Abstract
A major promise of genomic research is information that can transform health care and public health through earlier diagnosis, more effective prevention and treatment of disease, and avoidance of drug side effects. Although there is interest in the early adoption of emerging genomic applications in cancer prevention and treatment, there are substantial evidence gaps that are further compounded by the difficulties of designing adequately powered studies to generate this evidence, thus limiting the uptake of these tools into clinical practice. Comparative effectiveness research (CER) is intended to generate evidence on the “real-world” effectiveness compared with existing standards of care so informed decisions can be made to improve health care. Capitalizing on funding opportunities from the American Recovery and Reinvestment Act of 2009, the National Cancer Institute funded seven research teams to conduct CER in genomic and precision medicine and sponsored a workshop on CER on May 30, 2012, in Bethesda, Maryland. This report highlights research findings from those research teams, challenges to conducting CER, the barriers to implementation in clinical practice, and research priorities and opportunities in CER in genomic and precision medicine. Workshop participants strongly emphasized the need for conducting CER for promising molecularly targeted therapies, developing and supporting an integrated clinical network for open-access resources, supporting bioinformatics and computer science research, providing training and education programs in CER, and conducting research in economic and decision modeling.
Advances in genomic medicine are ushering in a new era of individualized cancer care and prevention. There are currently more than 1000 genomic tests available in clinical practice for an estimated 2500 conditions (1,2) with approximately 30% specific to oncology (3). Many genome-based tests are marketed to physicians, with some being marketed directly to consumers. However, with the accelerated pace of discoveries and the relatively unregulated availability of genome-based tools and markers for cancer care and prevention (4), there has been little research to determine the clinical utility of these applications. Health insurers have struggled to determine which emerging genomic applications merit coverage. To date, most of those applications have been deemed as having insufficient evidence of clinical utility by technology assessment groups (2,5,6). Because genomic information is accumulating rapidly, efficient empirical research and advanced, timely methods of systematic review, decision modeling, and practice-based research are required to assess clinical utility of genomics and personalized medicine applications (7,8).
Comparative effectiveness research (CER) is intended to generate evidence on the “real-world” effectiveness compared with existing standards of care so informed decisions can be made to improve health care (9–11). Because CER is by definition comparative, it can address questions on both the clinical utility of genomic tests and the added clinical value of these tests compared with standard care. Developing better evidence on which clinical interventions work best in subgroups of patients is essential in reducing health-care costs and achieving a more sustainable health-care delivery system (12).
Capitalizing on funding opportunities from the American Recovery and Reinvestment Act (ARRA) of 2009, the National Cancer Institute (NCI) supported a 2-year initiative to advance methods for analysis, synthesis, modeling, and evaluation of the clinical validity and utility of existing and emerging genomic medicine applications in cancer control and prevention. The NCI funded seven research teams to perform projects (Table 1) on CER in genomic and precision medicine to begin filling in this gap in knowledge, develop novel methodological approaches, and address necessary data infrastructure needs. In addition, investigators from these research teams formed four working groups (Table 2) to cover CER methodology, stakeholder engagement, evidence synthesis and horizon scanning, and infrastructure, with NCI sponsoring three in-person workshops.
Table 1.
Goals of each of the comparative effectiveness research (CER) in genomic and precision medicine projects by lead institution
| Project title | Lead institution | Project goal(s) |
|---|---|---|
| Building a Genome-Enabled Electronic Medical Record | University of Virginia | Enable efficient and accurate collection and integration into electronic medical records of personal, family, and genomic information for risk assessment and delivery of decision support to providers and patients |
| Comparative Effectiveness in Genomic and Personalized Medicine for Colon Cancer (CERGEN) | Kaiser Permanente | Conduct CER on the genomics of colorectal cancer (CRC); including evaluating the cost-effectiveness for genetic testing of Lynch Syndrome and KRAS testing in CRC treatment management |
| Programs in Clinical Effectiveness of Cancer Pharmacogenomics | Duke University | Develop biospecimen and data registries to support evidence generation and clinical effectiveness research for evaluating pharmacogenomic markers in lung and breast cancer |
| Developing Information Infrastructure Focused on Cancer Comparative Effectiveness | H. Lee Moffit Cancer Center | Develop an information infrastructure for CER, including CER metadata standards, and a comprehensive CER data dictionary to support the use of the emerging infrastructure |
| Comparative Effectiveness in Genomic Medicine (CEGeM) | University of Pennsylvania | Conduct four studies: pharmacogenomics of nicotine addiction; using single nucleotide polymorphism panels in breast cancer risk screening and prevention; personalized treatment for non–small cell lung cancer; and CDKN2A/p16 testing and adherence to melanoma prevention |
| Center for Comparative Effectiveness Research in Cancer Genomics (CANCERGEN) | Fred Hutchinson Cancer Center | Forge collaborations with external stakeholders to incorporate decision modeling, database linkage, ethics, policy, and clinical trial design to leverage the Soutwest Oncology Group clinical trials network |
| Clinical Validity and Utility of Genomic Targeted Chemoprevention of Prostate Cancer | Wake Forest University | Evaluate the clinical validity and utility of genomic targeted chemoprevention for prostate cancer |
Table 2.
