Abstract
Using a commercial protein expression system, we sought the crucial elements and conditions for the expression of proteins with genetically encoded unnatural amino acids. By identifying the most important translational components, we were able to increase suppression efficiency to 55% and to increase mutant protein yields to levels higher than achieved with wild type expression (120%), reaching over 500 µg/mL of translated protein (comprising 25 µg in 50 µL of reaction mixture). To our knowledge, these results are the highest obtained for both in vivo and in vitro systems. We also demonstrated that efficiency of nonsense suppression depends greatly on the nucleotide following the stop codon. Insights gained in this thorough analysis could prove useful for augmenting in vivo expression levels as well.
Introduction
The methodology based on unnatural amino acids (UAAs) incorporation into desired loci of the protein of interest is widely used for understanding protein structure-function relationships, investigating protein-based biological processes, and generating proteins and organisms with new properties [1]. Over the past two decades, the most established methods to site-specifically incorporate UAA in vivo were based on genetic code expansion. This is accomplished by supplying organisms with a non-endogenous aminoacyl-tRNA synthetase/tRNA pair, referred to as an orthogonal pair, that directs site-specific incorporation of UAA in response to a unique codon [2]. The orthogonal aminoacyl-tRNA synthetase (aaRS) aminoacylates a cognate orthogonal tRNA (but no other cellular tRNAs) with UAA. The orthogonal tRNA is a substrate for the orthogonal aaRS but is not aminoacylated by any endogenous aaRS [3]. The orthogonal aaRS/tRNA pair should, however, be compatible with the translational machinery of the host cell. The first orthogonal aaRS/tRNA pair used in Escherichia coli originated from the archaeon, Methanocaldococcus jannaschii, and was generated from the tyrosyl-tRNA synthetase and its cognate tRNA pair (MjTyrRS/tRNATyr) [4]. A unique codon is required to specify the UAA, and two main strategies are generally used: nonsense suppression, i.e. UAA incorporation in response to the least used stop codon recognized by a specific suppressor tRNA [5], [6]; and frame-shift suppression based on the application of four- or five-base extended codons and cognate suppressors [7]–[9].
Nowadays, genetic code expansion in E. coli using the amber suppression strategy and evolved variants of orthogonal M. jannaschii TyrRS/MjtRNATyr CUA is considered to be the most established and robust methodology to site-specifically incorporate UAAs. However, this methodology has not been reported to achieve high protein yields. Suppression efficiency depends greatly on the degree of orthogonal tRNA compatibility with the translational apparatus of host cell. The reasons for such an incompatibility are low affinity of the elongation factor Tu (EF-Tu) to UAA-charged MjtRNACUA derived from archeal tRNATyr [10] or its inability to recognize and deliver orthogonal tRNA to the ribosomal A-site [11]. On the other hand, suppression efficiency can be interrupted by the release factors, RF1 and RF2, which are responsible for the release of the growing polypeptide chain from the ribosome. RF functionality depends on the particular stop codon encoded in the mRNA sequence, as well as the fourth base following the stop codon [12], [13]. Recently, tremendous efforts were geared towards the production of greater yields of recombinant proteins containing UAAs, including design of specific vectors encoding multiple copies of MjtRNACUA [14], identification of newly optimized suppressor tRNACUA opt with modified T-stem [15]–[17], selection of the orthogonal ribosome [18], and selection and application of RF1-depleated E. coli variants [19], [20]. However, given the in vivo nature of the methodology, overall yield of recombinant protein with site-specifically incorporated UAA has never been reported to exceed 50% comparing to wild type.
Additionally, the evolution of cognate aaRS/tRNA pairs and the incorporation of UAAs into proteins in living organisms is not possible for certain amino acids. These include toxic UAAs, amino acids with extreme redox potentials or hydrophilicity that are not able to cross the cell membrane or UAAs which cannot be generated in large amounts, due to their difficult multi-step chemical synthesis. Moreover, site-directed UAAs incorporation into eukaryotic proteins expressed in mammalian cells is problematic, since no efficient system is available yet.
Herein we suggest complementing caveats that exist in the in vivo translational approach by using an efficient cell-free system for the different applications of this robust technology. The cell-free protein translation system can be considered as a good alternative for in vivo incorporation of UAAs [21], [22] due to several advantages over current in vivo processes, such as possibility to direct all of the cellular resources towards the production of a single protein [23]; to control the level of an orthogonal aaRS/tRNA pair and of the UAA employed in protein expression due to the absence of a cell wall. Another advantage of the in vitro approach is the possible use of aforementioned UAAs which can not be applied in vivo.
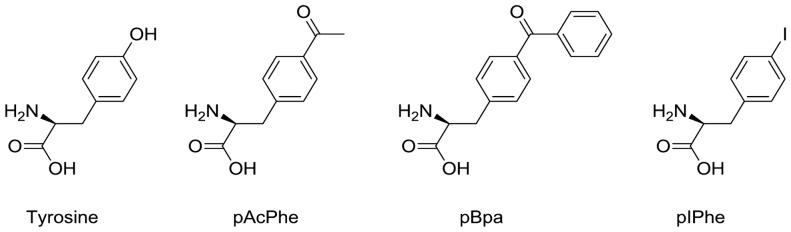
Here, we describe a general strategy based on the use of commercially available cell-free expression systems, combined with orthogonal M. jannaschii synthetases and cognate MjtRNACUA or synthetic tRNACUA Opt [4], [24], to obtain high yields of UAA-labeled proteins. This approach allowed us to incorporate tyrosine, as well as p-acetyl-L-phenylalanine (pAcPhe), p-benzoyl-L-phenylalanine (pBpa) and p-Iodo-L-phenylalanine (pIPhe; Fig. 1) into Green Fluorescent Protein (GFP), in response to the TAG stop codon with high fidelity and efficiency. The final yield of modified proteins varied widely and under optimal conditions reached roughly 50–120% of the wild type expression levels, depending on the type of suppressor tRNA, aaRS used and UAA used.
Figure 1. Structures of tyrosine and the three unnatural amino acids used in this study.
