects in only some tissues, such as the testes, with other tissues or organs that require BMP4 signaling, such as the limb and kidney, developing normally (11). It is conceivable that ligand produced by cleavage at NS and S2 sites does not affect the magnitude of signaling but rather the signaling between specific sets of cells. Consistent with this possibility, we found that although the S1 site is required for hBMP4 signaling, we could not detect a difference in the amount of signaling induced by mutations in S2 and NS, as well as hBMP4-R162Q in transfected cells (fig. S3), where the production or reception of a putative large hBMP4 ligand form may not occur, especially over the basal extent of autocrine signaling. Instead, it will be of greater interest to test for the effect of the CLP mutation hBMP4-R162Q on BMP signaling specifically in the orofacial primordia because this tissue may be most sensitive to a large form of the hBMP4 ligand or a prodomain-ligand complex with a shorter prodomain segment. BMP4 plays an instrumental role in the morphological differences in the beaks of Darwin’s finches, as well as in jaw development of the rapidly evolving species of cichlid fishes occupying Lakes Victoria, Malawi, and Tanganyika of the East African Rift valley (63–65). Although the existence of a large BMP4 ligand has yet to be determined, it is intriguing to consider that among the East African THMV (Tropheini, Haplochromine, Lake Malawi, and Lake Victoria) group of cichlids, the prodomain of BMP4 contains a high number of amino acid substitutions not seen in the signal peptide or ligand domain (66). Either the regulatory function of the prodomain in a BMP4 latent complex could be critical for modulating ligand activity during jaw development or amino acid changes in a putative large BMP4 ligand could alter its signaling abilities in orofacial development of cichlids.
In conclusion, our studies have identified a proconvertase cleavage site that is essential for the full activity of the Drosophila BMP5/6/7 ortholog Gbb, as well as for that of several human BMPs. Given the evolutionary and functional conservation of alternative processing in the N-terminal region of the proprotein, it is of interest to note that despite lower sequence conservation between BMP prodomains, they are thought to share a folded core domain (58). TGF-β and BMP family members that are processed to yield both large and small ligand forms may exhibit an expanded range of signaling activities because of differences in ligand structural motifs or interaction faces. Thus, the posttranslational regulation of BMP ligands is more intricately controlled than previously anticipated. A role for proconvertase processing as a mechanism to control both quantity and quality of ligand production is now more strongly supported by the identification of the NS cleavage site critical for signaling activity, by its evolutionary conservation, and by the fact that mutations in this site appear to disrupt human development (Fig. 3). Our results raise the possibility that context-dependent behaviors reported for various TGF-β and BMP ligands could be explained by the action of such alternate ligand forms.
MATERIALS AND METHODS
Drosophila strains
All Drosophila strains have been described previously (31, 36) or are in FlyBase and were obtained from Bloomington Stock Center (67). Crosses were performed at 25°C on standard sugar, cornmeal, and yeast extract media. UAS-gbb and UAS-gbb-CM were inserted at chromosomal location 86Fb with the φC31 system (37). The null allele gbb1 was recombined onto chromosomes bearing gbbR-gbb or gbbR-gbb-CM insertions at the φC31 landing site at 53B2. The presence of gbb1 in the resulting gbbR-gbb* gbb1 lines was verified by sequence analysis. Random clones expressing gbb, gbbmS1, and gbbmNS were generated by the FLPout system (68, 69) in hsFLP/+; AyGal4 UAS-GFP/+; UAS-gbb*/+ with 10 min heat shock at 37°C.
