Abstract
Alicyclobacillus are spoilage microbes of many juice products, but contamination of kiwi products by Alicyclobacillus is seldom reported. This study aims to investigate the whole production line of kiwi products in China to assess the potential risk of their contamination. A total of 401 samples from 18 commercial products, 1 processing plant and 16 raw material orchards were tested, and 76 samples were positive, from which 76 strains of microbes were isolated and identified as 4 species of Alicyclobacillus, including Alicyclobacillus acidoterrestris, Alicyclobacillus contaminans, Alicyclobacillus herbarius and Alicyclobacillus cycloheptanicus, and another 9 strains as 3 species of Bacillus by sequencing of their 16S rDNA. Through phylogenetic tree construction and RAPD-PCR amplification, it was found that there exist genotypic diversities to some extent among these isolates. Four test strains (each from one species of the 4 Alicyclobacillus species isolated in this study) could spoil pH adjusted kiwi fruit juice and some commercial kiwi fruit products with producing guaiacol (11–34 ppb).
Introduction
Alicyclobacillus species are a group of thermo-acidophilic, non-pathogenic rod-shaped, endospore-forming bacteria which were first isolated from Japanese hot springs in 1967 and can cause the spoilage of many fruit juice products [1]–[4]. Fruit juice are susceptible to the growth of yeasts, mycelial fungi and lactic acid bacteria due to their ability to grow in high-acid environments [5], but the pH of fruit juice had been thought to be too low for the spore-forming bacteria to grow [6]. In 1982, a large-scale spoilage incident caused by a new type of bacterium happened in Germany [7]. The most common characteristic of the spoilage is a medicinal, antiseptic offensive off-odor in commercial pasteurised apple juice [7]. The microbe causing this spoilage incident was a thermo-acidophilic, endospore-forming bacterium that was later identified as Alicyclobacillus acidoterrestris [2]. After this spoilage incident the potential dangers of Alicyclobacillus species were realized by the juice industries. During the next decade, after many this kind of thermo-acidophilic bacteria, which were distinct from the bacteria of the genus Bacillus, have been reported, they were allocated to a new genus, Alicyclobacillus, based on comparative sequence analysis of their 16S rRNA genes and the presence of ω-alicyclic fatty acids in their cell membrane [2]. To date, 19 species, 2 subspecies and 2 genomic species that belong to the genus Alicyclobacillus have been identified [8], [9].
The thermal processes to inactivate pathogenic foodborne microorganisms and vegetative non-heat resistant spoilage microorganisms in juice are insufficient to inactivate Alicyclobacillus [10]. Chang and Kang [11] also proved that the pasteurisation treatments applied to fruit products are not sufficient to inactivate Alicyclobacillus in 2004. A 1998 survey by the European National Food Processors Association showed that 35% of juice manufacturing participants experienced Alicyclobacillus spp. related problems. In North America, as well as in Europe, a similar survey in the same year conducted by the Grocery Manufacturers Association of the USA also showed that almost 1/3 of the responded companies had experienced spoilage incidents which may be caused by Alicyclobacillus [12]. In 1999, Eiroa et al. [5] found that up to 14.7% of 75 orange juice samples from 11 Brazilian companies to contain Alicyclobacillus. According to a survey by the European Fruit Juice Association (AIJN) in 2005, 45% of the 68 companies from the fruit processing industry experienced Alicyclobacillus related product contaminations during the three years prior to the survey, including 33% experiencing contaminations more than once [13]. These reports provide evidence to support that the problem caused by Alicyclobacillus spp. is a major and widespread microbial spoilage concern for the juice and beverage industries, which has not been thoroughly studied [12], [14].
Alicyclobacillus spp. are soil-borne bacteria [15], and there have been reports of Alicyclobacillus isolated from orchards [16], [17]. Alicyclobacillus have been isolated from many kinds of fruit juices products and concentrated fruit juices products, including apple [18]-[20], pear [16], [17], [14], orange [21], banana [22], watermelon [21], mango [23], lemon [24], grapefruit and blueberry [14] and so on with apple and pear are the most frequent isolation sources, but there are almost no reports about Alicyclobacillus contamination of kiwi fruit products. According to a 14-year survey in Argentina, 8556 samples from 7 Argentinean provinces of 19 different kinds of fruit and vegetable juices were analyzed for the presence of Alicyclobacillus, and the result showed that there was no Alicyclobacillus found in the only one sample of kiwi fruit [25].
Even so, we believe that there might be a potential risk of Alicyclobacillus contamination in kiwi fruit products because we have isolated 1 strain of Alicyclobacillus from a kiwi fruit product. As the origin source area of kiwi fruit and the largest production country in the world, China’s industry of kiwi fruit products (juice, vinegar, wine, fresh-cut slices) is developing very fast in recent years, and out of 60% of kiwi fruit production are from Shaanxi province [26], especially from Mei county and Zhouzhi county. Gocmen and Pettipher [27] [28] have proved that cell numbers between 105 and 106 CFU/mL of A. acidoterrestris produced sufficient guaiacol (2 ppb) to spoil orange and apple juices, but to our knowledge there is still no reports about the growth and taint (mainly guaiacol) production of Alicyclobacillus in kiwi fruit juice or commercial beverages related to kiwi fruit, although vanillin, which is a precursor of guaiacol is contained in kiwi fruit. This research aims to investigate the existence condition of Alicyclobacillus in the production line of kiwi products (orchards, processing factories and commercial products) in China's Shaanxi province and study the growth to and taint production of Alicyclobacillus in kiwi fruit juice and commercial kiwi fruit products to assess the potential risk of Alicyclobacillus contamination in kiwi fruit production line under the situation of a rapid development of this industry.
Materials and Methods
Sampling and Isolation
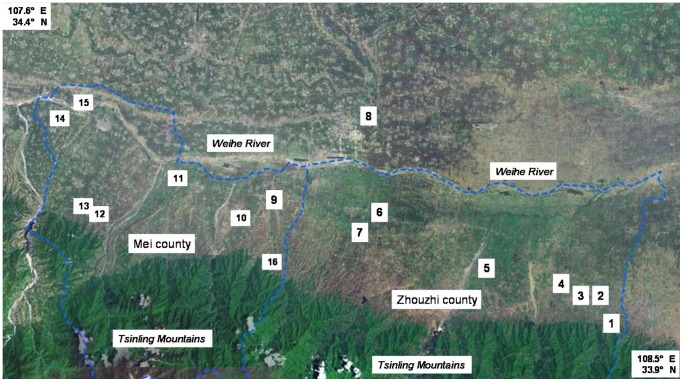
Samples were collected from three sources: eighteen samples were collected from commercial kiwi products bought from supermarkets, stores in Yangling or online, 102 samples were collected from a Hazard Analysis Critical Control Point accredited fresh-cut and frozen fruit and vegetable producer in Shaanxi province of China (the name of the authority who issued the permission: Baoji DuLe Food Co., Ltd), and 281 samples (fruits, soil and air) were collected from 16 orchards covering main regions of kiwi fruit production in Shaanxi province (Figure 1) (Table 1). The names of the authorities who issued the permissions for each sampling location are also shown in Table 1.
Figure 1. Sampling sites of 16 orchards.
The blue dash lines on the map are the administrative boundaries of Mei county and Zhouzhi county which located on the middle of Shaanxi province of China. The plain between the Weihe River and the Tsinling Mountains within these two counties, which is a part of the Guanzhong Plain, is the main region of kiwi fruit production in Shaanxi province.