Activities and Goals of comparative effectiveness research (CER) in genomic and precision medicine (GPM) working groups
| Working group | Objectives |
|---|---|
| Methodology | Define CER in GPM Catalog and share existing CER methods in collaboration with general CER groups, such as the Clinical & Translational Science Awards Consortium and Agency for Healthcare Research and Quality Explore development and application of new methodologies relevant to GPM |
| Stakeholder engagement | Catalog and share existing approaches to stakeholder engagement in GPM from other groups Meet with high-level stakeholders |
| Evidence synthesis and horizon scanning | Catalog and share existing methods for evidence synthesis Develop and apply new methods during next 2 years and perform evidence synthesis on topics under study Develop a framework for knowledge development in CER and GPM |
| Infrastructure | Catalog and share existing tools and resources Develop new resources |
The last workshop was held on May 30, 2012, in Bethesda, Maryland, and brought ARRA-funded investigators together to highlight their findings. Meeting participants discussed the scientific challenges they encountered and identified promising areas in CER genomic medicine. This report provides an overview of recent research efforts from the seven research teams, challenges to conducting CER, barriers to clinical practice implementation, and research priorities and opportunities in CER in genomic and precision medicine identified by the investigators during the workshop.
Knowledge Generation and Synthesis: Highlights From NCI-Sponsored CER Studies
Lyman proposed a framework of the CER process (7). He suggested that it is “a process for systematically identifying and synthesizing the totality of available evidence on the comparative effectiveness safety and overall value of competing strategies.” That includes incorporating the results from randomized controlled trials, meta-analyses, observational studies, and decision modeling to facilitate the development of clinical guidelines. This concept was expanded by Ginsburg and Kuderer (13), who suggested that to further CER—specifically in integrating genomic approaches to oncology—several strategies are needed: 1) “developing and applying timely systematic reviews and analytic tools”; 2) “establishing and utilizing disease-focused multidisciplinary research teams of translational clinical investigators, genomic experts, biostatisticians, and health outcome research methodologists to conduct and evaluate the data analysis and systematic reviews”; 3) “integrating the evidence synthesis with the evaluation of emerging data from the longitudinal registries, clinical trials, or pragmatic trials to guide the selection of genomic biomarkers for optimally designed Phase III confirmatory CER”; 4) “developing and evaluating clinical simulation models”; and 5) “providing clinical and policy recommendations through the formulation of evidence-based clinical practice guidelines.”
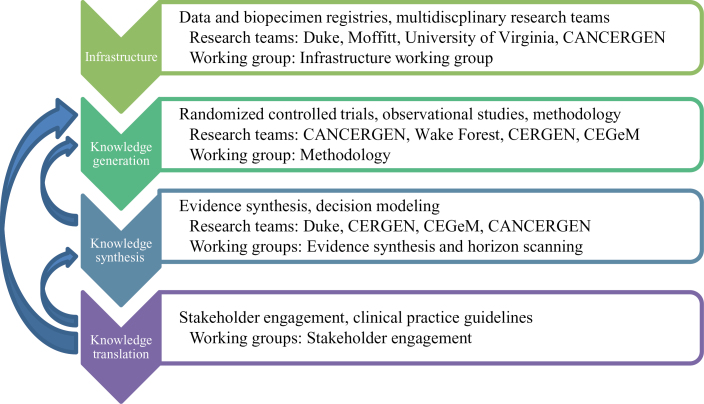
Figure 1 summarizes these strategies in four categories—infrastructure, knowledge generation, knowledge synthesis, and knowledge translation—and lists various methods and approaches associated with each. The ARRA-funded research institutions heavily invested in those categories are also provided in Figure 1. Highlights of the groups’ accomplishments presented at the workshop are described below.
Figure 1.