DNA constructs
The Gateway system (Invitrogen) was used to clone most DNA constructs. gbb and gbb cleavage mutants (gbb-CMs) were generated by first amplifying the gbb coding sequence (cds) with the entire 5′ untranslated region (UTR) (70) by polymerase chain reaction (PCR) with primers containing the attB1 sequence at the 5′ and attB2 at the 3′ end (table S2). The PCR product was cloned into pDONR221 (Invitrogen) with the BP reaction. Cleavage site mutations were generated in pDONR221-gbb by Quik-Change (Stratagene). After confirming the DNA sequence of pDONR221 clones, the insertions were transferred into pAW [Drosophila Genomics Resource Center (DGRC), http://dgrc.cgb.indiana.edu/] by the LR reaction to generate pAW-gbb and the various pAW-gbbCMs for Drosophila cell culture. gbb and gbb-CM were also amplified from pDONR221 constructs by PCR with primers containing an Eco RI site at the 5′ end and an Xba I site at the 3′ end (table S2) and were cloned into pUAST attB to make pUAS-attB-gbb and pUAS-attB-gbb-CMs for generating transgenic lines with the φC31 system (37). HA-tagged gbb constructs were generated by the introduction of either a single HA tag between Thr351 and Arg352 of the Gbb, GbbmS1, and GbbmNSmS1 ligand domain by PCR. The amino acid sequence of the junction is EPMEST(351)-GG-1x HA-GG-R(352) SCQMQ. A 3xHA tag was obtained from pBRacpA dpp 3xHA (71) and introduced between Ser353 and Cys354 of the Gbb ligand domain by PCR, resulting in MESTRS(353)-GGG-3xHA-GGG-C(354)QMQTL. A C-terminally GFP-tagged gbb with attB1 and attB2 sites was generated by PCR with pUAST-gbb-GFP (72) as a template DNA. All PCR products were cloned into pAW (DGRC) vector with the Invitrogen Gateway system as described above.
We had previously shown that a 6.8-kb Sal I genomic fragment contains all the necessary information for gbb expression and function during development (36). To generate a series of gbb rescue constructs (gbbR), we generated a genomic subclone lacking only the gbb coding sequences. First, a Not I site was engineered in place of the gbb coding region by PCR amplification of a 3.8-kb upstream (A) and 2.0-kb downstream (B) genomic region with the following primers: 5′-GCGACGAGCTCCTCTAGAACGTCTGGCTGGGATGGC-3′ (sense primer for A with Sac I–Xba I sites), 5′-CAGGCGGCCGCTTTGGACGGATCGGTTGC-3′ (antisense primer for A with Not I site), 5′-CAGGCGGCCGCTTTGGACGGATCGGTTGC-3′ (sense primer for B with Not I site), and 5′-GGACCTGGGTACCATCTAGATCCAGAACAGCATC-3′ (antisense primer for B with Xba I–Kpn I sites). A and B fragments were cloned into Sac I/Not I and Not I/Kpn I sites of pBluescript II SK+, resulting in pBS gbb AB. The AB fragment was excised with Xba I and subcloned into pattB (http://flyc31.frontiers-in-genetics.org/index.php) with the multiple cloning Not I site destroyed. Finally, the wild-type gbb or various gbb cleavage mutant sequences generated in pDONR221-gbb were transferred into pattB-gbb-AB with Not I to generate gbbR-gbb or gbbR-gbb-CMs.
A Flag-tagged form of sax was generated with pAWF (DGRC; an S2 cell expression vector with C-terminal 3 × Flag). The sax-RA (FlyBase, http://flybase.org/) transcript without the stop codon was amplified by PCR and cloned into pAWF. The coding sequence of human Bmp4 (hbmp4) was cloned with a 5′ primer containing attB1–Hind III–Kozak sequences and a 3′ primer with Xho I–attB2 sequences (table S2) to generate pDONR221-hbmp4. Mutations in the hBMP4 furin cleavage consensus sequences and the R162Q mutation were generated in pDONR221-hbmp4. Each insert was transferred into pcDNA3 (Invitrogen) for mammalian cell culture with digested hbmp4 Hind III and Xho I.
Cell-based signaling assays
BMP signaling assays were performed in Drosophila S2 cells or in HEK293 cells as described previously (29, 31, 73, 74). In brief, the S2 signaling assay uses a lacZ reporter construct under the control of Notch activation through Su(H) binding sites and BMP signaling–mediated repression through the brkSE (silencer element). Direct binding of phosphorylated Mad-Med-Shn to the brkSE and a quantitative response to BMP signaling have been demonstrated previously (29, 31). S2 cells were transfected (Effectene, Qiagen) with 100 ng of gbb and gbb-CM constructs and incubated at 25°C for 60 to 72 hours before lysis (~3 × 106 cells) to measure β-galactosidase activity. β-Galactosidase activity was measured with the Dual-Light system (Applied Biosystems). Differences in signaling activity between wild-type gbb, gbb-CMs, and control were tested for significance with the Tukey-Kramer HSD (honestly significant difference) procedure and Hsu’s MCB (multiple comparisons with the best mean) test for multiple comparisons at P < 0.05.