Table 1. The number of samples and positive samples of the 16 orchards and their geographic coordinate.
| Number of orchards | Locations of orchards | Geographic coordinate of orchards | authority who issued the permission | Number of samples | Positive samples |
| East | |||||
| 1 | Shaanxi Zhouzhi Bairui Kiwi fruit Experimental Base | 108.45°E, 34.05°N | Shaanxi Bairui Kiwi Fruit Research Institute Co. Ltd. | 14 | 5 |
| 2 | Kiwi fruit Experimental Base of Agricultural Science-technology Demonstration Park (Nanqianhu village in Zhouzhi county) | 108.45°E, 34.07°N | the government of Jiufeng township | 18 | 4 |
| 3 | Gengxi village, Jiufeng township, Zhouzhi county | 108.41°E, 34.07°N | p | 22 | 0 |
| 4 | Liujiabao village, Jixian township, Zhouzhi county | 108.38°E, 34.10°N | p | 16 | 4 |
| 5 | Erhezhuang village, Louguan township, Zhouzhi county | 108.27°E, 34.11°N | p | 15 | 6 |
| 6 | Shangtiantun village, Situn township, Zhouzhi county | 108.14°E, 34.17°N | p | 19 | 3 |
| 7 | Changdong village, Yabai township, Zhouzhi county | 108.10°E, 34.15°N | p | 14 | 6 |
| 8 | Xiajiagou village, Yangcun township, Yangling town | 108.11°E, 34.30°N | p | 21 | 7 |
| West | |||||
| 9 | Jinjia village, Hengqu town, Mei county | 107.99°E, 34.19°N | p | 22 | 5 |
| 10 | Tuling village, Tangyu town, Mei county | 107.93°E, 34.18°N | p | 18 | 9 |
| 11 | Taoyuan village, Huaiya town, Mei county(pollution free orchards) | 107.85°E, 34.21°N | p | 16 | 3 |
| 12 | Luoyukou village, Yingtou town, Mei county | 107.75°E, 34.16°N | p | 8 | 0 |
| 13 | Tongyu village, Yingtou town, Mei county | 107.74°E, 34.16°N | p | 7 | 2 |
| 14 | Yuechen village, Diwucun town, Mei county | 107.68°E, 34.28°N | p | 25 | 1 |
| 15 | Chenjiagou village, Diwucun town, Mei county | 107.71°E, 34.29°N | p | 26 | 0 |
| 16 | Kiwi fruit Experimental Station of Northwest A & F University (Qinghua Town) | 107.99°E, 34.12°N | Northwest A&F University | 20 | 2 |
| Total | 281 | 57 |
p: private land, and the owner of the land have given permission to conduct the study on this site.
As the most efficient isolation medium, the Bacillus acidoterrestris medium (BAM) [29] which can support nearly all species and all spoilage related species of Alicyclobacillus, was used as the medium for the enrichment of Alicyclobacillus in this study [9]. A hundred milliliter samples of kiwi products were mixed with 200 mL BAM broth in 500 mL flasks followed by a heat shock treatment at 80°C for 10 min [18] to inhibit the growth of yeast and fungi and promote the germination of Alicyclobacillus spores, after which the flasks were shaken at 50°C, 120 rpm in a shaker for 5 days to obtain an enrichment. Then 1 mL of these cultures were diluted appropriately in test tubes and obtain a same heat shock treatment and streaked on BAM plate and yeast starch glucose (YSG) [30] plates duplicately followed by a incubation at 45°C (BAM) or 60°C (YSG) for 4 days. After the enrichment, according to their morphology features, colonies which may be Alicyclobacillus species were selected randomly from plates and re-streaked to obtain pure colonies [9]. Then all pure colonies were streaked on pH 7.0 Luria-Bertani (LB) plates. The colonies which did not grow on LB plates were selected and observed with a microscope. Colonies of spore-forming rods were selected for further examination and stored at -40°C in corresponding broth supplemented with 30% sterile glycerol.
The number and the enrichment methods of the samples are shown in Table 2. Samples from the orchards were collected randomly into sterile sample bags from 2 sources: soil and fruits (both on trees and dropped), and the air samples were also collected with BAM plates. The samples were treated immediately after collection with 100 mL sterile water poured into the sample bags directly and then put in a shaker for 10 min at 120 rpm. After settlement the supernatant solution was mixed with 100 mL BAM broth and incubated at 50°C, 120 rpm for 5 days after a heat shock treatment at 80°C for 10 min to obtain an enrichment. The consequent isolation steps of samples from the plant and orchards were the same as the isolation steps of kiwi products samples. The plates containing air samples were incubated at 50°C for 5days directly followed by isolation steps similar to other samples.
Table 2. The number and the sampling and enrichment methods of the samples from the producer.
| Sample form and sampling sections | Number of samples | Sampling and enrichment methods | Positive samples |
| Orchards | |||
| Kiwi fruits before peeling | 10 | Samples were put into sterile sample bags and treated immediately after collection with 300 mL sterile water poured into the sample bags and then put in a shaker for 10 min at 120 rpm. After settlement 100 mL of the supernatant solution was mixed with 100 mL BAM broth and incubated at 50°C, 120 rpm for 5 days after a heat shock treatment at 80°C for 10 min to have an enrichment. | 2 |
| Kiwi fruits after peeling | 12 | s | 2 |
| Kiwi fruits after color protection treatment | 8 | s | 2 |
| Washing & fresh-cut shops | |||
| Kiwi fruits before washing | 7 | s | 1 |
| Kiwi fruits after washing | 9 | s | 1 |
| Kiwi fruit slices | 10 | s | 1 |
| Wash water | 5 | A total 2500 mL of wash water was collected from 5 sites of the washer randomly with five 500 mL sterile flasks. After delivery to the lab all the samples were mixed with BAM broth at the ratio of 1∶1 in 500 mL flasks and put in a shaker at 50°C, 120 rpm for 5 days after a heat shock treatment at 80°C for 10 min to have an enrichment. | 2 |
| Shop environment (walls) | 4 | Sampling were carried out with sterile cotton bud smearing on the surfaces of the shop environment and then put into test tubes containing 10 mL BAM broth. The tubes were incubated at 50°C for 5 days after a heat shock treatment at 80°C for 10 min as an enrichment. | 1 |
| Shop environment (floor) | 8 | s | 2 |
| Shop environment (raw material bins) | 9 | s | 3 |
| Shop environment (air) | 10 | s | 0 |
| Quick-freezing shop | |||
| Quick-frozen kiwi fruit slices | 10 | After melting at room temperature, the samples were treated by the same method as used for samples from the orchards. | 1 |
| Total | 102 | 18 |
s: same as above.
16S rRNA Gene Amplification and Sequencing
Selected colonies were grown in corresponding broth, and then their genomic DNA was extracted with the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Then a portion of their 16S rRNA gene was amplified using the primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′) (BGI, supplied by BGI, Beijing, China) [31].
PCR amplifications were performed under conditions as below: fifty microliter of a total reaction volume, 0.3 µL of 5 U/µL Taq DNA polymerase (Takara, supplied by Takara Biotechnology Co. Ltd., Dalian, China), 5 µL of 10× PCR reaction buffer, 3 µL of 25 mM MgCl2, 4 µL of deoxyribonucleoside triphosphate (dNTPs) mixture (Takara) with 2.5 mM each, 4 µL of each primers, 3 µL of template DNA. PCR reactions were done in an Alpha Unit Block Assembly for Peltier Thermal Cycler DNA Engine Systems (MJ RESEARCH Inc., Watertown, Massachusetts, USA) under conditions as below: initial denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 45 s, elongation at 72°C for 2 min, and final elongation at 72°C for 10 min [17].
The PCR products were checked by 1% agarose (Invitrogen, supplied by Invitrogen, Carlsbad, CA92008, USA) electrophoresis with an electrophoresis apparatus (Liuyi, Beijing Liuyi Instrument Factory, Beijing, China) to confirm they contained a 1.5 kb fragment each. Then these fragments were sent to BGI for purification and sequencing.
Phylogenetic Tree Construction and RAPD-PCR Amplification
The sequence data of 16S rDNA of these isolates were compared to sequences in GenBank using BLAST 2.2.27 and then submitted to GenBank using Sequin. Then these 16S rDNA sequences of isolates were aligned with CLUSTAL W to construct a phylogenetic tree with MEGA 5.10 using both the neighbour-joining method [32] based on the p-distance model and the maximum-parsimony method [33].