Materials and Methods
GFP Mutants
The X-ray crystallographic structure of template GFP (PDB accession number: 1EMA), encoded by the GFP control vector (RTS, 5 PRIME, Hamburg, Germany), was analyzed to choose sites for UAA incorporation. To examine the effect of the nucleotide following the stop codon on protein yields, we selected four amino acid residues on two external β-sheet of GFP and its adjacent loop. The selection of these residues for substitution by an UAA was chosen as not to obstruct proper GFP folding. The coding sequence of GFP was thus modified to replace the codons encoding tyrosine 39, lysine 41, leucine 42 or lysine 45 to an amber stop codon (TAG), with the nucleotide following the stop codon being G, C, A or T, respectively. The codons of amino acids at another β-sheet, i.e. histidine 148, asparagine 149 and valine 150, were substituted to TAG, such that A, G and T followed the stop codon, respectively. In order to generate mutation at permessive site of GFP the codon of tyrosine 151 was replaced to TAG, and the adjacent isoleucine 152 (ATC) was substituted by leucine (CTC), so that C has followed the amber stop codon. Mutagenesis was performed in the control vector containing the gene for GFP, by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit (Stratagen, Agilent Technologies, Santa Clara, CA) and the following primers:
QC-GFPY39 F (5'–GCATGGCAGGGGTTCAAATCCCCTCCGCCGGA–3') and QC-GFPY39 R (5'–TCCGGCGGAGGGGATTTGAACCCCTGCCATGC–3'); QC-GFPK41 F (5'–GGGTGAAGGTGATGCAACATACGGATAGCTTACCCTTAAATTTATTTGC–3') and QC-GFPK41 R (5'–GCAAATAAATTTAAGGGTAAGCTATCCGTATGTTGCATCACCTTCACCC–3'); QC-GFPL42 F (5'–GATGCAACATACGGAAAATAGACCCTTAAATTTATTTGCAC–3') and QC-GFPL42 R (5'–GTGCAAATAAATTTAAGGGTCTATTTTCCGTATGTTGCATC–3'); QC-GFPK45 F (5'–CATACGGAAAACTTACCCTTTAGTTTATTTGCACTACTGG–3') and QC-GFPK45 R (5'–CCAGTAGTGCAAATAAACTAAAGGGTAAGTTTTCCGTATG–3'); QC-GFPH148 F (5'–GGAATACAACTATAACTCATAGAATGTATACATCATGGCAG–3') and QC-GFPH148 R (5'–CTGCCATGATGTATACATTCTATGAGTTATAGTTGTATTCC–3'); QC-GFPN149 F (5'–CAACTATAACTCACACTAGGTATACATCATGGCAGAC–3') and QC-GFPN149 R (5'–GTCTGCCATGATGTATACCTAGTGTGAGTTATAGTTG–3'); QC-GFPV150 F (5'–GGAATACAACTATAACTCACACAATTAGTACATCATGCAGAC–3') and QC-GFPV150 R (5'–GTCTGCCATGATGTACTAATTGTGTGAGTTATAGTTGTATTCC–3'); QC-GFPY151 F (5'–CTATAACTCACACAATGTATAGCTCATGGCAGACAAACAAAAGAATGG–3') and QC-GFPY151 R (5'–CCATTCTTTTGTTTGTCTGCCATGAGCTATACATTGTGTGAGTTATAG–3'). Simultaneously, lysine 41, leucine 42 or lysine 45 codons were substituted to tyrosine TAC codon applying site-directed mutagenesis methodology and the following primers: QC-K41Y F (5'–GAAGGTGATGCAACATACGGATACCTTACCCTTAAATTTATTTGC–3') and QC-K41Y R (5'–GCAAATAAATTTAAGGGTAAGGTATCCGTATGTTGCATCACCTTC–3'); QC-L42Y F (5'–GGTGATGCAACATACGGAAAATACACCCTTAAATTTATTTGCACTAC–3') and QC-L42Y R (5'–GTAGTGCAAATAAATTTAAGGGTGTATTTTCCGTATGTTGCATCACC–3'); QC-K45Y F (5'–GCAACATACGGAAAACTTACCCTTTACTTTATTTGCACTACTGGAAAAC–3') and QC-K45Y R (5'–GTTTTCCAGTAGTGCAAATAAAGTAAAGGGTAAGTTTTCCGTATGTTGC–3').
UAA Incorporation Components
The plasmids pSup-MjmutTyrRS-6TRN encoding the orthogonal M. jannaschii tyrosyl-synthetase variants (M. jannaschii aaRS) for incorporation of tyrosine, pAcPhe and pBpa (Bachem, Bubendorf, Switzerland) and pIPhe (Sigma-Aldrich, St. Louis, MO) and suppressor tRNA were a kind gift from Dr. Peter Schultz (The Scripps Research Institute, La Jolla, CA). Coding sequences of M. jannaschii aaRS from plasmid pSup-pMjmutTyrRS-6TRN were amplified by PCR using primers Syn-Nde F (5' – TTACATATGGACGAATTTGAAATGATAAAGAGA –3') and Syn-Bam R (5' – CGGATCCTTATAATCTCTTTCTAATTGGCTCTAAAA –3'). The M. jannaschii aaRS cDNA copies were cloned directly into a pET-15b vector downstream the T7 promoter, RBS and His-tag regions for efficient expression and purification. Plasmid pET-MjmutTyrRS encoding mutated aaRS was transformed into E. coli BL21 (DE3) cells (Invitrogen, Grand Island, NY). All vector constructs were confirmed by DNA sequencing at the sequencing facility of the National Institute for Biotechnology in the Negev (NIBN), Ben-Gurion University of the Negev (BGU).
Preparation of MjtRNACUA, tRNACUA Opt
cDNA copies of all the RNA molecules under regulation of the T7 promoter were obtained by annealing the following two synthetic oligonucleotides: MjtRNACUA Forward 5'– TAATACGACTCACTATACCGGCGGTAGTTCAGCAGGGCAGAACGGCGGACTCTAAATCCGCATGGCGCTGGTTCAAATCCGGCCCGCCGGACCA–3' and MjtRNACUA Reverse 5'– TGGTCCGGCGGGCCGGATTTGAACCAGCGCCATGCGGATTTAGAGTCCGCCGTTCTGCCCTGCTGAACTACCGCCGGTATAGTGAGTCGTATTA –3'; and tRNACUA Opt Forward 5'–TAATACGACTCACTATACCGGCGGTAGTTCAGCAGGGCAGAACGGCGGACTCTAAATCCGCATGGCAGGGGTTCAAATCCCCTCCGCCGGACCA–3' and tRNACUA Opt Reverse 5'–TGGTCCGGCGGAGGGGATTTGAACCCCTGCCATGCGGATTTAGAGTCCGCCGTTCTGCCCTGCTGAACTACCGCCGGTATAGTGAGTCGTATTA–3'. Obtained products were used for in vitro RNA transcription by TranscriptAid High Yield Transcription Kit (Fermentas, Vilnius, Lithuania). Purified RNA molecules were heated to 90°C for 2 minutes, placed on ice for 2 minutes, and folded by addition of RNA Structuring Buffer (10 mM Tris-HCl, pH 7, 0.1 M KCl, 10 mM MgCl) and kept at 37°C for 20 minutes.