Methods and approaches used in comparative effectiveness research of genomics and personalized medicine in American Recovery and Reinvestment Act (ARRA)–funded research institutions. The framework depicts CER strategies grouped into four categories—infrastructure, knowledge generation, knowledge synthesis, and knowledge translation; ARRA-funded research institutions heavily invested in those categories are also provided. CANCERGEN = Center for Comparative Effectiveness Research in Cancer Genomics; CEGeM = Comparative Effectiveness in Genomic Medicine; CERGEN = Comparative Effectiveness in Genomic and Personalized Medicine for Colon Cancer.
Infrastructure
The research team at H. Lee Moffitt Cancer Center developed an information infrastructure for CER, within Total Cancer Care, which is a personalized cancer care initiative started in 2006 to collect tumor specimens and clinical data throughout a patient’s lifetime (14). Total Cancer Care, which also engages the patient to give them active feedback about their health and upcoming appointments, expanded electronic health record, biospecimen, cancer registry, epidemiologic, and genomics data management resources, integrated automated data extraction methods, and created interfaces to data for researchers and clinicians. This effort, guided by several CER studies on myelodysoplastic syndrome (15–17) led to the development of data quality standards, a comprehensive data dictionary, and an integrated metadata strategy to support CER and the development of new methodologies to integrate data from multiple sources (eg, electronic medical records, biospecimen databases, molecular data). The research team has, to date, received consent from more than 91 000 patients from 18 sites and has collected more than 33 500 tumors specimens, generating more than 17 000 high-quality gene expression profiles and 4500 massively parallel sequences (whole genome, whole exome, and targeted exome). They are also currently conducting clinical trial matching using patient stratification based on clinical and molecular data and enlisting a new statistical method (18) that will guide further enhancements to reduce time and cost for performing clinical trials based on the Total Cancer Care consortium.
The research team at the University of Virginia developed Health Heritage (19), which is a Web-based family health history tool, to facilitate the electronic collection of personal, family, and genomic data from the electronic medical record. Health Heritage also gives patients the ability to share electronically health information that has been sourced, in part, from their electronic medical record. This effort enables the efficient collection of more complete and accurate family health histories, develops a complete patient record for CER, and provides a mechanism to translate important research findings in cancer CER back to patients and providers through decision support logic within Health Heritage.
The Duke University team developed a comprehensive registry of biological specimens linked to an information database that integrates molecular data with patient-reported outcomes, clinical outcomes, and economic data for lung and breast cancer patients to support evidence generation and CER for evaluating cancer pharmacogenomics markers. Information incorporated into the registry included, but was not limited to, patient demographics, drug and treatment information, radiology and surgical procedures, outpatient medications, pathology, comorbidities, and outcome measures (eg, symptoms, quality-of-life, survival). As a first step, this information was used to evaluate the ability of novel gene expression biomarkers to direct treatments and predict toxicities and recurrence or other outcomes related to lung and breast cancer (20–22). Additional ongoing studies using the registry include developing second-generation genomic biomarkers in breast cancer, identifying genomic biomarkers for chemotherapy sensitivity in lung cancer using microarray gene expression analysis, and comparing these markers to usual care.
The consortium led by Fred Hutchinson Cancer Research Center created the Center for Comparative Effectiveness Research in Cancer Genomics (CANCERGEN), which consists of Fred Hutchinson Cancer Research Center, the University of Washington, the Center for Medical Technology Policy, and the Southwest Oncology Group (SWOG). CANCERGEN partners with an external stakeholder advisory group composed of health insurers; clinicians with expertise in genomics, oncology, and primary care; patient advocates; researchers; genomic test developers; and pharmaceutical representatives. The overall objective of CANCERGEN was to use a multistakeholder process to prioritize genomic tests for prospective clinical trials within the SWOG network. As part of this process, investigators conducted a landscape analysis of cancer genomic tests followed by a prioritization process to identify the highest priority tests. From these efforts, CANCERGEN and SWOG investigators have collaborated on two study concepts and anticipate incorporating comparative effectiveness endpoints (eg, quality of life related to testing, costs) into other clinical trials implemented through SWOG.
Knowledge Generation
As described in Figure 1, there are different CER methods and approaches to generate evidence; the advantages and limitations of each approach have been discussed extensively (9,23,24). A brief description of the research and any preliminary data highlighted at the workshop are presented below, organized by approach.
Randomized Controlled Trial.