RAPD-PCR amplifications were performed in 50 µL volume reactions containing 0.3 µL of 5 U/µL Taq DNA polymerase (Takara), 5 µL of 10× PCR reaction buffer, 3 µL of 25 mM MgCl2, 4 µL of dNTPs mixture (Takara) with 2.5 mM each, 4 µL of 2 different primers (A-01∶5′-CAGGCCCTTC-3′; AZ-14∶5′-CACGGCTTCC-3′) (BGI). The volume of template DNA was adjusted from 1 to 4 µL to get the clearest electrophoresis patterns. The PCR conditions were the same as above, except for annealing at 38°C for 45 s, and elongation at 72°C for 2 min 30 s. The results were checked by 1% agarose electrophoresis.
Enumeration of Growth and GC-MS Analysis for Taint Production
Four strains (C-ZJB-12-32, C-ZJB-12-35, C-ZJB-12-55 and C-ZJB-12-69, each 1 strain for one species of Alicyclobacillus isolated in this study) were selected and inoculated into BAM broth and incubated for 12 h to reach the log phase for activation, then these cultures were serially diluted (to make the number of CFU/mL of each sample was below 100 after inoculation) and each inoculated into 150 mL kiwi fruit juice (pH adjusted to 4.2 using 4 M NaOH) and 150 mL kiwi fruit juice (pH 2.5, without adjustment), then all 8 samples were incubated at 45°C for 21 days. Activated C-ZJB-12-35 was also inoculated into 150 mL of some commercial kiwi products mentioned in the sampling and isolation part, including 2 kinds of kiwi fruit juice (pH 3.5, soluble solids (SS) content 10°Brix, raw kiwi fruit juice content 60%), vinegar (pH 2.5, 5°Brix) and wine (pH 3.5, 7°Brix, alcohol 6%) (<100 CFU/mL after inoculation) and incubated at 45°C for 21 days. All 14 samples were plated onto BAM agar every day for the first week of incubation and every 3 days for the remainder of the experiment. Samples of kiwi fruit juice without pH adjustment obtained another test with a heat shock treatment of 80°C for 10 min before plating to check spores in them.
According to some previous reports [27] [28] [34] [35], guaiacol and 2 kinds of halophenols, 2,6-dibromophenol (2,6-DBP) and 2,6-dichlorophenol (2,6-DCP) were chosen as aim taint compounds. Standard solutions were prepared with guaiacol (Fluka Analytical, 2931 Soldier Springs Road, Laramie, WY, USA), 2,6-DBP (SUPELCO, 595 North Harrison Road, Bellefonte, PA, USA) and 2,6-DCP (SIGMA-ALDRICH, Co., 3050 Spruce Street, St. Louis MO, USA) in concentrations of 2.5, 5,7.5, 10 and 20 ppb in distilled water. A 5 ppb standard solution for guaiacol was also prepared in kiwi fruit juice as a contrast. All standard solutions were kept at 4°C in the dark until analysis (within 2 days). Ten milliliter of samples which could accumulate >106 CFU/mL after incubation and all standard solutions were transferred to 20 mL glass vials. After a 15-min equilibrium at 45°C, a solid phase microextraction (SPME) fiber (50/30mm DVB/Carboxen/PDMS; Supelco Co., Bellefonte, PA, USA) was exposed to the headspace of the vials at 45°C for 1 h to extract their volatile compounds.
After the extraction, the fiber was inserted into a Thermo-Finnigan Trace GC ultra/Trace DSQ (Thermo-Finnigan, San Jose, CA, USA) injection port using a 30 m ×0.25 mm i.d.×0.25 µm DB-Wax column (Agilent, Palo Alto, CA, USA). The GC-MS conditions were as follows: carrier gas He at 1 ml/min, splitless mode, injector temperature 250°C, starting temperature 50°C (2 min), final temperature 230°C (5 min), temperature rate 10°C /min, ion source temperature 230°C, scanning mass range 50–350 m/z. NIST 2002 fragmentation spectra database was used for identifications.
Nucleotide Sequence Accession Numbers
All the 16S rRNA gene (16S rDNA) sequences were deposited in GenBank (accession numbers KC193182 to KC193190 and KC354615 to KC354691, also see details in Figure 2).
Figure 2. Neighbour-joining phylogenetic tree of isolates and reference species.
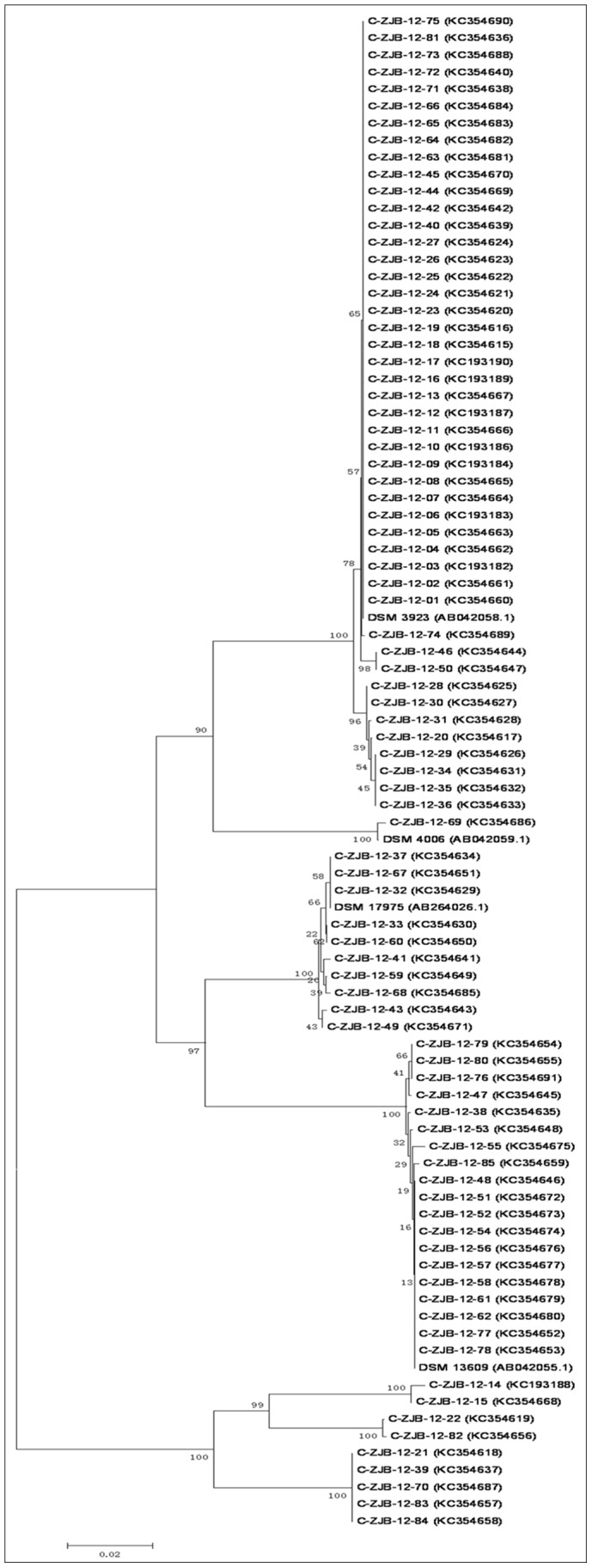
The 4 reference species are Alicyclobacillus acidoterrestris DSM 3923, Alicyclobacillus cycloheptanicus DSM 4006, Alicyclobacillus contaminans DSM 17975, and Alicyclobacillus herbarius DSM 13609. Construction is based on 16S rRNA gene sequence comparisons. Bootstrap percentages based on 1000 replicates are shown. Bar, p-distance (0.02 substitutions per nucleotide position).
Results
Positive Percentages and Properties of Isolates
A total of 401 samples were tested, and 76 samples were positive (Alicyclobacillus detected, 19.0%), from which 85 strains of microbes were isolated and identified. The total percentages of positive samples of the 16 orchards and all sampling sections of the producer were 20.3% and 17.6%, respectively (Table 1; Table 2), and only one sample (around 140 CFU/mL) among 18 commercial kiwi fruit products was detected as positive (5%), from which the one isolate was identified as Alicyclobacillus acidoterrestris later.
