Abstract
Background
Oncolytic adenoviral virotherapy (OV) is a highly promising approach for the treatment of glioblastoma multiforme (GBM). In practice, however, the approach is limited by poor viral distribution and spread throughout the tumor mass.
Methods
To enhance viral delivery, replication, and spread, we used a US Food and Drug Administration–approved neural stem cell line (NSC), HB1.F3.CD, which is currently employed in human clinical trials. HB1.F3.CD cells were loaded with an oncolytic adenovirus, CRAd-Survivin-pk7, and mice bearing various human-derived GBMs were assessed with regard to NSC migration, viral replication, and therapeutic efficacy. Survival curves were evaluated with Kaplan–Meier methods. All statistical tests were two-sided.
Results
Antiglioma activity of OV-loaded HB1.F3.CD cells was effective against clinically relevant human-derived glioma models as well as a glioma stem cell–enriched xenograft model. Median survival was prolonged by 34% to 50% compared with mice treated with OV alone (GBM43FL model median survival = 19.5 days, OV alone vs NSC + OV, hazard ratio of survival = 2.26, 95% confidence interval [CI] = 1.21 to 12.23, P = .02; GBM12 model median survival = 43.5 days, OV alone vs NSC + OV, hazard ratio of survival = 2.53, 95% CI = 1.21 to 10.38, P = .02). OV-loaded HB1.F3.CD cells were shown to effectively migrate to the contralateral hemisphere and hand off the therapeutic payload of OV to targeted glioma cells. In vivo distribution and migratory kinetics of the OV-loaded HB1.F3.CD cells were successfully monitored in real time by magnetic resonance imaging. OV-loaded NSCs retained their differentiation fate and were nontumorigenic in vivo.
Conclusions
HB1.F3.CD NSCs loaded with CRAd-Survivin-pk7 overcome major limitations of OV in vivo and warrant translation in a phase I human clinical trial for patients with GBM.
Neural stem cells (NSCs) are defined as multipotent progenitor cells that originate from the developing and adult central nervous system (1). NSCs display intrinsic tumor tropism that can be exploited for targeted anticancer drug delivery to invasive and metastatic cancer (2,3). In theory, the tumor-homing property of NSCs offers a substantial advantage over other targeted therapies, such as antibody-directed drug delivery, because of their ability to detect various cues generated by satellite tumor foci and respond to such cues by extravasating through complex tissue microenvironments and migrating to distant diseased areas (4). Glioblastoma multiforme (GBM) is the most common and aggressive primary central nervous system tumor in adults and is characterized by its propensity to infiltrate throughout the brain and cause relapses in patients because of the existence of an aberrant chemo- and radio-resistant glioma stem cell (GSC) population (5). Thus, a true cure for this formidable disease cannot arise from the application of traditional antineoplastic principles; it requires a dynamic agent capable of targeting scattered disease lesions as well as eliminating the tumor-initiating cancer stem cells effectively with minimal disruption of the existing delicate neural architecture (6).
Based on this, our lab has extensively evaluated NSCs as a cellular vehicle for the targeted delivery of glioma tropic oncolytic adenoviral virotherapy (OV) CRAd-S-pk7 (7–9). We have proposed that by combining NSCs’ unique tumor tropism with the OV’s ability to target chemo- and radio-resistant GSCs (6,10) we may overcome the deficiencies inherent to each approach deployed in isolation and can effectively target GBM. As a proof-of-principle we have previously demonstrated that 1) NSCs can be used as cellular vehicles for the in vivo delivery of an OV to intracranial gliomas (7); 2) intratumoral delivery of NSCs loaded with the CRAd-S-pk7, a glioma-tropic OV regulated by the tumor-specific survivin promoter (7,11), increased median survival by 50% compared with mice treated with OV alone in an orthotopic xenograft model of human glioma (8); and 3) NSCs demonstrated superior therapeutic efficacy when compared with mesenchymal stem cells as a cell carrier for OV in the context of intracranial gliomas (9). Because these previously published results argue in favor of using NSCs as targeted cellular delivery vehicles for antiglioma oncolytic virotherapy, we conducted the following critical translational studies to justify its application in a phase I clinical trial for patients with GBM: 1) identified an optimal NSC-based cell carrier for antiglioma oncolytic virotherapy; 2) tested the selected NSC-based cell carrier in several diverse and clinically relevant glioma xenograft models; 3) developed a noninvasive imaging protocol to monitor in vivo distribution and migratory activity of NSC-based cell carriers in real time; 4) examined the capacity for the NSC-based cell carrier to deliver antiglioma OV to a distant tumor burden in a glioma xenograft model; and 5) evaluated the therapeutic efficacy of NSC-based oncolytic virotherapy in a distance-delivery glioma xenograft model.
In this report, we provide a detailed evaluation of two immortalized NSC lines as cell carriers for targeted antiglioma therapy. Our results indicate that HB1.F3.CD, a US Food and Drug Administration–approved NSC line for human clinical trials (NCT01172964) is the most suitable NSC cell carrier for the future application of cell-based OV delivery in the clinical setting. We found that HB1.F3.CD cells can effectively hand off the viral therapeutic payload to distant tumor sites and substantially prolong median survival in diverse orthotopic models of human glioma. Thus, data presented in this study solidify the notion that NSCs can be used as cell carriers for the targeted delivery of antiglioma oncolytic viruses and serve as the foundation for an investigational new drug application for a human clinical trial involving newly diagnosed and recurrent patients with malignant gliomas.
Methods
Cell Culture and Establishment of Fluorescent-Labeled Cell Lines
The U87MG,U118MG, U251, and A172 human glioma cell lines were purchased from the AmericanType Culture Collection (Manassas, VA) and maintained according to vendor recommendations. Human primary brain tumor specimens (GBM43 and GBM12) were obtained from Dr David James (University of California at San Francisco) in accordance to a protocol approved by the institutional review board at UCSF. Tumor specimens were confirmed as World Health Organization grade IV malignant glioma by an attending neuropathologist. Human ReNcells (NSCs) were obtained from Millipore (Temecula, CA) and maintained according to the manufacturer’s protocol. Briefly, these NSCs were isolated from the cortical region of 14-week-old fetal tissue and immortalized by retroviral transduction and insertion of the c-myc gene. Cells were characterized according to the expression of nestin, SOX-2, CD133, and CD44 (data not shown) stem cell markers. Further details are available in the Supplementary Methods (available online).
Viral Vector
The replication competent adenoviral vector CRAd-S-pk7 contains the wild-type adenovirus replication protein E1A under the control of human survivin promoter. This vector has been created by homologous recombination using a shuttle plasmid containing the human survivin promoter upstream to the viral E1A gene. Shuttle plasmids containing these regions were further homologously recombined into adenoviral plasmids modified to contain a poly-lysine (pk7) incorporation into the C-terminus of the wild-type fiber protein (7,11).
