Abstract
Background
Trypanosoma brucei is the causative agent of African Sleeping Sickness in humans and contributes to the related veterinary disease, Nagana. T. brucei is segregated into three subspecies based on host specificity, geography and pathology. T. b. brucei is limited to animals (excluding some primates) throughout sub-Saharan Africa and is non-infective to humans due to trypanolytic factors found in human serum. T. b. gambiense and T. b. rhodesiense are human infective sub-species. T. b. gambiense is the more prevalent human, causing over 97% of human cases. Study of T. b. gambiense is complicated in that there are two distinct groups delineated by genetics and phenotype. The relationships between the two groups and local T. b. brucei are unclear and may have a bearing on the evolution of the human infectivity traits.
Methodology/Principal Findings
A collection of sympatric T. brucei isolates from Côte d’Ivoire, consisting of T. b. brucei and both groups of T. b. gambiense have previously been categorized by isoenzymes, RFLPs and Blood Incubation Infectivity Tests. These samples were further characterized using the group 1 specific marker, TgSGP, and seven microsatellites. The relationships between the T. b. brucei and T. b. gambiense isolates were determined using principal components analysis, neighbor-joining phylogenetics, STRUCTURE, FST, Hardy-Weinberg equilibrium and linkage disequilibrium.
Conclusions/Significance
Group 1 T. b. gambiense form a clonal genetic group, distinct from group 2 and T. b. brucei, whereas group 2 T. b. gambiense are genetically indistinguishable from local T. b. brucei. There is strong evidence for mating within and between group 2 T. b. gambiense and T. b. brucei. We found no evidence to support the hypothesis that group 2 T. b. gambiense are hybrids of group 1 and T. b. brucei, suggesting that human infectivity has evolved independently in groups 1 and 2 T. b. gambiense.
Introduction
The parasite Trypanosoma brucei is the causative agent of African Sleeping Sickness in humans and one of several pathogens that cause the veterinary disease Nagana. These diseases have a wide distribution across sub-Saharan Africa and affect some of the poorest areas of the world. T. brucei is traditionally segregated into three morphologically identical sub-species based on host, geography and pathology. T. b. brucei is limited to domestic and wild animals throughout sub-Saharan Africa and is non-infective to humans (and some primates) due to sensitivity to trypanosome lytic factors found in their serum [1]. T. b. gambiense and T. b. rhodesiense are human infective sub-species, named due to their relative geographic locations. While T. b. rhodesiense can infect humans due to possession of a serum resistance associated (SRA) gene [2], this gene is not present in the more prevalent West and central African trypanosome subspecies, T. b. gambiense [3]–[5], causative agent of over 95% of reported cases of sleeping sickness [6], [7]. How this sub-species is able to resist lysis by human trypanolytic factors is still unknown, although there is a decrease in uptake of lytic factor due to modification of the HpHbR cell surface receptor that contributes to resistance [8]–[12]. The study of T. b. gambiense is complicated in that there are two distinct groups within the sub-species that can be delineated by genetics and several phenotypic characteristics [13]. Group 1 T. b. gambiense causes a chronic infection, is invariably resistant to human serum and is by far the more prevalent group of the two. It appears to be largely a disease limited to humans, although some animal reservoirs have been described [14]–[19]. Conversely, group 2 T. b. gambiense are reported to be more virulent and exhibit a variable human serum resistance mechanism in a manner similar to T. b. rhodesiense [16], [19]. Unlike group 1 T. b. gambiense, group 2 T. b. gambiense does not possess the modification to the cell surface receptor HpHbR to avoid lysis by human serum and appears to utilize a different method to counter lytic factors that is distinct from group 1 T. b. gambiense or T. b. rhodesiense [20]. This group of T. b. gambiense has only been described in Côte d’Ivoire, Cameroon and Burkina Faso to date, and always in geographical areas where group 1 T. b. gambiense is also found [13], [21]. This geographical overlap of the two groups of T. b. gambiense raises interesting questions as to whether they share a close genetic relationship and possibly the ability to undergo sexual recombination.
Investigation of the population genetics of T. b. gambiense would reveal if such a relationship exists, however most research on T. b. gambiense field populations has focused on the more prevalent group 1. Studies with isoenzymes and RFLPs have indicated that group 1 T. b. gambiense populations exhibit low genetic variation and appear distinct from T. b. brucei populations [16], [19], [21]–[26] and recent comprehensive microsatellite genotyping techniques appear to have confirmed this for some populations [27], [28]. This is in contrast to high genetic variation found in T. b. brucei populations [16], [19], [22]–[26], [29]. It has also been shown that while group 1 T. b. gambiense are clonal within a disease focus, there are genetic differences between isolates from different geographic locations [27]. During the decades of investigation into the population structure of T. brucei, a second group of human infective T. b. gambiense from Côte d’Ivoire and Burkina Faso (formally Upper Volta) were identified that possess different isoenzyme and RFLP profiles from the classical group 1 T. b. gambiense isolates. These isolates also showed greater genetic variation and appeared more similar to local T. b. brucei [16], [19], [22]–[26], [30]. The greater variation in genetic markers and similarity to the T. b. brucei population suggested that this second group of T. b. gambiense might be genetically competent, in contrast to group 1. This has subsequently been confirmed by laboratory crosses between T. b. gambiense group 2 and both T. b. brucei and T. b. rhodesiense strains [31]–[33]. The possibility that different mating structures may exist in the two groups of T. b. gambiense has implications in evaluating the relationships between them and the evolution of several traits in the population, including human infectivity. It is possible that this trait has evolved twice in separate populations of T. b. gambiense or conversely evolved once but due to mating events has become invariant in group 1 and variable in group 2 T. b. gambiense.
A recent study of microsatellite multi-locus genotypes from many T. brucei isolates from across Africa has found that group 1 T. b. gambiense form a clade separate from T. b. brucei, T. b. rhodesiense and group 2 T. b. gambiense [29]. Group 2 T. b gambiense individuals formed a cluster positioned between T. b. brucei and group 1 T. b. gambiense, suggesting that group 2 T. b. gambiense may be a hybrid of group 1 and T. b. brucei and would perhaps share a mechanism for human infectivity. However, the T. b. brucei used in the study were mostly from East Africa and are not representative of West African T. b. brucei that are sympatric with the two T. b. gambiense groups. In order to resolve the relationships between the two groups of T. b gambiense and local T. b. brucei, we used a microsatellite genotyping approach to examine a collection of T. brucei isolates from Côte d’Ivoire [15], [16], [34]. The isolates have been characterized by the Blood Incubation Infectivity Test (BIIT) [35], [36] and defined as highly resistant (all of 5 rodents infected), intermediate or sub-resistant (1–4 rodents were infected) or sensitive (none of 5 rodents infected) (Table S1). They have also been typed as classical gambiense (group 1) or non-gambiense by isoenzyme or RFLP profiles [16], [21]. All of these samples were collected from a similar time period (1978–1983) and from a geographic area that contains both groups of T. b. gambiense. This collection of isolates allows the analysis of the sub-species genetics at this time point and the elucidation of the relationships between them, especially those between group 1 and 2 T. b. gambiense, which has implications for the evolution of human infectivity traits.
