Abstract
Background
The mechanism for the contribution of eosinophils (EOS) to asthma pathophysiology is not fully understood. Genome-wide expression analysis of airway EOS by microarrays has been limited by the ability to generate high quality RNA from sufficient numbers of airway EOS.
Objective
To identify, by genome-wide expression analyses, a compendium of expressed genes characteristic of airway EOS following an in vivo allergen challenge.
Methods
Atopic, mild asthmatic subjects were recruited for these studies. Induced sputum was obtained before and 48h after a whole lung allergen challenge (WLAC). Individuals also received a segmental bronchoprovocation with allergen (SBP-Ag) 1 month before and after administering a single dose of mepolizumab (anti-IL-5 monoclonal antibody) to reduce airway EOS. Bronchoalveolar lavage (BAL) was performed before and 48 h after SBP-Ag. Gene expression of sputum and BAL cells was analyzed by microarrays. The results were validated by qPCR in BAL cells and purified BAL EOS.
Results
A total of 299 transcripts were up-regulated by more than 2-fold in total BAL cells following SBP-Ag. Mepolizumab treatment resulted in a reduction of airway EOS by 54.5% and decreased expression of 99 of the 299 transcripts. 3 of 6 post-WLAC sputum samples showed increased expression of EOS-specific genes, along with the expression of 361 other genes. Finally, the intersection of the 3 groups of transcripts (increased in BAL post SBP-Ag (299), decreased after mepolizumab (99), and increased in sputum after WLAC (365)) was composed of 57 genes characterizing airway EOS gene expression.
Conclusion
We identified 57 genes that were highly expressed by BAL EOS compared to unseparated BAL cells after in vivo allergen challenge. 41 of these genes had not been previously described in EOS and are thus potential new candidates to elucidate EOS contribution to airway biology.
Introduction
Recruitment of EOS to the lung has been reproducibly reported in allergic asthma [1]. While airway eosinophilia is commonly associated with increased risk for asthma exacerbation, severity, and poor prognosis [2]–[4], the precise correlation of EOS to the pathophysiology of asthma remains controversial. Reduction of airway EOS is associated with decline of submucosal matrix protein deposition and airway smooth muscle hyperplasia [5], [6] suggesting that EOS contribute to airway remodeling. Through the production and release of pro-inflammatory mediators, EOS can amplify the expression of Th1, Th2, and Th17 cytokines and chemokines [7]–[9] indicating they play a role in the adaptive immune response. Recent trials of anti-IL-5 antibodies (mepolizumab and reslizumab) have shown benefits in asthma, particularly in reducing rates of exacerbations [10]–[12].
One approach to understanding the biology of EOS in asthma is gene expression analysis by microarrays. Initial GeneChip analysis, which was performed using in vitro IL-5-activated circulating EOS, identified 66 genes that were up-regulated by IL-5 and predicted to have functions in adhesion, recruitment, activation and survival [13]. A subsequent study performed by our group showed that the expression of more than 200 genes was increased in vitro in IL-5- and GM-CSF-activated EOS, including the anti-apoptotic serine/threonine protein kinase Pim-1 [14], [15]. During their egress to the airway, in response to allergen, the phenotype of peripheral blood EOS changes dramatically [16]–[18]; however, gene analysis with microarrays of airway EOS has not been explored.
We performed gene expression array analysis on sputum samples obtained following whole lung allergen challenge (WLAC), and on bronchoalveolar lavage (BAL) cells obtained following segmental bronchoprovocation with an allergen (SBP-Ag). These two in vivo allergen challenge models are well-established asthma models that lead to eosinophilic airway inflammation [19]–[21]. Typically SBP-Ag causes EOS to increase in the BAL from ∼0.5% at baseline to ∼70% after allergen challenge [9] while WLAC induces an increase of EOS in sputum from ∼3% to ∼10% [21]–[24]. Therefore, we anticipated that analysis of total BAL and sputum cells by microarrays after SBP-Ag and WLAC and purified BAL EOS would facilitate identification of genes specifically expressed by airway EOS.
Materials and Methods
Subjects
The study was approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board (IRB). Informed written consent was obtained from subjects prior to participation. Subjects had a history of mild atopic asthma as defined by at least one positive skin prick test and a history of allergen-induced asthma exacerbation with reversibility to albuterol >12% and/or a provocative concentration of methacholine producing a 20% fall in FEV1 (PC20) of <8 mg/ml. None of the subjects were using inhaled or oral corticosteroids. 10 subjects finished the SBP-Ag protocol and 2 were excluded for lower percentage of EOS in BAL after SBP-Ag (<67%) or low purity of BAL EOS (<99%). 18 subjects completed the sputum protocol and 12 were excluded due to having <500,000 cells, greater than 60% epithelial cells in the sputum, a greater than 2-fold difference in the percentage of epithelial cells pre- and post- WLAC, lack of RNA after extraction, or RNA integrity that was insufficient for microarrays.
Bronchoscopy, SBP-Ag, and Anti-IL-5 Administration
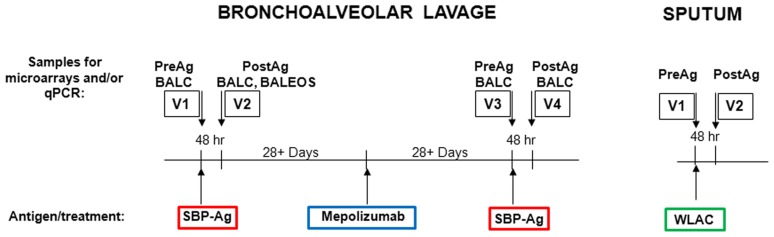
Detailed methods for bronchoscopy, SBP-Ag, and BAL cell preparation have previously been described [25]. A graded inhaled Ag challenge with Dermatophagoides farinae (Der p 1, dust mite), Felis catus domesticus (Fel d 1, cat) or Ambrosia artemisiifolia (Amb a 1, ragweed), obtained from Greer Laboratories (Lenoir, NC, USA) was performed to determine the participant’s AgPD20 (the provocative dose of Ag leading to a 20% fall in FEV1). One month later, a baseline bronchoscopy with BAL (4 × 40 ml aliquots of sterile 0.9% NaCl) was performed followed by administration of Ag at a dose of 20% of the AgPD20 into the lavaged segment (BAL V1, Figure 1). Bronchoscopy with BAL was repeated 48 h later (BAL V2, Figure 1). One month after the first SBP-Ag, a single dose of anti-IL-5 (750 mg, mepolizumab generously donated by GlaxoSmithKline) was administered intravenously to reduce the number of airway EOS. One month after mepolizumab treatment, the second SBP-Ag was performed with BAL immediately before (BAL V3) and 48 h later (BAL V4). Circulating EOS number was <100 per µl at the time of the second SBP-Ag (BAL V3). BAL cells were assessed by hemocytometer using Turk’s counting solution containing acetic acid and methylene blue and cell differentials were determined on cytospin preparations stained with the Wright-Giemsa-based Hema-3 (ThermoFisher/Thermo Scientific, Rockford, IL, USA). On BAL V2 (Figure 1), airway EOS were purified from BAL cells using a two-step Percoll gradient as previously described [8]. EOS were collected from the 1.085/1.100 g/ml interface.
Figure 1. Bronchoalveolar lavage and sputum timelines.
Atopic mild asthmatics underwent bronchoalveolar lavage (BAL) followed by segmental challenge with an allergen (SBP-Ag) on visit 1 (BAL V1). 48 h later on visit 2 (BAL V2), BAL was performed at the site of the challenged segment. One month later, a single dose of mepolizumab was administered. One month after dosing, BALs were repeated both before and after SBP-Ag. BAL cells (BALC) were prepared on visits 1, 2, 3, and 4 and airway EOS (BALEOS) were purified on visit 2. For the induced sputum study, sputum collection was performed followed by a whole lung allergen challenge (WLAC) on visit 1 (sputum V1). Collection of a second induced sputum was done 48 h after WLAC on visit 2 (sputum V2).