All isolates were Gram-positive, endospore-forming rods, and could form creamy white or yellowish, opaque colonies. Some isolates were motile. Among all 85 isolates, 1 was isolated from commercial products, 16 were isolated from the producer and 68 were isolated from the 16 orchards, including 26 from fruits samples (mainly from drop fruits), 37 from soil samples and 5 from air samples. The data revealed that Alicyclobacillus are soil-borne bacteria, as reported by Deinhard et al. in 1987 [15]. All samples from orchards’ air were negative except 2 air samples (formed 5, 7 colonies on plates respectively) from the orchard in Xiajiagou village, which might be because of the sprinkle on the day of sampling.
Identification of Isolates
As shown in Table 3, the similarity (max identity of BLAST results) of 16S rDNA among all isolates was beyond 99% except C-ZJB-12-85, and the number of nucleotides sequenced of all isolates were beyond 1400 (almost complete sequence of 16S rDNA) except C-ZJB-12-53 and C-ZJB-12-55. These mean that the isolates were very likely the same species as their nearest phylogenetic neighbours showed in Table 3. Forty six isolates from 20 sources (all sampling sections of the quick-frozen kiwi fruit slice producer and 7 orchards: the orchard in Xiajiagou village, Bairui Kiwi fruit Experimental Base, Changdong village, Liujiabao village, Kiwi fruit Experimental Station of Northwest A & F University, kiwi fruit Experimental Base of Agricultural Science-technology Demonstration Park, and Yuechen village) of total 85 isolates (54.1%) were identified as Alicyclobacillus acidoterrestris through the sequence data compared with nucleotides sequences in GenBank, which means that Alicyclobacillus acidoterrestris is the most widespread species of Alicyclobacillus.
Table 3. All isolated strains, their sources, number of nucleotides sequenced, nearest phylogenetic neighbour and similarity.
| Isolated strains | Source | No. of nucleotides sequenced | Nearest phylogenetic neighbour (GenBank accession number) | Similarity |
| C-ZJB-12-01 | Kiwi fruit wine (pH 3.5) | 1423 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-02 | Kiwi fruits before peeling | 1413 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-03 | s | 1424 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.6% |
| C-ZJB-12-04 | s | 1437 | Alicyclobacillus acidoterrestris (AJ133631.1) | 99.7% |
| C-ZJB-12-05 | s | 1428 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-06 | Kiwi fruits after peeling | 1428 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-07 | Kiwi fruits after color protection treatment | 1430 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-08 | Kiwi fruits before washing | 1430 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-09 | s | 1433 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-10 | Kiwi fruits after washing | 1438 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-11 | Wash water | 1432 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.7% |
| C-ZJB-12-12 | Shop environment (walls) | 1440 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.6% |
| C-ZJB-12-13 | Shop environment (floor) | 1422 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.9% |
| C-ZJB-12-14 | Shop environment (raw material bins) | 1470 | Bacillus coagulans (AB696800.1) | 98.8% |
| C-ZJB-12-15 | s | 1441 | Bacillus coagulans (AB696800.1) | 99.9% |
| C-ZJB-12-16 | s | 1441 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.2% |
| C-ZJB-12-17 | s | 1435 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.5% |
| C-ZJB-12-18 | Fruits from the orchard in Xiajiagou village | 1409 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-19 | s | 1434 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-20 | s | 1412 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.8% |
| C-ZJB-12-21 | Soil from the orchard in Xiajiagou village | 1450 | Bacillus fumarioli (AJ581126.1) | 99.7% |
| C-ZJB-12-22 | s | 1429 | Bacillus ginsengihumi (NR_041378.1) | 99.9% |
| C-ZJB-12-23 | Air from the orchard in Xiajiagou village | 1429 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.7% |
| C-ZJB-12-24 | s | 1422 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-25 | s | 1432 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-26 | s | 1409 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-27 | s | 1435 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-28 | Fruits from the orchard in Bairui Kiwi fruit Experimental Base | 1423 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.5% |
| C-ZJB-12-29 | s | 1417 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.6% |
| C-ZJB-12-30 | s | 1419 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.5% |
| C-ZJB-12-31 | s | 1423 | Alicyclobacillus acidoterrestris (AB682383.1) | 99.4% |
| C-ZJB-12-32 | s | 1430 | Alicyclobacillus contaminans (NR_041475.1) | 99.3% |
| C-ZJB-12-33 | s | 1418 | Alicyclobacillus contaminans (AB264027.1) | 99.4% |
| C-ZJB-12-34 | s | 1429 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.6% |
| C-ZJB-12-35 | s | 1412 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.6% |
| C-ZJB-12-36 | s | 1430 | Alicyclobacillus acidoterrestris (AB059675.1) | 99.5% |
| C-ZJB-12-37 | s | 1431 | Alicyclobacillus contaminans (NR_041475.1) | 99.5% |
| C-ZJB-12-38 | Soil from the orchard in Bairui Kiwi fruit Experimental Base | 1444 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-39 | s | 1433 | Bacillus fumarioli (J581126.1) | 99.8% |
| C-ZJB-12-40 | s | 1428 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-41 | s | 1451 | Alicyclobacillus contaminans (AB264027.1) | 99.2% |
| C-ZJB-12-42 | Fruits from the orchard in Changdong village | 1412 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-43 | s | 1437 | Alicyclobacillus contaminans (AB264027.1) | 99.5% |
| C-ZJB-12-44 | s | 1422 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-45 | s | 1424 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.7% |
| C-ZJB-12-46 | s | 1451 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.9% |
| C-ZJB-12-47 | s | 1455 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-48 | s | 1453 | Alicyclobacillus herbarius (AB681266.1) | 99.5% |
| C-ZJB-12-49 | s | 1437 | Alicyclobacillus contaminans (AB264027.1) | 99.6% |
| C-ZJB-12-50 | s | 1438 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.8% |
| C-ZJB-12-51 | Soil from the orchard in Changdong village | 1449 | Alicyclobacillus herbarius (AB681266.1) | 99.5% |
| C-ZJB-12-52 | s | 1426 | Alicyclobacillus herbarius (AB681266.1) | 99.4% |
| C-ZJB-12-53 | s | 993 | Alicyclobacillus herbarius (AB681266.1) | 99.7% |
| C-ZJB-12-54 | s | 1448 | Alicyclobacillus herbarius (AB681266.1) | 99.1% |
| C-ZJB-12-55 | s | 1003 | Alicyclobacillus herbarius (AB681266.1) | 99.0% |
| C-ZJB-12-56 | s | 1439 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-57 | s | 1429 | Alicyclobacillus herbarius (AB681266.1) | 99.4% |
| C-ZJB-12-58 | s | 1435 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-59 | Soil from the orchard in Liujiabao village | 1401 | Alicyclobacillus contaminans (AB264027.1) | 99.5% |
| C-ZJB-12-60 | s | 1439 | Alicyclobacillus contaminans (AB264027.1) | 99.0% |
| C-ZJB-12-61 | s | 1448 | Alicyclobacillus herbarius (AB681266.1) | 99.5% |
| C-ZJB-12-62 | s | 1449 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-63 | s | 1433 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-64 | s | 1426 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.5% |
| C-ZJB-12-65 | s | 1429 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.7% |
| C-ZJB-12-66 | s | 1431 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.6% |
| C-ZJB-12-67 | Fruits from the orchard in Shangtiantun village | 1440 | Alicyclobacillus contaminans (NR_041475.1) | 99.2% |
| C-ZJB-12-68 | s | 1452 | Alicyclobacillus contaminans (AB264027.1) | 99.5% |
| C-ZJB-12-69 | s | 1435 | Alicyclobacillus cycloheptanicus (AB680830.1) | 99.4% |
| C-ZJB-12-70 | Soil from the orchard in Shangtiantun village | 1442 | Bacillus fumarioli (AJ581126.1) | 99.9% |
| C-ZJB-12-71 | Soil from the orchard in Kiwi fruit Experimental Station of Northwest A & F University | 1422 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.8% |
| C-ZJB-12-72 | s | 1412 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-73 | Fruits from the orchard in kiwi fruit Experimental Base of Agricultural Science-technology Demonstration Park | 1434 | Alicyclobacillus acidoterrestris (AB682390.1) | 99.7% |
| C-ZJB-12-74 | Soil from the orchard in kiwi fruit Experimental Base of Agricultural Science-technology Demonstration Park | 1407 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.9% |
| C-ZJB-12-75 | s | 1428 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.6% |
| C-ZJB-12-76 | s | 1430 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-77 | s | 1452 | Alicyclobacillus herbarius (AB681266.1) | 99.3% |
| C-ZJB-12-78 | s | 1456 | Alicyclobacillus herbarius (AB681266.1) | 99.2% |
| C-ZJB-12-79 | s | 1451 | Alicyclobacillus herbarius (AB681266.1) | 99.0% |
| C-ZJB-12-80 | s | 1451 | Alicyclobacillus herbarius (AB681266.1) | 99.3% |
| C-ZJB-12-81 | Soil from the orchard in Yuechen village | 1420 | Alicyclobacillus acidoterrestris (NR_040844.1) | 99.7% |
| C-ZJB-12-82 | Soil from the orchard in Erhezhuang village | 1460 | Bacillus ginsengihumi (FJ357590.1) | 100% |
| C-ZJB-12-83 | s | 1459 | Bacillus fumarioli (AJ581126.1) | 99.9% |
| C-ZJB-12-84 | s | 1457 | Bacillus fumarioli (AJ581126.1) | 99.8% |
| C-ZJB-12-85 | Soil from the orchard in Tongyu village | 1447 | Alicyclobacillus herbarius (AB681266.1) | 98.8% |
s: same as above.