Analysis of Viral Replication
To detect the level of viral replication by quantitative polymerase chain reaction (PCR), NSCs were plated at a density of 2.5×104 cells/well in 24-well plastic tissue culture dishes. The next day, cells were infected with indicated IU/cell of CRAd-S-pk7. After a 1-hour incubation period, virus-containing media was removed, cells were washed with 1X phosphate-buffered saline (PBS), and a fresh portion of complete growth media was added. Infected cells were harvested at indicated time points. Total DNA was extracted from infected cells using a DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Gene expression was quantified by real-time quantitative PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and primers recognizing the viral E1A gene (7). In this process, DNA was amplified using an Opticon 2 system (Bio-Rad, Foster City, CA) and was detected by measuring the binding of the fluorescent dye SYBR green. Each sample was run in triplicates. Results were presented as the average number of E1A copies per nanogram of DNA.
We used the Adeno-X Rapid Titer Kit (Clontech, Mountain View, CA) according to the manufacturer’s protocol to titrate the levels of infectious viral progeny. Briefly, infected cells and media from each group were collected and subsequently subjected to three cycles of freezing and thawing. As a consequence, infectious progenies were released from infected cells. Cell lysates were then incubated with adherent HEK293 cells in serial 10-fold dilution. Forty-eight hours later, the amount of IU was calculated using the Adeno-X Rapid Titer Kit according to vendor recommendations. The titration units (IU) used by this protocol are similar to plaque-forming units.
Evaluation of NSC Migration and Viral Delivery In Vitro
To analyze the migratory capacity and OV delivery characteristics of HB1.F3-CD and ReNcells in vitro, we used a similar system to that described earlier (8), with a slight modification. Further details are available in the Supplementary Methods (available online).
Animal Studies
U87MG glioma cells were implanted by cranial guide screw. Briefly, mice were anesthetized with a ketamine/xylazine mixture (115/17mg/kg), and a burr hole was made. Stereotactic injection was carried out by using a 10 μL Hamilton syringe (Hamilton, Reno, NV) with a 30-gauge needle, which was inserted through the burr hole to a depth of 3mm mounted on a mice-exclusive stereotactic apparatus (Harvard Apparatus, Holliston, MA). Male athymic/nude mice were obtained from Charles River Laboratory (Wilmington, MA). Mice were cared for according to a study-specific animal protocol approved by the University of Chicago Institutional Animal Care and Use Committee. For detailed description of the methods used for all the in vivo experiments, please see the Supplementary Methods (available online).
Statistical Analysis
All statistical analyses were performed using Graphpad Prism 4 (GraphPad Software, San Diego CA). Data represent the results for assays performed in triplicate and repeated at least three different times, and error bars represent 95% confidence intervals (CIs). For continuous variables, comparisons between two groups were evaluated by statistical significance of difference of means in independent sample sets and determined using the Student t test; comparisons between more than two treatment groups were made using one-way analysis of variance or Kruskal–Wallis with Dunnett post hoc test. Survival curves were generated by the Kaplan–Meier method, and the log-rank test was used to compare the distributions of survival times. All reported P values were two-sided and were considered to be statistically significant at less than .05.
Results
Evaluation of CRAd-S-pk7 Progeny and Hand-Off From NSCs to Glioma Cells
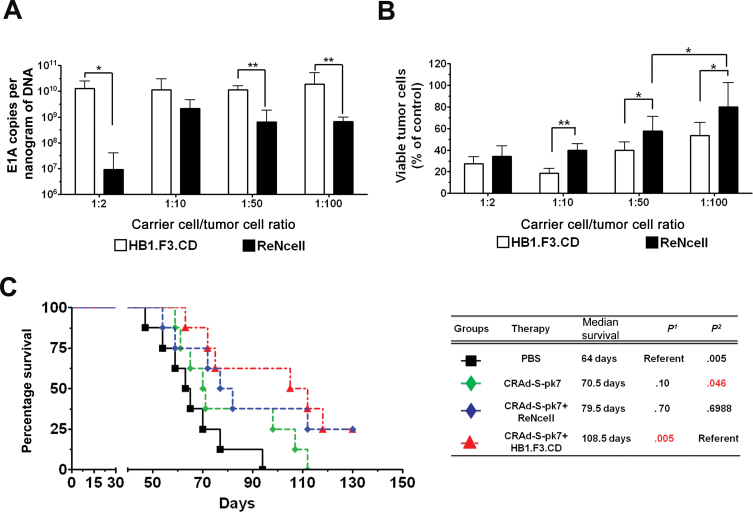
The initial characterization of both the HB1.F3.CD and ReNcell NSC lines and optimization of OV loading dose for the carrier cells is described in the Supplementary Results section and Supplementary Figure 1 (available online). Next, we examined in both NSC lines the capacity of released viral progeny to lyse targeted tumor cells. Both NSC lines were infected with 50 IU of OV per cell, the optimal OV loading dose according to our previous experiments [Supplementary Figure 1 and ref. (8)] and placed in the upper chambers of a transwell plate, whereas glioma cells were cultured in the lower chamber at the NSC/glioma cell ratios of 1:2, 1:10, 1:50, and 1:100. Viral E1A copies in the target glioma cells were measured by quantitative PCR from total isolated DNA. At the NSC/glioma cell ratio of 1:50 and 1:100, the recovered E1A copies from glioma cells were lower by approximately 1.3 (95% CI = 0.84 to 1.81; P = .002) and 1.4 (95% CI = 0.47 to 2.25; P = .02) orders of magnitude, respectively, in the cocultures containing ReNcells as compared with HB1.F3.CD cells (Figure 1A). Moreover, CRAd-S-pk7–loaded HB1.F3.CD cells demonstrated statistically significantly more killing of the targeted glioma cells than ReNcells at the NSC/glioma cell ratio of 1:10 (ReN vs HB1.F3.CD = 39.9% vs 18.7%, difference = 21.2%, 95% CI = 14.4% to 28.1%, P < 0.001), 1:50 (ReN vs HB1.F3.CD = 57.5% vs. 39.9%, difference = 17.6%, 95% CI = 3.2% to 32.0%, P = .02), and 1:100 (ReN vs HB1.F3.CD = 80.1% vs 53.6%, difference = 26.4%, 95% CI = 5.8% to 47.0%, P = 0.02) (Figure 1B).
Figure 1.
Permissiveness of neural stem cell (NSC) lines for adenovirus replication and efficacy in vitro/vivo. A) A transwell chamber assay was used to measure the viral progeny released from NSCs and subsequently capable of infecting glioma cells. HB1.F3.CD and ReNcell were infected with 50 IU per cell of CRAd-S-pk7 in the upper chambers of the transwell assay in the following ratio to glioma cells: 1:2, 1:10, 1:50, and 1:100. Glioma cells were placed on the bottom chambers of the transwell assay, and cells were harvested and infectivity was measured by quantitative real-time polymerase chain reaction for the viral E1A gene. Bars represent means from three independent experiments; error bars refer to 95% confidence intervals. Student t test was used. *P < .05; **P < .01. B) Cytotoxicity by trypan blue exclusion 96 hours after coculture. Bars represent means from five independent experiments; error bars refer to 95% confidence intervals. Student t test was used. *P < .05; **P < .01. C) CRAd-S-pk7 virus-loaded NSCs inhibit xenograft growth and prolong survival of mice with orthotopic glioblastoma. We stereotactically injected 2.5×105 U87MG cells into the right hemisphere of the brains of athymic nude mice (n = 8 per group). Three days after glioma establishment, both NSC lines were infected with 50 IU per cell of CRAd-S-pk7. Separate groups of mice received an injection of either 5×105 HB1.F3.CD or ReN cells loaded with CRAd-S-pk7 in a volume of 2.5 µL per mouse 2 to 3mm away from the original tumor site. Two additional groups of mice received either 2.5×107 IU of CRAd-S-pk7 alone or phosphate-buffered saline (PBS) in an identical volume and location in the brain. P 1 represents P value 1 in this table where PBS treated group served as reference and P 2 represents P value 2 where CRAd-S-pk7+HB1.F3.CD serve as reference. Survival curves were obtained by the Kaplan–Meier method, and overall survival time was compared between groups using log-rank test. All statistical tests were two-sided.