One of the aims of CANCERGEN is to conduct a CER study within a pragmatic clinical trial conducted by SWOG. The RxPONDER (Rx for Positive Node, Endocrine Responsive Breast Cancer) Trial (SEOG S1007) opened in January 2011, and researchers plan to enroll 4000 early-stage, hormone receptor–positive, HER2-negative breast cancer patients who have been found to have involvement of one to three lymph nodes with Oncotype DX recurrence scores of 25 or less (4). Patients are randomized to two treatment arms: chemotherapy plus endocrine therapy or endocrine therapy only. A goal of the trial is to assess whether patients with node-positive breast cancer who have low to intermediate recurrence scores benefit from adjuvant chemotherapy. The trial will also investigate whether there is an optimal cutpoint where chemotherapy should be recommended to this subset of breast cancer patients.
The research team at Wake Forest investigated whether using a genomic-based targeting approach has the potential to improve prostate cancer outcomes compared with a nontargeted approach. The investigators used an existing randomized clinical trial—the REDUCE (Reduction by Dutasteride of Prostate Cancer Events) Trial— to first demonstrate that single nucleotide polymorphisms identified from a genome-wide association study can be used to identify individuals who are at high risk for prostate cancer. The team also conducted a new trial in which men at risk for prostate cancer were identified from primary care clinics to assess the effectiveness of a genomic-targeted approach. The new trial randomized 700 men at risk for prostate cancer to one of four groups to determine the impact of genetic information on their perceived risk for prostate cancer and their intended and actual uptake of health-care recommendations made by their physicians. In addition, primary care physicians and urologists will be randomized to one of the four groups and surveyed to determine whether genetic information impacted their decision to recommend specific actions, such as screening, to their patients.
Observational Studies.
The research team at the University of Pennsylvania conducted a prospective cohort study of 613 patients with non–small cell lung cancer (NSCLC) to compare the effects of treatment guided by epidermal growth factor receptor (EGFR) and KRAS tumor mutation testing to standard therapy (platinating agents). Outcomes of interest include measuring feasibility (eg, tissue available for biopsy), degree to which use of mutation testing results alters therapeutic approach, patient acceptance, clinical outcomes (eg, response rate, progression-free survival, overall survival), and cost. To date, 497 enrolled patients have EGFR mutation results available. Preliminary results suggest improved survival among patients with an EGFR mutation who were treated with erlotinib.
Knowledge Synthesis
Evidence Synthesis.
The purpose of a systematic review or evidence synthesis is to evaluate, synthesize, and summarize the results of the best available research related to a given topic, and various groups (eg, Evaluation of Genomic Applications in Practice and Prevention [EGAPP], Institute of Medicine, Blue Cross Blue Shield Technology Evaluation Center) have developed processes for assessing evidence (5,6). Two sites conducted systematic reviews related to pharmacogenomics and clinical response. The Duke University research team conducted a systematic review of genomic studies predicting response to chemotherapy in breast cancer patients (25,26), whereas the Comparative Effectiveness Research in Genomics of Colorectal Cancer (CERGEN) team completed a systematic review of pharmacogenetic testing for predicting clinical benefit to anti-EGFR therapy in metastatic colorectal cancer (27). Both teams found that most studies were not of high quality. Some of the specific scientific challenges faced included limited data quality, large variation in genomic methodology, little or no demonstration of clinical utility for most genomic tests, infrequent independent validation, and difficulty in adjusting methods to match the rapid pace of developments in genomics.
Decision Modeling.
A major promise of genomic research is information that can transform health care through earlier diagnosis, more effective prevention and treatment of disease, and avoidance of drug side effects. Although there is interest in the early adoption of emerging genomic applications in cancer prevention and treatment, there are substantial evidence gaps that are further compounded by the difficulties of designing adequately powered studies to generate this evidence, thus limiting the uptake of these tools into clinical practice. By synthesizing evidence in a rigorous way, modeling can be an additional timely method to inform decision-makers about the clinical and economic value of these tests. As such, there is increasing interest in developing novel CER strategies to help funding groups maximize the value of their research portfolio when choosing among competing research projects.
Value-of-research (VOR) analysis is a quantitative method that can be used to determine whether more research is justified in regards to a medical intervention or technology (28). It applies economic methods and decision analysis to estimate the health impact and economic value of performing additional research. CANCERGEN conducted a VOR analysis to prioritize emerging cancer genomics technologies for further evaluation through the SWOG clinical trials network. They evaluated three different genomic tests that were identified to be top research priorities by a stakeholder group (29)—namely, EGFR testing for erlotinib maintenance therapy after first-line chemotherapy in advanced NSCLC patients, excision repair cross-complementing 1 (ERCC1) gene expression testing for platinum-based adjuvant therapy in resected early-stage NSCLC patients, and breast cancer tumor markers for detection of recurrence in breast cancer patients after primary therapy—to determine which test provides the highest net benefit. The results from the VOR analyses indicated that there would be substantial value from additional research on ERCC1 testing in early-stage NSCLC patients and breast cancer tumor marker testing in breast cancer patients, whereas there would be considerably less value for EGFR testing in advanced NSCLC patients.