In this study, 10 strains of Alicyclobacillus contaminans (4 orchards: the orchards in Bairui Kiwi fruit Experimental Base, Changdong village, Liujiabao village, and Shangtiantun village), 19 strains of Alicyclobacillus herbarius (5 orchards: the orchards in Bairui Kiwi fruit Experimental Base, Changdong village, Liujiabao village, kiwi fruit Experimental Base of Agricultural Science-technology Demonstration Park, and Tongyu village) and 1 strains of Alicyclobacillus cycloheptanicus (Shangtiantun village) were also isolated.
One thing to be noted was that there were 9 strains of thermo-acidophilic Bacillus isolated, including 2 strains of Bacillus coagulans from shop environment (raw material bins), 5 strains of Bacillus fumarioli from soil of the orchards in 4 locations (Xiajiagou village, Shangtiantun village, Bairui Kiwi fruit Experimental Base, Erhezhuang village), and 2 strains of Bacillus ginsengihumi from soil of the orchard in Erhezhuang village and Xiajiagou village.
Phylogenetic Positions and Genotypic Diversities of Isolates
From the neighbour-joining phylogenetic tree of the 85 isolates and 4 reference species (Figure 2) we can see that all strains fell into 7 clusters. All the strains of each of the 7 species (4 Alicyclobacillus species and 3 Bacillus species) clustered together. The biggest cluster (46 isolate strains and 1 reference strain) including all Alicyclobacillus acidoterrestris strains were mainly composed of 2 groups. The bootstrap values of the branches within these 2 groups and the Alicyclobacillus contaminans cluster and the Alicyclobacillus herbarius cluster were low (some bootstrap values were even below 30), which indicated that the differences among these strains were very small according to their 16S rDNA sequences.
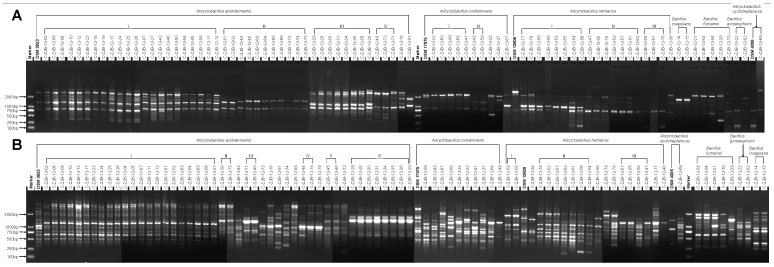
Genotypic diversities of all 85 isolates and 4 reference species were evaluated by RAPD-PCR (Figure 3). Generally, the RAPD-PCR with primer AZ-14 (pattern B in Figure 3) generated more bands after electrophoresis than with primer A-01 (pattern A in Figure 3), which might be caused by the relatively higher affinity of primer AZ-14 to the genomic DNA of Alicyclobacillus. Therefore, the genotypic diversity revealed through RAPD-PCR with primer AZ-14 was bigger than with primer A-01. In detail, in pattern A, within the cluster of Alicyclobacillus acidoterrestris, there were 4 groups and 2 unique strains which were different from each other, as well as from any strains of the 4 groups. Whereas in pattern B, there were 6 groups and 7 unique strains in the same cluster. But the biggest 2 groups in this cluster were similar. Group I was composed of 22 strains in both pattern A and B, and 16 strains of them were the same, while group III in pattern A and group VI in pattern B were composed of exactly the same 8 strains, which agreed with the result of the phylogenetic tree’s corresponding part precisely. Within the cluster Alicyclobacillus contaminans there were 2 groups and 3 unique strains in pattern A, but each strain including the reference strain DSM 17975 belong to this cluster in pattern B was different from each other, which suggested there might exist great genotypic diversity within this species of Alicyclobacillus in the kiwi fruit production area in Shaanxi province of China. Although there were 3 groups within the cluster Alicyclobacillus herbarius in both 2 patterns, but pattern B still showed bigger genotypic diversity, with 6 unique strains, whereas there was only 1 unique strain in pattern A. But the 2 patterns also shared some similarity. There were 2, 5, and 2 same strains between group I, II and III, respectively.
Figure 3. Electrophoresis patterns of RAPD-PCR of the genomic DNA of isolates and reference species.
The 4 reference species are Alicyclobacillus acidoterrestris DSM 3923, Alicyclobacillus cycloheptanicus DSM 4006, Alicyclobacillus contaminans DSM 17975, Alicyclobacillus herbarius DSM 13609. I–VI indicate groups within a certain species. Marker, DNA marker DL2000 (BGI). A, Primer A-01; B, Primer AZ-14.
Growth and Taint Production
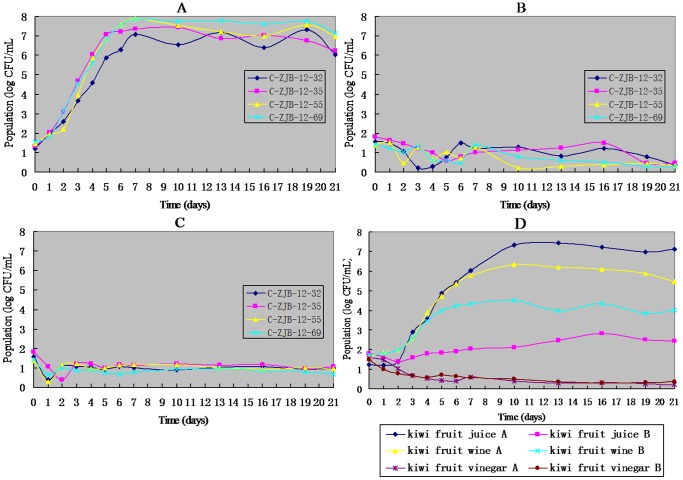
Changes in microbial populations over incubation are shown in Figure 4. Replicate uninoculated control samples of 2 kinds of kiwi fruit juice and 6 commercial kiwi fruit products had undetectable Alicyclobacillus populations (<30 CFU/mL) over incubation (data not shown). Microbial population levels of all pH adjusted kiwi fruit juice samples reached >107 CFU/mL within 7 days of incubation, and the growth curves of all 4 test strains were similar. All curves in Figure 4B did not seem extending regularly, which might be caused by the testing errors (with a inoculation volume of 100 µL, there usually were only <5 CFUs on a plate). But it was obvious that there was hardly any growth in all kiwi fruit juice samples without pH adjustment. In all these samples a certain number of spores kept their abilities of reproduction throughout the whole incubation time (Figure 4C). One juice sample of the six commercial kiwi fruit products samples reached population levels >107 CFU/mL within 10 days, and the 2 wine samples accumulated Alicyclobacillus population levels 106 and 104, respectively. Alicyclobacillus did not grow obviously in the rest samples of this group.