Preparation of M. jannaschii Tyrosyl-tRNA Synthetase and its Evolved Derivatives
E. coli BL21 (DE3) cells transformed with one of the above plasmids were grown to an OD600 of 0.5–0.7 in 1 L Luria-Bertani (LB) medium. Isopropyl-ß-D-thiogalactoside (IPTG) was added to a final concentration of 1 mM and the cells were grown for additional 4–5 h at 37°C. Cells were harvested at 8,000 g for 10 min at 4°C. The cell pellet was resuspended in 4 mL of lysis buffer (300 mM NaCl, 10 mM imidazole, 50 mM NaH2PO4, pH 8.0) per gram of cell paste. A cell lysate was prepared using BugBuster Protein Extraction Reagent (Novagen, Darmstadt, Germany) with addition of benzonase nuclease (Novagen) and Protease Inhibitor Cocktail Set III (Merck, Darmstadt, Germany). The lysate was centrifuged at 16,000 g at 4°C for 30 minutes. The His-tagged synthetase was then purified using Ni-NTA agarose (Qiagen). The Ni-NTA agarose beads were washed twice with wash buffer (300 mM NaCl, 20 mM imidazole, 50 mM NaH2PO4, pH 8.0) and the protein was eluted with elution buffer (300 mM NaCl, 250 mM imidazole, 50 mM NaH2PO4, pH 8.0). The eluate was dialyzed against sterile PBS buffer pH 7.4 three times and concentrated using an Amicon Ultra-centrifuge device 10 kDa MWCO (Millipore, Beverly, MA).
Coupled in vitro Transcription/translation Reaction
Cell-free protein expression was performed using the RTS 100 E. coli HY Kit (5 PRIME, Hamburg, Germany) at 30°C for 6 h in 10 (for Western Blot) or in 50 µL (for protein purification and MS analysis) of reaction mixture. Expression of GFP with an incorporated UAA was achieved by mixing the RTS 100 E. coli HY Kit reaction mixture containing 0.5 µg of modified control vector GFP with purified M. jannaschii aaRS derivatives (100–450 µg/mL, final concentration) and orthogonal suppressor either MjtRNACUA or tRNACUA Opt (480–600 µg/mL) in the absence or presence of the corresponding UAA (1 mM).
Proteins Quantitative Analysis and Purification
Following UAA incorporation using the cell-free expression system, 5 µL of reaction solution were precipitated with acetone to avoid protein aggregation. The protein pellet was resuspended in an equal volume of LDS loading buffer (Invitrogen). The resulting solution was incubated at 70°C for 10 minutes and resolved by SDS-PAGE. Proteins of interest were visualized by Coomassie staining using SimplyBlue SafeStain (Invitrogen). For Western blot analysis, proteins were transferred to a nitrocellulose membrane using Immuno-Blot PVDF membrane apparatus (Bio-Rad, Hercules, CA). To visualize proteins in the Western blot, we used primary mouse monoclonal IgG directed against His-tag (Santa Cruz Biotechnology, Santa Cruz, CA) and horseradish peroxidase-conjugated goat polyclonal secondary antibodies to mouse IgG1 heavy chain (Abcam, Cambridge, UK). Chemiluminescence was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL). Absolute quantities of expressed proteins were estimated by purification of WT and mutated GFP by Ni-NTA agarose (Qiagene), concentration by Amicon Ultra-centrifuge device 10 kDa MWCO (Millipore) and resolving by SDS-PAGE. Proteins of interest were visualized by Coomassie staining using SimplyBlue SafeStain (Invitrogen), the desired proteins bands were cut from the gel and eluted using Model 422 Electro-Eluter (Bio-Rad). The concentration of proteins were measured by Implen Nanophotometer (Labfish, Germany). The relative quantity of expressed proteins was analyzed by densitometry using GeneTools software (SynGene, Cambridge, UK). All of the proteins expressed using the RTS 100 E. coli HY Kit were purified according to the manufacturer’s manual using Ni-NTA Spin Columns (Qiagen, Hilden, Germany).
Proteolysis and Mass Spectrometry
The trypsin cleavage sites in WT GFP were predicted by PeptideCutter (http://web.expasy.org/peptide_cutter/). In-gel digestion of purified GFP containing UAA, was performed using Trypsin Gold (Promega, Madison, WI) according to a modified manufacturer’s procedure. Briefly, the gel bands corresponding in size to GFP were finely sliced, washed for 5 minutes with water, 15 minutes with acetonitrile (ACN), and dried in a vacuum centrifuge. After reduction (10 mM dithiothreitol in 100 mM NH4HCO3) and alkylation (10% iodoacetamide in 100 mM NH4HCO3), dried gel slices were incubated on ice for 15–30 minutes with Trypsin gold (12.5 ng/µL) in digestion buffer (50 mM NH4HCO3, 5 mM CaCl2). Excess buffer was removed and 20–30 µL of digestion buffer without Trypsin Gold was added to the gel slices followed by incubation at 37°C overnight. Peptides were extracted from the gel slices by washing once with 25 mM NH4HCO3, ACN and 1% formic acid (FA). The samples were then dried in a vacuum centrifuge. The extracted peptides were purified and concentrated using ZipTip pipette tips (Millipore), following the manufacturer’s instruction.
ESI-MS/MS analysis was performed using reverse phase nano-LC (Agilent Technology) connected directly to the LTQ XL Orbitrap EDT mass spectrometer (Thermo Electron, Wien, Austria) at the Analytical Research Services & Instrumentation Unit, BGU. The peptides were eluted with an increasing ACN gradient (Solvent A, 0.1% FA, 5% ACN; Solvent B, 0.1% FA, 80% ACN) over a period of 70 min. MS/MS spectra were acquired in a data-dependent fashion. Instrument control was performed using the Xcalibur software package (Thermo Electron).
Theoretical monoisotopic masses for the peptides generated by trypsin digestion of WT GFP were predicted with PeptideMass (http://web.expasy.org/peptide_mass/), while fragmentation of the FSVSGEGEGDATYGK peptide and theoretical molecular masses of the peptide species were calculated with the MS-Product software at the ProteinProspector web service (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct). Theoretical molecular masses for peptides containing UAA were adjusted manually.