Induced Sputum
Induced sputum was obtained as previously described [22] on visit 1 (sputum V1, Figure 1) and 48 h after a graded WLAC leading to a 20% fall in FEV1 (sputum V2). Briefly, patients inhaled nebulized 3% saline for 5 minutes, rinsed out their mouth and then coughed with collection of the sputum produced. This was repeated a total of three times. The sputum was diluted 1∶1 with PBS. After centrifugation, cytospins were prepared and stained with Giemsa to determine cell distributions. Cells were stored in Trizol solution for RNA purification.
RNA Preparation and Microarray Hybridization
Total RNA was extracted from BAL cells or purified BAL EOS using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), treated with DNase (RNase-free DNase kit, Qiagen), and RNA from BAL cells were submitted to the University of Wisconsin-Madison Gene Expression Center (Madison, WI) for labeling and hybridization. RNA quality and integrity was evaluated via Agilent 2100 Bioanalyzer platform (Agilent Technologies). Sense-strand cDNA was generated from total RNA using the Ambion Whole Transcript (WT) Target Expression Kit (Life Technologies, Grand Island, NY, USA). Fragmentation and labeling of the single-stranded cDNA was performed using the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA). The labeled cDNAs were hybridized to Human Gene ST 1.0 GeneChips (Affymetrix) according to the manufacturer’s protocols.
Induced sputum samples were shipped to Miltenyi Biotech for RNA extraction using standard RNA Trizol extraction protocols. Sample integrity was evaluated via Agilent 2100 Bioanalyzer platform (Agilent Technologies). 50 ng of RNA was amplified and labeled using the Aglient Low Input Quick Amp Labeling Kit (AgilentTechnologies). Hybridization was performed using the Agilent Gene Expression Hybridization Kit to Agilent Whole Human Genome Oligo Microarrays 8X60K for microarray analyses. The Agilent Feature Extraction Software was used to read out and process the microarray image files.
Microarray Data Analysis
For BAL cells, raw data (CEL files) were uploaded into ArrayStar software version 4.0 (DNASTAR Inc., Madison, WI, USA) for normalization and statistical analysis. The robust multichip analysis (RMA) algorithm was used for background correction, quantile normalization and median polish summarization. Microarrays are deposited in the Gene Expression Omnibus repository (accession number: GSE46159). Four pairs of data comparisons that might reveal genes up-regulated in BAL cells after allergen challenge with or without mepolizumab were performed. The transcripts were filtered on the basis of ≥2-fold difference. The 99 genes up-regulated by SBP-Ag and decreased 1.5-fold by mepolizumab (Table S5) were determined using both the direct fold change between BAL V4 and V2 and the fold change between BAL V2 and V1 divided by the fold change between BAL V4 and V3 (see Figure 1).
For induced sputum samples, raw data files were analyzed using the Agilent Feature Extraction Software (FES) (Agilent Technologies). The software was used to determine feature intensities, perform background subtraction, reject outliers, and calculate statistical confidences. Microarrays are deposited in the Gene Expression Omnibus repository (accession number: GSE46238).To determine differential gene expression, the data outputted from FES was then analyzed using the Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware). Ratios were calculated as control vs. sample, or before and after allergen challenge, by division of sample intensity through control signal intensity.
Real-time qPCR
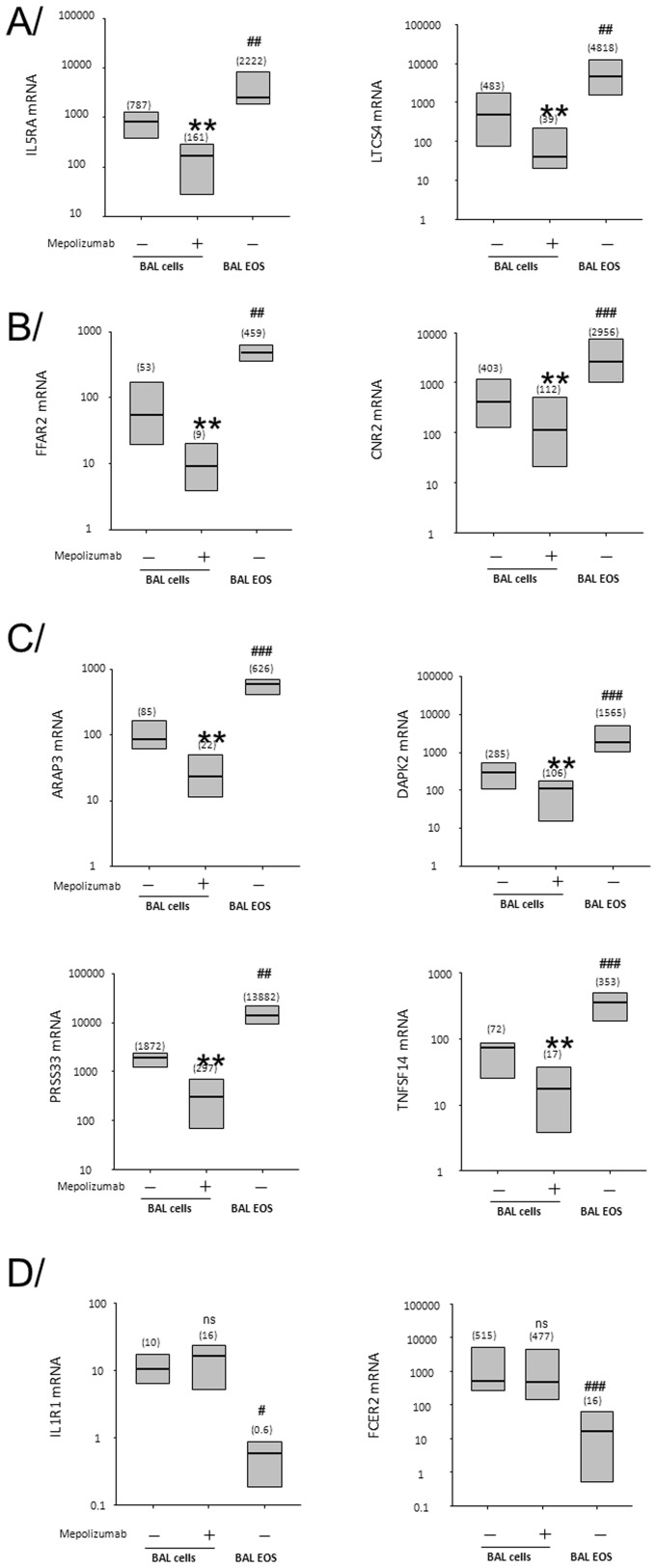
The reverse transcription reaction was performed using the Superscript III system (Invitrogen/Life Technologies, Grand Island, NY, USA). mRNA expression was determined by qPCR using SYBR Green Master Mix (SABiosciences, Frederick, MD, USA) and human IL5RA, LTC4S, FFAR2, CNR2, ARAP3, DAPK2, PRSS33, TNFSF14, IL1R1 and FCER2 (CD23ß) forward and reverse specific primers (see Table S1 for primer sequences) were designed using Primer Express 3.0 (Applied Biosystems, Carlsbad, CA, USA) and blasted against the human genome to determine specificity using http://www.ncbi.nlm.nih.gov/tools/primer-blast. The reference gene, ß-glucuronidase ((GUSB), forward: caggacctgcgcacaagag, reverse: tcgcacagctggggtaag), was used to normalize the samples. Standard curves were performed and efficiencies were determined for each set of primers. Efficiencies ranged between 91 and 96%. Data are expressed as fold change using the comparative cycle threshold (ΔΔCT) method as described previously [9]. The values presented in Figure 4 are fold change = (2−ΔΔCt) compared to expression in BAL cells at V1 (before SBP-Ag).