Materials and Methods
Ethical Statement
Ethical approval for the human derived isolates in this study have been previously published [16], [17].
Isolate Library
The collection of 43 T. brucei isolates used for population analysis was collected from a disease focus in Côte d’Ivoire that encompasses the townships of Gagnoa, Vavoua, Daloa and Bouaflé [16], [21] from a range of hosts, including humans, between 1978 and 1983 (Table 1) The townships are approximately 50km to 90km apart. Samples were obtained as purified DNA or blood spots from stabilates on FTA cards that were washed and prepared as per the manufacturer’s instructions before analysis (Whatman). The collection comprises both classical group 1 T. b. gambiense and group 2 T. b. gambiense as identified by isoenzymes and RFLPs [35], [36], and also non-human infective T. b. brucei determined by BIIT. A 5 rodent BIIT [37] was used to assess whether each isolate possessed the potential for human infectivity [11]. In addition to the main sample collection, the following reference isolates were also included: the group 1 T. b. gambiense genome strain MHOM/CI/86/DAL972 [27], [38], the 2 T. b gambiense strain STIB386 (MHOM/CI/78/TH114/78E) (also included in the main collection [39]) used in the T. b. gambiense genetic map [27], [38] and the comprehensively studied group 1 T. b. gambiense strain MHOM/CI/52/ELIANE were also included.
Table 1. Details of the isolates used in the study, including the isolate name, the host each strain was isolated from, the serum resistance profile resulting from a 5 rodent BIIT (R = highly resistant, I = intermediate or sub-resistant and S = sensitive) and the geographic location of the isolate when collected within Côte d’Ivoire.
| Isolate Name | Host | Serum Resistance | Location |
| MHOM/CI/78/TH112 | Human | S | Bouafle |
| MHOM/CI/78/TH114 | Human | R | Bouafle |
| MHOM/CI/78/TH126 | Human | I | Bouafle |
| MSUS/CI/78/TSW065 | Pig | S | Bouafle |
| MSUS/CI/78/TSW100 | Pig | I | Bouafle |
| MSUS/CI/78/TSW113 | Pig | I | Bouafle |
| MSUS/CI/78/TSW115A | Pig | R | Bouafle |
| MSUS/CI/78/TSW168 | Pig | I | Bouafle |
| MSUS/CI/78/TSW175 | Pig | R | Bouafle |
| MSUS/CI/78/TSW178 | Pig | S | Bouafle |
| MSUS/CI/78/TSW18 | Pig | I | Bouafle |
| MSUS/CI/78/TSW182 | Pig | I | Bouafle |
| MSUS/CI/78/TSW19 | Pig | I | Bouafle |
| MSUS/CI/78/TSW190 | Pig | I | Bouafle |
| MSUS/CI/78/TSW196 | Pig | S | Bouafle |
| MSUS/CI/78/TSW209 | Pig | I | Bouafle |
| MSUS/CI/78/TSW251 | Pig | S | Bouafle |
| MSUS/CI/78/TSW308 | Pig | I | Bouafle |
| MSUS/CI/78/TSW332 | Pig | S | Bouafle |
| MSUS/CI/78/TSW38 | Pig | S | Bouafle |
| MSUS/CI/78/TSW390 | Pig | S | Bouafle |
| MSUS/CI/78/TSW65 | Pig | S | Bouafle |
| MSUS/CI/78/TSW77 | Pig | I | Bouafle |
| MHOM/83/DAL587 | Human | R | Bouafle |
| MHOM/83/DAL598 | Human | R | Daloa |
| MHOM/83/DAL642 | Human | R | Daloa |
| MHOM/83/DAL645 | Human | R | Daloa |
| MHOM/83/DAL403 | Human | R | Daloa |
| MHOM/83/DAL633 | Human | R | Daloa |
| MHOM/CI/86/DAL972 | Human | R | Daloa |
| MHOM/CI/82/DAL654 | Human | R | Daloa |
| MHOM/CI/83/DAL596 | Human | R | Daloa |
| MHOM/CI/83/DAL607 | Human | R | Daloa |
| MHOM/CI/78/DAL069 | Human | R | Daloa |
| MHOM/CI/78/DAL072A | Human | R | Daloa |
| MHOM/83/DAL595 | Human | R | Gagnoa |
| MHOM/CI/83/DAL543 | Human | R | Vavoua |
| MHOM/CI/78/TH1 | Human | S | Vavoua |
| MSUS/CI/78/TSW155 | Pig | S | Vavoua |
| MSUS/CI/78/TSW158 | Pig | I | Vavoua |
| MSUS/CI/78/TSW187 | Pig | I | Vavoua |
| MHOM/83/DAL542 | Human | R | Vavoua |
| MHOM/84/DAL740 | Human | R | Vavoua |
| MHOM/CI/82/DAL494 | Human | R | Vavoua |
| MHOM/CI/52/ELIANE | Human | R | Côte d’Ivoire |
Genotyping
Samples were genotyped for population analysis using previously published nested PCR primers for eight microsatellite markers; Ch1/18, Ch1/D2/7, Ch2/PLC, Ch2/5, Ch5/JS2, Ch11/110, Ch3/IJ15/1 and Ch4/M12C12 [40]. The Ch4/M12C12 marker proved to be monomorphic for these samples and was excluded. Markers on the same chromosome are at opposite ends of the chromosome to each other and are unlikely to be linked [27]. All of the primers used to amplify the microsatellites have been previously identified and verified for both T. b. brucei and T. b. gambiense populations [41]. The PCR conditions for each nested reaction were 95°C 50 secs, 55°C 50 secs and 65°C 60 secs, for 30 cycles. PCR was performed in a total volume of 30 µl with the primers at a final concentration of 10 µM each and Taq polymerase final concentration of 0.25 units/µl (Thermo Scientific). If a sample was homozygous at a marker, it was repeated, otherwise all reactions were performed once. To determine allele size, one of the second round primers was tagged using either AFAM or HEX fluorescent dye to allow accurate sizing on a capillary ABI sequencer against ROX labeled size standards (Dundee Sequencing Unit). The size of the tagged PCR product was measured to within 2bp using the Peak Scanner® software package (Applied Biosystems). Each distinct allele size was given a number and the two alleles for each locus were catalogued for each isolate. This allowed the creation of a multi-locus genotype (MLG) for each isolate. In addition to the microsatellite MLG, the samples were further characterized using the previously published diagnostic PCR for the presence/absence of the TgSGP gene [42], [43] and an improved set of nested TgSGP primers that span the 5′ and 3′ regions of the gene [44]. The PCR conditions used for these primers were 95°C 50 secs, 55°C 50 secs and 65°C 120 secs, for 35 cycles. TgSGP PCR was performed in a total volume of 10 µl.