In HEK293 cells, the Id1 promoter luciferase reporter (BRE-luciferase) and simian virus 40 (SV40)–Renilla luciferase were used to examine BMP signaling activity and transfection efficiency, respectively. To measure BMP signaling activities of each hBMP4-CM and hBMP4R162Q, we transiently transfected 293 cells with 1.1 μg of each hBMP4 construct and 250 ng of BRE-luciferase and SV40-Renilla luciferase. After 48 hours of incubation, cells were lysed and BMP signaling activities were measured with the Dual-Luciferase Reporter Assay System (Promega).
Sample preparation, coimmunoprecipitation, and Western analysis
S2 cells transfected with 2 μg of pAW gbb or pAW gbb-CM were grown for 4 days at 25°C. Either the entire cell culture was recovered and used as “total fraction” or it was centrifuged at 2000 rpm for 5 min at room temperature to separate the pellet (“cell fraction”) and supernatant fractions. The supernatant fraction was recentrifuged at 13,000 rpm for 5 min at room temperature to completely remove debris and used as “conditioned media fraction.” To assess BMP signaling by measuring phosphorylated Mad, we transfected S2 cells with pAc-Mad-Flag (a gift of M. O’Connor) and incubated with either 1× or 1/10× conditioned medium (prepared as described above) from pAc-, pAW-gbb–, and gbbmS1-transfected S2 cells for 3 hours at room temperature. After incubation, cells were collected by centrifugation at 2000 rpm for 5 min at room temperature and were lysed in SDS sample buffer. yw, gbb1/gbb2, and gbb1/gbb3 third instar larvae or dissected tissues were homogenized in SDS sample buffer.
Protein samples were analyzed by Western blot with ECL-Plus (GE Healthcare Life Sciences). Monoclonal antibody 3D6-24 against Gbb peptide 424-440 was generated at the UAB Epitope Recognition and Immunoreagent Core Facility. Anti-Gbb (1:1000), anti-actin (1:10,000; Millipore), anti-Flag (M2, 1:2000, Sigma), anti-pSmad3 [EP823Y, 1:2000, Epitomics (51)], and horseradish peroxidase–conjugated secondary antibodies (1:10,000; Jackson ImmunoResearch) were used for Western blot analyses.
S2 cells were transfected with 1 μg of pAW gbb-HA, pAW gbbmS1-HA, or pAW gbbmNSmS1-HA and 500 ng of pAWF-sax and grown for 4 days at 25°C. Cultures were solubilized in PBST (phosphate-buffered saline containing 1% Triton X-100, pH 7.0) and incubated with Dynabeads proteins G (Invitrogen)–conjugated mouse anti-Flag antibody (M2, Sigma) for 20 hours at 4°C with rotation. Proteins bound to Flag antibody were washed with TBST (50 mM tris-HCl, 150 mM NaCl, and 0.2% Triton X-100, pH 7.4) and eluted with SDS sample buffer. Then, protein samples were analyzed by Western blots with mouse anti-Flag (1:1000, M2, Sigma) and rat anti-HA antibodies (1:1000, 12CA5, Roche).
Immunohistochemistry
Immunostaining of wing discs was conducted as described previously (17) with anti-pSmad3 [EP823Y, 1:1,000, Epitomics (51)], anti-Sal [1:200 (75)] and Alexa Fluor secondary antibodies (1:500, Invitrogen). Images of wing discs were collected on either a Leica SP2 or a Zeiss 510 Meta confocal microscope as Z stacks of 1-μm sections. For each experiment, the same confocal settings were used during image capture. Except where noted, fluorescence intensity values were obtained from a fixed segment across the dorsal wing pouch (20% offset from the dorsal-ventral boundary) of six SUMed confocal slices with Plot Profile (Fiji). The average intensity profile was plotted from more than three discs with the SE.
