Abstract
National Surgical Adjuvant Breast and Bowel Project protocol C-08 tested the worth of adding 1 year of bevacizumab to oxaliplatin-based standard adjuvant chemotherapy regimen in the treatment of stage II/III colon cancer. Although the overall result was negative, the possibility that a molecularly defined subset could benefit from bevacizumab cannot be ruled out. We performed post hoc Cox regression analyses to test for marker-by-treatment interactions for standard pathological features and survival analyses using the Kaplan–Meier method. All statistical tests were two-sided and considered statistically significant at the .05 level. Patients diagnosed with mismatch repair defective (dMMR) tumors derived statistically significant survival benefit from the addition of bevacizumab (hazard ratio [HR] = 0.52; 95% confidence interval [CI] = 0.29 to 0.94; P = .02) in contrast with no benefit in patients diagnosed with mismatch repair proficient tumors (HR = 1.03; 95% CI = 0.84 to 1.27; p = .78; P interaction = .04). Although a post hoc finding, this data suggests that a molecularly defined subset of colon cancer patients may derive clinical benefit from antiangiogenesis agents and underscores the need for independent validation in other clinical trials.
Although the anti-VEGF antibody bevacizumab showed promise for the treatment of stage IV colon cancer (1–4), it failed to improve clinical outcome of patients diagnosed with stage II/III colon cancer when added to adjuvant chemotherapy.
The C-08 protocol conducted by the National Surgical Adjuvant Breast and Bowel Project (NSABP) randomly assigned 2710 patients diagnosed with stage II/III colon adenocarcinoma to receive either oxaliplatin-based chemotherapy (mFOLFOX6) or mFOLFOX6 plus bevacizumab for 12 months (5). According to the primary endpoint analysis after median follow up of 35.6 months, the addition of bevacizumab to mFOLFOX6 did not result in a statistically significant increase in disease-free survival (hazard ratio [HR] = 0.89; 95% confidence interval [CI] = 0.76 to 1.04; P = .15). Tests for a potential interaction of the effect of bevacizumab with sex, age, and nodal status were not statistically significant. However, mismatch repair (MMR) status was not examined at that time.
We have updated the analysis of C-08 with the inclusion of MMR status and longer follow-up. MMR status was determined by immunohistochemistry (IHC) with mutL homolog 1 (MLH1) and mutS homolog 2 (MSH2) proteins as described by Lindor (6). Any patients that showed negative staining of one of the two proteins in the tumor cells in the presence of positive staining in the surrounding normal cells were classified as MMR deficient (dMMR), whereas others were classified as MMR proficient (pMMR). These two immunohistochemistry markers provide both a sensitive and specific alternative to microsatellite instability in detecting DNA MMR defects (6).
The C-08 correlative study was conducted with approvals from institutional review boards for NSABP Biospecimen Bank and Biostatistics Center. Informed consent was required for participation. Formalin-fixed paraffin-embedded tumor blocks were available from 2100 of 2710 randomized patients. Patient characteristics of the MMR study subset were not different from the original trial cohort (Supplementary Table 1, available online). MMR status could be determined in 1993 patients. There were 107 case subjects with either assay failures with no staining in the normal cells or tissue detachment during the staining procedure. There were 252 case subjects (12.6%) classified as dMMR. In the set of patients with known MMR status, 25% were stage II, and median follow-up was 5.7 years (range = 0.2–7.4 years).
We also examined the V600E BRAF mutation based on its association with dMMR and worse overall survival (7). V600E mutation was determined using a primer extension assay as previously reported (n = 1764)(8).
Formal statistical tests for marker-by-bevacizumab interaction were performed in a Cox regression model including indicator variables for the marker, bevacizumab treatment, and the interaction term for the following variables: age (<65 vs ≥65 years; n = 2159), sex (n = 2159), T stage (high vs low; n = 2145), N stage (N0, N1, N2 with a 2-degree of freedom interaction test; n = 2159), MMR defects defined by two immunohistochemistry markers (MLH1 and MSH2; deficient, proficient; n = 1993), and V600E BRAF mutation (n = 1764) (Table 1). Survival was estimated by the Kaplan–Meier method. No correction for multiple comparisons was made. All statistical tests were two-sided and considered statistically significant at the .05 level.