Population Analysis
The multi-locus genotype (MLG) of each isolate studied was used to create a dendrogram to infer relationships within the Côte d’Ivoire T. brucei population. Clustering calculator (http://www.biology.ualberta.ca/jbrzusto/cluster.php) was used to create a Phylip Drawtree string (analysed using neighbor-joining clustering and Canberra Distances). A bootstrap with 100 iterations was also generated. This Drawtree string was then converted into a dendrogram using Figtree software (http://tree.bio.ed.ac.uk/). Heterozygosity, Nei’s genetic distance and FST were calculated using the GenAlEx software package for Microsoft Excel [41]. Hardy-Weinberg calculations were also performed using GenAlEx and linkage disequilibrium calculations were performed using GenePop v4.0.10 [45]–[47] and LIAN v3.5 [47] software packages. The likelihood of replicated genotypes occurring due to sexual recombination was examined using MLGsim 2.0 [48], usingP sex values calculated for each replicated MLG compared to 10,000 simulations for a population of the same size. Additionally, a principal component analysis (PCA) of the genetic distance was performed on the three populations using the GenAlEx software package [41]. k-means clustering was calculated from the eigenvalues from the PCA using the kmean() function of the R software package [49]. Finally, a sub-structuring modeling analysis was performed on the MLG using the STRUCTURE software package [50]. The models were formulated assuming an admixture model and correlated allele frequencies and a burn-in value of 50,000 and replication value of 250,000 [27], [28]. Several estimates of population size (K) were investigated for K = 2 to 12. The most likely estimate for K was inferred using both the posterior probability of 5 repeat runs for each value of K [40], [51], [52] and an estimate of delta K [40].
Results
All studies of the population structure of group 1 T. b. gambiense to date have revealed foci to be largely clonal and relatively homogeneous [40]. Isoenzymes and RFLP analyses suggest that group 1 T. b. gambiense is genetically distinct from sympatric T. b. brucei and from group 2 T. b. gambiense. Here we have examined parasites isolated from both human patients and animals in four neighboring foci of infection in the Côte d’Ivoire in the late 1978 and 1983. The samples can be segregated into three different classes based on previous isoenzyme and RFLP studies and by the ability to resist lysis by human serum measured using the BIIT. Isolates that are invariably resistant to lysis and possess the classical isoenzymes of T. b. gambiense are classified as group 1 T. b. gambiense. A second group of parasites that also show resistance (either by BIIT or due to being isolated from a human host) but lack the isoenzyme markers characteristic of group 1 T. b. gambiense, are defined as non-gambiense or group 2 T. b. gambiense. Isoenzyme profiles of these isolates were highly variable [11]. The third group of isolates were sensitive to human serum in the BIIT and were not isolated from human patients; they were therefore classified as T. b. brucei and the isoenzyme profiles of these isolates were also highly variable [11].
TgSGP Genotyping
The cell surface receptor TgSGP has been proposed as a diagnostic marker of group 1 T. b. gambiense [52] and PCR targeting the 3′ end of the gene was recommended to test for the presence of the gene as a diagnostic for T. b. gambiense group 1 [52]. Unfortunately, several group 2 T. b. gambiense are also positive for the published PCR diagnostic test, limiting its use in foci with both groups of T. b. gambiense [40]. Further work has now shown that the 3′ end of the TgSGP gene is closely related to the VSG Tb10.v4.0178 and this VSG is likely the progenitor of this section of the TgSGP gene [47]. Tb10.v4.0178 is present in several strains, including T. b brucei, T. b. rhodesiense and group 2 T. b. gambiense isolates [27], [28], so there is a possibility of false positives when using the originally proposed primers. In our analysis using the original primers targeted to the 3′ end of the TgSGP gene [27], [28], 21 isolates were positive for the TgSGP gene diagnostic, of which, 18 were previously characterized as group 1 T. b. gambiense on the basis of isoenzymes and RFLPs. However, two isolates (MSUS/CI/78/TSW209 and MSUS/CI/78/TSW158) that would be putatively identified as group 2 T. b. gambiense on the basis of human serum resistance phenotype in the BIIT but non-classical isoenzyme profiles, also showed positive for the 3′ end of the TgSGP gene. Additionally, a serum sensitive isolate (MSUS/CI/78/TSW308) was also positive. However, when using nested PCR primers that span both the 5′ and 3′ regions of the gene, the TgSGP gene was only shown to be present in the 18 isolates previously identified as group 1 T. b. gambiense (Figure 1a). This would suggest that the three non-group 1 T. b. gambiense isolates are false positives and possess either VSG Tb10.v4.0178 or a closely related VSG. To investigate the relationships between the isolates from this disease focus, the isolates were further characterized using microsatellite genotyping.
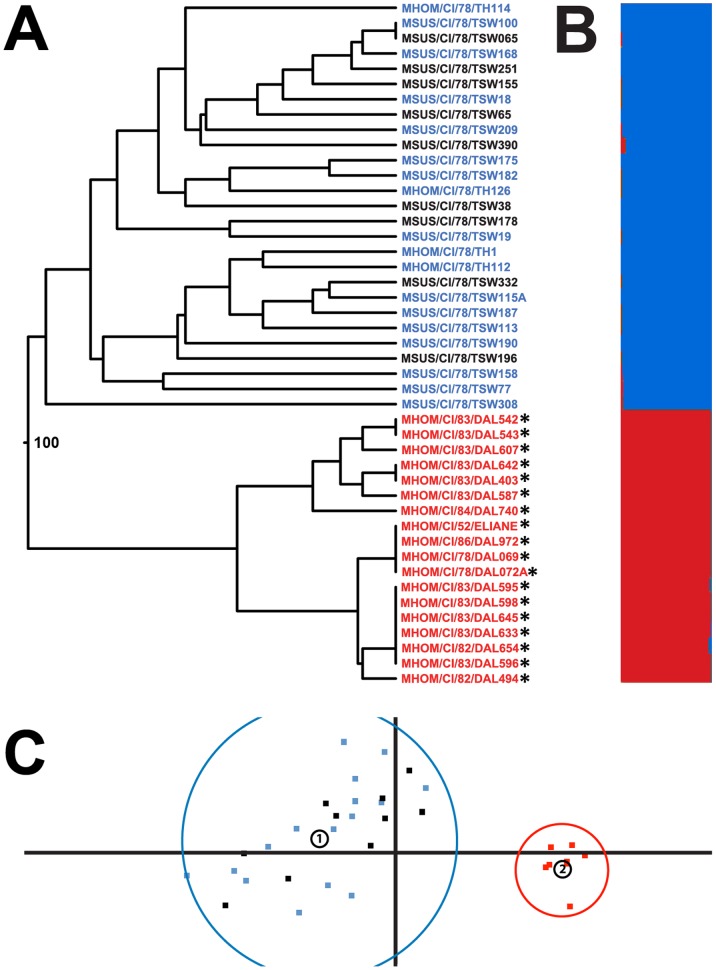
Figure 1. Genetic analysis of the Trypanosoma brucei population at the Côte d’ivoire focus.