Next, to evaluate the therapeutic efficacy of different NSC-based antiglioma virotherapy in vivo, 2.5×104 cells of U87MG were implanted into the right hemisphere of athymic nude (nu/nu) mice. Three days after implantation of the glioma xenograft, a single injection of CRAd-S-pk7 (50 IU of pk7 per NSC × 5×1 05 NSCs per mouse = 2.5×107 IU of OV total) or 5×105 NSCs loaded with 50 IU per cell of CRAd-S-pk7 was administered ipsilaterally 2 to 3mm away from the original tumor implantation site. The mice were monitored for survival. The median survival for the PBS-treated control group was 64 days compared with 70.5 days for the CRAd-S-pk7 group (P= .10), 79.5 days for the OV-loaded ReNcell group (P = .06), and 108.5 days for the OV-loaded HB1.F3.CD group (P = .005) (Figure 1C). Thus, the group of animals bearing glioma xenografts treated with CRAd-S-pk7–loaded HB1 survived 29 days longer than the group treated with CRAd-S-pk7–loaded ReNcell. Based on this, we concluded that the HB1.F3.CD cell line functions more effectively as a cell carrier for the OV CRAd-S-pk7 both in vitro and in vivo and used it as our OV carrier cell in the remainder of the studies.
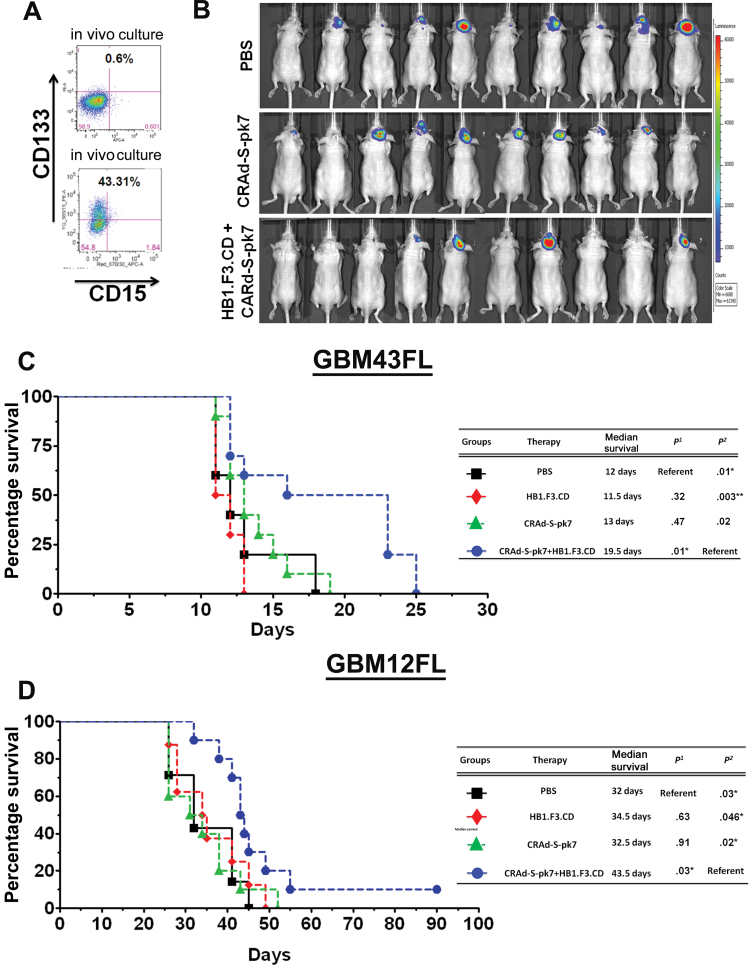
Evaluation of HB1.F3.CD as a Cell Carrier in Multiple Clinically Relevant GBM Patient-Derived Glioma Xenograft Models
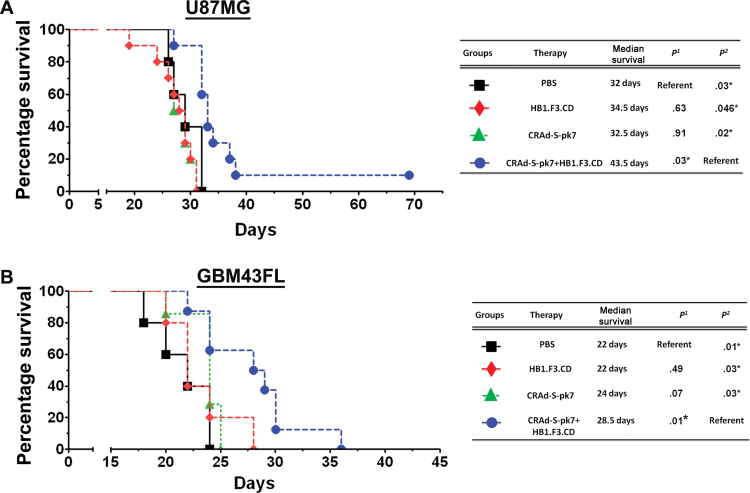
Next, we tested our OV-NSC system in two patient-derived orthotopic models: GBM43FL and GBM12FL. To maintain their intrinsic patient GBM properties, both cell lines were serially propagated in the flank of nude mice (12). As shown in Figure 2A, through a Fluorescent activated cell sorting (FACS) assay, after 2 weeks, GBM43FL cells grown in vitro expressed only 0.6% of the CD133+ GSC population, whereas 43.31% of GBM43FL propagated in the flanks of nude mice were CD133+. We implanted 2.5×104 GBM43FL cells in the right hemisphere of the brain and kept the HB1.F3.CD loaded with the same amount of virus as the CRAd-S-pk7 injection protocol described in Figure 1C, with the exception that the therapy was delivered in an ipsilateral intratumoral fashion. Through bioluminescence monitoring, we showed that four of 10 mice in the group treated with CRAd-S-pk7 loaded in HB1.F3.CD cells had tumors as compared to eight of 10 in control PBS and the OV alone group (Figure 2B). Median survival was prolonged by 34% to 50% compared with mice treated with OV alone; the median survival of group treated with CRAd-S-pk7–loaded HB1.F3.CD cells was 19.5 days, which represented a 6.5 day median survival increase over the OV group (median survival = 13 days; P = .02) and an 8-day median survival increase over the HB1.F3.CD treated group (median survival = 11.5 days; P = .003) (Figure 2C) (hazard ratio of survival = 2.26, 95% CI = 1.21 to 12.23, P = .02). In the GBM12FL model, an identical injection protocol was used as GBM43FL line for Figure 2C. The median survival was 34.5 days (P = .046) for the mice treated with HB1.F3.CD alone, 32.5 days (P = .02) for the mice treated with CRAd-S-pk7, and 43.5 days for the mice treated with HB1.F3.CD–loaded CRAd-S-pk7 (Figure 2D). The hazard ratio of survival was 2.53 (95% CI = 1.21 to 10.38, P = .02).