Results
Site-specific Incorporation of Tyrosine into GFP in Response to a UAG-stop Codon in a Cell-free Expression System
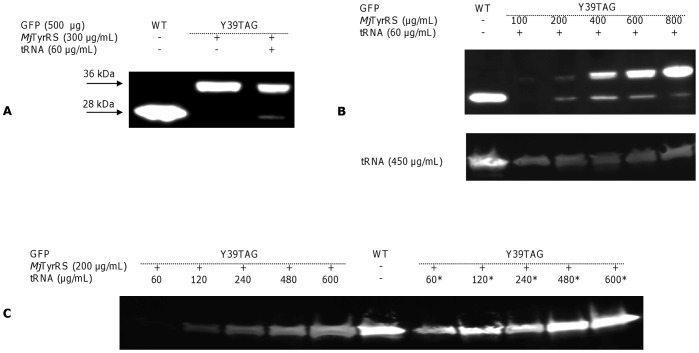
To incorporate tyrosine in response to TAG stop codon we expressed plasmid GFP Y39TAG obtained by site-directed mutagenesis in the RTS 100 E. coli HY Kit mixture supplied with external components, purified MjTyrRS and a suppressor MjtRNACUA; we employed GFP, encoded by a control vector of the kit, as a reporter protein. Western blot with anti His-antibodies enables to visualize full-length and not truncated GFP at a size of 28 kDa, as well as MjTyrRS of 36 kDa (Fig. 2A). The addition of purified MjTyrRS and synthetic MjtRNACUA to the reaction mixture permitted site-specific incorporation of tyrosine in response to the stop codon at GFP Y39TAG-mutated proteins, while no bands corresponding in size to GFP were detected in the reaction mixture supplied only with MjTyrRS, indicating orthogonality of M. jannaschii synthetase to endogenous tRNA molecules. Since estimated band intensity corresponding to GFP Y39TAG did not exceed 10% of the WT expression level, we further adjusted MjTyrRS and MjtRNACUA concentrations in the reaction mixture. The synthetase concentration required for maximal suppression efficiency was found to vary widely and depended on the concentration of MjtRNACUA (Fig. 2B). At a MjTyrRS concentration equal to 400–450 µg/mL and in the presence of 60 µg/mL MjtRNACUA, the expression level of Y39TAG GFP reached the maximum possible under these conditions and was roughly 20% of WT expression level, while in the presence of 450 µg/mL MjtRNACUA, the maximal expression level reached 35% of WT expression level and required only 100 µg/mL of MjTyrRS. To determine whether the suppressor tRNA structure is limiting, we sought another suppressor. We then adjusted the concentration of both MjtRNACUA and T-stem-modified tRNACUA Opt for optimal tyrosine incorporation. Using both the original and alternate suppressor, the expression of full-length GFP was demonstrated to depend greatly on the nonsense suppressor concentration (Fig. 2C). A maximum yield of Y39TAG GFP constituting 55% and 115% of the WT expression level was achieved at MjtRNACUA concentration of 600 µg/mL and of a tRNACUA Opt concentration of 480 µg/mL, respectively. This experiment revealed that the nature of suppressor tRNA significantly affects the efficiency of TAG nonsense suppression in recombinant proteins, while application of tRNACUA Opt allows one to reach at least similar protein yields as that of WT protein.
Figure 2. Western Blot of WT GFP and GFP Y39TAG mutant expression in a cell-free translation system.
Synthesis of WT GFP and the GFP Y39TAG mutant was performed using the RTS E. coli HY Kit, to which the corresponding plasmid (500 µg/mL), purified MjTyrRS and cognate suppressor MjtRNACUA (tRNA) or T-stem modified tRNACUA Opt (denoted as *) were added. (A) Expression of WT GFP and the GFP Y39TAG mutant in the presence of MjTyrRS (300 µg/mL) and synthetic MjtRNACUA (60 µg/mL). The band at 28 kDa corresponds to full-length GFP. (B) Western blot analysis demonstrates enhanced GFP Y39TAG protein expression as a function of increased MjTyrRS concentrations in a cell-free reaction medium supplied with MjtRNACUA (60 µg/mL – top panel and 450 µg/mL – bottom panel). (C) Dependence of GFP Y39TAG yield on the type and concentration of nonsense suppressor, as visualized by Western blot.
The Effect of the Nucleotide following the Stop Codon on the Expression of UAA-containing Proteins
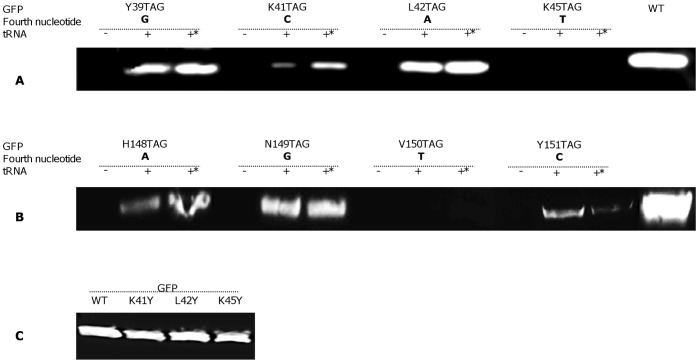
As it is mentioned above, the TAG stop codon specifying the desired position for UAA incorporation into a recombinant protein could be recognized by either RF1 or by the cognate suppressor tRNA. It has been shown that the identity of the nucleotide following nonsense codon impinge on the selection rate of RF1 [12], i.e. a low rate of stop signal recognition by RF1 means that mRNA interaction with near-cognate aminoacyl-tRNA or frame-shifting occurs faster than does RF1 binding to the ribosomal A-site [12]. Based on the literature screened [17], [19], [22], [25], [26], we suggested that the forth nucleotide in tetra-nucleotide stop signal could affect the rate of nonsense suppression [12], [13]. To verify this notion, we selected several amino acids in two different GFP β-sheets and their adjacent loop that were substituted to TAG, the location was selected in a way that afforded that the immediate nucleotide downstream from the stop codon differed from one location to another. Thus, we constructed plasmids encoding GFP Y39TAG, K41TAG, L42TAG and K45TAG mutants, where the amber codon was followed by a guanine (G), a cytosine (C), an adenine (A) and a thymine (T), respectively; and GFP H148TAG, N149TAG, V150TAG and Y151TAG were stop codon was followed by A, G, T and C, respectively. All these constructs were expressed using the cell-free expression kit supplemented by MjTyrRS (150 µg/mL) in the absence or presence of either MjtRNACUA or tRNACUA Opt (480 µg/mL). Western blot analysis (Fig. 3A, 3B) revealed that the expression level of full-length GFP depended on the specific nucleotide following TAG stop codon in the cell-free protein translation system based on an E. coli lysate. The fourth nucleotide hierarchy for efficient suppression was demonstrated to be A≈G>C>T for both variants of nonsense suppressor tRNAs. A literature screen revealed that E. coli-based cell-free protein synthesis had been successfully employed for UAA incorporation into proteins when the TAG stop codon was followed by G [22], [26], A [19], [25], and C [22]. To the best of our knowledge, no expression of protein containing UAA has been reported when T followed the stop codon. To demonstrate that indeed the “following base” and not position change in the protein affects the rate of suppression efficiency, we have substituted K41Y, L42Y and K45Y to tyrosine and tested GFP expression level for these mutants relative to the WT. It should be noted that, the fourth nucleotides after the tyrosine codon remained as in the native sequence: G after the codon in the native protein GFP39Y, C after GFP K41Y, A after GFP L42Y and T after K45Y. Western blot analysis of these control mutants revealed that expression levels did not differ from those of WT and from one another (Fig. 3C), verifying that indeed the effects that we have observed imply context dependence.