a. Dendrogram of multi-locus genotypes (MLG) for the T. brucei isolates collected from several townships in Côte d’Ivoire, over the period of time 1978–1983 in addition to DAL972 and ELIANE. Bootstrap values from 100 iterations are indicated for branch nodes with a bootstrap value above 10. The presence of TgSGP using primers spanning the 5′ and 3′ ends is indicated by *. Isolates that displayed human serum resistance, the classical T. b. gambiense isoenzyme profile and possess TgSGP can be inferred to be group 1 T. b. gambiense (Red). Isolates that display a degree of resistance or were isolated from humans but did not possess the classical isoenzyme profile or TgSGP were determined to be group 2 T. b gambiense (Blue). Strains that exhibited no human serum resistance and were isolated from animals are most likely T. b. brucei (Black). b. Predicted structure of the Côte d'Ivoire T. brucei focus for the most likely population number (K = 2). The proportion of each population that an isolate is a member of is indicated by red and blue in the histogram. c. Principal component analysis (PCA) of the Côte d’Ivoire T. brucei isolates using a pair-wise genetic distance comparison between each isolates MLG. The x-axis explains 56.45% of the variability in the populations and the y-axis 13.98%, for a total of 70.45%. Isolates are colored as outlined in 1a. The circled numbers indicate the centroids of the two clusters identified by k-means analysis. The limits of these clusters are also indicated.
Microsatellite Genotyping
To investigate the relationships between the group 1 and 2 T. b. gambiense and T. b. brucei populations, the genotype of each of the 45 samples was determined using seven microsatellite markers. All seven markers are polymorphic, featuring several alleles at each locus (Table S1). For the entire sampled population, the majority of markers display a level of heterozygosity that is in in agreement with Hardy-Weinberg distribution (Table 2). Using these multi-locus genotypes (MLG), a dendrogram of relatedness was created to visualize the relationship between isolates in the study (figure 1a). The dendrogram revealed a discrete population containing only the 18 isolates characterized as group 1 T. b. gambiense by isoenzymes, RFLPs, BIIT and the presence of TgSGP. This clade also included the genome reference strain MHOM/CI/86/DAL972 and the well characterized group 1 strain MHOM/CI/52/ELIANE. The bootstrap value (100) indicates a high degree of support for the node that separates this group. The remaining 27 isolates formed a separate cluster distinct from the group 1 T. b. gambiense, containing both serum sensitive T. b. brucei (n = 9) and serum resistant group 2 T. b. gambiense isolates (n = 18). There is little bootstrap support for any of the branching nodes within this clade, suggesting this cluster represents a single population and that T. b. brucei and group 2 T. b. gambiense are largely indistinguishable from each other.
Table 2. Polymorphisms and heterozygosity of T. brucei at the Côte d’Ivoire focus (N = 45).
| Locus | Alleles | ObservedHeterozygosity | Expected Heterozygosity |
| Ch5/JS2 | 8 | 0.667 | 0.784 |
| Ch11/110 | 3 | 0.200 | 0.182 |
| Ch11/51 | 3 | 0.356 | 0.299 |
| Ch1/18 | 8 | 0.867 | 0.656 |
| Ch1/D2/7 | 8 | 0.267 | 0.709 |
| Ch2/PLC | 5 | 0.200 | 0.424 |
| Ch3/IJ15/1 | 6 | 0.359 | 0.363 |
Principal components analysis (PCA) was performed using the genetic distance of the isolates to create a multi-dimensional comparison (figure 1c). PCA identifies the independent factors from a data set that explain the maximum amount of correlation. Plotting these factors allows the structure of a population to be visualized. Further structure could be inferred by utilizing k-mean clustering and the eigenvalues from the PCA. This identified two distinct clusters; one containing T. b. brucei and group 2 T. b. gambiense populations and a second containing only isolates previously identified as group 1 T. b. gambiense. When analyzed by village, there is no evidence for any sub-structuring in the group 2 and T. b. brucei populations based on geography (data not shown) suggesting that the parasites within the townships are part of the same transmission cycle. Also, while not included in the analysis due to low sample numbers, similar genotypes are present in group 2 T. b. gambiense and T. b. brucei isolates from neighboring Burkina Faso, suggesting that there is one meta-population spanning the two countries (Table S1).
In order to further examine the population structure of the Côte d’Ivoire focus, STRUCTURE modeling was performed using the MLG data set. The most likely population number (K) was found to be 2 when estimated using both posterior probabilities and delta K (figure 1b) [47]. Isolates are either predominantly a member of a population containing only group 1 T. b. gambiense or a population containing group 2 T. b. gambiense and T. b. brucei. The removal of the group 1 isolates and then re-modeling with STRUCTURE did not indicate any further structure in the T. b. brucei/group 2 T. b. gambiense isolates (up to K = 12, data not shown).
Population Analysis
In order to determine the population structure within each clearly distinct cluster as defined by the dendrogram, PCA and STRUCTURE, each cluster was examined separately. Cluster 1 contained the 18 isolates that had previously been identified as group 1 T. b. gambiense isolates and have subsequently been shown to all possess TgSGP. Cluster 2 contained all other isolates (n = 27), i.e. serum sensitive T. b. brucei and the non-classical gambiense isolates that showed any degree of serum resistance and could be termed group 2 T. b. gambiense. To further examine the differences between the sub-species and groups, cluster 2 was sub-divided into those isolates that previously showed any degree of resistance in a BIIT (putative group 2 T. b. gambiense, n = 18) and those that were highly sensitive and could be described as T. b. brucei isolates (n = 9).
The relationship between these populations was analyzed using two statistical tests, Nei’s genetic distance and FST. Both Nei’s genetic distance and the FST statistic indicate that both populations within cluster 2 are distinct from cluster 1 and that within cluster 2, there is very little genetic difference between T. b. brucei and group 2 T. b. gambiense populations (Tables 3 & 4). As there is so little difference between populations, subsequent analysis considered the cluster 2 as a single population.