The CERGEN team conducted two comparative effectiveness studies: one on Lynch syndrome screening and the second on KRAS and BRAF testing. For Lynch syndrome, microsimulation was used to estimate the impact of alternative screening strategies recommended by the EGAPP Working Group. Based on that analysis, microsatellite instability testing was the preferred option compared with immunohistochemistry staining, BRAF, and germline testing. Similarly, microsimulation was used to estimate the impact of KRAS and BRAF testing. The investigators found that screening for KRAS and BRAF mutation improves the cost-effectiveness of anti-EGFR therapy; however, the incremental cost-effectiveness ratio was above the generally accepted threshold of $100 000/quality-adjusted life-year (30).
Knowledge Translation
Stakeholder Engagement.
The importance of stakeholder engagement in CER is widely recognized because CER’s purpose is to produce useful information for decision making (9,24). The Stakeholder Engagement Working Group developed a meeting agenda and selected clinical case studies for an NCI-sponsored workshop held on January 11, 2011, in Bethesda, Maryland. A diverse group of stakeholders—patient advocates, payers, health-care providers, industry, regulators, and researchers—met to discuss how evidence should be interpreted and what level of evidence is needed before a genomic test is adopted into clinical practice. The 22 stakeholders were asked to complete a survey before and during the meeting to decide whether they would recommend for or against clinical use of each test. Deverka et al. (31) found that, although there is a willingness by stakeholders to accept indirect evidence of clinical utility, they still rely on evidence reviews and recommendations from clinical guidelines committees to assess the appropriateness of genomic tests for clinical implementation.
Challenges and Barriers to Clinical Implementation
The workshop attendees noted a number of barriers to incorporating genomic tests into clinical practice beyond the evidence gaps already noted. One barrier is the divergence between the evidence requirements set by regulators for regulatory approval and those set by payers for reimbursement. The current regulatory structure focuses on analytical accuracy rather than clinical validity or utility and creates the potential that genomic tests will be introduced into clinical practice without sufficient evidence supporting their value compared with standard care or that tests that provide high clinical utility and value will not be used or reimbursed in practice. These regulatory uncertainties and relative lack of regulatory requirements make it challenging for test developers to design studies to meet the needs of both stakeholders. Other groups, such as the Institute of Medicine’s Roundtable on Translating Genomic-Based Research for Health, have hosted workshops to examine potential solutions to this issue (32). In addition, patients need to properly consent and be assured that appropriate safeguards are in place to protect personal health information when providing medical records data linked to genomic information.
Workshop participants also repeatedly raised the challenges of creating an infrastructure to support CER in genomic medicine. Not only are there technical challenges in linking clinical and genomic data stored in different databases—standardization of data, interoperability, electronic compatibility, harmonization across patient identifiers to link across multiple data sources into an analyzable record—but there are also proprietary and data security concerns that serve as additional barriers in creating such a data network. Furthermore, data critical to conducting CER are often not captured (eg, lack of outcomes data, difficulty in getting physicians to enter data, the lack of current procedural terminology codes for most genomic tests) by existing databases or are incomplete, potentially limiting the quality of the studies that can be conducted. Researchers also have little control over how data are collected and stored in electronic medical records, which adds to the problem. These problems are also compounded by the differing expectations of clinical researchers, informatics team, and data analysts.
There are also many challenges to generating useful evidence for decision making in cancer genomic medicine using CER (23); these include: the rapid pace of innovation, lack of regulation, and variable definitions and evidence thresholds for clinical and personal utility. In addition to the technical challenges noted earlier, other hurdles to conducting well-designed CER studies include high costs, large sample size needs, and data on cancer-related endpoints (eg, cancer recurrence, survival). Innovative approaches that are rapid, timely, efficient, and responsive are needed so relevant information can be generated before the evidence becomes outdated.
To facilitate this area of research and ultimately the translation of innovative genomic tests into clinical practice, the current workforce and any future demand for education and training will need to be evaluated. The promise of genomic and precision medicine has created new and greater demands for both those providing the scientific expertise—clinician-scientists, bioinformatics specialists, statisticians, health economists, molecular biologists—needed to conduct CER and those delivering clinical care (eg, physicians, genetic counselors) to the patients. The exacting needs of research and, to some extent, clinical practice where health-care professionals will need to be able to interpret more complex genomic data will require a degree of education and training that is currently not being met.