Figure 2.
The efficacy of HB1.F3.CD neural stem cells (NSCs) as a cell carrier for CRAd-S-pk7 virus in human-derived glioma xenografts. A) To demonstrate a major difference between glioma cells maintained in culture versus in vivo, in vitro–cultured or in vivo–cultured GBM43 cells were harvested at 2 weeks and stained for CD133 and CD15 (markers of glioma stem cells) and analyzed by fluorescent-activated cell sorting (FACS). B) To test the HB1.F3.CD cell line as a cell carrier for adenovirus against the GBM43FL glioma xenograft, 5×104 cells were implanted in the right hemisphere of nude mice. Three days after implantation, mice received intratumoral therapy of either 5×105 HB1.F3.CD cells infected with 50 IU per cell of CRAd-S-pk7, CRAd-S-pk7 alone (2.5×107 IU), HB1.F3.CD cells alone, or phosphate-buffered saline (PBS). Mice were monitored for tumor volume by bioluminescence imaging at 14 days post-therapy. C) Overall survival of mice bearing GBM43 human glioma xenografts (n = 7 per group). Survival curves were obtained by the Kaplan–Meier method, and overall survival time was compared between groups using log-rank test. All statistical tests were two-sided. P1 represents P value #1 in this table where PBS treated group served as reference and P2 represents P value #2 where CRAd-S-pk7+HB1.F3.CD serves as reference. D) The same injection strategy was used for mice bearing GBM12 human glioma xenografts (n = 10 per group). NSCs significantly increased the efficacy of CRAd-S-pk7 in both GBM43 and GBM12 models, as shown by the survival increase between the CRAd-S-pk7 group and the CRAd-S-pk7–loaded HB1.F3.CD group (P = .02 for both GBM43 and GBM12 models). P 1 represents P value 1 in this table where PBS treated group served as reference and P 2 represents P value 2 where CRAd-S-pk7+HB1.F3.CD serve as reference.
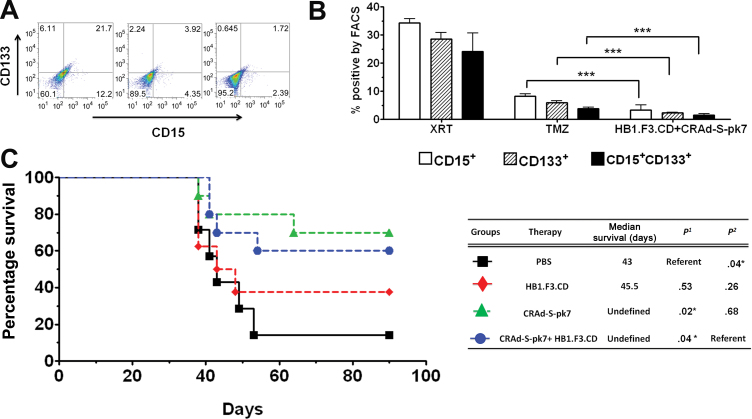
Evaluation of NSC-Based Antiglioma Oncolytic Virotherapy in a GSC-Derived Xenograft Model
To test whether our system is effective at targeting GSCs, we first performed an in vitro coculture experiment with CRAd-S-pk7–loaded HB1.F3.CD cells fluorescently labeled with green fluorescent protein (GFP) and GBM43FL cells. After 72 hours of coculture, cells were harvested and analyzed for the presence of GSCs (GFP−CD15+, GFP−CD133+, or GFP−CD15+CD133+) by FACS. As shown in Figure 3, A and B, the CRAd-S-pk7–loaded HB1.F3.CD cells statistically significantly decreased the CD15+ population approximately 10- and 2.5-fold (P < .001), the CD133+ population approximately 12- and 2.6-fold (P < .001), and the CD15+CD133+ population approximately 15- and 2.4-fold, compared with radiotherapy (XRT) (2 Gy) and temozolomide (TMZ) (50 μM), respectively (P < .001). To evaluate the therapeutic efficacy of OV-loaded HB1.F3.CD cells against GSCs in vivo, the CD133+ GSC population of cells was isolated from GBM43FL by FACS sorting (Figure 2A), and 5×103 CD133+ cells were implanted intracranially in the right hemisphere of nude mice. Three days after GSC tumor implantation, mice were divided into four groups and treated with intratumoral injections of either PBS, HB1.F3.CD alone, CRAd-S-pk7 alone, or CRAd-S-pk7–loaded HB1.F3.CD cells (as described in Figure 1C). As shown in Figure 3C, approximately 60% of mice treated with OV-loaded HB1.F3.CD cells and approximately 65% of mice treated with OV alone survived for more than 90 days, indicating that NSC-based oncolytic virotherapy can be effective in suppressing GSC-driven tumor growth in an orthotopic human glioma xenograft model.
Figure 3.
The efficacy of HB1.F3.CD neural stem cells (NSCs) as a cell carrier for CRAd-S-pk7 virus in a glioma stem cell–derived xenograft model. A) GBM43FL glioma cells were cultured and exposed to a total of 10 Gy radiotherapy (XRT) treatment (5 days × 2 Gy), temozolomide (50 uM), or cocultured with HB1.F3.CD-GFP+ cells loaded with CRAd-S-pk7 (50 IU per cell). After 72 hours of incubation, cells were collected and stained for glioma stem cell (GSC) markers CD133 and CD15 and subjected to fluoresecent-activated cell sorting (FACS) analysis. Representative FACS plots show the percentage of CD15+, CD133+, or CD15+CD133+ GBM43FL cells. B) The percentage of positive populations of GSCs in the three treatment groups. All three populations of GSCs were statistically significantly reduced in oncolytic adenoviral virotherapy (OV)–loaded NSC treatment group compared with the XRT or chemotherapy (TMZ) treatment groups (P < .001), compared using Student t test. Bars represent means from three independent experiments; error bars refer to 95% confidence intervals. C) The OV-loaded HB1.F3.CD therapy was tested in vivo. GBM43FL cells were FACS sorted, and 5×103 CD133+ cells were intracranially implanted in the brains of nude mice (n = 7 per group). Three days after tumor implantation, mice were treated either with phosphate-buffered saline (PBS), 5×105 HB1.F3.CD cells, 5×105 HB1.F3.CD cells loaded with 50 IU per cell of CRAd-S-pk7, or 2.5×107 IU of CRAd-S-pk7 intratumorally. P 1 represents P value 1 in this table where PBS treated group served as reference and P 2 represents P value 2 where CRAd-S-pk7+HB1.F3.CD serve as reference. Survival curves were obtained by the Kaplan–Meier method, and overall survival time was compared between groups using the log-rank test. All statistical tests were two-sided. ***P < .001; **P < .01; *P < .05.