Table 3. Nei’s genetic distance between the populations of group 1 and 2 T. b. gambiense and T. b. brucei at the Côte d’Ivoire focus.
| T. b. brucei (N = 9) | Gp 2 T. b. gambiense (N = 16) | Gp 1 T. b. gambiense (N = 20) | |
| T. b. brucei | 0.000 | – | – |
| Gp 2 T. b. gambiense | 0.026 | 0.000 | – |
| Gp 1 T. b. gambiense | 0.357 | 0.321 | 0.000 |
Table 4. Fst proportion indicating genetic distance between the populations of group 1 and 2 T. b. gambiense and T. b. brucei at the Côte d’Ivoire focus.
| T. b. brucei (N = 9) | Gp 2 T. b. gambiense (N = 16) | Gp 1 T. b. gambiense (N = 20) | |
| T. b. brucei | 0.000 | – | – |
| Gp 2 T. b. gambiense | 0.138 | 0.000 | – |
| Gp 1 T. b. gambiense | 0.277 | 0.264 | 0.000 |
If random mating is occurring within the Côte d’Ivoire population then the number of heterozygous genotypes within the population should be a predictable proportion based on the number and frequency of alleles present in the population, in agreement with Hardy-Weinberg expectations. Analysis of the genotype frequencies in cluster 2 (the combined T. b. brucei and group 2 T. b. gambiense isolates) revealed that none of the seven markers significantly deviated from Hardy-Weinberg equilibrium (Table 5). These data are consistent with the hypothesis that some degree of mating is occurring within this cluster i.e. within and between the group 2 T. b. gambiense and T. b. brucei. In contrast, analysis of markers for cluster 1 shows that the group 1 T. b. gambiense population display a strong deviation from Hardy-Weinberg, with markers being either monomorphic or possessing heterozygote excess strongly disagreeing with the Hardy-Weinberg hypothesis (Table 6).
Table 5. Hardy-Weinberg analysis for the combined population of group 2 T. b. gambiense and T. b. brucei at the Côte d’Ivoire focus (N = 25).
| Locus | DF | ?2 | Probability | Significance |
| Ch5/JS2 | 15 | 21.021 | 0.136 | Not Significant |
| Ch11/110 | 3 | 1.080 | 0.782 | Not Significant |
| Ch11/51 | 3 | 0.173 | 0.982 | Not Significant |
| Ch1/18 | 15 | 18.798 | 0.223 | Not Significant |
| Ch1/D2/7 | 6 | 6.762 | 0.343 | Not Significant |
| Ch2/PLC | 10 | 15.693 | 0.109 | Not Significant |
| Ch3/IJ15/1 | 6 | 6.832 | 0.559 | Not Significant |
Table 6. Hardy-Weinberg analysis of group 1 T. b. gambiense isolates at the Côte d’Ivoire focus (N = 20).
| Locus | DF | ?2 | Probability | Significance |
| Ch5/JS2 | 3 | 18.000 | 0.000 | P<0.001 |
| Ch11/110 | Monomorphic | – | – | – |
| Ch11/51 | 1 | 4.500 | 0.034 | P<0.05 |
| Ch1/18 | 1 | 18.000 | 0.000 | P<0.001 |
| Ch1/D2/7 | 6 | 18.147 | 0.006 | P<0.001 |
| Ch2/PLC | 1 | 0.015 | 0.904 | Not Significant |
| Ch3/IJ15/1 | Monomorphic | – | – | – |
A second method to assess the degree of mating within cluster 2 is to estimate the amount of linkage disequilibrium in allele and genotype frequencies. The presence of mating in a population will cause alleles at unlinked loci to be inherited in a randomly assorted manner, leading to linkage equilibrium, while a population with little mating will exhibit linkage disequilibrium. Analysis of linkage equilibrium revealed that the overwhelming majority of allele combinations do not show significant linkage disequilibrium in cluster 2 (Table 7). Additionally, both the T. b. brucei and group 2 T. b. gambiense populations exhibit a standardized Index of Association (IA) that tends towards zero. IA s a function of the rate of recombination and is zero for complete linkage equilibrium, so our data are consistent with linkage equilibrium existing in these populations (Table 8). Linkage equilibrium is also exhibited when the T. b. brucei and group 2 populations are analyzed together, suggesting linkage equilibrium between the populations. These data, combined with the Hardy-Weinberg analysis suggest that there is a degree of mating and re-assortment both between and within the T. b. brucei and group 2 T. b. gambiense populations. The group 1 T. b. gambiense alleles are largely monomorphic or uninformative so a linkage disequilibrium study is not possible with this population. However the low number of alleles and limited variability correlates with studies of other foci, suggesting that group 1 T. b. gambiense possesses a clonal population structure that is separate from that of the group 2 T. b. gambiense and T. b. brucei population [27], [28]. The low variability in genotypes of the group 1 T. b. gambiense isolates were tested for clonality using MLGsim. This indicated that all observed P sex values for each MLG expansion were highly significant at the p<0.01 threshold, suggesting that all of the samples are clonal. Conversely, the same analysis on the T. b. brucei and group 2 T. b. gambiense populations shows that most genotypes have likely arisen due to sexual recombination and only two of the samples possess possible clonal genotypes at the p<0.01 significance threshold, MSUS/CI/78/TSW158 and MSUS/CI/78/TSW175.
Table 7. Linkage disequilibrium for each genetic marker (statistically significant disequilibrium highlighted in bold) between pair wise polymorphic loci in the combined T. b. brucei and group 2 T. b. gambiense population at the Côte d’Ivoire focus (N = 25).
| Locus 1 | Locus 2 | Probability | Standard Error |
| Ch5/JS2 | Ch11/110 | 0.345 | 0.014 |
| Ch5/JS2 | Ch11/51 | 0.053 | 0.006 |
| Ch11/110 | Ch11/51 | 1.000 | 0.000 |
| Ch5/JS2 | Ch1/18 | 0.299 | 0.029 |
| Ch11/110 | Ch1/18 | 0.136 | 0.008 |
| Ch11/51 | Ch1/18 | 0.410 | 0.012 |
| Ch5/JS2 | Ch1/D2/7 | 0.172 | 0.019 |
| Ch11/110 | Ch1/D2/7 | 0.351 | 0.009 |
| Ch11/51 | Ch1/D2/7 | 1.000 | 0.000 |
| Ch1/18 | Ch1/D2/7 | 0.040 | 0.008 |
| Ch5/JS2 | Ch2/PLC | 0.081 | 0.012 |
| Ch11/110 | Ch2/PLC | 0.648 | 0.003 |
| Ch11/51 | Ch2/PLC | 0.573 | 0.018 |
| Ch1/18 | Ch2/PLC | 0.007 | 0.032 |
| Ch1/D2/7 | Ch2/PLC | 0.293 | 0.010 |
| Ch5/JS2 | Ch3/IJ15/1 | 0.373 | 0.015 |
| Ch11/110 | Ch3/IJ15/1 | 0.237 | 0.022 |
| Ch11/51 | Ch3/IJ15/1 | 0.363 | 0.021 |
| Ch1/18 | Ch3/IJ15/1 | 0.721 | 0.023 |
| Ch1/D2/7 | Ch3/IJ15/1 | 0.274 | 0.011 |
| Ch2/PLC | Ch3/IJ15/1 | 0.525 | 0.021 |
The data were analyzed using the Genepop 4.0 software package.