Priority Areas and Future Direction
Each of the ARRA-funded research teams was asked to identify research opportunities and priority areas in CER, which were then compiled and synthesized into nine priority areas. Each research team was asked to weight the priorities to further refine the list. The priorities identified are not specific to any of the institutions represented at the meeting but rather serve to summarize the collective deliberations of the group and their opinions of what is needed to advance CER in genomic medicine. Here we summarize the top five priority areas identified during the priority-setting session, which have been categorized into knowledge generation, infrastructure, and knowledge synthesis-based recommendations.
Knowledge Generation
CER for Targeted Therapies.
Workshop participants strongly emphasized the critical need for CER of promising targeted therapies to generate data that can inform clinical practice. These include clinical utility studies that compare the effectiveness of genomic testing to usual care and other genomic applications (eg, Oncotype DX vs Mammaprint) in preventing and treating cancer for which promising genomic applications exist, validating promising biomarkers in heterogeneous populations, supporting studies of genomic markers for toxicity to chemotherapeutic agents, and incorporating clinical trial matching of patients to therapies based on their genomic and molecular profiles.
Infrastructure
Integrated Population-Based Clinical Network for Open-Access Resources.
To fully realize the promise of CER in cancer genomic medicine, workshop participants emphasized the need for the development and support of an integrated network to provide open-access to key resources. The ultimate goal of such a network would be to provide the necessary data and biospecimens to allow researchers and clinicians to conduct “real-world” studies so evidence-based clinical guidelines that provide specific decision algorithms for the selection of effective treatments or combinations of treatments based on the molecular and clinical characteristics of each patient’s cancer disease can be developed. This network should be representative of the underlying population and include detailed prospective data on treatments, outcomes, diagnosis and staging, risk factors, and comorbidities collected on hundreds of thousands of cancer patients who agree to release their medical data and provide their germline DNA and tissue samples for a number of research purposes. Such a population resource would allow researchers to not only conduct CER studies of new genomic technologies but also molecularly characterize the patients’ cancers, understand their responses to different cancer therapies based on clinical and genomic information, identify molecular targets that can be used for the development of new treatments, and match patients to specific molecularly based clinical trials. To create this network would require leveraging partnerships with patients, clinicians, industry, academia, and public health. Two proprietary models that are ongoing include those established by the Moffitt Cancer Center, whose Total Cancer Care program is partnering with patients, community, clinicians, industry, academy, and 17 hospitals (14), and the Rapid-Learning Personalized Health Care Model being put in place at Duke University and other institutes (33). The need to develop an adequate infrastructure to support CER has also been articulated by Tunis et al. (24) so good quality evidence can be generated on relevant genomic applications before they become outdated.
Methods Research.
Because of the overwhelming volume of genomic data being generated, solutions are needed to better collate, store, access, analyze, and exchange research and clinical data. Workshop participants noted that there is a great need to support bioinformatics/computer science research so patient and biospecimen data can be integrated to advance CER and cancer genomic medicine. This need was also highlighted at a recent Institute of Medicine workshop that focused specifically on informatics needs and challenges in cancer research (34).
Training.
Another priority that was repeatedly mentioned by workshop participants was the lack of opportunities for scientists to be trained in CER methodology. Training programs and K (career) awards for CER, such as those awarded the Agency for Healthcare Research and Quality (35), were suggested resource investments.
Knowledge Synthesis
Economic/Decision Modeling Research.
Workshop participants also emphasized the need for methods research in economic and decision modeling. For example, decision-analytic frameworks are needed to assist stakeholders in making informed decisions and weighing the risks and benefits of various genomic biomarkers and genomic sequence data for use in clinical practice. Methods to develop such frameworks may accelerate utilization and practice-based evidence development of genomic tests that pose low risk and offer plausible clinical benefit while discouraging premature use of tests that provide little benefit or pose considerable health risks compared with usual care. Examples of this type of work are demonstrated by a risk–benefit framework focused on pharmacogenomic tests developed by Veenstra et al. (36) and the categorization of genetic variants identified in the course of whole genome sequencing into predetermined clinically relevant “bins,” defined by the strength of the evidence for the use of these variants in clinical practice, developed by Berg et al. (37).
Another CER approach where more methods work would be beneficial is cost-effectiveness analysis (38). This approach formally assesses the incremental value of technologies and can incorporate different outcomes—such as clinical outcomes or health-care costs—although, as noted by Goddard et al. (23), the value placed on genetic information is difficult to measure and incorporate into policy decisions.