Figure 4. Validation by qPCR of the 57 genes associated with airway EOS after airway allergen challenge.
qPCR was performed on BAL cells before SBP-Ag (BAL V1 as shown on Figure 1), 48 h after allergen with (V4) or without mepolizumab treatment (V2), and on purified BAL EOS (BALEOS) after allergen challenge (no mepolizumab, V2). Fold changes compared to expression in BAL cells before allergen challenge and mepolizumab (V1) are presented. Box plots depict the median and the interquartile range between the 25th and the 75th percentiles. 6 subjects are included in each group and means are presented in parentheses. ** indicates that mRNA level in BAL cells is significantly decreased by mepolizumab compared to BAL cells on V2 (p<0.01). # indicates that mRNA level in BAL EOS at V2 is significantly higher or lower compared to total BAL cells on V2 (#, p<0.05; ##, P<0.01: ###, P<0.001).
Statistical Analyses
To compare expression of genes in total BAL cells and purified BAL EOS by qPCR, data were log transformed and analyzed using the Student’s paired t-test. Statistical analyses were performed using SigmaPlot 11.0 software package.
Results
Gene Expression Analysis in BAL Cells 48 h after SBP-Ag by Microarrays
While less than 1% of BAL cells were EOS before SBP-Ag, they increased to 73.9% ±4.2 (n = 8) of the total BAL cell population after SBP-Ag. After mepolizumab, SBP-Ag elicited a significant but much attenuated increase in EOS counts compared to before mepolizumab (34% ±9 versus 73.9% ±4.2; p<0.001, paired t test, n = 8) [26]. 2 of 8 subjects (Figure 2) were randomly chosen for gene expression analysis using a whole human genome transcript microarray. For these 2 subjects, EOS constituted 72 and 74% of the BAL cells after SBP-Ag and were reduced to 24 and 39% of the BAL when SBP-Ag was performed after mepolizumab.
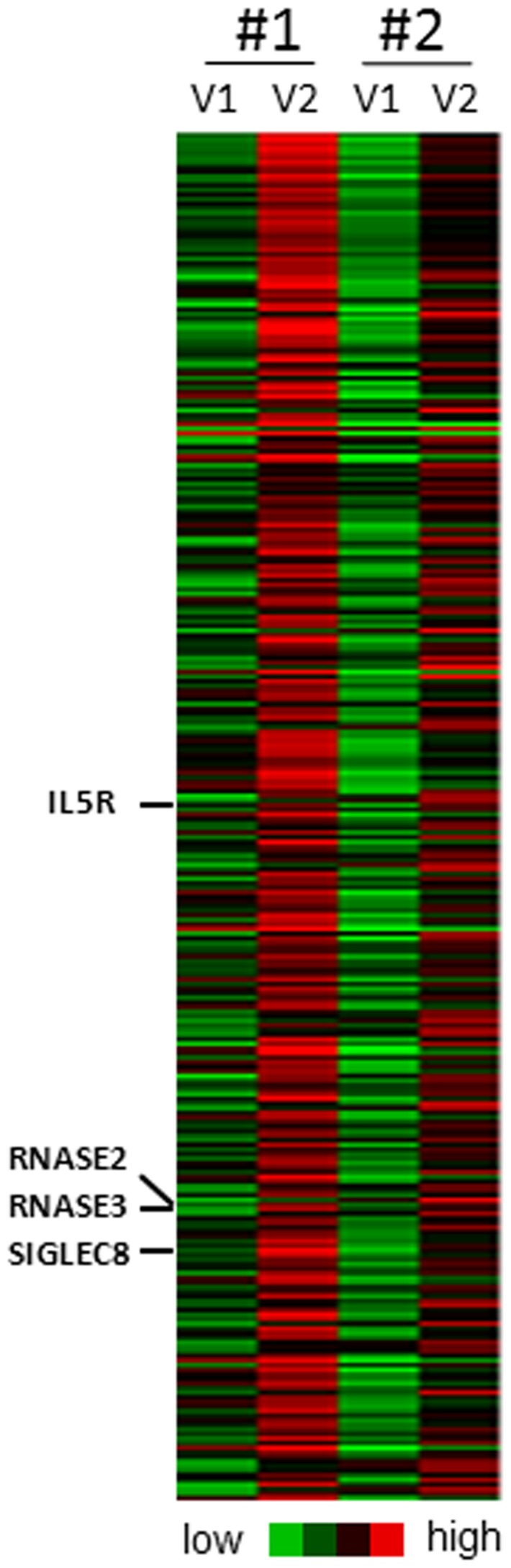
Figure 2. Heat map generated using Cluster 3.0 and presenting the 299 genes up-regulated by more than 2-fold in BAL cells from both subjects #1 and #2, 48 h after SBP-Ag (BAL V2 as shown in Figure 1) compared to before SBP-Ag (V1).
Eosinophil-markers (IL5RA, RNASE2, RNASE3 and SIGLEC8) are highlighted. The full list of gene names is presented on Table S2.
In total BAL cells, expression of 299 genes was up-regulated by more than 2-fold 48 h after SBP-Ag (Figure 2 and Table S2). In both subjects, 22 transcripts were increased by more than 5-fold (Table S3) and 4 transcripts by more than 10-fold (ALOX15, CD1A, CD24 and FAM101B). The 299 genes up-regulated after SBP-Ag included marker genes for EOS (IL5RA, RNASE2, RNASE3, and SIGLEC8), and other genes well-known to be expressed by EOS (ADAM8, CCR3, CD69, CDA, GATA1, HRH4, ICAM3, IL1RL1, LTC4S, OXER1, P2RY2, PADI4, and SIGLEC10). Genes associated with lung remodeling were also found such as metalloproteinase genes (MMP12, MMP9), as well as some IL-13-induced chemokines (CCL13, CCL2) and Th2-associated genes (CCL22, FCER2, and IL1R2) [27]–[29]. While expression of genes coding for soluble mediators (e.g. cytokines) were unchanged, receptors for several mediators known to participate in lung fibrosis were up-regulated (CCR2, FGFR2, FLT1, IGF1R, IL1R1 and IL6R) [27], [29], [30]. 19 transcripts mostly expressed in dendritic cells, lymphocytes or mast cells (Table S4), Swissprot research at http://www.uniprot.org/) were also included in the 299 transcripts. Further analysis using DAVID Bioinformatics Resources indicated that 31 genes have been previously associated with asthma, allergy or a pulmonary disease (DAVID Bioinformatics Resources 6.7; National Institute of Allergy and Infectious Diseases (NIAID), NIH) (Table S5).
99 (33.1%) of the 299 transcripts elevated by more than 2-fold in BAL cells after SBP-Ag were decreased by more than 1.5-fold after reduction of EOS by mepolizumab (Table S6). Of note, 1.5- versus 2-fold reduction was chosen because mepolizumab decreased EOS percentage in BAL cells by ∼50%. Among the 99 genes whose expression was down-regulated after SBP-Ag are the well-known EOS-specific genes (IL5RA, RNASE2, RNASE3 and SIGLEC8) and all the other transcripts mentioned above as previously known to be expressed by EOS (ADAM8, CD69, GATA1, HRH4, ICAM3, IL1RL1, LTC4S, OXER1, P2RY2, PADI4 and SIGLEC10) except for CCR3. In addition, other genes that are not known to be expressed by EOS were down-regulated by mepolizumab. Whether these genes are expressed by other cell types (lymphocytes, dendritic cells, macrophages, or mast cells) that are affected directly or indirectly by the lack of IL-5 or the reduction of EOS or directly expressed by EOS is not known.