Table 8. Standardised indices of association (IA) for the populations of group 1 and 2 T. b. gambiense and T. b. brucei at the Côte d’Ivoire focus.
| Population analyzed | VD- | VE- | IA |
| Total Population (N = 45) | 2.739 | 1.476 | 0.143 |
| Combined T. b. brucei & gp 2 T. b. gambiense (N = 25) | 1.686 | 1.311 | 0.048 |
| Gp 2 T. b. gambiense (N = 16) | 1.716 | 1.227 | 0.067 |
| T. b. brucei (N = 9) | 1.441 | 1.360 | 0.099 |
| Gp 1 T. b. gambiense (N = 20) | – | – | ∞ |
Data were analyzed using the LIAN software package to test the null hypothesis of linkage equilibrium. This null hypothesis is that the variance of loci that do not show linkage (VD –) is equal to the expected variance of loci modelled under linkage equilibrium (VE –).
It would appear from these data that the T. b. brucei and group 2 T. b. gambiense populations exhibit some degree of mating, both within and between sub-species. The group 1 T. b. gambiense population is clonal and distinct from the sympatric T. b. brucei and group 2 T. b gambiense populations.
Discussion
These data reinforce studies from other foci that populations of group 1 T. b. gambiense are likely clonal [27], [28]. The reasons for the clonality of group 1 T. b. gambiense are unclear as the genes necessary for meiosis are present [37] and are expressed during tsetse infection (Peacock and Gibson, unpublished data). Another possible reason for the low frequency of mating observed in group 1 T. b. gambiense is that, due to the limited number of genotypes present, the chances of a mixed genotype infection occurring in the tsetse vector is unlikely. This precludes any mating other than selfing, which has been observed in laboratory crosses for T. b. brucei [53]. However a high frequency of selfing would result in a population with an excess of homozygotes rather than the observed excess of heterozygotes, and is therefore unlikely.
The observed homogeneous population of group 1 T. b. gambiense could also be due to a bottleneck effect that occurred in the past or if group 1 T. b. gambiense underwent clonal expansion, possibly after it gained the human infectivity trait. Despite questions as to how the clonal population structure has developed, these data show that the group 1 T. b. gambiense genome reference strain MHOM/CI/86/DAL972 is clearly a good representative of group 1 strains from this area. Previous studies have shown that the MLG in the populations of various group 1 T. b. gambiense foci are significantly different from each other [27] and examination of the microsatellite alleles in this study with previous studies indicates that there are few shared alleles at each marker. Indeed, the necessity to include new markers specific to this study reinforces the fact that the focus possesses a different clonal population of group 1 T. b. gambiense to that found in other foci.
This study has also shown that the STIB386 strain (MHOM/CI/78/TH114) used in genetic mapping studies [54] is a good representative for Côte d’Ivoire group 2 T. b. gambiense in that it is distinct from the group 1 T. b. gambiense population and clusters with other group 2 T. b. gambiense strains. Previous studies have suggested that group 2 T. b. gambiense are more varied than group 1 and more similar to T. b. brucei [16], [19], [22]–[26]. The data presented here show that not only was there a large and genetically varied breeding population of group 2 T. b. gambiense at the Côte d’Ivoire focus, that these individuals were almost certainly breeding with local T. b. brucei. The T. b. brucei and group 2 T. b. gambiense populations are very similar, suggesting that they form one breeding population. There is a suggestion that this breeding population extends to neighboring Burkina Faso, as similar microsatellite alleles are found in both countries. However, this may be due to the seasonal, economic migration of the Mossi people between the two nations [55] that transports parasites rather then a large contiguous population. Nevertheless, group 2 T. b. gambiense can be seen as an extended host range variant T. b. brucei.
The degree to which mating occurs in a population is an important consideration due to the process acting as a major source of variation. While the human infectivity trait present in group 2 T. b. gambiense may have evolved once, it is now present in a wide range of genetic backgrounds. This is in contrast to group 1 T. b. gambiense which has limited genetic diversity at each discrete disease focus and hence a lower capacity to evolve. The genetic backgrounds of the group 2 T. b. gambiense infectivity trait may be in constant flux due to evidence suggesting the presence of mating within the population. This may go someway to explaining the emergence of a “new” group of T. b. gambiense in Côte d’Ivoire [56]. This new human infective group has been claimed as distinct from both group 1 T. b. gambiense and group 2 T. b. gambiense from the early 1980s. However, based on the heterogeneity and propensity for mating displayed by group 2 in our study, it is likely that this new group belongs to group 2, with the group 2 human serum resistance mechanism on a new genetic background. Unfortunately, the human serum resistance profile of the new group has not yet been described and it is unknown whether it also displays a variable, non-SRA mediated resistance [56]. Elucidation of the resistance mechanism for group 2 T. b. gambiense would also allow this hypothesis to be investigated.
A previous microsatellite study has indicated that although group 2 T. b. gambiense from Côte d’Ivoire is related to T. b. brucei, it also clusters close to group 1 T. b. gambiense [29]. This has led to the suggestion it is a hybrid of T. b. brucei and group 1 T. b. gambiense. However, the T. b. brucei used in the study were mostly from East Africa and are not be representative of T. b. brucei in Côte d’Ivoire. Our data clearly shows that the West African T. b. brucei and group 2 T. b. gambiense populations are largely indistinguishable and that group 2 is not closer to group 1 T. b. gambiense than local T. b. brucei. There is no evidence to support the hypothesis that group 2 T. b. gambiense is a hybrid of group 1 T. b. gambiense and T. b. brucei and that this sub-species has evolved separately. We have also shown that TgSGP is not present in the local group 2 T. b. gambiense or T. b. brucei populations by using nested primers that span the 5′ and 3′ ends of the gene. While this is counter to other published data [40], the primers described in the original diagnostic PCR are likely to show false positives due to similarity to the 3′ end of TgSGP and the VSG Tb10.v4.0178 [52]. This observation adds further weight to the hypothesis that there is no mating between group 1 or group 2 T. b. gambiense, and that group 2 has not emerged as a result of hybridizing event.
The study presented here, in conjunction with other published data, suggests that human infectivity has arisen on at least three occasions (with some evidence of a fourth variant). In East Africa the evolution of SRA and the high levels of mating occurring there have allowed the human infectivity trait to spread through the T. brucei population, generating the T. b. rhodesiense sub-species [57], [58]. However, there are also human infective trypanosomes in this area that do not possess SRA, suggesting a second novel mechanism [3], [59]. In West and Central Africa, group 1 T. b. gambiense is the dominant cause of human African sleeping sickness. This group of parasites has evolved a constitutively expressed human serum resistance mechanism that does not depend on SRA. After evolution of this trait, the sub-species appears to have expanded clonally in the human population. Just as in East Africa, there is also a human infective T. brucei population separate from the dominant form that appears to have a novel resistance mechanism. This group 2 T. b. gambiense population appears to be a host range variant of T. b. brucei and the human infectivity trait it possesses is freely transmitted among the local T. b. brucei population. The presence of mating amongst group 2 T. b. gambiense and T. b. brucei strains makes evolution of new traits more likely and may allow the human infectivity trait to be combined with new phenotypes that may increase its virulence. It also allows the human resistance trait to persist in the local T. brucei animal reservoir through genetic transfer. Understanding whether the inter-sub-species mating and transfer of human infectivity traits exhibited in Côte d’Ivoire can also occur in other foci is becoming more pressing. In Uganda, the T. b. gambiense and T. b. rhodesiense foci are moving closer together and their ranges will begin to overlap [60], [61]. Fortunately the possibility of mating between group 1 T. b. gambiense and T. b. rhodesiense appears unlikely due to the clonal nature of group 1 T. b gambiense [27], [28], and while T. b. rhodesiense mating with group 2 T. b. gambiense is certainly possible [33], the ranges of these two diseases are not converging at present. Taken together, all of these data indicate that T. brucei has a high zoonotic potential despite specific trypanolytic countermeasures that have been inherited by humans.