Additional methods research is needed in VOR modeling for emerging genomic applications in cancer prevention and treatment. In a world of limited research funds, this approach can be useful to decision makers who must try to maximize the value of their research portfolio when choosing among competing research projects.
Summary
Despite the advances being made in precision medicine, the translation of genomic discoveries into clinical practice has been hindered by insufficient evidence of clinical utility of promising applications. Furthermore, because of the rapid pace of innovation, more advanced and timely methods of systematic review, decision modeling, and practice-based research are needed to fill in this evidence gap.
Particularly in light of the concerns over the growing costs of health care, which are currently estimated to be greater than 17% of growth domestic product in the United States (39), answering questions on the clinical utility of genomic tests and the added clinical value of those tests will make CER a practical necessity.
Funding
The National Cancer Institute sponsored the 2012 workshop.
The authors report no conflicts of interest. The study sponsor(s) had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
References
- 1. Gwinn M, Grossniklaus DA, Yu W, et al. Horizon scanning for new genomic tests. Genet Med. 2011;13(2):161–165 [DOI] [PubMed] [Google Scholar]
- 2. United Healthcare Personalized medicine: trends and prospects for the new science of genetic testing and molecular diagnostics. United Healthcare Working Paper. 2012.
- 3. Centers for Disease Control and Prevention CancerGEM KB (Cancer Genomic Evidence-based Medicine Knowledge Base): An Integrated, Searchable Knowledge Base of Human Genome Epidemiology and Genomic Applications in Cancer Care and Prevention. Atlanta: CDC; 2012. [Google Scholar]
- 4. Ramsey SD, Veenstra D, Tunis SR, Garrison L, Crowley JJ, Baker LH. How comparative effectiveness research can help advance “personalized medicine” in cancer treatment. Health Aff. 2011;30(12):2259–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blue Cross and Blue Shield Blue Cross and Blue Shield Technology Evaluation Center Criteria. 2012. Available at: http://www.bcbs.com/blueresources/tec/tec-criteria.html Accessed November 26, 2012. [Google Scholar]
- 6. Evaluation of Genomic Applications in Practice and Prevention Evaluation of Genomic Applications in Practice and Prevention. 2012. Available at: http://www.egappreviews.org/recommendations/index.htm Accessed November 26, 2012. [Google Scholar]
- 7. Lyman GH. Comparative effectiveness research in oncology: the need for clarity, transparency and vision. Cancer Invest. 2009;27(6):593–597 [DOI] [PubMed] [Google Scholar]
- 8. Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(suppl):1–32 [DOI] [PubMed] [Google Scholar]
- 9. Insitute of Medicine Initial National Priorities for Comparative Effectiveness Research. Washington, DC: National Academies Press; 2009. [Google Scholar]
- 10. Rich EC. The policy debate over public investment in comparative effectiveness research. J Gen Intern Med. 2009;24(6):752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Congressional Budget Office Research on the Comparative Effectiveness of Medical Treatments. 2007. Available at: http://www.cbo.gov/ftpdocs/88xx/doc8891-12-18-ComparativeEffectiveness.pdf Accessed January 7, 2013. [Google Scholar]
- 12. Orszag P. Health costs are the real deficit threat. Wall Street Journal. May 15, 2009. [Google Scholar]
- 13. Ginsburg GS, Kuderer NM. Comparative effectiveness research in oncology: the need for clarity, transparency and vision. J Clin Oncol. 2012;30(34):4233–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. 2011;17(6):528–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cogle CR, Craig BM, Rollison DE, List AF. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117(26):7121–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Craig BM, Rollison DE, List AF, Cogle CR. Diagnostic testing, treatment, cost of care, and survival among registered and non-registered patients with myelodysplastic syndromes. Leuk Res. 2011;35(11):1453–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig BM, Rollison DE, List AF, Cogle CR. Underreporting of myeloid malignancies by United States cancer registries. Cancer Epidemiol Biomarkers Prev. 2012;21(3):474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han G, Schell MJ, Kim J. Comparing two exponential distributions using the exact likelihood ratio test. Stat Biopharm Res. 2012;4(4):348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cohn WF, Ropka ME, Pelletier SL, et al. Health Heritage: a web-based tool for the collection and assessment of family health history: initial user experience and analytic validity. Public Health Genomics. 2010;13(7–8):477–491 [DOI] [PubMed] [Google Scholar]
- 20. Kuderer NM, Culakova E, Huang M. et al. , eds. Quality appraisal of clinical validation studies for multigene prediction assays of chemotherapy response in early-stage breast cancer. Paper presented at the annual meeting of theAmerican Society of Clinical Oncology; June 2011; Chicago, IL
- 21. Culakova E, Poniewierski MS, Huang M. et al. , eds. Assessment of genomic prognostic signatures as predictors of response to neoadjuvant chemotherapy in patients with early stage breast cancer. Paper presented at the American Association for Cancer Research–San Antonio Breast Cancer Symposium; November 2011; San Antonio, TX.