Identification of EOS-associated Genes by Microarray Using Induced Sputum after WLAC
Sputum cells from 6 subjects were analyzed by a whole genome transcript microarray. WLAC increased the percentage of EOS in sputum from 2.0 to 8.2% (p<0.001). Marker genes for EOS (RNASE2, RNASE3, IL5RA and SIGLEC8) were increased by more than 2-fold after WLAC in 3 of the 6 subjects involved in this analysis. Sputum samples from these 3 individuals allowed us to establish a list of 365 EOS-associated genes whose expression was increased by more than 1.5-fold after WLAC (Table 1). In addition to the original EOS markers (RNASE2, RNASE3, IL5RA and SIGLEC8), the 365 transcripts (Table 1) included some other well-known genes expressed by EOS. In fact, 11 of the 13 genes (ADAM8, CCR3, CD69, CDA, HRH4, ICAM3, IL1RL1, LTC4S, P2RY2, PADI4 and SIGLEC10) known to be expressed by EOS and up-regulated in BAL cells after SBP-Ag (Table S2) were also seen in sputum after WLAC.
Table 1. EOS-associated genes in sputum after WLAC.
| AATK | CD48 | DNAJB5 | GPR56 | KCNK5 | NKX6-2 | RAB37 | ST6GAL1 |
| ABCB1 | CD69 | DNASE1L3 | GPR84 | KCNMB4 | NOV | RAB3D | ST8SIA4 |
| ABLIM2 | CD93 | DUSP2 | GPR97 | KCNN4 | NPDC1 | RASAL1 | STAB1 |
| ACAP1 | CDA | DYSF | GPT | KCTD15 | NRARP | RASSF2 | SULF2 |
| ACOX2 | CDKN1C | ECE1 | GRASP | KIF21B | NRG1 | RASSF5 | SYNE1 |
| ACPP | CDRT8 | EGR3 | GRB10 | KLF10 | NTRK1 | RD3 | SYNE2 |
| ADAM19 | CECR6 | EGR4 | GSDMA | KRTAP19-1 | ODF4 | RGAG4 | TAC4 |
| ADAM28 | CHI3L1 | EIF2C2 | HAS1 | LBH | OLIG1 | RGL4 | TAGAP |
| ADAM8 | CHST15 | EMB | HBA2 | LGALS12 | OLIG2 | RGS16 | TAS2R50 |
| ADAMTS10 | CHST2 | EMR4P | HBB | LGALS2 | OSBPL3 | RGS2 | TBC1D3B |
| ADORA2A | CHSY1 | ENHO | HBD | LIMK2 | OSM | RHOH | TESC |
| AGER | CLC | ENPP1 | HCG27 | LPPR2 | P2RY14 | RNASE2 | TET2 |
| AGPAT9 | CLEC10A | ENPP2 | HDAC4 | LRG1 | P2RY2 | RNASE3 | TGM2 |
| ALPL | CLEC4F | EPHB3 | HDC | LTC4S | P2RY6 | RNASE6 | THBS4 |
| AMPD2 | CLEC4G | ETV3 | HGF | LTF | PADI2 | RNF19B | TIAM2 |
| ANKH | CLEC5A | F13A1 | HHIP | LY9 | PADI4 | RUFY4 | TIMP1 |
| APOBEC3B | CMTM2 | FAM101B | HIC1 | MARCKSL1 | PAK1 | RUNX2 | TMEM154 |
| ARAP3 | CNR2 | FAM198B | HIF1A | MAST4 | PALLD | RYBP | TMEM156 |
| AREG | COL18A1 | FAM20A | HIP1R | MATK | PARM1 | S100B | TMEM71 |
| ASB2 | COMP | FAM46B | HIST1H1D | MBOAT7 | PDCD1 | S1PR1 | TNFRSF10C |
| ASGR2 | CORO1A | FAM65B | HIST1H1E | MCTP2 | PDE2A | SATB1 | TNFSF14 |
| ATHL1 | CREB3L3 | FAM83A | HIST1H2AC | MECOM | PGLYRP1 | SCHIP1 | TPCN1 |
| ATL2 | CREB5 | FCAR | HIST1H2AE | MGAM | PHF19 | SCNN1D | TREML2 |
| ATP2A3 | CSGALNACT2 | FCER1A | HIST1H2BO | MGC24103 | PHLDB1 | SDS | TRERF1 |
| ATP8B4 | CST7 | FCER2 | HIST1H4A | MMP1 | PIK3R5 | SELL | TRPM6 |
| B3GNT8 | CTTNBP2 | FCGR2B | HIST2H2BE | MMP10 | PIK3R6 | SEMA4B | TSPAN18 |
| B4GALNT3 | CX3CR1 | FCN1 | HSPA6 | MMP12 | PIP5K1B | SEMA7A | TTYH2 |
| BIRC3 | CXCR1 | FFAR2 | ICAM3 | MMP25 | PLAUR | SIGLEC10 | VDR |
| CACNA1E | CXCR2 | FFAR3 | IFITM1 | MMP7 | PLD4 | SIGLEC8 | VEGFA |
| CACNA2D3 | CXCR2P1 | FGF11 | IGFL1 | MPZL3 | PLEKHG2 | SLC16A10 | VENTX |
| CALCRL | CYP1B1 | FHL3 | IL10 | MRC2 | PLUNC | SLC24A3 | VNN2 |
| CAMK1 | CYP4F12 | FLJ11710 | IL18R1 | MRVI1 | PPP1R14A | SLC26A8 | VSTM1 |
| CASS4 | CYSLTR2 | FNDC1 | IL1R1 | MSRB3 | PPP2R2C | SLC2A13 | WNT5A |
| CBFA2T3 | CYTIP | FRY | IL1R2 | MST4 | PRDM1 | SLC3A1 | WTAP |
| CCL17 | DAB2IP | FZD2 | IL1RAP | MT1G | PRKCB | SLC40A1 | XYLT1 |
| CCL26 | DACH1 | GADD45A | IL1RL1 | MTVR2 | PRSS33 | SLC6A10P | YPEL3 |
| CCR3 | DAPK2 | GAGE7 | IL21R | MXD1 | PSTPIP1 | SLC6A8 | ZBTB46 |
| CD177 | DCHS1 | GAS6 | IL4 | NCAM2 | PTGIR | SLC7A5 | ZBTB46 |
| CD1A | DEFB122 | GATA2 | IL5RA | NCRNA00085 | PTGS2 | SNAI1 | ZDHHC18 |
| CD1B | DEFB130 | GCOM1 | IL6R | NCRNA00152 | PTP4A3 | SNHG3 | ZDHHC8 |
| CD1C | DENND3 | GFRA2 | INSIG1 | NCRNA00287 | PTPN7 | SOCS2 | ZNF395 |
| CD1E | DGKA | GGT5 | IRX6 | NDE1 | PVRL2 | SORBS1 | ZNF469 |
| CD207 | DGKD | GLYCTK | ITGB1BP2 | NDEL1 | PXN | SORL1 | ZNF507 |
| CD209 | DIRAS1 | GPR160 | JHDM1D | NECAB2 | PYROXD2 | SPATA9 | |
| CD226 | DNAH17 | GPR183 | KCNH2 | NHSL2 | QPCT | SPNS3 | |
| CD300LB | DNAJB1 | GPR35 | KCNJ15 | NKD1 | RAB33A | SPOCK1 |
365 genes upregulated by more than 1.5 in each of the 3 subjects with high levels of EOS-characterizing genes (IL5RA, RNASE2, RNASE3 and SIGLEC8) after WLAC.