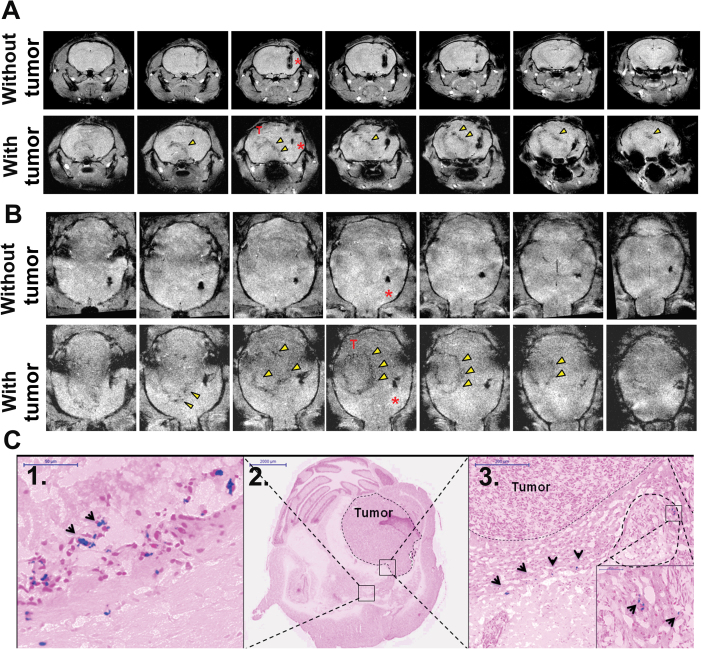
Magnetic Resonance Imaging (MRI) Monitoring of NSC Migration In Vivo
To evaluate whether OV-loaded HB1.F3.CD cells migrate to distant tumor foci in animal brains, microparticles of iron oxide (MPIO)–labeled HB1.F3.CD cells [please refer to Supplementary Methods and Supplementary Figure 3, available online, for labeling protocol and ref. (13)] with or without OV (50 IU/cell) were stereotactically implanted in the left hemisphere 10 days after U87 glioma cell or PBS injection in the right hemisphere. The MPIO-labeled HB1.F3.CD grafts resulted in a hypointense area surrounding the injection sites (Figure 4A). Figure 4, A and B, show serial continuous slices of coronal and axial T1 weighted images, respectively, from representative mice (n = 4) at 3 days after implantation of the MPIO-labeled HB1.F3.CD cells loaded with CRAd-S-pk7 in the contralateral hemisphere of the animal brain without (Figure 4, A and B, 1st row) or with (Figure 4, A and B, 2nd row) a U87 xenograft tumor. A hypointense stream directed toward the side of the implanted U87 tumor extended gradually from the HB1.F3.CD cell graft over time (Figure 4A, arrowheads), suggesting the migration of OV-loaded HB1.F3.CD cells toward the tumor site (Figure 4, A and B, image rows with tumor). In contrast, no obvious signal changes were observed along the corpus callosum in the animal brain without tumor (Figure 4, A and B, image rows without tumor). Immunohistological verification with Prussian Blue staining of the same mouse brain with tumor as shown in Figure 4, A and B, is shown in Figure 4C and revealed the presence of iron from MPIO-labeled HB1.F3.CD (arrowheads) at the implanted site (Figure 4C, 1), at the edges of the tumor mass (dotted line) (Figure 4C, 2), and inside the tumor (Figure 4C, 3). These results confirm that OV-loaded HB1.F3.CD cells retain their tropism for tumor as detected by MRI in vivo and that MRI tracking of NSC migration is a feasible and valuable strategy moving forward from preclinical studies.
Figure 4.
Magnetic resonance imaging of tumor-tropic migration of oncolytic adenoviral virotherapy (OV)–HB1.F3.CD neural stem cells (NSCs) in vivo. Microparticles of iron oxide (MPIO)–labeled HB1.F3.CD cells loaded with CRAd-S-pk7 (50 IU per cell) were implanted in the left hemisphere of control mice (no tumor) or mice bearing U87 xenografts in the contralateral hemisphere. A) Serial axial T1 weighted images of a mouse brain 3 days after implantation without tumor (top panel) and with tumor (bottom panel). Arrowheads point to the area of hypointense signal extending from the site of NSC injection toward the tumor graft. B) Serial coronal T1 weighted images show an alternative view of oncolytic adenovirus–loaded NSCs without tumor (top panel) or with tumor (bottom panel). C) Prussian blue staining of the corresponding animal brains confirmed the presence of iron MPIOs at the (C1) NSC implantation site, (C2) at the tumor border, and (C3) inside the tumor mass. Scale bars, (size).
In Vivo Differentiation Status of Implanted OV-Loaded HB1.F3.CD Cells
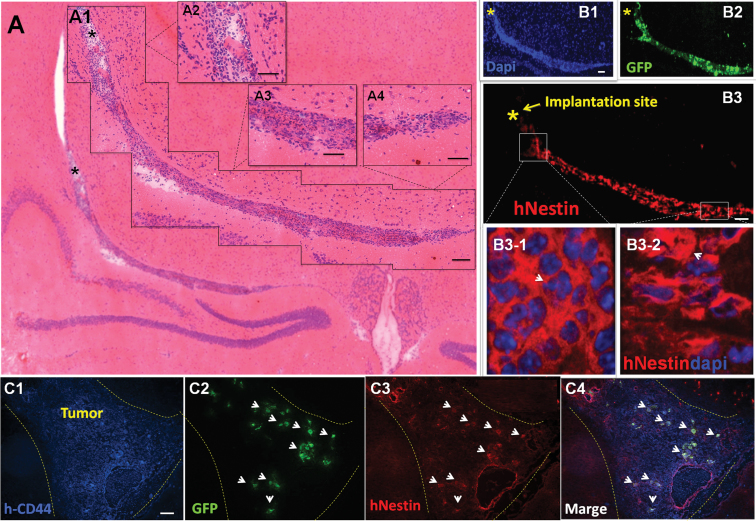
The in vivo differentiation status of CRAd-S-pk7–loaded HB1.F3.CD cells after implantation is an important issue to consider for both safety (nononcogenic) and tumor-homing properties of loaded NSCs (14). To examine the differentiation status of transplanted HB1.F3.CD cells loaded with OV in vivo, HB1.F3.CD-GFP+ cells infected with OV were implanted in the contralateral hemisphere of nude mouse brains containing U87 glioma xenografts established 10 days earlier. At 24 and 72 hours after NSC implantation, the mice were killed and their brains were subjected to immunohistochemical analysis. Within 24 hours of implantation, the GFP-tagged HB1.F3.CD cells began to cross the midline of the brain and migrate toward the implanted tumor in the contralateral hemisphere. Figure 5A shows the hematoxylin and eosin staining of the migratory path of implanted HB1.F3.CD OV-loaded cells, which is validated by the immunofluorescence staining by both 4'6-diamidino-2-phenylindole (DAPI) (Figure 5, B1) and GFP (Figure 5, B2) antibodies on the same animal brain. Next, to evaluate the stemness of implanted HB1.F3.CD cells loaded with CRAd-S-pk7, we counterstained the slides with an antihuman-specific nestin antibody and observed that the majority of the GFP+ HB1.F3.CD cells were also nestin+ (Figure 5, B3, B3-1, and B3-2). When we examined the implanted xenograft tumor in the contralateral hemisphere 72 hours after NSC implantation, we observed the presence of GFP+ cells (Figure 5, C2) inside human CD44+ tumor foci (Figure 5, C1). Additionally, the HB1.F3.CD cells that migrated and reached the tumor site (white dotted area) also stained positive for human nestin (Figure 5, C3 and C4). Furthermore, as shown in Supplementary Figure 2 (available online), even though 24 hours after OV infection of HB1.F3.CD cells in vitro the NSC markers Sox2 and Oct4 as well as the differentiation markers Galc (oligodendrocyte marker), GFAP (astrocyte), and beta-III tubulin (neuronal) mRNA levels decreased, by 72 hours gene expression normalized and was non-statistically significantly different from the uninfected control HB1.F3.CD cells (Supplementary Figure 3, available online). Most important, OV-loaded HB1.F3.CD cells implanted intracranially into nude mice did not show any signs that implanted NSCs became tumorigenic as demonstrated by complete animal survival and immunohistochemistry (data not shown). Taken together, these data indicate that implanted HB1.F3.CD cells loaded with CRAd-S-pk7 retained their NSC differentiation fate and displayed substantial pathotropism in a mouse model of glioma.