Sequence and phylogenetic analysis
Potential proconvertase cleavage sites were predicted with ProP (76). Phylogeny.fr (http://www.phylogeny.fr/version2_cgi/index.cgi) was used to build a phylogenetic tree. BMP and TGF-β protein sequences were aligned with MUSCLE. The tree was calculated with the maximum likelihood method of PhyML with WAG substitution model.
Supplementary Material
Acknowledgments
We thank B. Hogan for hbmp4, R. Barrio for Sal antibody, Q. Chen and R. Freiman for assistance with mammalian cell culture, and the Bloomington Stock Center for Drosophila stocks. We thank C. Kelsey for technical assistance and appreciate the efforts of J. deLeon, K. Murray, and V. Le in generating transgenic lines. We thank C. Dunn for advice on the phylogenetic analysis, M. A. Accavitti-Loper for assistance with monoclonal generation (University of Alabama at Birmingham Epitope Recognition and Immunoreagent Core Facility, supported by NIH P30 AR48311), and members of the Wharton lab for discussions and helpful comments.
Funding: This work was supported by a grant from NIH GM068118 to K.A.W.
Footnotes
Author contributions: T.A. and K.A.W. designed the experiments and analyzed the data; T.A. performed most of the experiments; G.M. generated the Gbb antibody; and K.A.W. performed the phylogenic analysis, made the figures, and wrote the paper.
Competing interests: The authors declare that they have no competing interests.
www.sciencesignaling.org/cgi/content/full/5/218/ra28/DC1
Fig. S1. Signaling activity of the HA-tagged Gbb constructs.
Fig. S2. Gbb38 and Gbb15 bind to the Sax type I receptor.
Fig. S3. Signaling activity of the hBMP4 and hBMP4 cleavage mutants.
Fig. S4. Failure to cleave Gbb at the NS results in wing defects.
Fig. S5. The gbb double cleavage mutant exhibits dominant-negative behavior when expressed in dpp-expressing cells of the wing imaginal disc.
Fig. S6. Wild-type gbb (Gbb38 and Gbb15) and gbbmS1 (Gbb38) exhibit nonautonomous signaling.
Table S1. Macrochaetae and leg defects associated with misexpression of gbb and gbb cleavage mutants in dpp-expressing cells.
Table S2. Primers used in this study.
References
- 1.Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 3.Wu MY, Hill CS. TGF-β superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Affolter M, Basler K. The Decapentaplegic morphogen gradient: From pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 5.Bangi E, Wharton K. Dpp and Gbb exhibit different effective ranges in the establishment of the BMP activity gradient critical for Drosophila wing patterning. Dev Biol. 2006;295:178–193. doi: 10.1016/j.ydbio.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Dansereau DA, Lasko P. The development of germline stem cells in Drosophila. Methods Mol Biol. 2008;450:3–26. doi: 10.1007/978-1-60327-214-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: A critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Hackenmiller R, Berg L, Jean F, Nakayama T, Thomas G, Christian JL. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15:2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sopory S, Nelsen SM, Degnin C, Wong C, Christian JL. Regulation of bone morphogenetic protein-4 activity by sequence elements within the prodomain. J Biol Chem. 2006;281:34021–34031. doi: 10.1074/jbc.M605330200. [DOI] [PubMed] [Google Scholar]
- 11.Goldman DC, Hackenmiller R, Nakayama T, Sopory S, Wong C, Kulessa H, Christian JL. Mutation of an upstream cleavage site in the BMP4 prodomain leads to tissue-specific loss of activity. Development. 2006;133:1933–1942. doi: 10.1242/dev.02368. [DOI] [PubMed] [Google Scholar]