Identification of Airway EOS Gene Expression
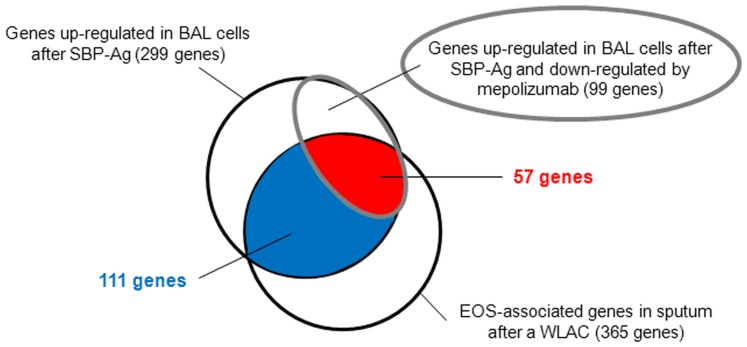
We have utilized 3 airway models that either augmented or reduced the EOS population, and defined genes concomitantly expressed with markers of EOS genes in the context of allergen challenges in vivo. Figure 3 shows that 168 (111+57) (46%) of the 365 EOS-associated genes in sputum were also identified in the 299 genes up-regulated in BAL cells. This suggests that despite distinct anatomical sites and provocative protocols, infiltrating EOS express a consistent group of transcripts. Furthermore, the intersection of these 2 sets with the genes decreased by mepolizumab identified a group of 57 genes (Figure 3 and Table 2). We propose that this intersection represents a stringent selection for gene expression by airway EOS. A literature search in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) indicates that only 16 of the 57 genes (28%) have been previously linked to EOS. Therefore, 41 genes (>70%) in this subset have not previously been associated with EOS.
Figure 3. Venn diagram identifying 57 genes highly associated with airway EOS after an allergen challenge.
57 genes defined at the intersection of genes up-regulated in BAL cells after SPB-Ag, down-regulated when SBP-Ag was performed following mepolizumab, and part of the EOS-associated genes in the sputum following WLAC.
Table 2. 57 genes upregulated in BAL cells after SBP-AG, down-regulated when SBP-Ag was performed following mepolizumab, and part of the EOS-associated genes in sputum after WLAC.
| ADAM28 | CNR2 * | FFAR3 | LGALS12 | PRSS33 * | SPNS3 |
| ADAM8 | CORO1A | FHL3 | LTC4S * | RAB37 | TESC |
| ARAP3 * | CYP4F12 | GGT5 | MCTP2 | RAB3D | TNFSF14 * |
| ASB2 | DACH1 | GPR56 | MMP25 | RD3 | TREML2 |
| ATP2A3 | DAPK2 * | GPR97 | NHSL2 | RNASE2 | TRPM6 |
| CASS4 | DGKD | ICAM3 | P2RY2 | RNASE3 | TSPAN18 |
| CD300LB | EMR4P | IFITM1 | PADI2 | SIGLEC10 | VSTM1 |
| CD69 | FAM101B | IL1RL1 | PADI4 | SIGLEC8 | |
| CDA | FAM65B | IL5RA * | PGLYRP1 | SLC24A3 | |
| CHST15 | FFAR2 * | KIF21B | PIK3R6 | SORL1 |
genes chosen for validation of the list by qPCR.
IL5RA, RNASE2, RNASE3 and SIGLEC8 are known markers of EOS.
Validation of the Airway EOS-associated Genes
To validate the expression of these genes by airway EOS, we performed qPCR for 8 of the 57 genes (Table 2). The samples from the 6 subjects not used in the microarray analyses were analyzed by qPCR. Gene expression in highly purified BAL EOS (BAL V2, see Figure 1) and total BAL cells after SBP-Ag before (BAL V2) and after mepolizumab infusion (BAL V4) are presented as fold change (2−ΔΔCt method) compared to gene expression in BAL cells before SBP-Ag (BAL V1). Initially, 2 established genes expressed by EOS were chosen as controls (IL5RA and LTCS4). As shown in Figure 4A, expression of both genes was highly up-regulated by SBP-Ag, and strongly decreased (>80%) by mepolizumab. Both genes displayed very high levels in pure BAL EOS compared to total BAL cells, suggesting that EOS are likely the main source of IL5RA and LTCS4 in BAL cells obtained after SBP-Ag. In Figure 4B, FFAR2 and CNR2 were selected as genes that have been connected with asthma and/or allergy and have been reported to be expressed by EOS [31], [32]. Both genes exhibited the same profile as IL5RA and LTCS4, indicating that EOS are also an important source of FFAR2 and CNR2 in BAL cells. In addition, 4 of the 41 genes for which expression in EOS has not previously been described were randomly selected. As shown in Figure 4C, all 4 genes (ARAP3, DAPK2, PRSS33 and TNFSF14) presented the same profile of expression as IL5RA, LTCS4, FFAR2 and CNR2 demonstrating that these transcripts are highly expressed in airway EOS. IL1R1 and FCER2 (CD23ß) were chosen as negative controls as they were up-regulated in unseparated BAL cells after SBP-Ag (Table S2) and part of the EOS-associated genes as established in sputum (Table 2), but were not affected by mepolizumab (Table S6). Conversely to IL5RA, LTCS4, FFAR2, CNR2, ARAP3, DAPK2, PRSS33 and TNFSF14, IL1R1 and FCER2 displayed very low expression in BAL EOS compared to total BAL cells (Figure 4D). Figure 4 validates the data obtained by microarrays and demonstrates that BAL EOS are likely the source of the 57 genes that increased post SBP-Ag in BAL cells. Using DAVID Bioinformatics Resources, genes were clustered according to their known functions. Six genes have been associated with a respiratory disease, 7 genes produce secreted proteins and 27 genes code for plasma membrane protein. Some other important clusters included cell adhesion (7 genes), immune and defense response (6 genes), and receptors for a variety of stimuli such as carbohydrates and nucleotides (15 genes) (Table S7).
Discussion
We have used a unique approach combining 3 whole genome expression microarrays (sputum cells after WLAC, BAL cells after SBP-Ag, and BAL cells after mepolizumab and SBP-Ag) to identify 57 genes highly expressed by airway EOS after an allergen challenge. We demonstrated that EOS are likely the main source of these 57 genes expressed by total BAL cells. The 57 genes included genes such as CD69, IL5RA, LTC4S, RNASE2, RNASE3, SIGLEC8, ADAM8, CDA, ICAM3, IL1RL1 (ST2), and P2RY2 that are known to be expressed in EOS, and other genes not previously linked to EOS. Cluster analysis indicated these genes might have an impact on EOS migration and activation, as well as the innate and adaptive immune responses.