Figure 3. Cell-free expression of WT GFP and tyrosine-incorporating mutant GFP, as visualized by Western blot.
(A and B) Co-translational incorporation of tyrosine at different positions in response to the amber stop codon was achieved by adding purified MjTyrRS (200 µg/mL) and two types of suppressor tRNA (480 µg/mL) to the reaction mixture (tRNA denotes synthetic MjtRNACUA, *– tRNACUA Opt). (C) Western blot visualization of the expression level of GFP WT and tyrosine-substituted proteins.
Genetic Incorporation of UAA in Response to the Amber Stop Codon
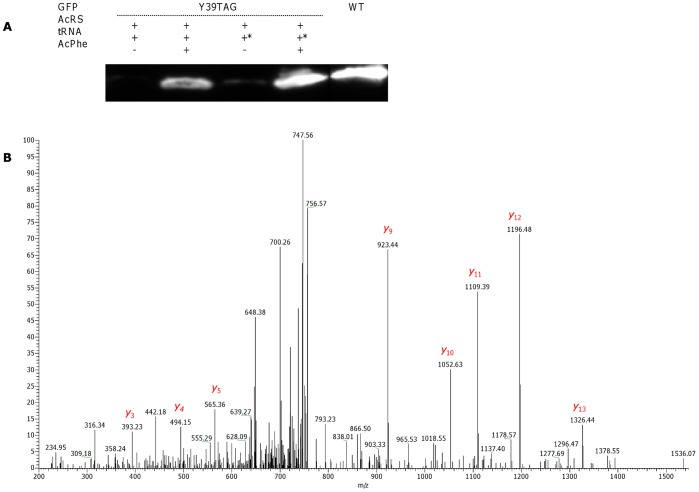
To test the generality of the developed platform, we examined its ability to incorporate diverse UAAs at position 39 of GFP in response to the TAG stop codon, applying both types of suppressor tRNAs and three variants of MjTyrRS derivatives. The three evolved variants of M. jannaschii aaRS, i.e. AcRS [27], BpaRS [28] and IPheRS [29], were tested for the ability to suppress the amber stop codon in GFP Y39TAG mutants together with either MjtRNACUA or tRNACUA Opt in the absence or presence of their cognate UAA in a cell-free translation system. The expression of full-length GFP Y39TAG was shown (Fig. 4A and 5A) to depend on the presence of pBpa and pIPhe. GFP expression was not detected in the absence of pBpa and pIPhe. Although AcRS has been widely used for site-specific protein labeling in vivo [17], [30], [31], its application in cell-free reaction medium led to background suppression in the absence of pAcPhe (Fig. 6A). The reason for background suppression in vivo is from mis-acylation of the suppressor tRNA molecules by the evolved synthetase with an endogenous amino acid, such as tyrosine or phenylalanine, in the rich media [17]. The overall level of background suppression was estimated to be less than 2 and 4.5% of GFP WT expression level for MjtRNACUA and tRNACUA Opt, respectively; however, since the main disadvantage of using previously reported eukaryotic-based cell-free systems for UAA incorporation was a high degree of mis-acylation with endogenous amino acids [21], site-specifically modified GFP Y39TAG were further characterized by mass spectrometry (MS).
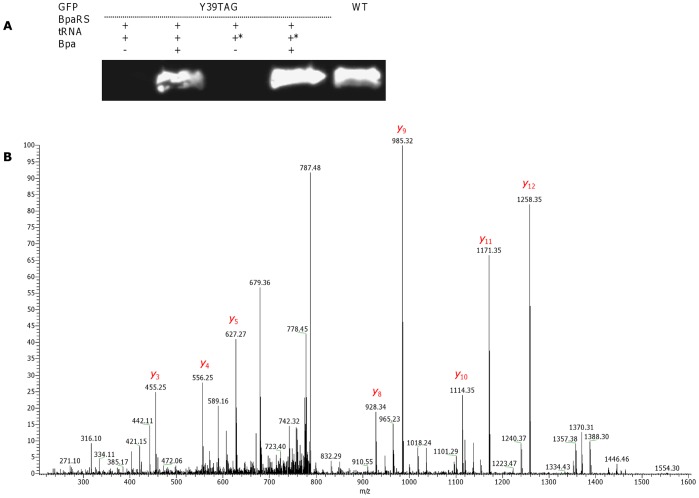
Figure 4. Site-specific Bpa incorporation into GFP in a cell-free expression system.
(A) Western blot visualization of WT GFP and Bpa-incorporating GFP Y39TAG. The synthesis of GFP was performed using an in vitro translation kit. Co-translational incorporation of Bpa was achieved by addition of M. jannaschii BpaRS (100 µg/mL), different types of suppressor tRNA (480 µg/mL) and Bpa (1 mM) to the reaction medium. tRNA denotes synthetic MjtRNACUA, Opt – tRNACUA Opt. (B) Annotated MS/MS spectrum of the FSVSGEGEGDATY*GK peptide from GFP Y39Bpa. The mass shift of 88 Da between Bpa in GFP Y39Bpa and tyrosine in WT GFP is clearly observed. For clarity, only the most abundant “y” ions are assigned.