Figure 4. Changes in microbial populations during the 45°C incubation.
A, pH adjusted kiwi fruit juice inoculated with 4 strains of Alicyclobacillus; B, kiwi fruit juice inoculated with 4 strains of Alicyclobacillus; C, kiwi fruit juice inoculated with 4 strains of Alicyclobacillus (with a heat shock treatment); D, six kinds commercial kiwi fruit products inoculated with C-ZJB-12-35.G.
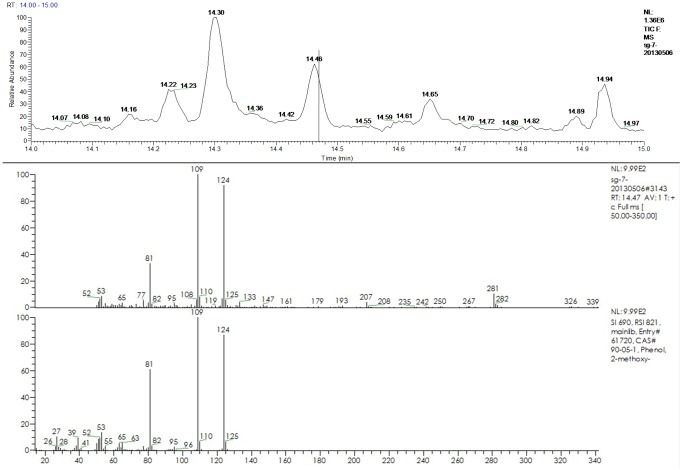
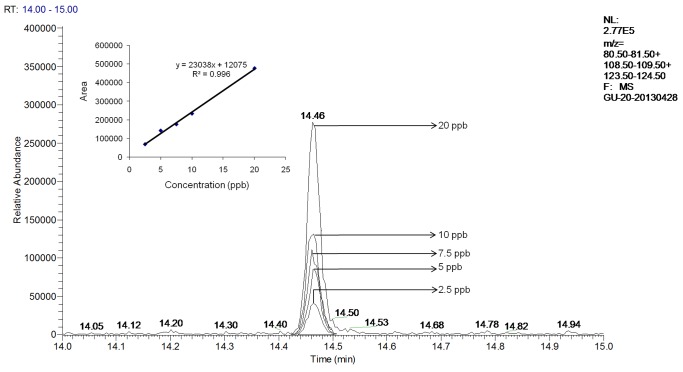
A GC–MS chromatogram, a mass spectrum from the peak thought to be guaiacol in the sample of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-35 and a mass spectrum of standard guaiacol are shown in Figure 5. The compound peaked at 14.47 min should be guaiacol based on the values of SI and RSI given by the NIST 2002 library and comparison with the retention time of standard solutions. Three major mass fragments of guaiacol (mass to charge, m/z, 81,109, 124) can be seen in Figure 5. The GC–MS chromatograms of other chosen samples were similar to the above one, according to which guaiacol were produced by all chosen samples. No 2,6-DBP and 2,6-DCP were detected. A GC–MS chromatogram of all standard solutions and a standard curve generated by Excel are shown in Figure 6. The mass spectrums of the peaks at 14.47 min in the TIC chromatograms of the contrast 5 ppb standard solution for guaiacol in kiwi fruit juice and the sample of commercial kiwi fruit wine A show low SI and RSI values of many compounds through NIST library. The reason might be there were several compounds detected at this retention time or the limit of the instrument precision, and changing a more suitable column or optimizing the conditions of microextraction and GC-MS might obtain better results. The peak areas of guaiacol in samples and the calculated concentrations of guaiacol are shown in Table 4 (see Chromatogram S1-S6 for their raw data files). According to the standard curve all the samples accumulated guaiacol between 11–34 ppb after the 21 days incubation.
Figure 5. A GC–MS chromatogram of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-35.
Mass spectrums from the peak (14.47 min) thought to be guaiacol in this sample and from standard guaiacol are also shown. The compound peaked at 281 m/z may come from the SPME fiber, because it exist in the peak around 14.47min in all samples. Because the scanning mass range was 50–350 m/z, peaks below 50 m/z were not included. Both of these may reduce the SI and RSI values.
Figure 6. A GC–MS chromatogram of all standard solutions and a standard curve generated by Excel.
For comparison purposes only peaks between the mass range of 81, 109, 124 m/z are displayed.
Table 4. The peak areas of guaiacol of samples and the calculated concentrations.
| Samples | Start Retention Time (min) | End Retention Time (min) | Peak area | Concentration (ppb) |
| C-ZJB-12-32 in A | 14.44 | 14.54 | 280389.50 | 11.65 |
| C-ZJB-12-35 in A | 14.44 | 14.53 | 435151.81 | 18.36 |
| C-ZJB-12-55 in A | 14.43 | 14.52 | 678012.37 | 28.91 |
| C-ZJB-12-69 in A | 14.44 | 14.50 | 812313.79 | 34.74 |
| C-ZJB-12-35 in B | 14.43 | 14.50 | 650602.09 | 27.72 |
According to the standard curve, concentrations were calculated based on the peak areas (mass range: 81, 109, 124 m/z).
A: pH adjusted kiwi fruit juice; B: commercial kiwi fruit juice A.
Discussion
The decline of percentages of positive samples from orchards to producer shops and commercial products reveals the distribution rule of microbes in a whole production line. There seem no clear relations among the percentages of positive samples of the 16 orchards, and the percentages of some orchards (Gengxi village, Luoyukou village, Chenjiagou village) are even 0%, despite their number of samples. This may be because of the sampling locations. Positive samples are likely from drop fruits and the soil around them (data not shown). Therefore, there are probably other species of Alicyclobacillus except the 4 species found in this study because of the randomness and limited quantity of sampling.
The result that Alicyclobacillus acidoterrestris is the most widespread species of Alicyclobacillus in this study is similar to many previous reports [36], [17], [14]. Alicyclobacillus acidoterrestris is the most important spoilage microbe of juice products and off-odour (mainly guaiacol) producer [18], [37], therefore there will be a potential risk of spoilage when more and more kiwi fruits from these orchards are used as raw materials for juice, wine and other products in the future. Except 2 isolates, all the isolates from all sampling sections of the producer, including the raw material orchard, processing shops and final products, are identified as Alicyclobacillus acidoterrestris. This result indicates that the potential risk mentioned above truly exists, which is also proved by the fact that the only one isolate from commercial kiwi fruit products is identified as Alicyclobacillus acidoterrestris too. If more producers’ shops are tested, other species of Alicyclobacillus may be found.
To our knowledge, this is the first report on the isolation of Alicyclobacillus contaminans and Alicyclobacillus herbarius from China’s orchards. Alicyclobacillus herbarius and Alicyclobacillus cycloheptanicus has also been proved as taint producers [27] [36]. Therefore, kiwi fruit products coming from these orchards may be contaminated by these non-acidoterrestris Alicyclobacillus.
Alicyclobacillus acidoterrestris and Alicyclobacillus contaminans were widely distributed among various fruit orchards in Japan [36]. According to the results of this study, it is thought that Alicyclobacillus acidoterrestris, Alicyclobacillus contaminans, and Alicyclobacillus herbarius might be the predominant species of Alicyclobacillus in kiwi fruit orchards in Shaanxi province of China.