Of the 57 transcripts associated with airway EOS, 16 have been previously detected in circulating EOS, including IL1RL1, CNR2, and FFAR2. IL1RL1 (also known as ST2) is the receptor for IL-33, a cytokine that is released following epithelial damage during an allergic reaction. IL-33 can activate CD4+ T cells, type-2 innate lymphoid cells, and mast cells, as well as EOS [33]. The function of IL-33 on circulating EOS includes induction of degranulation, cytokine release, and increased survival. In vivo, blockade of the IL-33/IL1RL1 pathway reduces allergic inflammation and airway responsiveness [34]. Our study suggests that EOS might be the main source of IL-33 receptor among the airway inflammatory cells present after SBP-Ag, and EOS could contribute to IL-33-mediated effects in allergic asthma. CNR2 (also called CB2 receptor) is the cannabinoid receptor expressed in lymphoid organs [35]. CNR2 is known to be expressed by circulating EOS, and an endogenous ligand (2-arachidonoylglycerol) induces EOS migration [36]. In mice, genetic deletion of Cnr2 exacerbated contact allergic inflammation and influenza-induced excessive airway injury possibly via induction of pro-inflammatory and pro-remodeling mediators (IL-17, IL-13, TNF-α, GM-CSF) [37], [38]. CNR2 agonists reduced bronchoconstriction, mast cell degranulation and pulmonary inflammation in guinea pigs [39]. Another endogenous CNR2 ligand, anandamide is increased in BAL fluids after SBP-Ag, and correlated with the number of EOS in the airway [40]. Finally, FFAR2 expression by EOS has only been reported once [32]. FFAR2 (also known as GPR43) is a lipid G-protein coupled receptor for short-chain fatty acids that is produced by bacteria after fermentation of dietary fibers. Interestingly, the lack of FFAR2 led to unresolved inflammation in several animal models of inflammation, including allergen-induced airway inflammation [32], suggesting FFAR2 activation by short-chain fatty acids would be beneficial to reduce asthma.
One example of potentially novel EOS-derived mediators is TNFSF14 (also called LIGHT), a membrane-expressed or secreted protein that interacts with TNFRSF14 membrane receptor (HVEM), and was characterized as a co-stimulatory ligand for lymphoid cells [41]. As most of the co-stimulatory members of the TNF/TNFR superfamily, TNFSF14 has been studied for its impact on autoimmune disease and transplant rejection [41]. More recently, LIGHT has been implicated in induction of airway fibrosis and smooth muscle hyperplasia via TGF-ß and IL-13 production in an allergic animal model [42]. Interestingly, in human sputum, LIGHT protein level has been associated with lower lung function (FEV1% predicted) [43].
Another example of novel mediators in human EOS is PGLYRP1 or PGRP-S (also called Tag7), which recognizes bacterial peptidoglycan and has an antibacterial function [44]. PGLYRP1 is a 21Kd protein with a signal peptide for secretion. While PGLYRP1 has not been described in human EOS, bovine and mouse EOS are known to express and store the bovine and murine ortholog of PGLYRP1 [45], [46]. In addition, to its antimicrobial function, several mouse models using PGLYRP1 knock-out, have demonstrated both an anti-inflammatory role for PGLYRP1 that down-regulated the production of type-1 cytokines and chemokines [47], and a pro-inflammatory role in atopic dermatitis, contact dermatitis, and pulmonary inflammation models promoting the Th1, Th2 and Th17 response [48], [49]. Also, through its interaction with Hsp70 on cytotoxic lymphocytes, PGLYRP1 possesses anti-tumor cell activity [50].
Another intriguing gene is PRSS33 (also called Protease EOS). This member of the trypsin-like serine proteases has been described to be predominantly produced by macrophages [51]. However, an international patent has described PRSS33 as expressed by EOS as well (Pub. No.: WO/2001/016290). Proteases have crucial roles in maintaining homeostasis and regulating inflammation and immune reactions, thus their expression in EOS is under active investigation. For instance, PRSS21 (esp-1) has been found in EOS but not in neutrophils [52], and we have shown very recently that EOS could generate high amounts of another protease, MMP-9 [53]. Yet, the production of PRSS33 by human EOS remains uncertain.
There is little overlap between airway EOS genes described in the present study and previous microarrays using in vitro activated blood EOS. In vitro, IL-5 induced the up-regulation of CD69 by blood EOS [13], [14]. Surprisingly, CD69 was the only common gene between our 57 EOS-associated genes and the genes up-regulated by IL-5 or GM-CSF in these 2 previous studies. This suggests that a short-term in vitro activation using IL-5 or GM-CSF might not be an adequate model to reflect in vivo EOS activation after an allergen challenge. In addition, the present study analyzed the presence of genes associated with eosinophilia while the 2 previous studies analyzed genes that are cytokine-induced. Thus, in addition to their exposure to cytokines, airway EOS likely become activated as a result of their transmigration out of the blood and through the tissues. Future studies will be required to determine if the 57 genes are inducible or constitutively expressed by airway EOS. Comparison of gene expression in airway EOS after allergen challenge with blood EOS obtained from individuals who do not receive challenge would provide information on the inducible versus constitutive expression of these genes.
In a study by Nakajima et al. [54], microarray analysis examined EOS-specific transcripts by comparison to other nucleated peripheral blood cells. Among the 30 EOS-specific genes, 3 were present in our list of 57 genes (RNASE2, IL5RA and CNR2). 11 of the 30 genes were also up-regulated in our BAL cells after SBP-Ag including GPR44, ALOX15, P2RY14, IDO1, ADORA3, CAMK1, CCR3 and P2RY10; while 10 of the 30 genes were part of the EOS-associated genes in induced sputum (CLC, PLAUR, MARCKSL1, P2RY14, CAMK1, CCR3, and OLIG2).
Gene expression analysis by microarrays has been previously performed on bronchial biopsies from asthmatic patients and on airway epithelial cells [55], [56] but microarray analysis using BAL cells after in vivo challenge has not been reported. In our whole genome expression analysis, 299 genes were up-regulated after SBP-Ag in BAL cells. Cluster analysis by DAVID Bioinformatics Resources identified 36 genes involved in defense, immune, or wound-healing responses, 32 genes implicated in cell adhesion or locomotion, 23 genes that regulate transcription, 21 genes associated with apoptosis and 12 genes that are part of the second-messenger-mediated signaling. Importantly, the EOS markers (RNASE2 and RNASE3, also called EDN and ECP) as well as 15 other well-known EOS-associated genes were up-regulated concomitantly with EOS accumulation in the airway following SBP-Ag. Of these 17 genes, all but CCR3 were decreased by the mepolizumab infusion. Among the 13 genes up-regulated by SBP-Ag and related to lung remodeling (CCL2, CCL13, CCL22, CCR2, FCER2, FGFR2, FLT1, IGF1R, IL1R1, IL1R2, IL6R, MMP9 and MMP12), 11 of them were unchanged following mepolizumab. This suggests that in this model mepolizumab did not impact expression of remodeling-associated genes.
Potential limitations of this study include the inability to assess gene expression in non-hematopoietic airway cells (including epithelial cells) or to detect by microarray transcripts, such as cytokines, that are in low abundance. Unlike microarray analysis, qPCR allowed detection of IL-13 and IL-17A in BAL after SBP-Ag [9]. This suggests that some genes might have been excluded from the final eosinophilic genes and that the list of 57 genes is not an exhaustive inventory. Finally, it is important to note that a portion of the genes that are not affected by mepolizumab may in fact represent genes associated with a distinct phenotype of eosinophils that were recruited to or differentiated in the airway following administration of mepolizumab.
In conclusion, we have identified 57 genes highly expressed in vivo by airway EOS (BAL and sputum). Most of the 57 genes have not been previously studied in EOS and are important candidates to understand tissue EOS biology and to find new targets for therapies associated with an excessive eosinophilic response.
Supporting Information
Primer sequences used for real-time PCR.
(DOCX)
299 Genes upregulated in BAL cells 48 h after segmental allergen challenge.
(DOCX)
22 Genes upregulated by more than 5 fold in both subjects in BAL cells 48 h after segmental allergen challenge.
(DOCX)
Main cellular sources (non-eosinophilic) of the transcripts up-regulated in BAL cells after SBP-Ag.
(DOCX)
Among the 299 genes upregulated in BAL cells after SBP-Ag, 31 have been previously genetically associated with asthma, allergy or a pulmonary disease. DAVID Bioinformatics Resources 6.7; National Institute of Allergy and Infectious Diseases (NIAID), NIH; http://david.abcc.ncifcrf.gov/.
