Abstract
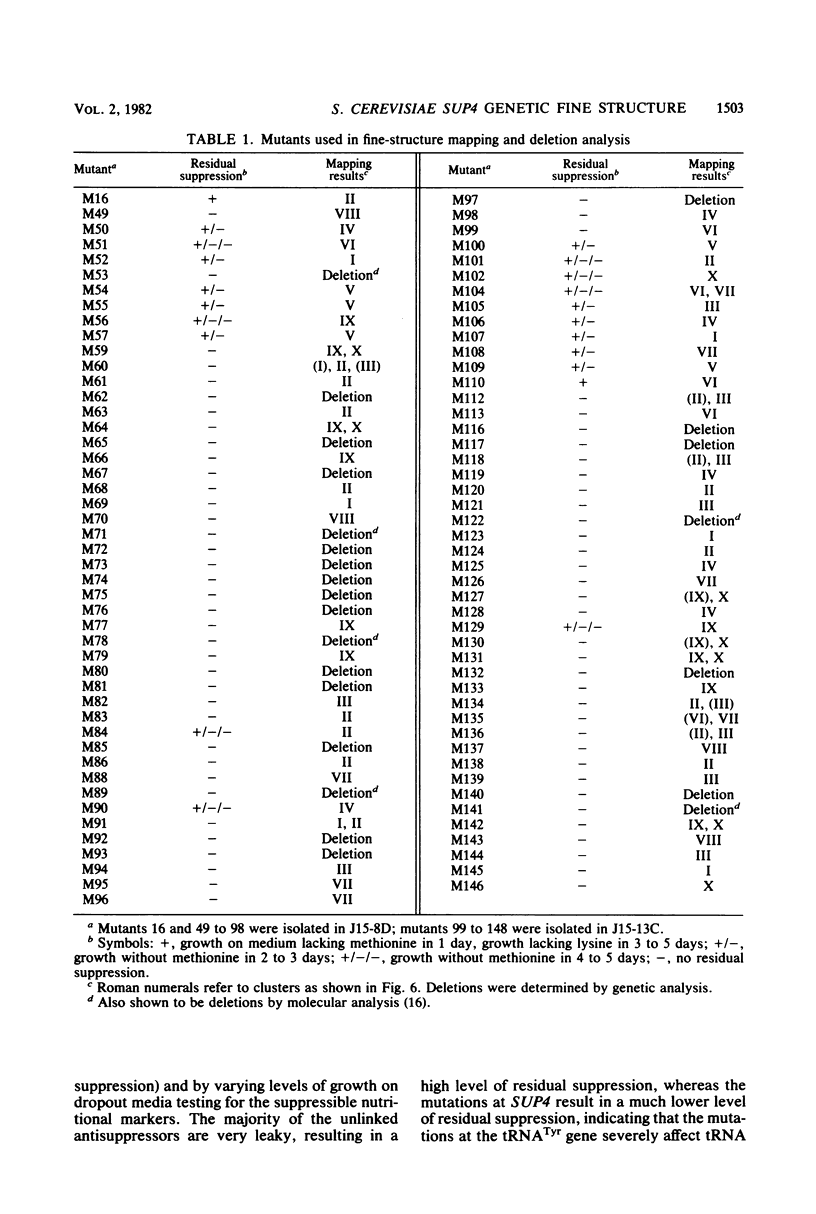
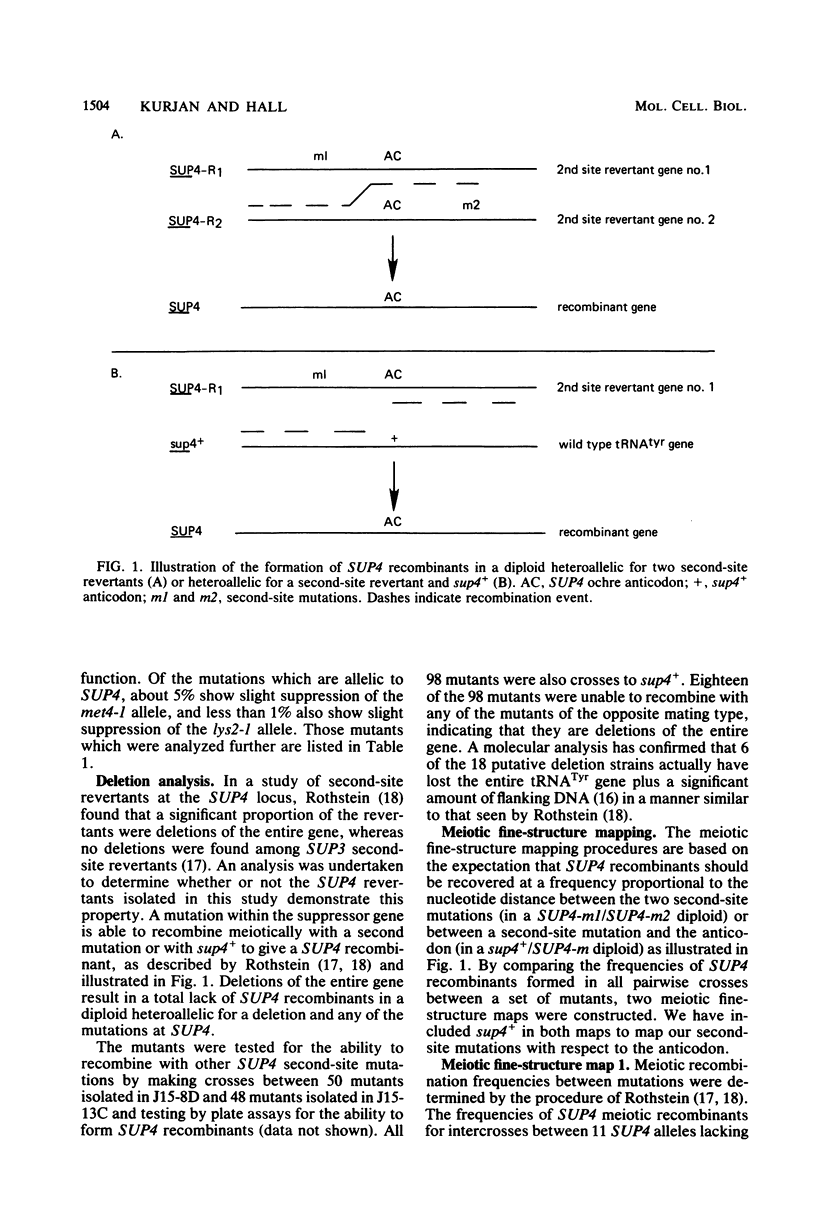
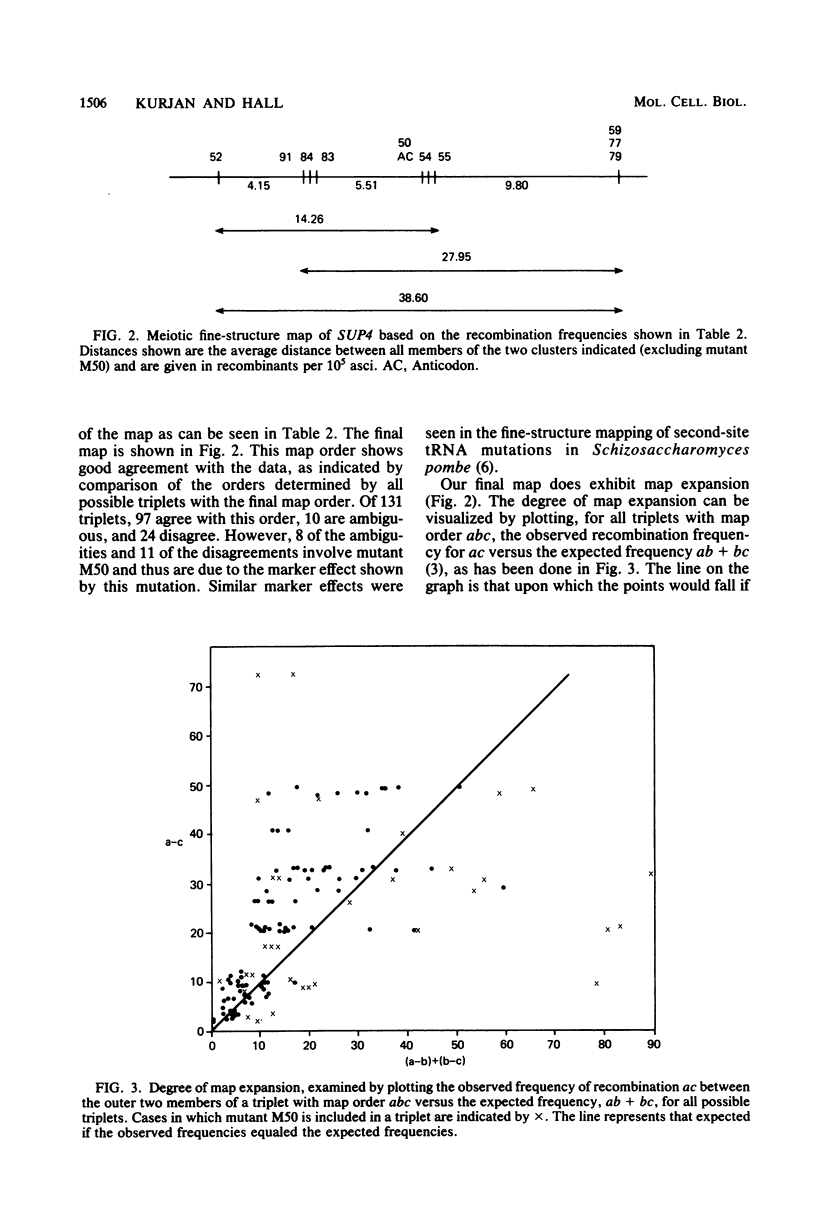
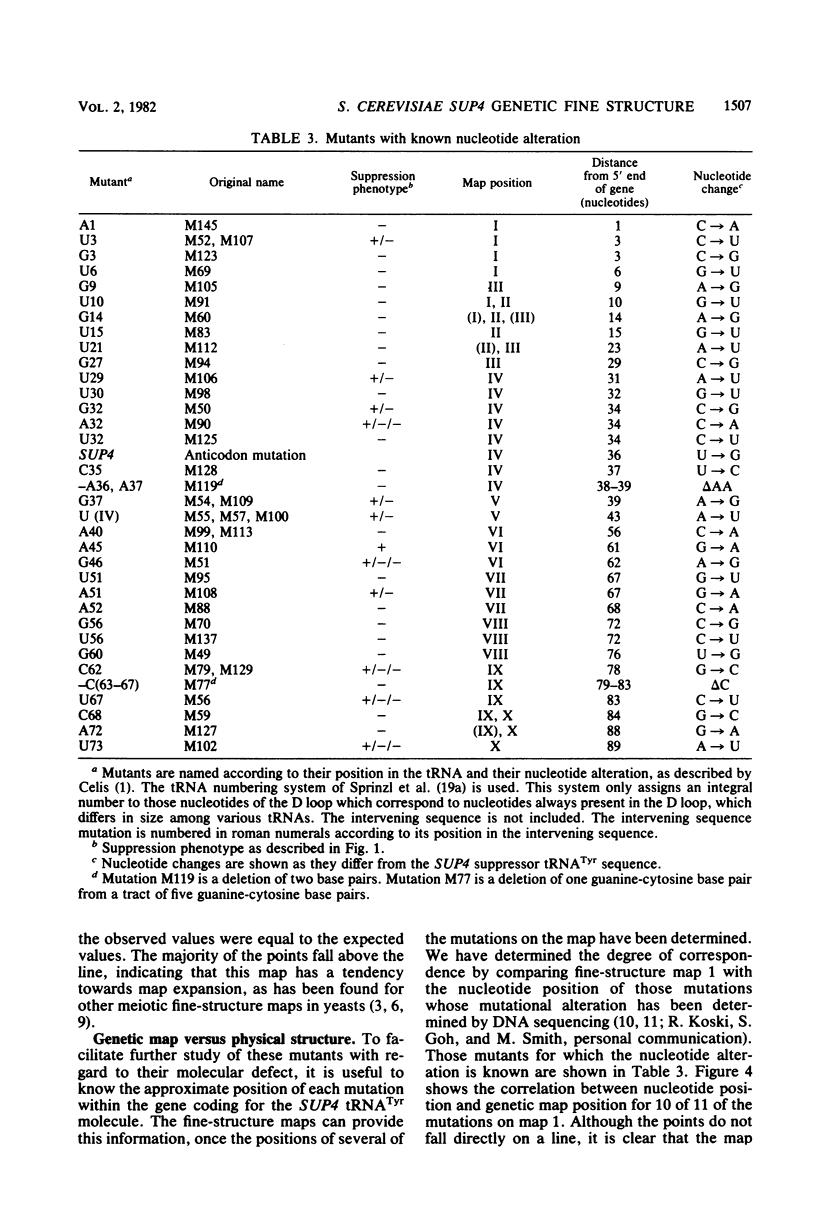
The SUP4 tRNATyr locus in Saccharomyces cerevisiae has been studied by the isolation and characterization of mutations at the SUP4 gene which result in the loss of suppressor function. Most of the mutations act as single-site mutations, whereas about a third of the mutations are deletions of the entire gene. Two meiotic fine-structure maps of the gene were made. The first mapping technique placed 10 mutations plus the sup4+ anticodon on a map by a measurement of levels of recombination between pairs of mutations. The second map utilized a more qualitative estimate of recombination frequency, allowing 69 mutations and the sup4+ anticodon to be mapped. The maps were compared with the physical structure of the gene for the 34 mutations whose nucleotide alteration has been determined by DNA sequencing (Koski et al., Cell 22:415-425, 1980; Kurjan et al., Cell 20:701-709, 1980). Both maps show a good correlation with the physical structure of the gene, even though certain properties of genetic fine-structure maps, such as marker effects and “map expansion,” were seen.