- 22. Kuderer NM. Prognostic markers in estrogen receptor–positive breast cancer: a cross-platform comparison of genomic signatures and identification of novel prognostic genes and pathways. Paper presented at the American Association for Cancer Research–San Antonio Breast Cancer Symposium; November 2012; San Antonio, TX.
- 23. Goddard KA, Knaus WA, Whitlock E, et al. Building the evidence base for decision making in cancer genomic medicine using comparative effectiveness research. Genet Med. 2012;14(7):633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tunis SR, Benner J, McClellan M. Comparative effectiveness research: plicy context, methods development and research infrastructure. Stat Med. 2010;29(19):1963–1976 [DOI] [PubMed] [Google Scholar]
- 25. Lyman GH, Culakova E, Poniewierski MS. et al. , eds. Multigene signature assays in patients with early-stage breast cancer receiving neoadjuvant chemotherapy: a systematic review and evidence summary of predictive performance. Paper presented at the American Association for Cancer Research–San Antonio Breast Cancer Symposium; November 2011; San Antonio, TX.
- 26. Lyman GH. Ki67 as a predictive markers of response to neoadjuvant chemotherapy in patients with early-stage breast cancer: a systematic review and evidence summary. Paper presented at the American Assocation for Cancer Research–San Antonio Breast Cancer Symposium; November 2012; San Antonio, TX.
- 27. Lin JS, Webber EM, Senger CA, Holmes RS, Whitlock EP. Systematic review of pharmacogenetic testing for predicting clinical benefit to anti-EGFR therapy in metastatic colorectal cancer. Am J Cancer Res. 2011;1(5):650–662 [PMC free article] [PubMed] [Google Scholar]
- 28. Claxton KP, Schulper M. J. Using value of information analysis to prioritise health research. Pharamacoeconomics. 2006;24(11):1055–1068 [DOI] [PubMed] [Google Scholar]
- 29. Thariani R, Wong W, Carlson JJ, et al. Prioritization in comparative effectiveness research: the CANCERGEN Experience. Med Care. 2012;50(5):388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behl AS, Goddard KA, Flottemesch TJ, et al. Cost-effectiveness analysis of screening for KRAS and BRAF . mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012;104(23):1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deverka PA, Schully SD, Ishibe N, et al. Stakeholder assessment of the evidence for cancer genomic tests: insights from three case studies. Genet Med. 2012;14(7):656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Institute of Medicine Genome-Based Diagnostics: Clarifying Pathways to Clinical Use—Workshop Summary. Washington, DC: National Academies Press; 2012. [PubMed] [Google Scholar]
- 33. Ginsburg GS, Staples J, Abernethy AP. Academic medical centers: ripe for rapid-learning personalized health care. Sci Transl Med. 2011;3(101):101cm27 [DOI] [PubMed] [Google Scholar]
- 34. Institute of Medicine Informatics Needs and Challenges in Cancer Research: Workshop Summary. Washington, DC: National Academies Press; 2012. [PubMed] [Google Scholar]
- 35. Agency for Healthcare Research and Quality AHRQ’s Patient-Centered Outcomes Research (PCOR) Institutional Award (K12) Fact Sheet. 2012. http://www.ahrq.gov/fund/training/pcork12.htm Accessed January 7, 2013 [Google Scholar]
- 36. Veenstra DL, Roth JA, Garrison LP, Jr, Ramsey SD, Burke W. A formal risk-benefit framework for genomic tests: facilitating the appropriate translation of genomics into clinical practice. Genet Med. 2010;12(11):686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13(6):499–504 [DOI] [PubMed] [Google Scholar]
- 38. Gold MR, Siegel JE, Russell LB, Weinstein MC. Cost-Effectiveness in Health and Medicine. New York: Oxford University Press; 1996. [Google Scholar]
- 39. World Bank Health Expenditure, Total (% of GDP). http://data.worldbank.org/indicator/SH.XPD.TOTL.ZS Accessed April 12, 2013