Table 1.
Variables examined and their interaction with bevacizumab*
| Time to recurrence | Overall survival | |||||
|---|---|---|---|---|---|---|
| Variables | No. | Recurrences, No. (%) | Marker-by-bevacizumab interaction P† | No. | Deaths, No. (%) | Marker-by-bevacizumab interaction P† |
| Age, years | ||||||
| <65 | 1556 | 342 (21.0) | .44 | 1556 | 272 (17.5) | .42 |
| ≥65 | 603 | 129 (21.4) | 603 | 161 (26.7) | ||
| Sex | ||||||
| Female | 1067 | 224 (21.0) | .29 | 1067 | 192 (18.0) | .42 |
| Male | 1092 | 247 (22.6) | 1092 | 241 (22.1) | ||
| T stage‡ | ||||||
| Low | 713 | 66 (9.3) | .498 | 713 | 77 (10.8) | .51 |
| High | 1432 | 403 (28.1) | 1432 | 354 (24.7) | ||
| N stage | ||||||
| N0 | 530 | 45 (8.5) | .25 | 530 | 53 (10.0) | .21 |
| N1 | 990 | 178 (18.0) | 990 | 166 (16.8) | ||
| N2 | 639 | 248 (38.8) | 639 | 214 (33.5) | ||
| MMR status | ||||||
| Proficient | 1741 | 394 (22.6) | .08 | 1741 | 349 (20.1) | .03 |
| Deficient | 252 | 32 (12.7) | 252 | 49 (19.4) | ||
| BRAF | ||||||
| Not mutated | 1563 | 352 (22.5) | .28 | 1563 | 307 (19.6) | .37 |
| Mutated | 201 | 43 (21.4) | 201 | 54 (26.9) | ||
* MMR = mismatch repair.
† P is for the interaction in a Cox model containing bevacizumab, the variable, and the variable–bevacizumab interaction.
‡ T-stage category is defined as “low” for stage II T3 and stage III T1&T2 and “high” for stage II T4 and stage III T3 & T4.
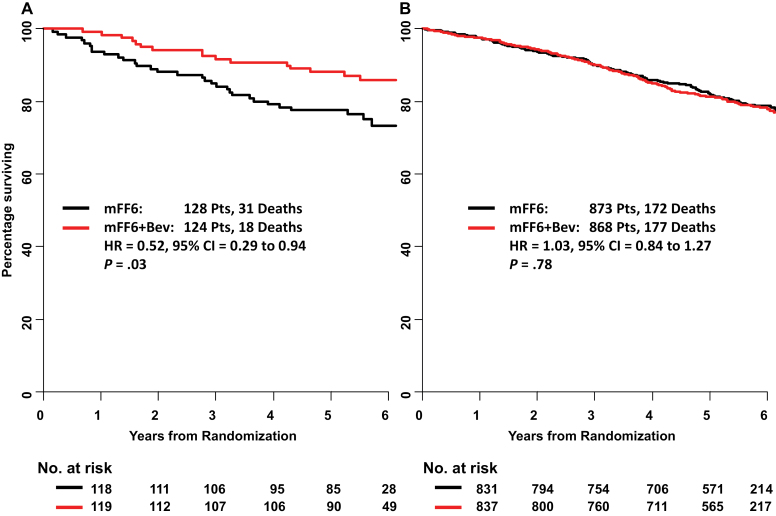
For the overall survival endpoint, only MMR status showed statistically significant interaction with bevacizumab (P = .03; not significant if corrected for multiple comparisons), with a decrease in mortality observed only in patients with dMMR tumors. Although 31 of 128 patients with dMMR tumors treated with chemotherapy died, only 18 of 124 patients who received bevacizumab in addition to chemotherapy died during the same follow-up period (HR = 0.52; 95% CI = 0.29 to 0.94; P = .03) (Figure 1A). In contrast, there was no difference in mortality between the control arm and bevacizumab arm in those who were diagnosed with pMMR tumors. One hundred seventy-two of 873 pMMR patients treated with chemotherapy died, whereas 177 of 868 pMMR patients treated with bevacizumab died during the same follow-up period (HR = 1.03; 95% CI = 0.84 to 1.27; P = .78) (Figure 1B). For time to recurrence, there was a trend for interaction in the same direction, but it was not statistically significant (P interaction = .08).