- 12.Künnapuu J, Björkgren I, Shimmi O. The Drosophila DPP signal is produced by cleavage of its proprotein at evolutionary diversified furin-recognition sites. Proc Natl Acad Sci USA. 2009;106:8501–8506. doi: 10.1073/pnas.0809885106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sopory S, Kwon S, Wehrli M, Christian JL. Regulation of Dpp activity by tissue-specific cleavage of an upstream site within the prodomain. Dev Biol. 2010;346:102–112. doi: 10.1016/j.ydbio.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Constam DB, Robertson EJ. Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J Cell Biol. 1999;144:139–149. doi: 10.1083/jcb.144.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian J, Andrée B, Jones CM, Sampath K. The pro-domain of the zebrafish Nodal-related protein Cyclops regulates its signaling activities. Development. 2008;135:2649–2658. doi: 10.1242/dev.019794. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Schier AF. The zebrafish Nodal signal Squint functions as a morphogen. Nature. 2001;411:607–610. doi: 10.1038/35079121. [DOI] [PubMed] [Google Scholar]
- 17.Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalsa O, Yoon JW, Torres-Schumann S, Wharton KA. TGF-β/BMP superfamily members, Gbb-60A and Dpp, cooperate to provide pattern information and establish cell identity in the Drosophila wing. Development. 1998;125:2723–2734. doi: 10.1242/dev.125.14.2723. [DOI] [PubMed] [Google Scholar]
- 19.Ray RP, Wharton KA. Context-dependent relationships between the BMPs gbb and dpp during development of the Drosophila wing imaginal disc. Development. 2001;128:3913–3925. doi: 10.1242/dev.128.20.3913. [DOI] [PubMed] [Google Scholar]
- 20.Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-β family in Drosophila. Development. 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- 21.Lander AD, Nie Q, Wan FY. Do morphogen gradients arise by diffusion? Dev Cell. 2002;2:785–796. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- 22.Kicheva A, Gonzalez-Gaitan M. The Decapentaplegic morphogen gradient: A precise definition. Curr Opin Cell Biol. 2008;20:137–143. doi: 10.1016/j.ceb.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Schwank G, Dalessi S, Yang SF, Yagi R, de Lachapelle AM, Affolter M, Bergmann S, Basler K. Formation of the long range Dpp morphogen gradient. PLoS Biol. 2011;9:e1001111. doi: 10.1371/journal.pbio.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlunegger MP, Grutter MG. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-β2. Nature. 1992;358:430–434. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 25.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-β2: An unusual fold for the superfamily. Science. 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 26.Imbeaud S, Carre-Eusebe D, Rey R, Belville C, Josso N, Picard JY. Molecular genetics of the persistent Müllerian duct syndrome: A study of 19 families. Hum Mol Genet. 1994;3:125–131. doi: 10.1093/hmg/3.1.125. [DOI] [PubMed] [Google Scholar]
- 27.Dixit H, Rao LK, Padmalatha VV, Kanakavalli M, Deenadayal M, Gupta N, Chakrabarty B, Singh L. Missense mutations in the BMP15 gene are associated with ovarian failure. Hum Genet. 2006;119:408–415. doi: 10.1007/s00439-006-0150-0. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, Natsume N, Shimozato K, Ohbayashi N, Suzuki Y, Niimi T, Minami K, Yamamoto M, Altannamar TJ, Erkhembaatar T, Furukawa H, Daack-Hirsch S, L’Heureux J, Brandon CA, Weinberg SM, Neiswanger K, Deleyiannis FW, de Salamanca JE, Vieira AR, Lidral AC, Martin JF, Murray JC. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 30.Pyrowolakis G, Hartmann B, Müller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Bangi E, Wharton K. Dual function of the Drosophila Alk1/Alk2 ortholog Saxophone shapes the BMP activity gradient in the wing imaginal disc. Development. 2006;133:3295–3303. doi: 10.1242/dev.02513. [DOI] [PubMed] [Google Scholar]