(DOCX)
99 genes up-regulated in BAL cells by SBP-Ag and down-regulated after mepolizumab.
(DOCX)
Genes up-regulated in BAL cells after allergen challenge, down-regulated by mepolizumab and part of the EOS-associated genes in the sputum: functional annotation clustering (DAVID Bioinformatics Resources 6.7, National Institute of Allergy and Infectious Diseases, NIH).
(DOCX)
Acknowledgments
The authors thank Mary Jo Jackson, RN, BSN, Holly Eversoll, RN, BSN, Michele Wolff, RN, MSN, and Evelyn Falibene, BS, for patient recruitment and screening; the Laboratory core that recruited and screened subjects and purified blood eosinophils; Paul Fichtinger, BS, for eosinophil purification; and the UW-Asthma Program Project Grant group for helpful suggestions.
Funding Statement
This work was supported in part by a Program Project Grant (NIH HL088594), the University of Wisconsin General Clinical Research Center grant (NIH M01RR03186), and the University of Wisconsin Institute for Clinical and Translational Research (NCRR/NIH 1UL1RR025011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bousquet J, Chanez P, Lacoste JY, Barnéon G, Ghavanian N, et al. (1990) Eosinophilic inflammation in asthma. New Engl J Med 323: 1033–1039. [DOI] [PubMed] [Google Scholar]
- 2.Broekema M, Timens W, Vonk JM, Volbeda F, Lodewijk ME, et al.. (2010) Persisting Remodeling and Less Airway Wall Eosinophil Activation in Complete Remission of Asthma. Am J Respir Crit Care Med. [DOI] [PubMed]
- 3. Broekema M, Volbeda F, Timens W, Dijkstra A, Lee NA, et al. (2010) Airway eosinophilia in remission and progression of asthma: accumulation with a fast decline of FEV(1). Respir Med 104: 1254–1262. [DOI] [PubMed] [Google Scholar]
- 4. Busse WW, Ring J, Huss-Marp J, Kahn JE (2010) A review of treatment with mepolizumab, an anti-IL-5 mAb, in hypereosinophilic syndromes and asthma. J Allergy Clin Immunol 125: 803–813. [DOI] [PubMed] [Google Scholar]
- 5. Flood-Page P, Menzies-Gow A, Phipps S, Ying S, Wangoo A, et al. (2003) Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest 112: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, et al. (2004) A critical role for eosinophils in allergic airways remodeling. Science 305: 1776–1779. [DOI] [PubMed] [Google Scholar]
- 7. Spencer LA, Szela CT, Perez SA, Kirchhoffer CL, Neves JS, et al. (2009) Human eosinophils constitutively express multiple Th1, Th2, and immunoregulatory cytokines that are secreted rapidly and differentially. J Leukoc Biol 85: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu LY, Mathur SK, Sedgwick JB, Jarjour NN, Busse WW, et al. (2006) Human airway and peripheral blood eosinophils enhance Th1 and Th2 cytokine secretion. Allergy 61: 589–598. [DOI] [PubMed] [Google Scholar]
- 9. Esnault S, Kelly EA, Nettenstrom LM, Cook EB, Seroogy CM, et al. (2012) Human eosinophils release IL-1ß and increase expression of IL-17A in activated CD4(+) T lymphocytes. Clin Exp Allergy 42: 1756–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro M, Mathur S, Hargreave F, Boulet LP, Xie F, et al.. (2011) Reslizumab for Poorly Controlled, Eosinophilic Asthma: A Randomized, Placebo-Controlled Study. Am J Respir Crit Care Med. [DOI] [PubMed]
- 11. Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, et al. (2009) Mepolizumab and exacerbations of refractory eosinophilic asthma. New Engl J Med 360: 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, et al. (2009) Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med 360: 985–993. [DOI] [PubMed] [Google Scholar]
- 13. Temple R, Allen E, Fordham J, Phipps S, Schneider HC, et al. (2001) Microarray analysis of eosinophils reveals a number of candidate survival and apoptosis genes. Am J Respir Cell Mol Biol 25: 425–433. [DOI] [PubMed] [Google Scholar]
- 14. Bates ME, Liu LY, Esnault S, Stout BA, Fonkem E, et al. (2004) Expression of IL-5- and GM-CSF-responsive genes in blood and airway eosinophils. Am J Respir Cell Mol Biol 30: 736–743. [DOI] [PubMed] [Google Scholar]
- 15. Stout BA, Bates ME, Liu LY, Farrington NN, Bertics PJ (2004) IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol 173: 6409–6417. [DOI] [PubMed] [Google Scholar]
- 16. Bates ME, Sedgwick JB, Zhu Y, Liu LY, Heuser RG, et al. (2010) Human airway eosinophils respond to chemoattractants with greater eosinophil-derived neurotoxin release, adherence to fibronectin, and activation of the Ras-ERK pathway when compared with blood eosinophils. J Immunol 184: 7125–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, et al. (2002) Decreased expression of membrane IL-5R alpha on human eosinophils: I. Loss of membrane IL-5 alpha on eosinophils and increased soluble IL-5R alpha in the airway after antigen challenge. J Immunol 169: 6452–6458. [DOI] [PubMed] [Google Scholar]
- 18. Johansson MW, Kelly EA, Busse WW, Jarjour NN, Mosher DF (2008) Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J Immunol 180: 7622–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calhoun WJ, Jarjour NN, Gleich GJ, Stevens CA, Busse WW (1993) Increased airway inflammation with segmental versus aerosol antigen challenge. Am Rev Respir Dis 147: 1465–1471. [DOI] [PubMed] [Google Scholar]
- 20. Jarjour NN, Calhoun WJ, Kelly EA, Gleich GJ, Schwartz LB, et al. (1997) The immediate and late allergic response to segmental bronchopulmonary provocation in asthma. Am J Respir Crit Care Med 155: 1515–1521. [DOI] [PubMed] [Google Scholar]
- 21. Boulay ME, Boulet LP (2002) Lower airway inflammatory responses to repeated very-low-dose allergen challenge in allergic rhinitis and asthma. Clin Exp Allergy 32: 1441–1447. [DOI] [PubMed] [Google Scholar]
- 22. Liu LY, Swenson CA, Kita H, Kelly EAB, Busse WW (2000) The relationship of airway eosinophilia and sputum cell generation of IL-5 following antigen challenge. J Allergy Clin Immunol 106: 1063–1069. [DOI] [PubMed] [Google Scholar]
- 23. Lopuhaa CE, Out TA, Jansen HM, Aalberse RC, van der Zee JS (2002) Allergen-induced bronchial inflammation in house dust mite-allergic patients with or without asthma. Clin Exp Allergy 32: 1720–1727. [DOI] [PubMed] [Google Scholar]
- 24. Pin I, Freitag AP, O'Byrne PM, Girgis-Gabardo A, Watson RM, et al. (1992) Changes in the cellular profile of induced sputum after allergen-induced asthmatic responses. Am Rev Respir Dis 145: 1265–1269. [DOI] [PubMed] [Google Scholar]
- 25. Liu LY, Bates ME, Jarjour NN, Busse WW, Bertics PJ, et al. (2007) Generation of Th1 and Th2 chemokines by human eosinophils: evidence for a critical role of TNF-alpha. J Immunol 179: 4840–4848. [DOI] [PubMed] [Google Scholar]
- 26. Johansson MW, Gunderson KA, Kelly EA, Denlinger LC, Jarjour NN, et al. (2012) Anti-IL-5 attenuates activation and surface density of ß2-integrins on circulating eosinophils after segmental antigen challenge. Clin Exp Allergy 43: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Royce SG, Cheng V, Samuel CS, Tang ML (2012) The regulation of fibrosis in airway remodeling in asthma. Mol Cell Endocrinol 351: 167–175. [DOI] [PubMed] [Google Scholar]
- 28. Homer RJ, Elias JA, Lee CG, Herzog E (2011) Modern concepts on the role of inflammation in pulmonary fibrosis. Arch Pathol Lab Med 135: 780–788. [DOI] [PubMed] [Google Scholar]