Figure 5.
In vivo differentiation of CRAd-S-pk7–loaded HB1.F3.CD cells. To follow the differentiation status of oncolytic adenovirus–loaded HB1.F3.CD cells implanted into the brains of mice, HB1.F3.CD-GFP+ loaded with CRAd-S-pk7 (50 IU per cell) were implanted into the contralateral hemisphere of mice bearing U87 glioma xenografts. Mice were killed 24 and 72 hours after neural stem cell (NSC) implantation, and brains were prepared for immunohistochemical analysis. A) Hematoxylin and eosin staining of the migratory path of NSCs from the injection site represented by the (*) to the tumor site (A1). Magnified views of the migration path: injection site (A2), center of migration path (A3), end of migration site or tumor (A4). B) Hematoxylin and eosin staining was confirmed by 4'6-diamidino-2-phenylindole (DAPI). (B1) and green fluorescent protein (GFP) (B2) staining. HB1.F3.CD-GFP+ cells (B3) were also positive for human nestin along the migratory path. (B3-2) corresponds to the slightly elongated shape seen in (A4) along the migratory path where nestin staining is spread out (arrowhead) as opposed to (B3-1), a bunched (arrowhead) shape that corresponds to (A2) or NSC implantation site. C) Mice were also sacrificed at 72 hours after NSC implantation. (C2) shows the borders of the tumor (represented by dotted line) and HB1.F3.CD-GFP+ positive cells inside human CD44+ tumor foci (C1). (C3) HB1.F3.CD-GFP+ cells that colocalize with CD44+ cells also stained positive for nestin. C4) Merge. Sizing of scale bars: Figure 5 A1 (200 μm); Figure 5 A2-A4 (50 μm); Figure 5 B1-B2 (200 μm); Figure 5 C1 (100 μm).
In Vivo Hand-off and Expansion of CRAd-S-pk7 at Distant Tumor Sites by HB1.F3.CD Cell Carrier
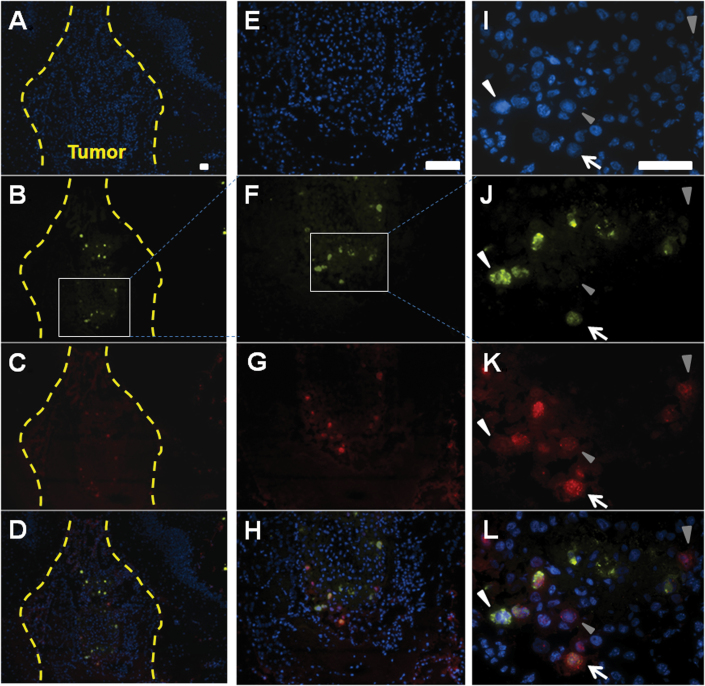
To investigate the hand-off and amplification of CRAd-S-pk7 at the targeted tumor site, we implanted OV-loaded HB1.F3.CD cells in the contralateral hemisphere of nude mice bearing U87 xenograft tumors (10 days after tumor implantation). Seventy-two hours after NSC implantation, mouse brains were harvested and subjected to immunohistochemical analysis for the expression of the adenoviral early gene E1A (Figure 6, B, F, and J) and the late hexon gene (Figure 6, C, G, and K). Figure 6 illustrates viral hand-off by the presence of newly infected tumor cells as designated by positive staining for E1A. Tumor cells transitioning from an early to late phase of infection are represented by E1A + hexon+ markers (Figure 6, J, K, and L, white arrow). Cells that are hexon+ denote a late phase of infection (Figure 6 J, K, and L, gray arrowhead). Based on these data, we conclude that OV-loaded HB1.F3.CD cells implanted in the contralateral hemisphere of glioma-bearing mice can home to, amplify, and hand off their therapeutic payload at distant tumor sites.
Figure 6.
Neural stem cells (NSCs) hand off and expand oncolytic adenoviral virotherapy (OV) therapeutic payload at distant tumor foci in vivo. OV-loaded HB1.F3.CD cells were implanted in the contralateral hemisphere of mice bearing U87 xenograft tumors. Mice were killed 72 hours after NSC implantation, and brains were preserved and prepared for immunohistochemical analysis. A, E, I) stained with 4’6-diamidino-2-phenylindole (DAPI) only. Scale bars: A–D = 200 μm, E–H=100 μm I–L = 50 μm. B, F, J) Early stages of viral replication represented by the positive staining for E1A (gray arrowhead) inside the tumor border (dotted line). C, G, K) Intermediate stages of viral replication denoted by the costaining of E1A and hexon. D, H, L) (white arrow) Hexon positive staining represents the late phases of viral infection (white arrowhead). Sizing of scale bars: Figure 6 A-D (200 μm); Figure 6 E-H (100 μm); Figure 6 I-L (50 μm).