Supporting Information
Overview of sample origin, host, human serum resistance phenotype and alleles present for each microsatellite marker. The presence absence of TgSGP is also indicated, whether it possesses the full gene (1) or the progenitor VSG (2).
(XLSX)
Funding Statement
PC was a BBSRC research student, CD was a Wellcome funded research student. MT, AM and AC were funded by a Wellcome Project Grant. The Wellcome Trust Centre for Molecular Parasitology is supported by core funding from the Wellcome Trust [085349]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pays E, Vanhollebeke B (2009) Human innate immunity against African trypanosomes. Current Opinion in Immunology 21: 493–498. [DOI] [PubMed] [Google Scholar]
- 2. Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, et al. (1998) A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense . Cell 95: 839–846. [DOI] [PubMed] [Google Scholar]
- 3. De Greef C, Chimfwembe E, Kihang'a Wabacha J, Bajyana Songa E, Hamers R (1992) Only the serum-resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann Soc Belg Med Trop 72 Suppl 113–21. [PubMed] [Google Scholar]
- 4. Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, et al. (2002) The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense . Am J Trop Med Hyg 67: 684–690. [DOI] [PubMed] [Google Scholar]
- 5. Turner C, McLellan S, Lindergard L, Bisoni L, Tait A, et al. (2004) Human infectivity trait in Trypanosoma brucei: stability, heritability and relationship to sra expression. Parasitology 129: 445–454. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization (2006) Weekly epidemiological record: relevé épidémiologique hebdomadaire.
- 7. Simarro PP, Diarra A, Postigo JAR, Franco JR, Jannin JG (2011) The Human African Trypanosomiasis Control and Surveillance Programme of the World Health Organization 2000–2009: The Way Forward. PLoS Negl Trop Dis 5: e1007 doi:10.1371/journal.pntd.0001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kieft R, Capewell P, TURNER CMR, Veitch NJ, MacLeod A, et al. (2010) Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proceedings of the National Academy of Sciences 107: 16137–16141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeJesus E, Kieft R, Albright B, Stephens NA, Hajduk SL (n.d.) A Single Amino Acid Substitution in the Trypanosoma brucei gambiense Haptoglobin-Hemoglobin Receptor Abolishes TLF-1 Binding. PLoS Pathogens: e1003317. [DOI] [PMC free article] [PubMed]
- 10.Higgins MK, Tkachenko O, Brown A (2013) Structure of the trypanosome haptoglobin–hemoglobin receptor and implications for nutrient uptake and innate immunity. Proceedings of the National Academy of Sciences. doi:−10.1073/pnas.1214943110/−/DCSupplemental [DOI] [PMC free article] [PubMed]
- 11. Bullard W, Kieft R, Capewell P, Veitch NJ, Macleod A, et al. (2012) Haptoglobin-hemoglobin receptor independent killing of African trypanosomes by human serum. Virulence 3: 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symula RE, Beadell JS, Sistrom M (2012) Trypanosoma brucei gambiense Group 1 Is Distinguished by a Unique Amino Acid Substitution in the HpHb Receptor Implicated in Human Serum Resistance. PLoS Neglected … 6: e1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gibson W (1986) Will the real Trypanosoma b. gambiense please stand up. Parasitol Today 2: 255–257. [DOI] [PubMed] [Google Scholar]
- 14. Gibson W, Mehlitz D, Lanham SM (1978) The identification of Trypanosoma brucei gambiense in Liberian pigs and dogs by isoenzymes and by resistance to human plasma. Tropenmedizin und Parasitologie 29: 335–345. [PubMed] [Google Scholar]
- 15. Felgner P, Brinkmann U, Zillmann U, Mehlitz D (1981) Epidemiological studies on the animal reservoir of gambiense sleeping sickness. Part II. Parasitological and immunodiagnostic examination of the human population. Tropenmedizin und Parasitologie 32: 134–140. [PubMed] [Google Scholar]
- 16. Mehlitz D, Zillmann U, Scott CM (1982) Epidemiological studies on the animal reservoir of Gambiense sleeping sickness. Part III. Characterization of trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmedizin und Parasitologie 33: 113–118. [PubMed] [Google Scholar]
- 17.Mehlitz D (1986) Le réservoir animal de la maladie du sommeil á Trypanosoma brucei gambiense. Études et Synthèses de I'IEMVT, Deutsche Gesellschaft für Technische Zusammenarbeit (GTZ).