- 29. Al Muhsen S, Johnson JR, Hamid Q (2011) Remodeling in asthma. J Allergy Clin Immunol 128: 451–462. [DOI] [PubMed] [Google Scholar]
- 30. Elias JA, Zhu Z, Chupp G, Homer RJ (1999) Airway remodeling in asthma. J Clin Invest 104: 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kishimoto S, Oka S, Gokoh M, Sugiura T (2006) Chemotaxis of human peripheral blood eosinophils to 2-arachidonoylglycerol: comparison with other eosinophil chemoattractants. Int Arch Allergy Immunol 140 Suppl 13–7. [DOI] [PubMed] [Google Scholar]
- 32. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461: 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H (2008) A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol 121: 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kearley J, Buckland KF, Mathie SA, Lloyd CM (2009) Resolution of allergic inflammation and airway hyperreactivity is dependent upon disruption of the T1/ST2-IL-33 pathway. Am J Respir Crit Care Med 179: 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munro S, Thomas KL, Abu-Shaar M (1993) Molecular characterization of a peripheral receptor for cannabinoids. Nature 365: 61–65. [DOI] [PubMed] [Google Scholar]
- 36. Oka S, Ikeda S, Kishimoto S, Gokoh M, Yanagimoto S, et al. (2004) 2-arachidonoylglycerol, an endogenous cannabinoid receptor ligand, induces the migration of EoL-1 human eosinophilic leukemia cells and human peripheral blood eosinophils. J Leukoc Biol 76: 1002–1009. [DOI] [PubMed] [Google Scholar]
- 37. Karsak M, Gaffal E, Date R, Wang-Eckhardt L, Rehnelt J, et al. (2007) Attenuation of allergic contact dermatitis through the endocannabinoid system. Science 316: 1494–1497. [DOI] [PubMed] [Google Scholar]
- 38. Karmaus PW, Chen W, Crawford RB, Harkema JR, Kaplan BL, et al. (2011) Deletion of cannabinoid receptors 1 and 2 exacerbates APC function to increase inflammation and cellular immunity during influenza infection. J Leukoc Biol 90: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giannini L, Nistri S, Mastroianni R, Cinci L, Vannacci A, et al. (2008) Activation of cannabinoid receptors prevents antigen-induced asthma-like reaction in guinea pigs. J Cell Mol Med 12: 2381–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zoerner AA, Stichtenoth DO, Engeli S, Batkai S, Winkler C, et al. (2011) Allergen challenge increases anandamide in bronchoalveolar fluid of patients with allergic asthma. Clin Pharmacol Ther 90: 388–391. [DOI] [PubMed] [Google Scholar]
- 41. del Rio ML, Lucas CL, Buhler L, Rayat G, Rodriguez-Barbosa JI (2010) HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J Leukoc Biol 87: 223–235. [DOI] [PubMed] [Google Scholar]
- 42. Doherty TA, Soroosh P, Khorram N, Fukuyama S, Rosenthal P, et al. (2011) The tumor necrosis factor family member LIGHT is a target for asthmatic airway remodeling. Nat Med 17: 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, et al. (2010) Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol 125: 1028–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osanai A, Sashinami H, Asano K, Li SJ, Hu DL, et al. (2011) Mouse peptidoglycan recognition protein PGLYRP-1 plays a role in the host innate immune response against Listeria monocytogenes infection. Infect Immun 79: 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tydell CC, Yount N, Tran D, Yuan J, Selsted ME (2002) Isolation, characterization, and antimicrobial properties of bovine oligosaccharide-binding protein. A microbicidal granule protein of eosinophils and neutrophils. J Biol Chem 277: 19658–19664. [DOI] [PubMed] [Google Scholar]
- 46.Park SY, Jing X, Gupta D, Dziarski R (2013) Peptidoglycan Recognition Protein 1 Enhances Experimental Asthma by Promoting Th2 and Th17 and Limiting Regulatory T Cell and Plasmacytoid Dendritic Cell Responses. J Immunol. [DOI] [PMC free article] [PubMed]
- 47. Saha S, Jing X, Park SY, Wang S, Li X, et al. (2010) Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe 8: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park SY, Gupta D, Kim CH, Dziarski R (2011) Differential effects of peptidoglycan recognition proteins on experimental atopic and contact dermatitis mediated by Treg and Th17 cells. PLoS ONE 6: e24961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park SY, Jing X, Gupta D, Dziarski R (2013) Peptidoglycan Recognition Protein 1 Enhances Experimental Asthma by Promoting Th2 and Th17 and Limiting Regulatory T Cell and Plasmacytoid Dendritic Cell Responses. J Immunol. [DOI] [PMC free article] [PubMed]
- 50. Sashchenko LP, Dukhanina EA, Shatalov YV, Yashin DV, Lukyanova TI, et al. (2007) Cytotoxic T lymphocytes carrying a pattern recognition protein Tag7 can detect evasive, HLA-negative but Hsp70-exposing tumor cells, thereby ensuring FasL/Fas-mediated contact killing. Blood 110: 1997–2004. [DOI] [PubMed] [Google Scholar]
- 51. Chen C, Darrow AL, Qi JS, D'Andrea MR, Andrade-Gordon P (2003) A novel serine protease predominately expressed in macrophages. Biochem J 374: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inoue M, Kanbe N, Kurosawa M, Kido H (1998) Cloning and tissue distribution of a novel serine protease esp-1 from human eosinophils. Biochem Biophys Res Commun 252: 307–312. [DOI] [PubMed] [Google Scholar]
- 53. Kelly EA, Liu LY, Esnault S, Quinchia-Rios BH, Jarjour NN (2012) Potent synergistic effect of IL-3 and TNF on matrix metalloproteinase 9 generation by human eosinophils. Cytokine 58: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakajima T, Matsumoto K, Suto H, Tanaka K, Ebisawa M, et al. (2001) Gene expression screening of human mast cells and eosinophils using high-density oligonucleotide probe arrays: abundant expression of major basic protein in mast cells. Blood 98: 1127–1134. [DOI] [PubMed] [Google Scholar]
- 55. Laprise C, Sladek R, Ponton A, Bernier MC, Hudson TJ, et al. (2004) Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics 5: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA (2005) Effects of allergen challenge on airway epithelial cell gene expression. Am J Respir Crit Care Med 171: 579–586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for real-time PCR.
(DOCX)
299 Genes upregulated in BAL cells 48 h after segmental allergen challenge.
(DOCX)
22 Genes upregulated by more than 5 fold in both subjects in BAL cells 48 h after segmental allergen challenge.
(DOCX)
Main cellular sources (non-eosinophilic) of the transcripts up-regulated in BAL cells after SBP-Ag.
(DOCX)
Among the 299 genes upregulated in BAL cells after SBP-Ag, 31 have been previously genetically associated with asthma, allergy or a pulmonary disease. DAVID Bioinformatics Resources 6.7; National Institute of Allergy and Infectious Diseases (NIAID), NIH; http://david.abcc.ncifcrf.gov/.
(DOCX)
99 genes up-regulated in BAL cells by SBP-Ag and down-regulated after mepolizumab.
(DOCX)
Genes up-regulated in BAL cells after allergen challenge, down-regulated by mepolizumab and part of the EOS-associated genes in the sputum: functional annotation clustering (DAVID Bioinformatics Resources 6.7, National Institute of Allergy and Infectious Diseases, NIH).
(DOCX)