In Vivo Evaluation of the Therapeutic Efficacy of Distantly Delivered NSC-Based Oncolytic Virotherapy
Finally, we set out to evaluate the therapeutic potential of the OV-loaded HB1.F3.CD cell carrier when delivered at a site distant from the primary tumor mass. We stereotactically implanted 5×105 OV-loaded (50 IU/cell) HB1.F3.CD cells in the left hemisphere of the cerebral cortex in nude mice bearing U87 or GBM43FL human glioma xenografts in the contralateral hemisphere (5×103 glioma cells were implanted 3 days before OV-loaded HB1.F3.CD cell implantation). In the U87 model, median survival of the group treated with CRAd-S-pk7–loaded HB1.F3.CD cells was 33 days, which represented an 18% increase in median survival over the OV group (median survival = 28 days; P < .001) and a 16% increase over the HB1.F3.CD–treated group (median survival = 28.5 days; P < .001) (Figure 7A). In the GBM43FL model, an identical injection protocol was used as described in Figure 7A. The median survival was 22 days (P = .03) for the mice treated with HB1.F3.CD alone, 24 days (P = .03) for the CRAd-S-pk7–treated group, and 28.5 days for the HB1.F3.CD–loaded CRAd-S-pk7 group (Figure 7B). These data indicate that NSC carrier-based OV therapy can be effective even when implanted in the contralateral hemisphere of the targeted glioma xenograft.
Figure 7.
Contralateral delivery of oncolytic adenoviral virotherapy (OV)–loaded HB1.F3.CD cells show therapeutic efficacy in a mouse model of glioma. To examine distance delivery, either 5×103 U87 or GBM43FL cells were implanted into the right hemisphere of nude mice (n = 7 per group). After 3 days, mice were treated with an injection of either phosphate-buffered saline (PBS), 5×105 HB1.F3.CD cells, 5×105 HB1.F3.CD cells loaded with 50 IU per cell of CRAd-S-pk7, or 2.5×107 IU of CRAd-S-pk7 into the contralateral left hemisphere of the brain. A) Overall survival of U87MG bearing mice. B) Overall survival of GBM43FL-bearing mice. CRAd-S-pk7–loaded HB1.F3.CD extended the efficacy of OV alone in both U87MG and GBM43FL models (P < .001, P = .03, respectively). P 1 represents P value 1 in this table where PBS treated group served as reference and P 2 represents P value 2 where CRAd-S-pk7+HB1.F3.CD serve as reference. Survival curves were obtained by the Kaplan–Meier method, and overall survival time was compared between groups using the log-rank test. All statistical tests were two-sided. *P < .05, **P < .01, ***P < .001
Discussion
NSC carrier-based oncolytic virotherapy holds great promise as an alternative and complimentary treatment modality for glioblastoma. In this study, we sought to evaluate the relative efficacy of two immortalized NSC lines as a cell carrier for antiglioma oncolytic virotherapy and observed that 1) the HB1.F3.CD cells were more efficient in supporting CRAd-S-pk7 OV replication as well as killing glioma cells both in vitro and in vivo compared with ReNcells; 2) the antiglioma activity of OV-loaded HB1.F3.CD cells was effective against clinically relevant human-derived glioma models as well as a GSC-enriched xenograft model; 3) OV-loaded HB1.F3.CD cells can effectively migrate to the contralateral hemisphere and hand off their therapeutic payload of oncolytic viruses to targeted glioma cells; 4) in vivo distribution and migratory kinetics of the OV-loaded HB1.F3.CD cells can be monitored by MRI and are reported in detail; and 5) distance delivery of OV-loaded HB1.F3.CD cells can prolong median survival in orthotopic mouse models with human glioma xenografts. These results along with our previously published data (8,9,15,16) argue in favor of NSC-based cell carriers for the targeted delivery of oncolytic virus against human glioma. We believe that the methods and protocols as well as preclinical data generated during this study will permit us to bridge the gap between preclinical animal studies and human clinical trials and will lead to the development of human clinical trial protocols in the future.
Despite these promising results, certain important points of emphasis deserve further attention. First, we witnessed variability in the therapeutic efficacy of NSC-based oncolytic virotherapy between different glioma models. The models in our study were selected to represent the interpatient and intratumor heterogeneity widely observed among GBMs. On a molecular level, the GBM43 xenograft model expresses p53 mutant and PTEN wild type, is negative for epidermal growth factor receptor, and expresses elevated levels of the chemo-resistance gene O6-methyltransferase (MGMT) (12,17). The GBM12 model expresses p53 null status and wild-type PTEN (12,17) but expresses epidermal growth factor receptor. On the other hand, U87 contains wild-type p53, PTEN mutant, expresses low levels of wild-type epidermal growth factor receptor, and is MGMT negative (18). With respect to their pathological features, the U87 cell line forms very localized noninfiltrative tumors, whereas GBM43 tumors are mildly invasive, and GBM12 tumors are extremely infiltrative in the rodent brain. It has been well documented that the genetic phenotype of tumor cells is one of the key determining factors of antitumor activity of an oncolytic virus (19). For example, wild-type p53 status in tumor cells can facilitate an efficient killing of these tumor cells by OVs (11,12,20). This is consistent with the observation in our study that demonstrated the most pronounced efficacy in the p53 wild-type U87 glioma model.
Furthermore, the data presented in this report convincingly demonstrate that within 24 hours after implantation, OV-loaded HB1.F3.CD cells are capable of migrating 4 to 6mm to the contralateral hemisphere and handing off therapeutic virus to targeted tumor cells in a rodent model of glioma. However, the smaller-sized brain makes it difficult to speculate on the migratory capacity of HB1.F3.CD cells in a larger-sized human brain with glioma. Fortunately, to mitigate the uncertainty behind this possible pitfall, we were able to demonstrate and optimize a valuable protocol where NSCs can be labeled with MPIOs and subsequently tracked by MRI imaging; therefore this technique may help serve as a key tool for the dynamic optimization of NSC delivery protocols in the clinical setting.
The fate of the NSC-based cell carrier is critical for maintaining tumor pathotropism (21). A number of recent studies reported that pluripotency and the differentiation status of the NSC are altered upon viral infection (22). Our data indicate that upon loading with the OV CRAd-S-pk7, the NSC-specific markers (nestin, Sox2 and Oct4) or different differentiation markers (Gal C, beta III tubulin, or GFAP) at the mRNA level remain unchanged. Moreover, the fate of the transplanted NSC in the mammalian brain has been shown to be influenced by the extrinsic factors that can drive their differentiation into either neurons or glia (23). The diseased brain seems to create an environment that can stimulate endogenous or exogenous stem cells to differentiate into specific cells types. However, our data, along with other published data, indicate that therapeutic stem cells implanted into the brains of tumor-bearing mice remain in an undifferentiated state (Figure 5) (2,24,25). These results strongly suggest that the tumor microenvironment may be deficient in factors necessary for stem cell differentiation.
Last, NSC allorejection could be detrimental to the success of NSC-based antiglioma oncolytic virotherapy in the clinic. Despite the poor immunogenic characteristics of NSCs, in vitro NSC allorejection by peripheral lymphocytes has been reported (14,26). The ideal solution to this possible pitfall would be using autologous NSCs, but the currently available technology for isolating and expanding autologous NSCs is in its infancy. With recent developments in the field of induced pluripotent stem cells, one might imagine a future in which medical centers can offer highly customized, patient-focused approaches to NSC-based antiglioma therapy by using personalized induced pluripotent stem cells or induced NSCs (27) catered toward each patient with unique genetic backgrounds (27–29).