- 32.Degnin C, Jean F, Thomas G, Christian JL. Cleavages within the prodomain direct intracellular trafficking and degradation of mature bone morphogenetic protein-4. Mol Biol Cell. 2004;15:5012–5020. doi: 10.1091/mbc.E04-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haerry TE. The interaction between two TGF-β type I receptors plays important roles in ligand binding, SMAD activation, and gradient formation. Mech Dev. 2010;127:358–370. doi: 10.1016/j.mod.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly AC, Randall RA, Hill CS. Transforming growth factor β-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol. 2008;28:6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wharton KA, Cook JM, Torres-Schumann S, de Castro K, Borod E, Phillips DA. Genetic analysis of the bone morphogenetic protein-related gene, gbb, identifies multiple requirements during Drosophila development. Genetics. 1999;152:629–640. doi: 10.1093/genetics/152.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez AR, Cook J, Deininger PL, Derynck R. Dominant negative mutants of transforming growth factor-β1 inhibit the secretion of different transforming growth factor-β isoforms. Mol Cell Biol. 1992;12:1674–1679. doi: 10.1128/mcb.12.4.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittbrodt J, Rosa FM. Disruption of mesoderm and axis formation in fish by ectopic expression of activin variants: The role of maternal activin. Genes Dev. 1994;8:1448–1462. doi: 10.1101/gad.8.12.1448. [DOI] [PubMed] [Google Scholar]
- 40.Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- 41.Osada SI, Wright CV. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development. 1999;126:3229–3240. doi: 10.1242/dev.126.14.3229. [DOI] [PubMed] [Google Scholar]
- 42.Sun BI, Bush SM, Collins-Racie LA, LaVallie ER, DiBlasio-Smith EA, Wolfman NM, McCoy JM, Sive HL. derrière: A TGF-β family member required for posterior development in Xenopus. Development. 1999;126:1467–1482. doi: 10.1242/dev.126.7.1467. [DOI] [PubMed] [Google Scholar]
- 43.Joseph EM, Melton DA. Mutant Vg1 ligands disrupt endoderm and mesoderm formation in Xenopus embryos. Development. 1998;125:2677–2685. doi: 10.1242/dev.125.14.2677. [DOI] [PubMed] [Google Scholar]
- 44.Eimon PM, Harland RM. Effects of heterodimerization and proteolytic processing on Derrière and Nodal activity: Implications for mesoderm induction in Xenopus. Development. 2002;129:3089–3103. doi: 10.1242/dev.129.13.3089. [DOI] [PubMed] [Google Scholar]
- 45.Gelbart WM. The decapentaplegic gene: A TGF-β homologue controlling pattern formation in Drosophila. Development. 1989;107(Suppl):65–74. doi: 10.1242/dev.107.Supplement.65. [DOI] [PubMed] [Google Scholar]
- 46.Tomoyasu Y, Nakamura M, Ueno N. Role of dpp signalling in prepattern formation of the dorsocentral mechanosensory organ in Drosophila melanogaster. Development. 1998;125:4215–4224. doi: 10.1242/dev.125.21.4215. [DOI] [PubMed] [Google Scholar]
- 47.Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- 48.Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, Julicher F, Gonzalez-Gaitan M. Dynamics of Dpp signaling and proliferation control. Science. 2011;331:1154–1159. doi: 10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- 49.Hamaratoglu F, de Lachapelle AM, Pyrowolakis G, Bergmann S, Affolter M. Dpp signaling activity requires Pentagone to scale with tissue size in the growing Drosophila wing imaginal disc. PLoS Biol. 2011;9:e1001182. doi: 10.1371/journal.pbio.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- 51.Dejima K, Kanai MI, Akiyama T, Levings DC, Nakato H. Novel contact-dependent bone morphogenetic protein (BMP) signaling mediated by heparan sulfate proteoglycans. J Biol Chem. 2011;286:17103–17111. doi: 10.1074/jbc.M110.208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang MS, Liang LF, Wang S, Ratovitski T, Holmstrom J, Barker C, Stotish R. Characterization and identification of the inhibitory domain of GDF-8 propeptide. Biochem Biophys Res Commun. 2004;315:525–531. doi: 10.1016/j.bbrc.2004.01.085. [DOI] [PubMed] [Google Scholar]