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Celis J. E. Collection of mutant tRNA sequences. Nucleic Acids Res. 1979 Jan;6(1):r21–r-27. doi: 10.1093/nar/6.1.419-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lam C., Turoscy V. Structural analysis of the dur loci in S. cerevisiae: two domains of a single multifunctional gene. Genetics. 1980 Mar;94(3):555–580. doi: 10.1093/genetics/94.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarprio L., Hastings P. J. Gene conversion and intragenic recombination at the SUP6 locus and the surrounding region in Saccharomyces cerevisiae. Genetics. 1976 Dec;84(4):697–721. doi: 10.1093/genetics/84.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M. S. X-ray and meiotic fine structure mapping of the adenine-8 locus in Saccharomyces cerevisiae. Genetics. 1968 Apr;58(4):507–527. doi: 10.1093/genetics/58.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S., Mortimer R. K. Informational transfer in meiotic gene conversion. Proc Natl Acad Sci U S A. 1969 Jan;62(1):96–103. doi: 10.1073/pnas.62.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M., Mousset M., Wiame J. M., Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta. 1966 Oct 31;127(2):325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Kurjan J. tRNA synthesis: identification of in vivo precursor tRNAs from parental and mutant yeast strains. Nucleic Acids Res. 1981 Feb 25;9(4):1019–1029. doi: 10.1093/nar/9.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Moore C. W., Sherman F. Role of DNA sequences in genetic recombination in the iso-1-cytochrome c gene of yeast. I. Discrepancies between physical distances and genetic distances determined by five mapping procedures. Genetics. 1975 Mar;79(3):397–418. doi: 10.1093/genetics/79.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., Sherman F. Role of DNA sequences in genetic recombination in the iso-1-cytochrome c gene of yeast. II. Comparison of mutants altered at the same and nearby base pairs. Genetics. 1977 Jan;85(1):1–22. doi: 10.1093/genetics/85.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse-Messenguy F., Fink G. R. Temperature-sensitive nonsense suppressors in yeast. Genetics. 1973 Nov;75(3):459–464. doi: 10.1093/genetics/75.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. A genetic fine structure analysis of the suppressor 3 locus in Saccharomyces. Genetics. 1977 Jan;85(1):55–64. doi: 10.1093/genetics/85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J., Esposito R. E., Esposito M. S. The effect of ochre suppression on meiosis and ascospore formation in Saccharomyces. Genetics. 1977 Jan;85(1):35–54. doi: 10.1093/genetics/85.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Deletions of a tyrosine tRNA gene in S. cerevisiae. Cell. 1979 May;17(1):185–190. doi: 10.1016/0092-8674(79)90306-4. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Grueter F., Spelzhaus A., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1980 Jan 11;8(1):r1–r22. [PMC free article] [PubMed] [Google Scholar]
- YANOFSKY C., CARLTON B. C., GUEST J. R., HELINSKI D. R., HENNING U. ON THE COLINEARITY OF GENE STRUCTURE AND PROTEIN STRUCTURE. Proc Natl Acad Sci U S A. 1964 Feb;51:266–272. doi: 10.1073/pnas.51.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]