Figure 1.
The effect of bevacizumab (Bev) treatment on overall survival by mismatch repair (MMR) status for colon cancer: NSABP C-08. A) MMR deficient. B) MMR proficient. In each panel, the survival estimates are derived by the Kaplan–Meier method, and the hazard ratio (HR), confidence intervals (CIs), and P value come from a Cox regression model containing only an indicator variable for treatment. The MMR–treatment interaction test (P = 0.04) is from a Cox regression model including indicator variables for MMR, bevacizumab treatment, and the interaction term. All statistical tests were two-sided. mFF6 = modified FOLFOX6.
Although BRAF did not show statistically significant interaction, because there was an association between MMR status and BRAF mutation (P < .0001), we examined whether a combination of the two markers could further define the subset that benefited from bevacizumab in an exploratory analysis. We found that a small subset of patients (n = 51 with 16 deaths) defined by BRAF mutation and dMMR derived the most benefit, with a hazard ratio of 0.27 (95% CI = 0.08 to 0.94; P = .03) (data not shown).
Because we defined dMMR based on two immunohistochemistry markers (MLH1 and MSH2), we would have misclassified about 25% of hypermutated tumors (with mutations in MLH3, MSH3, MSH6, PMS2, and POLE) as pMMR based on data from The Cancer Genome Atlas Network (9). It would be important to examine whether patients diagnosed with hypermutated tumors with the mutations in the latter genes also derived statistically significant clinical benefit from bevacizumab.
Because this was a finding from a retrospective analysis without an a priori hypothesis based on a mechanistic study, the results presented should be regarded only as hypothesis generating. According to published exome capture sequencing data from The Cancer Genome Atlas Network, dMMR tumors are hypermutated, with median number of nonsilent mutation of 728 compared with 58 in pMMR or nonhypermutated tumors (9). Unlike pMMR tumors that are poorly immunogenic, dMMR tumors are highly immunogenic because of the generation of mutated proteins, including those with frame-shift mutations (10,11). Therefore, dMMR tumor cells at the micrometastatic sites have to evade attack from the immune system in order to progress. VEGF-A is speculated to be one of the main tumor-derived soluble factors that act as a chemo-attractant for immature myeloid cells from the marrow to the tumor site and suppress dendritic cell maturation, creating an immune suppressive microenvironment (12–15). Furthermore, VEGF-A directly induces regulatory T-cell proliferation in tumor-bearing mice through VEGFR-2 (16). Intriguingly, blocking VEGF-A alone was sufficient to inhibit regulatory T-cell accumulation in tumor-bearing mice but not in tumor-naive mice (16). More important, adding bevacizumab to chemotherapy resulted in a substantial reduction in the proportion of regulatory T-cells in the peripheral blood of colon cancer patients (16). Thus we hypothesize that bevacizumab may be particularly effective in dMMR patients because of its disruption of the immunosuppressive microenvironment associated with these hypermutated and highly immunogenic tumors.
In conclusion, our data suggest that there may be a subset of colon cancer patients who may derive significant clinical benefit from the addition of bevacizumab to standard adjuvant chemotherapy. Although only hypothesis generating, this data warrants independent validation in other randomized clinical trials.