- 54.Varga J, Pasche B. Antitransforming growth factor-β therapy in fibrosis: Recent progress and implications for systemic sclerosis. Curr Opin Rheumatol. 2008;20:720–728. doi: 10.1097/BOR.0b013e32830e48e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hauburger A, von Einem S, Schwaerzer GK, Buttstedt A, Zebisch M, Schraml M, Hortschansky P, Knaus P, Schwarz E. The pro-form of BMP-2 interferes with BMP-2 signalling by competing with BMP-2 for IA receptor binding. FEBS J. 2009;276:6386–6398. doi: 10.1111/j.1742-4658.2009.07361.x. [DOI] [PubMed] [Google Scholar]
- 56.Sengle G, Ono RN, Lyons KM, Bachinger HP, Sakai LY. A new model for growth factor activation: Type II receptors compete with the prodomain for BMP-7. J Mol Biol. 2008;381:1025–1039. doi: 10.1016/j.jmb.2008.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.di Clemente N, Jamin SP, Lugovskoy A, Carmillo P, Ehrenfels C, Picard JY, Whitty A, Josso N, Pepinsky RB, Cate RL. Processing of anti-Müllerian hormone regulates receptor activation by a mechanism distinct from TGF-β. Mol Endocrinol. 2010;24:2193–2206. doi: 10.1210/me.2010-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhfahl S, Hauburger A, Thieme T, Groppe J, Ihling C, Tomic S, Schutkowski M, Sinz A, Schwarz E. Identification of a core domain within the proregion of bone morphogenetic proteins that interacts with the dimeric, mature domain. Biochem Biophys Res Commun. 2011;408:300–305. doi: 10.1016/j.bbrc.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 59.Ogiso Y, Tsuneizumi K, Masuda N, Sato M, Tabata T. Robustness of the Dpp morphogen activity gradient depends on negative feedback regulation by the inhibitory Smad, Dad. Dev Growth Differ. 2011;53:668–678. doi: 10.1111/j.1440-169X.2011.01274.x. [DOI] [PubMed] [Google Scholar]
- 60.Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 61.Hogan BLM. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 62.Derynck R, Miyazono K, editors. The TGF-β Family. Vol. 50 Cold Spring Harbor Laboratory Press; New York: 2008. [Google Scholar]
- 63.Wu P, Jiang TX, Suksaweang S, Widelitz RB, Chuong CM. Molecular shaping of the beak. Science. 2004;305:1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 65.Albertson RC, Kocher TD. Genetic and developmental basis of cichlid trophic diversity. Heredity. 2006;97:211–221. doi: 10.1038/sj.hdy.6800864. [DOI] [PubMed] [Google Scholar]
- 66.Terai Y, Morikawa N, Okada N. The evolution of the pro-domain of bone morphogenetic protein 4 (Bmp4) in an explosively speciated lineage of East African cichlid fishes. Mol Biol Evol. 2002;19:1628–1632. doi: 10.1093/oxfordjournals.molbev.a004225. [DOI] [PubMed] [Google Scholar]
- 67.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H Flybase Consortium. FlyBase: Enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 69.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 70.Wharton KA, Thomsen GH, Gelbart WM. Drosophila 60A gene, another transforming growth factor β family member, is closely related to human bone morphogenetic proteins. Proc Natl Acad Sci USA. 1991;88:9214–9218. doi: 10.1073/pnas.88.20.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O’Connor MB, Marsh JL. Twisted gastrulation is a conserved extracellular BMP antagonist. Nature. 2001;410:479–483. doi: 10.1038/35068578. [DOI] [PubMed] [Google Scholar]
- 72.Nahm M, Long AA, Paik SK, Kim S, Bae YC, Broadie K, Lee S. The Cdc42-selective GAP rich regulates postsynaptic development and retrograde BMP transsynaptic signaling. J Cell Biol. 2010;191:661–675. doi: 10.1083/jcb.201007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 74.Zilberberg L, ten Dijke P, Sakai LY, Rifkin DB. A rapid and sensitive bioassay to measure bone morphogenetic protein activity. BMC Cell Biol. 2007;8:41. doi: 10.1186/1471-2121-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barrio R, de Celis JF. Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc Natl Acad Sci USA. 2004;101:6021–6026. doi: 10.1073/pnas.0401590101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.