The D80°C of Bacillus coagulans is 40 min in a double concentrated tomato paste media according to Sandoval et al. [38], and Bacillus fumarioli grows optimally at pH 5.5 and 50°C [39], while Bacillus ginsengihumi can also grow in acid media at temperature up to 50°C [40]. These reports proved that these Bacillus can be an interference to the detection of Alicyclobacillus from samples and cause false positive results, although there are no reports relating them with spoilage of juice products and guaiacol production.
Through the analysis of the 2 patterns it was thought the division of genotypic diversities among a relatively large number of samples (as in this study) though PAPD-PCR with different primers may be different because of the different affinity between the primer and the sample strain’s genomic DNA. There seems no large possibility of RAPD-PCR as a traceability method of Alicyclobacillus contamination, because the RAPD-PCR results showed that one certain genotypic group within a cluster of a certain species could be isolated from different sources (such as group I of the cluster of Alicyclobacillus acidoterrestris, which included strains from 3 sources in pattern A and 4 sources in pattern B, with the plant of the producer of frozen kiwi fruit slice as one source), and strains from one source could be divided into different groups within a cluster of a certain species (such as strains from Xiajiagou village [C-ZJB-12-18 ∼ C-ZJB-12-27] which belonged to Alicyclobacillus acidoterrestris were in 2 groups in pattern A and 3 groups in pattern B). These results show some differences from Groenewald’s [17] report.
From Figure 4A,B and C we can see that the main reason for Alicyclobacillus to unable to grow in raw kiwi fruit juice is not its containing some bacteriostatic factors, but its relatively low pH (2.5) compared with other fruit juices (apple and orange, pH 3.5–4.0), and their spores can retain viability in raw kiwi fruit juice and grow and produce taint like in apple and orange juices when the pH rises while dilution or mixing with other relatively high pH juices to make palatable commercial products. Figure 4D proves this conclusion further. The growth curve in the commercial kiwi fruit juice A products are similar to the ones in pH adjusted kiwi fruit juice, only showing longer lag phase and longer time (about 8–10 days) to reach the population levels of 107 CFU/mL, which may be because of the lower pH and different media contents compared with pH adjusted kiwi fruit juice. The reason why C-ZJB-12–35 is unable to grow in the 2 vinegar samples may lie in their low pH (2.5). The alcohol in the 2 wine samples might have negative influence on the growth of Alicyclobacillus, but according to the GC-MS chromatogram of wine A, a small amount of guaiacol should exist in the sample, although guaiacol might not be separated well with other compounds. As for another commercial juice which unable to support the growth of C-ZJB-12-35, it is inferred that it does not contain enough raw kiwi fruit juice as described on its packaging (no antiseptics found).
Guaiacol has been found in all 5 samples proved that it is the most important taint compound, which is similar to Pettipher’s report [28]. It has been proved that the best estimated threshold (BET) value for odour of guaiacol was around 2 ppb in apple juice [28], and similar threshold values for odour of guaiacol in apple juice were even lower in other reports [41] [42]. Although there are still no reports about such values in kiwi fruit juice, it is thought they might not be much higher than in apple juice, therefore the guaiacol levels in this study are high enough to make a consumer aware of the spoilage. The different amount of guaiacol production among the 5 samples might be attributed to the differences among species, cell numbers. Goto et al. [36] has proved Alicyclobacillus herbarius to produce guaiacol, although in the same study, they did not detect guaiacol from Alicyclobacillus contaminans, which may be because of the limit of the test method and the culture media. In this study, we first proved that Alicyclobacillus contaminans could produce guaiacol (11.65 ppb) in pH adjusted kiwi fruit juice. There were no halophenols (2,6-DBP and 2,6-DCP) detected in all samples in this study, although Alicyclobacillus acidoterrestris has been proved to produce guaiacol and 2,6-DBP, and Alicyclobacillus cycloheptanicus has been proved to produce guaiacol and both 2,6-DBP and 2,6-DCP in Gocmen’s study [27]. This might be attributed to the media used in this study.
Although there was only one Alicyclobacillus strain isolated from commercial kiwi fruit products, Alicyclobacillus strains were isolated from kiwi fruit product processing plant and raw material orchards, and there existed genotypic diversities to some extent. Furthermore, the test strains from all 4 species isolated in this study can spoil the pH adjusted kiwi fruit juice and some commercial products, and their spores can retain viability in raw kiwi fruit juice. Therefore, there do exist a potential risk of Alicyclobacillus contamination, even spoilage incidents, when a large quantity of kiwi fruits were used as raw materials to make juice or wine or other products, although there is no report about such incidents yet in China. Because of the similarity of 16S rDNA between one species and its subspecies could beyond 99% in genus Alicyclobacillus (e.g. between some strains of Alicyclobacillus acidocaldarius and Alicyclobacillus acidocaldarius subsp. Rittmannii after searching through NCBI), the genotypic diversities (through RAPD-PCR) of these isolates indicate that there might be some new subspecies among them. Therefore, further research will be focused on the growth conditions and off-odour production of all isolated Alicyclobacillus in kiwi fruit juice and commercial products to further assess their capability of contamination, and looking for new subspecies to expand our knowledge about Alicyclobacillus.
Supporting Information
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-32.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-55.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-69.
(RAW)
The GC-MS raw data file of commercial kiwi fruit juice A inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of commercial kiwi fruit wine A inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of 5-ppb standard solution for guaiacol in kiwi fruit juice.
(RAW)
Funding Statement
This study was financially supported by the China State “12th-Five-Year Plan” Scientific and Technological Schemes Support (2012BAK17B06) (http://www.most.gov.cn/), and the National Natural Science Foundation of China (31071550, 31171721)(http://www.nsfc.gov.cn/).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Uhino F, Doi S (1967) Acido-thermophilic bacteria from thermal waters. Agricultural Biology and Chemistry 31: 817–822. [Google Scholar]
- 2. Wisotzkey JD, Jurtshuk P, Fox GE, Deinhard G, Poralla K (1992) Comparative sequence analysis on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus. Alicyclobacillus gen. nov. International Journal of Systematic Bacteriology 42: 263–269. [DOI] [PubMed] [Google Scholar]
- 3. Borlinghaus A, Engel R (1997) Alicyclobacillus incidence in commercial apple juice concentrate (AJC) supplies –method development and validation. Fruit Processing 7: 262–266. [Google Scholar]
- 4. Walls I, Chuyate R (2000) Spoilage of fruit juices by Alicyclobacillus acidoterrestris . Food Australia 52: 286–288. [PubMed] [Google Scholar]
- 5. Eiroa MNU, Junqueira VCA, Schmidt FL (1999) Alicyclobacillus in orange juice: occurrence and heat resistance of spores. Journal of Food Protection 62: 883–886. [DOI] [PubMed] [Google Scholar]
- 6. Blocher JC, Busta FF (1983) Bacterial spore resistance to acid. Food Technology 37: 87–99. [Google Scholar]
- 7. Cerny G, Hennlich W, Poralla K (1984) Spoilage of fruit juice by bacilli: isolation and characterisation of the spoiling microorganism. Zeitschrift feur Lebensmittel Untersuchung und Forsuchung 179: 224–227. [DOI] [PubMed] [Google Scholar]
- 8. Steyn CE, Cameron M, Witthuhn RC (2011) Occurrence of Alicyclobacillus in the fruit processing environment–A review. International Journal of Food Microbiology 147: 1–11. [DOI] [PubMed] [Google Scholar]
- 9. Smit Y, Cameron M, Venter P, Witthuhn RC (2011) Alicyclobacillus spoilage and isolation–A review. Food Microbiology 28: 331–349. [DOI] [PubMed] [Google Scholar]
- 10. Splittstoesser DF, Churey JJ, Lee CY (1994) Growth characteristics of aciduric spore forming bacilli isolated from fruit juices. Journal of Food Protection 57: 1080–1083. [DOI] [PubMed] [Google Scholar]
- 11. Chang SS, Kang DH (2004) Alicyclobacillus spp. in the fruit juice industry: history, characteristics, and current isolation/detection procedures. Critical Reviews in Microbiology 30: 55–74. [DOI] [PubMed] [Google Scholar]
- 12. Walker M, Phillips CA (2008) Alicyclobacillus acidoterrestris: an increasing threat to the fruit juice industry? International Journal of Food Science and Technology 43: 250–260. [Google Scholar]
- 13.Howard I (2006) ACB Workshop October 2005–Review. European Quality Control System (EQCS) Workshop 2006. Available: http://www.eqcs.org/download/Workshop2006/07_ACBworkshop2005.pdf.