- 18. Mehlitz D (1977) The behaviour in the blood incubation infectivity test of four Trypanozoon strains isolated from pigs in Liberia. Trans R Soc Trop Med Hyg 71: 86. [DOI] [PubMed] [Google Scholar]
- 19. Zillmann U, Mehlitz D, Sachs R (1984) Identity of Trypanozoon stocks isolated from man and a domestic dog in Liberia. Tropenmedizin und Parasitologie 35: 105–108. [PubMed] [Google Scholar]
- 20. Capewell P, Veitch NJ, Turner CMR, Raper J, Berriman M, et al. (2011) Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Negl Trop Dis 5: e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paindavoine P, Zampetti-Bosseler F, Coquelet H, Pays E, Steinert M (1989) Different allele frequencies in Trypanosoma brucei brucei and Trypanosoma brucei gambiense populations. Mol Biochem Parasitol 32: 61–71. [DOI] [PubMed] [Google Scholar]
- 22. Stevens JR, Tibyrenc M (1996) Trypanosoma brucei sl: evolution, linkage and the clonality debate. Parasitology 112: 481–488. [DOI] [PubMed] [Google Scholar]
- 23. Mathieu-Daude F, Bicart-See A (1994) Identification of Trypanosoma brucei gambiense group I by a specific kinetoplast DNA probe. Am J Trop Med Hyg 50: 13–19. [DOI] [PubMed] [Google Scholar]
- 24. Tait A, Babiker EA, Le Ray D (1984) Enzyme variation in Trypanosoma brucei ssp. I Evidence for the sub-speciation of Trypanosoma brucei gambiense . Parasitology 89: 311–326. [DOI] [PubMed] [Google Scholar]
- 25. Godfrey DG, Kilgour V (1976) Enzyme electrophoresis in characterizing the causative organism of Gambian trypanosomiasis. Trans R Soc Trop Med Hyg 70: 219–224. [DOI] [PubMed] [Google Scholar]
- 26. Gibson W, Marshall DC, Godfrey DG (1980) Numerical Analysis of Enzyme Polymorphism: A New Approach to the Epidemiology and Taxonomy of Trypanosomes of the Subgenus Trypanozoon . Advances in Parasitology 18: 175–246. [DOI] [PubMed] [Google Scholar]
- 27. Morrison LJ, Tait A, McCormack G, Sweeney L, Black A, et al. (2008) Trypanosoma brucei gambiense Type 1 populations from human patients are clonal and display geographical genetic differentiation. Infection, Genetics and Evolution 8: 847–854. [DOI] [PubMed] [Google Scholar]
- 28. Koffi M, De Meeûs T, Bucheton B, Solano P, Camara M, et al. (2009) Population genetics of Trypanosoma brucei gambiense, the agent of sleeping sickness in Western Africa Vol. 106: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balmer O, Beadell JS, Gibson W, Caccone A (2011) Phylogeography and taxonomy of Trypanosoma brucei . PLoS Negl Trop Dis 5: e961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hide G, Cattand P, LeRay D, Barry JD, Tait A (1990) The identification of Trypanosoma brucei subspecies using repetitive DNA sequences. Mol Biochem Parasitol 39: 213–225. [DOI] [PubMed] [Google Scholar]
- 31. Turner CMR, Sternberg J, Buchanan N, Smith E, Hide G, et al. (1990) Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology 106: 209–214. [DOI] [PubMed] [Google Scholar]
- 32. Jenni L, Marti S, Schweizer J, Betschart B, Le Page RW, et al. (1986) Hybrid formation between African trypanosomes during cyclical transmission. Nature 322: 173–175. [DOI] [PubMed] [Google Scholar]
- 33. Gibson W, Winters K, Mizen G, Kearns J, Bailey M (1997) Intraclonal mating in Trypanosoma brucei is associated with out-crossing. Microbiology 143: 909–920. [DOI] [PubMed] [Google Scholar]
- 34. Mehlitz D, Brinkmann U, Haller L (1981) Epidemiological studies on the animal reservoir of Gambiense sleeping sickness. Part I. Review of literature and description of the study areas. Tropenmedizin und Parasitologie 32: 129–133. [PubMed] [Google Scholar]
- 35. Rickman LR, Robson J (1970) The blood incubation infectivity test: a simple test which may serve to distinguish Trypanosoma brucei from T. rhodesiense . Bull World Health Organ 42: 650–651. [PMC free article] [PubMed] [Google Scholar]
- 36.Mehlitz D (1978) Untersuchungen zur Empfänglichkeit von Mastomys natalensis für Trypanosoma (Trypanozoon) brucei gambiense. Tropenmed Parasit: 101–107. [PubMed]
- 37. Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, et al. (2010) The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl Trop Dis 4: e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Macleod A, Tweedie A, McLellan S, Taylor S, Cooper A, et al. (2005) Allelic segregation and independent assortment in T. brucei crosses: Proof that the genetic system is Mendelian and involves meiosis. Mol Biochem Parasitol 143: 12–19. [DOI] [PubMed] [Google Scholar]
- 39. MacLeod A (2005) The genetic map and comparative analysis with the physical map of Trypanosoma brucei . Nucleic Acids Res 33: 6688–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radwanska M, Claes F, Magez S, Magnus E, Pérez-Morga D, et al. (2002) Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense . Am J Trop Med Hyg 67: 289–295. [DOI] [PubMed] [Google Scholar]
- 41. Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- 43. Rousset F (2008) genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8: 103–106. [DOI] [PubMed] [Google Scholar]
- 44. Haubold B, Hudson RR (2000) LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- 45. Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stenberg P, Lundmark M, Saura A (2003) mlgsim: a program for detecting clones using a simulation approach. Mol Ecol Notes 3: 329–331 doi:10.1046/j.1471-8286.2003.00408.x [Google Scholar]
- 49. Ihaka R, Gentleman R (1996) R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics 5: 299–314 doi:10.1080/10618600.1996.10474713 [Google Scholar]
- 50. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 51. Berberof M, Pérez-Morga D, Pays E (2001) A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense . Mol Biochem Parasitol 113: 127–138. [DOI] [PubMed] [Google Scholar]
- 52. Gibson W, Nemetschke L, Ndung'u J (2010) Conserved sequence of the TgSGP gene in Group 1 Trypanosoma brucei gambiense . Infection, Genetics and Evolution 10: 453–458. [DOI] [PubMed] [Google Scholar]
- 53. Peacock L, Ferris V, Bailey M, Gibson W (2008) Fly transmission and mating of Trypanosoma brucei brucei strain 427. Mol Biochem Parasitol 160: 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cooper A, Tait A, Sweeney L, Tweedie A, Morrison L, et al. (2008) Genetic analysis of the human infective trypanosome Trypanosoma brucei gambiense: chromosomal segregation, crossing over, and the construction of a genetic map. Genome Biol 9: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell SS, Jacobsen K, Stanley WD (1990) International migration and development in Sub-Saharan Africa. Cambridge, MA, USA: World Bank Discussion Papers.
- 56. Jamonneau V, Ravel S, Garcia A, Koffi M (2004) Characterization of Trypanosoma brucei sl infecting asymptomatic sleeping-sickness patients in Cote d'Ivoire: a new genetic group? Ann Trop Med Parasitol 98: 329–337. [DOI] [PubMed] [Google Scholar]
- 57. MacLeod A, Tweedie A, Welburn SC, Maudlin I, Turner CM, et al. (2000) Minisatellite marker analysis of Trypanosoma brucei: reconciliation of clonal, panmictic, and epidemic population genetic structures. Proceedings of the National Academy of Sciences 97: 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tait A, Barry JD, Wink R, Sanderson A (1985) Enzyme variation in T. brucei ssp II. Evidence for T. b. rhodesiense being a set of variants of T. b. brucei . … 90: 89–100. [DOI] [PubMed] [Google Scholar]
- 59. Enyaru JCK, Matovu E, Nerima B, Akol M, Sebikali C (2006) Detection of T.b. rhodesiense trypanosomes in humans and domestic animals in south east Uganda by amplification of serum resistance-associated gene. Ann N Y Acad Sci 1081: 311–319. [DOI] [PubMed] [Google Scholar]
- 60. Picozzi K (2005) Sleeping sickness in Uganda: a thin line between two fatal diseases. BMJ 331: 1238–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fèvre EM, Picozzi K, Fyfe J, Waiswa C, Odiit M, et al. (2005) A burgeoning epidemic of sleeping sickness in Uganda. Lancet 366: 745–747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of sample origin, host, human serum resistance phenotype and alleles present for each microsatellite marker. The presence absence of TgSGP is also indicated, whether it possesses the full gene (1) or the progenitor VSG (2).
(XLSX)