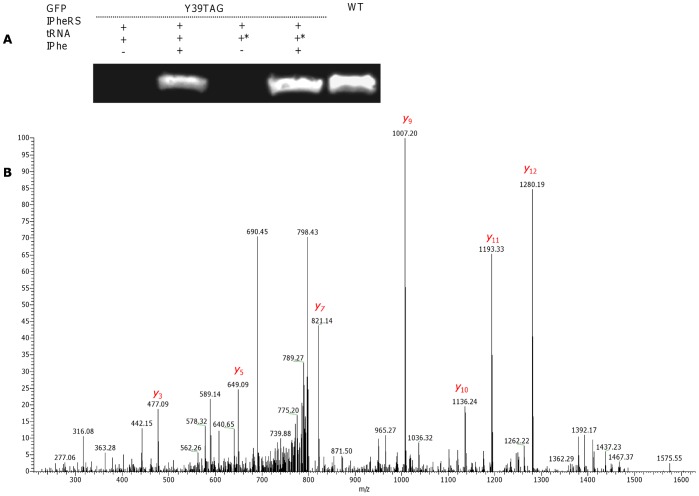
Figure 5. Site-specific incorporation of pIPhe into GFP in a cell-free expression system.
(A) Western blot visualization of WT GFP and pIPhe-incorporated GFP Y39TAG. Co-translational incorporation of pIPhe was achieved by addition of M. jannaschii IPheRS (100 µg/mL), different types of suppressor tRNA (480 µg/mL), and pIPhe (1 mM) to the reaction medium. tRNA denotes synthetic MjtRNACUA, Opt – tRNACUA Opt. (B) Annotated MS/MS spectrum of the FSVSGEGEGDATY*GK peptide from GFP Y39IPhe. The mass shift between pIPhe and tyrosine was calculated to be 109.9 Da, corresponding to the observed “y” ions.
Figure 6. Site-specific pAcPhe incorporation into GFP in a cell-free expression system.
(A) Western blot visualization of WT GFP and pAcPhe-incorporating GFP Y39TAG. The synthesis of GFP was performed using an in vitro translation kit. Co-translational incorporation of pAcPhe was achieved upon addition of the corresponding plasmid (500 µg/mL), M. jannaschii pAcPheRS (100 µg/mL), different types of suppressor tRNA (480 µg/mL), and pAcPhe (1 mM) to the reaction medium. tRNA denotes synthetic MjtRNACUA, Opt – tRNACUA Opt. (B) Annotated average MS/MS spectrum of peptides with m/z 765.84 corresponding to pAcPhe-incorporated FSVSGEGEGDATY*GK peptide of GFP Y39TAG. The Y* ion corresponding to pAcPhe (calculated m/z, 393.24; observed m/z, 393.23) can be easily detected. The distinguishing mass shift of 26 Da can be observed for most abundant “y” ions. For clarity, only the most abundant “y” ions are assigned.
The relative yields of GFP Y39TAG protein with site-specifically incorporated pIPhe and pAcPhe were measured to be 45–52% for the reactions utilizing MjtRNACUA, and an approximately 85% for tRNACUA Opt (Fig. 5A and 6A). The highest efficiency of UAA incorporation was obtained by applying BpaRS to the cell-free reaction mixture; the expression level of GFP Y39-mutated proteins comprised an approximately 80 and 120% of GFP WT level for MjtRNACUA and tRNACUA Opt, respectively (Fig. 4A).
Mass Spectrometry Analysis to Determine Specificity and Fidelity of Incorporation
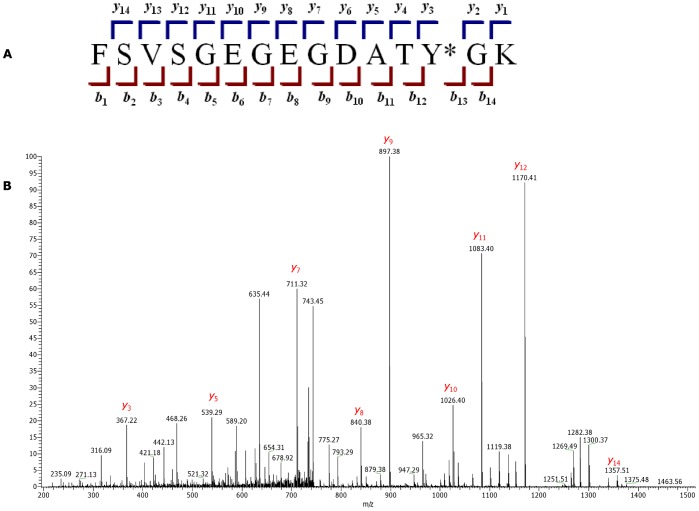
To examine the specificity and fidelity of M. jannaschii aaRS derivatives, WT GFP and GFP Y39TAG were expressed using a cell-free translation system, purified and separated by SDS-PAGE. The bands corresponding in size to GFP, as visualized by Coomassie staining, were excised, trypsin-digested and analyzed with a LTQ XL Orbitrap EDT mass spectrometer. The peptide of interest was predicted by the PeptideCutter software to be FSVSGEGEGDATY*GK, where Y* denotes either tyrosine in WT GFP or UAA in GFP Y39TAG. The monoisotopic mass calculated by PeptideMass software for this peptide generated by trypsin digest of the WT protein was 1503.6597 Da; the mass of the same peptide from GFP Y39TAG was adjusted manually (depending on the incorporated UAA mass). MS analysis of WT GFP identified the desired peptide, showing a doubly charged ion [M+2] peak of m/z 752.829, in a good agreement with the calculated mass. Peaks observed in the average MS/MS spectrum (Fig. 7) were also in a good agreement with predicted masses (calculated with the ProteinProspector MS-Product software) of “y” and “b” ions generated by FSVSGEGEGDATY*GK peptide fragmentation (Table S1). MS analysis of GFP Y39TAG revealed peaks with doubly charged ion masses of 765.84 (calculated mass, 765.85), 807.79 (calculated mass, 807.78) and 796.85 (calculated mass, 796.88) for FSVSGEGEGDATY*GK peptides containing pAcPhe, pBpa and pIPhe, respectively. Although Western blot revealed some degree of background suppression of amber stop codon of GFP Y39TAG obtained by using AcRS in the absence of pAcPhe, no peptide signals corresponding to GFP sequence containing tyrosine or phenylalanine at position 39 were detected, indicating that similarly to the observed in vivo process [17], [27] mis-acylation of suppressor tRNA by near-cognate endogenous amino acids was inhibited by addition of cognate UAA. Addition of higher concentrations of the UAA to the reaction, eliminate the competition fully, as no molecules with m/z equivalent to trypsin digested WT GFP were found for proteins obtained by addition of UAAs to the cell-free reaction. Moreover, no peptides containing canonical amino acids from near-cognate suppression of stop codon were detected, confirming good selectivity and fidelity of the analyzed M. jannaschii aaRSs. MS/MS analysis of GFP Y39TAG peptides (Fig. 4B, 5B and 6B) demonstrated a characteristic mass shift of 88.11, 109.9 and 26.04 Da relative to WT GFP, values that exactly match the differences between tyrosine and pBpa, pIPhe and pAcPhe, respectively. Mass shifts were also detected for peaks corresponding to “y” ions (from y3 to y14), as well as for b13 ion, indicating the site of UAAs incorporation to be position 39 of GFP. In general, we did not detect any differences in the GFP Y39TAG proteins obtained by utilization of either MjtRNACUA or tRNACUA Opt, apart from the different protein yields.