In conclusion, our study shows that NSC-based cell carriers can effectively deliver antiglioma OV to distant tumor sites, release the therapeutic payload at the target sites, and increase median survival in a diverse range of orthotropic human glioma xenograft models that stand to recapitulate the heterogeneity of the human disease. The importance of this point cannot be emphasized enough, because after all, the name “glioblastoma multiforme” does indeed mean “to have many forms, of multiple shapes,” which perfectly captures the essence of the multifaceted nature of the disease and the challenge it presents to developing an effective therapy. Thus, any novel antiglioma therapy must stand to address this crucial issue. In this study, we have demonstrated that the NSC OV platform has the ability to extend survival in a multitude of invasive models of human glioma and target the therapy-resistant and disease-reinitiating GSC population, thus fulfilling two critical considerations for successful clinical translation, and may serve as a future therapy that can complement the existing standard of care for glioblastoma.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke (U01NS069997, R01NS077388); National Cancer Institute (R01CA122930, K99CA160775); and University of Chicago IRI Pilot Grant.
Supplementary Material
We would like to thank Ms Shirley Bond from the Integrated Microscopy Core Facility and MRI Core Facility at the University of Chicago for her microscopy support. We are grateful to Dr. David James for providing patient derived GBM line GBM43 and GBM12.
References
- 1. Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci. 2010;11(3):176–187 [DOI] [PubMed] [Google Scholar]
- 2. Aboody KS, Brown A, Rainov NG, et al. Neural stem cells display extensive tropism for pathology in adult brain: evidence from intracranial gliomas. Proc Natl Acad Sci U S A. 2000;97(23):12846–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benedetti S, Pirola B, Pollo B, et al. Gene therapy of experimental brain tumors using neural progenitor cells. Nat Med. 2000;6(4):447–450 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed AU, Lesniak MS. Glioblastoma multiforme: can neural stem cells deliver the therapeutic payload and fulfill the clinical promise? Expert Rev Neurother. 2011;11(6):775–777 [DOI] [PubMed] [Google Scholar]
- 5. Nicholas MK, Lukas RV, Chmura S, Yamini B, Lesniak M, Pytel P. Molecular heterogeneity in glioblastoma: therapeutic opportunities and challenges. Semin Oncol. 2011;38(2):243–253 [DOI] [PubMed] [Google Scholar]
- 6. Alonso MM, Jiang H, Gomez-Manzano C, Fueyo J. Targeting brain tumor stem cells with oncolytic adenoviruses. Methods Mol Biol. 2012;797 111–125. 10.1007/978-1-61779-340-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tyler MA, Ulasov IV, Sonabend AM, et al. Neural stem cells target intracranial glioma to deliver an oncolytic adenovirus in vivo. Gene Ther. 2009;16(2):262–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmed AU, Thaci B, Alexiades NG, et al. Neural stem cell-based cell carriers enhance therapeutic efficacy of an oncolytic adenovirus in an orthotopic mouse model of human glioblastoma. Mol Ther. 2011;19(9):1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ahmed AU, Tyler MA, Thaci B, et al. A comparative study of neural and mesenchymal stem cell-based carriers for oncolytic adenovirus in a model of malignant glioma. Mol Pharm. 2011;8(5):1559–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanai R, Rabkin SD, Yip S, et al. Oncolytic virus-mediated manipulation of DNA damage responses: synergy with chemotherapy in killing glioblastoma stem cells. J Natl Cancer Inst. 2012;104(1):42–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ulasov IV, Zhu ZB, Tyler MA, et al. Survivin-driven and fiber-modified oncolytic adenovirus exhibits potent antitumor activity in established intracranial glioma. Hum Gene Ther. 2007;18(7):589–602 [DOI] [PubMed] [Google Scholar]
- 12. Sarkaria JN, Carlson BL, Schroeder MA, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12(7 Pt 1):2264–2271 [DOI] [PubMed] [Google Scholar]
- 13. Thu MS, Najbauer J, Kendall SE, et al. Iron labeling and pre-clinical MRI visualization of therapeutic human neural stem cells in a murine glioma model. PLoS One. 2009;4(9):e7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed AU, Alexiades NG, Lesniak MS. The use of neural stem cells in cancer gene therapy: predicting the path to the clinic. Curr Opin Mol Ther. 2010;12(5):546–552 [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed AU, Ulasov IV, Mercer RW, Lesniak MS. Maintaining and loading neural stem cells for delivery of oncolytic adenovirus to brain tumors. Methods Mol Biol. 2012;797 97–109. 10.1007/978-1-61779-340-0_8. [DOI] [PubMed] [Google Scholar]
- 16. Thaci B, Ahmed AU, Ulasov IV, et al. Pharmacokinetic study of neural stem cell-based cell carrier for oncolytic virotherapy: targeted delivery of the therapeutic payload in an orthotopic brain tumor model. Cancer Gene Ther. 2012;19(6):431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kitange GJ, Carlson BL, Schroeder MA, et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009;11(3):281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chahal M, Xu Y, Lesniak D, et al. MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro Oncol. 2010;12(8):822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto M, Curiel DT. Current issues and future directions of oncolytic adenoviruses. Mol Ther. 2010;18(2):243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Beusechem VW, van den Doel PB, Gerritsen WR. Conditionally replicative adenovirus expressing degradation-resistant p53 for enhanced oncolysis of human cancer cells overexpressing murine double minute 2. Mol Cancer Ther. 2005;4(6):1013–1018 [DOI] [PubMed] [Google Scholar]
- 21. Carney BJ, Shah K. Migration and fate of therapeutic stem cells in different brain disease models. Neuroscience. 2011;197 37–47. 10.1016/ j.neuroscience.2011.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das S, Basu A. Viral infection and neural stem/progenitor cell’s fate: implications in brain development and neurological disorders. Neurochem Int. 2011;59(3):357–366 [DOI] [PubMed] [Google Scholar]
- 23. Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438 [DOI] [PubMed] [Google Scholar]
- 24. Miletic H, Fischer Y, Litwak S, et al. Bystander killing of malignant glioma by bone marrow-derived tumor-infiltrating progenitor cells expressing a suicide gene. Mol Ther. 2007;15(7):1373–1381 [DOI] [PubMed] [Google Scholar]
- 25. Shah K, Bureau E, Kim DE, et al. Glioma therapy and real-time imaging of neural precursor cell migration and tumor regression. Ann Neurol. 2005;57(1):34–41 [DOI] [PubMed] [Google Scholar]
- 26. Ubiali F, Nava S, Nessi V, et al. Allorecognition of human neural stem cells by peripheral blood lymphocytes despite low expression of MHC molecules: role of TGF-beta in modulating proliferation. Int Immunol. 2007;19(9):1063–1074 [DOI] [PubMed] [Google Scholar]
- 27. Ring KL, Tong LM, Balestra ME, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11(1):100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izpisua Belmonte JC, Ellis J, Hochedlinger K, Yamanaka S. Induced pluripotent stem cells and reprogramming: seeing the science through the hype. Nat Rev Genet. 2009;10(12):878–883 [DOI] [PubMed] [Google Scholar]
- 29. Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.