Funding
This work was supported in part by Public Health Service Grants U10-CA-37377, U10-CA- 69974, U10-CA-12027, U10-CA-69651, and U24- CA-114732 from the National Cancer Institute, Department of Health and Human Services . It was also supported by Genentech Inc and Sanofi-Synthelabo Inc and funded in part under a grant from the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Supplementary Material
S. Paik had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflicts of interest.
We thank Melanie Finnigan for data and tissue block management; William Hiller and Teresa Oeler for histology; Wendy L. Rea, Christine I. Rudock, and Barbara Good for manuscript editing and preparation; Teresa A. Bradley, Ethan Barry, and Joyce Mull for regulatory affairs related to this manuscript; and Barbara Harkins and Francine Fonzi for protocol development. We also thank NSABP members who contributed tissue blocks as well as patients who enrolled in the study.
Clinical trials registration NSABP C-08: NCT00096278.
References
- 1. Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–2342 [DOI] [PubMed] [Google Scholar]
- 2. Hurwitz HI, Fehrenbacher L, Hainsworth JD, et al. Bevacizumab in combination with fluorouracil and leucovorin: an active regimen for first-line metastatic colorectal cancer. J Clin Oncol. 2005;23(15):3502–3508 [DOI] [PubMed] [Google Scholar]
- 3. Kabbinavar F, Hurwitz HI, Fehrenbacher L, et al. Phase II, randomized trial comparing bevacizumab plus fluorouracil (FU)/leucovorin (LV) with FU/LV alone in patients with metastatic colorectal cancer. J Clin Oncol. 2003;21(1):60–65 [DOI] [PubMed] [Google Scholar]
- 4. Kabbinavar FF, Hambleton J, Mass RD, et al. Combined analysis of efficacy: the addition of bevacizumab to fluorouracil/leucovorin improves survival for patients with metastatic colorectal cancer. J Clin Oncol. 2005;23(16):3706–3712 [DOI] [PubMed] [Google Scholar]
- 5. Allegra CJ, Yothers G, O’Connell MJ, et al. Phase III trial assessing bevacizumab in stages II and III carcinoma of the colon: results of NSABP protocol C-08. J Clin Oncol. 2011;29(1):11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20(4):1043–1048 [DOI] [PubMed] [Google Scholar]
- 7. Gavin PG, Yothers G, Tanaka N, et al. BRAF mutation. Clin Cancer Res. 2012;18(23): 6531–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost- effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saeterdal I, Bjorheim J, Lislerud K, et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci U S A. 2001;98(23):13255–13260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banejea A, Hands RE, Powar MP, et al. Microsatellite and choromosomal stable colorectal cancers demonstrate poor immunogenicity and early disease recurrence. Colorectal Dis. 2009;11(6):601–608 [DOI] [PubMed] [Google Scholar]
- 12. Bellamy WT, Richter L, Sirjani D, et al. Vascular endothelial cell growth factor is an autocrine promoter of abnormal localized immature myeloid precursors and leukemia progenitor formation in myelodysplastic syndromes. Blood. 2001;97(5):1427–1434 [DOI] [PubMed] [Google Scholar]
- 13. Gabrilovich DI, Chen HL, Girgis KR, et al. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat Med. 1996;2(10):1096–1103 [DOI] [PubMed] [Google Scholar]
- 14. Ohm JE, Gabrilovich DI, Sempowski GD, et al. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood. 2003;101(12):4878–4886 [DOI] [PubMed] [Google Scholar]
- 15. Oyama T, Ran S, Ishida T, et al. Vascular endothelial growth factor affects dendritic cell maturation through the inhibition of nuclear factor-kappa B activation in hemopoietic progenitor cells. J Immunol. 1998;160(3):1224–1232 [PubMed] [Google Scholar]
- 16. Terme M, Pernot S, Marcheteau E, et al. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. 2013;73(2):539–549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.