Figure 7. Annotated tandem MS spectra of the FSVSGEGEGDATY*GK peptide from WT GFP.
(A) “y”- and “b”-type ions generated during fragmentation of the FSVSGEGEGDATY*GK peptide. Y* denotes tyrosine in WT GFP or UAA in the GFP Y39TAG mutants. (B) MS/MS spectrum of FSVSGEGEGDATY*GK from WT GFP.
Discussion
Since it has been shown that protein synthesis is a ribosome-mediated process that does not require cell integrity, cell-free protein expression systems have been used for a variety of purposes, including site-specific UAA incorporation into recombinant proteins [21], [26]. In our efforts to develop an E. coli-based cell-free protein expression system to produce high yields of UAA containing recombinant proteins, we employed the most established M. janaschii orthogonal synthetases and nonsense suppressor molecules. In contrast to previously described cell-free expression systems adapted for protein synthesis with encoded UAAs, we employed purified MjTyrRS and its derivatives along with suppressor tRNA as additional components of defined reaction mixture allowing for control of the protein synthesis environment. In this manner, we were able to produce GFP Y39TAG with tyrosine with an absolute yield of 270 µg/mL and a suppression efficiency of 55% by adjusting MjTyrRS and MjtRNACUA concentrations. Although suppressor tRNA concentration is the major factor limiting production of proteins containing UAAs, further augmentation of the proportion of MjtRNACUA in the reaction medium (exceeding a final concentration of 600 µg/mL) led to the inhibition of recombinant protein biosynthesis, presumably because of translational apparatus overloading with non-endogenous elements. Replacement of the orthogonal MjtRNACUA suppressor by T-stem-modified tRNACUA Opt optimized for efficient recognition and binding by E. coli EF-Tu factor resulted in further enhancement of recombinant protein yields, which were estimated to be 120% of WT GFP expression. It was previously shown that the degree of improvement in suppression efficiency of evolved tRNACUA Opt varied, depending on the specific aaRS and cognate UAA used [17], [24]. We have also observed the ability of modified tRNACUA Opt to suppress amber stop codon in vitro with different efficiencies, depending on the UAA being incorporated. The application of optimized suppressors in a cell-free reaction medium for most aaRSs resulted in an enhanced protein yield, ranging from 85 to 110% of WT levels. Still, we cannot exclude the possibility that some of the evolved MjTyrRSs could have lower affinity to such a suppressor. For both tRNA molecules, the high fidelity of M. jannaschii aaRS derivatives in the cell-free expression system was confirmed by mass spectrometry.
The efficiency of UAAs incorporation in response to a stop codon is known to depend on the position of the mutation site and the nature of the recombinant protein, in particular the encoded amino acids and corresponding codon surrounding the amber codon. Although this phenomenon is usually taken into consideration when designing experiments, the precise reason and mechanism leading to this observation was not reported. It is well known that the identity the of nucleotide following stop codon influence the efficiency of translation termination. Depending on the nucleotide downstream the certain stop codon, the decoding site of 16S ribosomal RNA (rRNA) favors either translational termination by binding to the RF-factors, “read-through” of stop codon by augmentation of near-cognate aminoacyl-tRNA binding or a “frame-shift” [12], [32]. We hypothesized that the effectiveness of suppressor tRNA binding to its cognate nonsense codon would also depend on the following nucleotide. The expression of GFP Y39TAG, K41TAG, L42TAG and K45TAG mutants (where the fourth nucleotide was G, C, A, and T, respectively), and of GFP H148TAG, N149TAG, V150TAG and Y151TAG (where the fourth nucleotide was – A, G, T and C, respectively) demonstrated that the identity of the base following the amber codon determined the efficiency of UAA-charged suppressor MjtRNACUA or tRNACUA Opt interaction with UAG stop codon and, as a result, overall protein yields. According to our study, the strength of UAG stop codon selection and interaction was predicted by the fourth base hierarchy to be A≈G>C>T for both MjtRNACUA and tRNACUA Opt. The purines as the forth nucleotide improve the termination in the presence of RF1, however, we suggest that in the cell-free reaction medium prepared by us concentration of suppressor tRNA exceeds the concentration of RF1 many times rendering RF1 unavailable to the ribosomes; under these conditions, a purine nucleotide downstream the stop codon would augment UAG interaction with its cognate suppressor tRNA over the possibilities of mis-acylation or frame-shift. Analysis of the literature failed to find an example of the incorporation of any UAA in response to the amber stop codon followed by a T. Instead, the base following the stop codon was either G or A for the majority of UAA-containing proteins produced in vitro and in vivo that we have screened. These findings indirectly confirm our observation concerning the effect of the nucleotide downstream of the stop codon on the efficiency of UAA incorporation. Nonetheless, the actual mechanism by which the fourth base modulates the efficiency of stop codon suppression has yet to be revealed. In addition we would like to stress that a statistical analysis was not performed, hence we base our conclusions only on experimental evidence in our study combined with a literature screen.
It is also important to note that all of the reported protein yields are not absolute quantities and are reported as relative values and as percent of the WT expression levels, which were set to 100% in this study.
Conclusions
The cell-free translation system, modified as reported here, to genetically encode proteins containing UAAs resulted in increased amounts of recombinant proteins with very good fidelity. Concentrations of added UAAs with cognate RSs that have shown lower fidelity, can be tuned in this system in a controled manner, thus eliminating possible competition of incorporation of natural amino acids. Compettition of supressor tRNA with RF1 can be reduced significantly by using controled and higher concentrations of suppressor tRNA, thus affording higher supression efficiencies. The ability to control the concentrations of the different orthogonal components in this system afford reduced competition from natural components in the translational machinery.
The major advantage of the methodology reported here is its generality. Due to the availability of commercial cell-free translation systems with variety of modifications, it is possible to produce both prokaryotic and eukaryotic UAA-encoded proteins. The nature of the in vitro approach enables one to incorporate UAAs into nascent polypeptides that are not available for living organisms, provided that the right aaRS is available. It is also our belief that through this approach, more than one UAA could be incorporated into a protein with only a small loss in protein yield.