- 14. Durak MZ, Churey JJ, Danyluk MD, Worobo RW (2010) Identification and haplotype distribution of Alicyclobacillus spp. from different juices and beverages. International Journal of Food Microbiology 142: 286–291. [DOI] [PubMed] [Google Scholar]
- 15. Deinhard G, Blanz P, Poralla K, Altan E (1987) Bacillus acidoterrestris sp. nov., a new thermotolerant acidophile isolated from different soils. Systematic and Applied Microbiology 10: 47–53. [Google Scholar]
- 16. Wisse CA, Parish ME (1998) Isolation and enumeration of sporeforming, thermoacidophilic, rod-shaped bacteria from citrus processing environments. Diary, Food and Environmental Sanitation 18: 504–509. [Google Scholar]
- 17. Groenewald WH, Gouws PA, Witthuhn RC (2009) Isolation, identification and typification of Alicyclobacillus acidoterrestris and Alicyclobacillus acidocaldarius strains from orchard soil and the fruit processing environment in South Africa. Food Microbiology 26: 71–76. [DOI] [PubMed] [Google Scholar]
- 18. Walls I, Chuyate R (1998) Alicyclobacillus – historical perspective and preliminary characterization study. Dairy, Food and Environmental Sanitation 18: 99–503. [Google Scholar]
- 19. Previdi MP, Lusardi E, Vicini E (1999) Resistance of Alicyclobacillus spp. spores to a disinfectant. Industria conserve 74: 231–236. [Google Scholar]
- 20. Chen S, Tang Q, Zhang X, Zhao G, Hu X, et al. (2006) Isolation and characterization of thermo-acidophilic endosporeforming bacteria from the concentrated apple juice-processing environment. Food Microbiology 23: 439–445. [DOI] [PubMed] [Google Scholar]
- 21. Goto K, Mochida K, Kato Y, Asahara M, Ozawa C, et al. (2006) Diversity of Alicyclobacillus isolated from fruit juices and their raw materials, and emended description of Alicyclobacillus acidocaldarius . Microbiological Culture Collections 22: 1–14. [Google Scholar]
- 22. Baumgart J, Menje S (2000) The impact of Alicyclobacillus acidoterrestris on the quality of juices and soft drinks. Fruit Processing 10: 251–254. [Google Scholar]
- 23. Gouws PA, Gie L, Pretorius A, Dhansay N (2005) Isolation and identification of Alicyclobacillus acidocaldarius by 16S rDNA from mango juice and concentrate. International Journal of Food Science and Technology 40: 789–792. [Google Scholar]
- 24. Pinhatti MEMC, Variane S, Eguchi SY, Manfio GP (1997) Detection of acidothermophilic Bacilli in industrialized fruit juices. Fruit Processing 9: 350–353. [Google Scholar]
- 25. Oteiza JM, Ares G, Sant'Ana AS, Soto S, Giannuzzi L (2011) Use of a multivariate approach to assess the incidence of Alicyclobacillus spp. in concentrate fruit juices marketed in Argentina: Results of a 14-year survey. International Journal of Food Microbiology 151: 229–234. [DOI] [PubMed] [Google Scholar]
- 26. Huang H, Ferguson AR (2001) Review: Kiwifruit in China. New Zealand Journal of Crop and Horticultural Science 29: 1–14. [Google Scholar]
- 27. Gocmen D, Elston A, Williams T, Parish M, Rouseff RL (2005) Identification of medicinal off-flavours generated by Alicyclobacillus species in orange juice using GC-olfactometry and GC-MS. Letters in Applied Microbiology 40: 172–177. [DOI] [PubMed] [Google Scholar]
- 28. Pettipher GL, Osmundson ME, Murphy JM (1997) Methods for the detection and enumeration of Alicyclobacillus acidoterrestris and investigation of growth and production of taint in fruit juice and fruit juice-containing drinks. Letters in Applied Microbiology 24: 185–189. [DOI] [PubMed] [Google Scholar]
- 29.International Federation of Fruit Juice Producers (IFU) (2007) Method on the Detection of Taint Producing Alicyclobacillus in Fruit Juices. IFU Method No. 12. IFU, Paris, 1–11. [Google Scholar]
- 30. Goto K, Tanimoto Y, Tamura T, Mochida K, Arai D, et al. (2002) Identification of thermoacidophilic bacteria and a new Alicyclobacillus genomic species isolated from acidic environments in Japan. Extremophiles 6: 333–340. [DOI] [PubMed] [Google Scholar]
- 31. Jiang CY, Liu Y, Liu YY, You XY, Guo X, et al. (2008) Alicyclobacillus ferrooxydans sp. nov., a ferrous-oxidizing bacterium from solfataric soil. International Journal of Systematic and Evolutionary Microbiology 58: 2898–2903. [DOI] [PubMed] [Google Scholar]
- 32. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 33. Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 34. Danyluk MD, Friedrich LM, Jouquand C, Goodrich-Schneider R, Parish ME, et al. (2011) Prevalence, concentration, spoilage, and mitigation of Alicyclobacillus spp. in tropical and subtropical fruit juice concentrates. Food Microbiology 28: 472–477. [DOI] [PubMed] [Google Scholar]
- 35. Zierler B, Siegmund B, Pfannhauser W (2004) Determination of off-flavour compounds in apple juice caused by microorganisms using headspace solid phase microextraction–gas chromatography–mass spectrometry. Analytica Chimica Acta 520: 3–11. [Google Scholar]
- 36. Goto K, Nishibori A, Wasada Y, Furuhata K, Fukuyama M, et al. (2008) Identification of thermo-acidophilic bacteria isolated from the soil of several Japanese fruit orchards. Letters in Applied Microbiology 46: 289–294. [DOI] [PubMed] [Google Scholar]
- 37. Jensen N, Whitfield FB (2003) Role of Alicyclobacillus acidoterrestris in the development of a disinfectant taint in shelf-stable fruit juice. Letters in Applied Microbiology 36: 9–14. [DOI] [PubMed] [Google Scholar]
- 38. Sandoval AJ, Barreiro JA, Mendoza S (1992) Thermal Resistance of Bacillus coagulans in Double Concentrated Tomato Paste. Journal of Food Science 57: 1369–1370. [Google Scholar]
- 39. Clerck ED, Gevers D, Sergeant K, Rodríguez-Díaz M, Herman L, et al. (2004) Genomic and phenotypic comparison of Bacillus fumarioli isolates from geothermal Antarctic soil and gelatine. Research in Microbiology 155: 483–490. [DOI] [PubMed] [Google Scholar]
- 40. Ten LN, Im WT, Baek SH, Lee JS, Oh HM, et al. (2006) Bacillus ginsengihumi sp nov., a novel species isolated from soil of a ginseng field in Pocheon Province, South Korea. Journal of Microbiology and Biotechnology 16: 1554–1560. [Google Scholar]
- 41. Eisele TA, Semon MJ (2005) Best estimated aroma and taste detection threshold for guaiacol in water and apple juice. Journal of Food Science 70: 267–269. [Google Scholar]
- 42. Siegmund B, Pöllinger-Zierler B (2006) Odor thresholds of microbially induced off flavor compounds in apple juice. Journal of Agricultural and Food Chemistry 54: 5984–5989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-32.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-55.
(RAW)
The GC-MS raw data file of pH adjusted kiwi fruit juice inoculated with C-ZJB-12-69.
(RAW)
The GC-MS raw data file of commercial kiwi fruit juice A inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of commercial kiwi fruit wine A inoculated with C-ZJB-12-35.
(RAW)
The GC-MS raw data file of 5-ppb standard solution for guaiacol in kiwi fruit juice.
(RAW)