Supporting Information
Calculation of “y” and “b” ions of the FSVSGEGEGDATY*GK fragment (Y* denotes either tyrosine in WT GFP or UAA in the GFP Y39TAG mutants). Masses for the WT GFP-derived FSVSGEGEGDATY*GK peptide fragmentation were predicted by the MS-Product software of the ProteinProspector web service, while masses for GFP Y39TAG mutants were adjusted manually.
(DOC)
Acknowledgments
Mr. Ziv Roth and Dr. Isam Khalaila are thanked for their help in analyzing the MS data. Prof. Peter G. Schultz is gratefully acknowledged for generously supplying us with the pSup plasmid.
Funding Statement
The research leading to these results received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 260647 and from the Edmond J. Safra Center for the Design and Engineering of Functional Biopolymers. LA holds the Elaine S. and Alvin W. Wene Career Development Chair in Biotechnology Engineering. SS acknowledges the Harbour Foundation for a Fay and Bert Harbour Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1. Wang Q, Parrish AR, Wang L (2009) Expanding the genetic code for biological studies. Chem Biol 16: 323–336 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie J, Schultz PG (2006) A chemical toolkit for proteins - an expanded genetic code. Nat Rev Mol Cell Biol 7: 775–782 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 3. Liu DR, Schultz PG (1999) Progress toward the evolution of an organism with an expanded genetic code. Proc Natl Acad Sci U S A 96: 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang L, Magliery TJ, Liu DR, Schultz PG (2000) A new functional suppressor tRNA/Aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J Am Chem Soc 122: 5010–5011 10.1021/ja000595y. Available: http://dx.doi.org/10.1021/ja000595y via the Internet. [Google Scholar]
- 5. Bain JD, Diala ES, Glabe CG, Dix TA, Chamberlin AR (1989) Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J Am Chem Soc 111: 8013–8014 10.1021/ja00202a052 Available: http://dx.doi.org/10.1021/ja00202a052. [Google Scholar]
- 6. Noren C, Anthony-Cahill S, Griffith M, Schultz P (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244: 182–188 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 7. Hohsaka T, Ashizuka Y, Murakami H, Sisido M (2001) Five-base codons for incorporation of nonnatural amino acids into proteins. Nucleic Acids Res 29: 3646–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hohsaka T, Ashizuka Y, Murakami H, Sisido M (1996) Incorporation of nonnatural amino acids into streptavidin through in vitro frame-shift suppression. J Am Chem Soc 118: 9778–9779 10.1021/ja9614225 Available: http://dx.doi.org/10.1021/ja9614225. [Google Scholar]
- 9. Sisido M, Hohsaka T (2001) Introduction of specialty functions by the position-specific incorporation of nonnatural amino acids into proteins through four-base codon/anticodon pairs. Appl Microbiol Biotechnol 57: 274–281. [DOI] [PubMed] [Google Scholar]
- 10. LaRiviere FJ, Wolfson AD, Uhlenbeck OC (2001) Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 11. Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, et al. (2011) Expanding the genetic code of Escherichia coli with phosphoserine. Science 333: 1151–1154 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poole ES, Brown CM, Tate WP (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli . EMBO J 14: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poole ES, Major LL, Mannering SA, Tate WP (1998) Translational termination in Escherichia coli: Three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res 26: 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ryu Y, Schultz PG (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli . Nat Methods 3: 263–265 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 15. Schrader JM, Chapman SJ, Uhlenbeck OC (2009) Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol 386: 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eargle J, Black AA, Sethi A, Trabuco LG, Luthey-Schulten Z (2008) Dynamics of recognition between tRNA and elongation factor Tu. J Mol Biol 377: 1382–1405 10.1016/j.jmb.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young TS, Ahmad I, Yin JA, Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli . J Mol Biol 395: 361–374. [DOI] [PubMed] [Google Scholar]
- 18. Wang K, Neumann H, Peak-Chew SY, Chin JW (2007) Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol 25: 770–777 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 19. Short GF,3rd, Golovine SY, Hecht SM (1999) Effects of release factor 1 on in vitro protein translation and the elaboration of proteins containing unnatural amino acids. Biochemistry 38: 8808–8819 10.1021/bi990281r. [DOI] [PubMed] [Google Scholar]
- 20. Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, et al. (2011) RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol 7: 779–786 10.1038/nchembio.657; 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, et al. (2002) An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc Natl Acad Sci U S A 99: 9715–9720 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goerke AR, Swartz JR (2009) High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol Bioeng 102: 400–416 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- 23. Jewett MC, Swartz JR (2004) Rapid expression and purification of 100 nmol quantities of active protein using cell-free protein synthesis. Biotechnol Prog 20: 102–109 10.1021/bp0341693. [DOI] [PubMed] [Google Scholar]
- 24. Guo J, Melancon CE 3rd, Lee HS, Groff D, Schultz PG (2009) Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew Chem Int Ed Engl 48: 9148–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agafonov DE, Huang Y, Grote M, Sprinzl M (2005) Efficient suppression of the amber codon in E. coli in vitro translation system. FEBS Lett 579: 2156–2160. [DOI] [PubMed] [Google Scholar]
- 26. Sando S, Ogawa A, Nishi T, Hayami M, Aoyama Y (2007) In vitro selection of RNA aptamer against Escherichia coli release factor 1. Bioorg Med Chem Lett 17: 1216–1220 10.1016/j.bmcl.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 27. Wang L, Zhang Z, Brock A, Schultz PG (2003) Addition of the keto functional group to the genetic code of Escherichia coli . Proc Natl Acad Sci U S A 100: 56–61 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chin JW, Martin AB, King DS, Wang L, Schultz PG (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli . Proc Natl Acad Sci U S A 99: 11020–11024 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie J, Wang L, Wu N, Brock A, Spraggon G, et al. (2004) The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol 22: 1297–1301 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 30. Ye S, Kohrer C, Huber T, Kazmi M, Sachdev P, et al. (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem 283: 1525–1533 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 31. Liu CC, Schultz PG (2010) Adding new chemistries to the genetic code. Annu Rev Biochem 79: 413–444 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 32. Pape T, Wintermeyer W, Rodnina MV (2000) Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol 7: 104–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Calculation of “y” and “b” ions of the FSVSGEGEGDATY*GK fragment (Y* denotes either tyrosine in WT GFP or UAA in the GFP Y39TAG mutants). Masses for the WT GFP-derived FSVSGEGEGDATY*GK peptide fragmentation were predicted by the MS-Product software of the ProteinProspector web service, while masses for GFP Y39TAG mutants were adjusted manually.
(DOC)