Abstract
The nucleosome is the first level of genome organization and regulation in eukaryotes where negatively charged DNA is wrapped around largely positively charged histone proteins. Interaction between nucleosomes is dominated by electrostatics at long range and guided by specific contacts at short range, particularly involving their flexible histone tails. We have thus quantified how internucleosome interactions are modulated by salts (KCl, MgCl2) and histone tail deletions (H3, H4 N-terminal), using small-angle x-ray scattering and theoretical modeling. We found that measured effective charges at low salts are ∼1/5th of the theoretically predicted renormalized charges and that H4 tail deletion suppresses the attraction at high salts to a larger extent than H3 tail deletion.
Introduction
Highly charged DNA is packaged into nucleosome core particles (NCP) in eukaryotic cells by wrapping every 147 per ∼180 basepairs (bp) of DNA around a histone octamer. The octamer is a protein complex comprising two copies of each of the four histone proteins (named H2A, H2B, H3, and H4). Each histone has one C-terminal and one N-terminal tail that protrude out from the octameric core. Linked by ∼20–60 bp DNA like beads on a string, NCPs are further packaged into chromatin whose conformation and dynamics underpins gene maintenance and regulation. Thus, there has been long-standing interest in physically understanding nucleosome structure and interaction fundamental to chromatin function.
Polyelectrolyte behavior and histone tail modulation are among the key determinants of the multifactorial packaging of nucleosomes (1). One NCP carries a net negative charge of ∼150 e with −294 e from DNA and +144 e from histone proteins. Given its diameter of ∼10 nm and height of ∼6 nm, this leads to an average charge density of −0.43 e/nm2, about half of the −1 e/nm2 of DNA. The expected polyelectrolyte behavior of NCP is demonstrated by cation-modulated unfolding and compaction of NCP arrays (2,3). Notably, divalent cations go beyond screening the electrostatic repulsion between NCPs and can mediate inter-NCP attraction, e.g., Mg2+- or Ca2+-induced condensation of mono- and polynucleosomes (4). In contrast, these divalent cations are unable to condense DNA. This suggests significant roles of the complex charge distribution of NCP furnished by the histones (5). In particular, the tails of histones (∼30% of the histone mass), being highly positively charged and flexible, are known to be crucial in modulating nucleosome interaction and chromatin assembly (1).
There have been extensive experimental studies aiming to elucidate the electrostatics of nucleosomes and the roles of histone tails. The effects of cations of varied valences on the nucleosome assembly have been well characterized (1–3,6), e.g., by measuring the sedimentation coefficients of nucleosome arrays or the fraction of condensed nucleosomes (4,7). Among the 16 histone tails, the eight N-terminal tails hold the most positive charges and are considered more significant, e.g., NCPs with all N-terminal tails deleted remain soluble in high divalent salts (8). In an effort to dissect the contribution from each N-terminal tail, all 16 possible tail-deletion NCP mutants have been studied and it was found that all N-terminal tails contribute to the condensation of oligo-NCP arrays nonspecifically and additively (8). Still, the histone tails vary in length and charge and thus differ in strength in mediating nucleosome interactions, e.g., H3 and H4 tails, studied herein, were observed to be more important than H2A and H2B tails (8,9).
Systematic measurements have promoted widespread theoretical interests in mechanistically understanding the structure and interaction of nucleosomes (10,11). At the atomic level, enabled by the availability of high resolution crystal structures (12), molecular dynamics simulations have described the ion and solvent atmospheres of nucleosome and revealed rugged electrostatic surfaces (13,14). The flexible histone tails have been analyzed in detail and shown to display multistate conformational dynamics (15–17). At the coarse-grain level, both nucleosome arrays and chromatin have been modeled to recapture the conformational changes under varied conditions such as salt and mechanical stretching (18,19). However, to apply the detailed NCP structures and energetics predicted by theoretical approaches, stringent experimental tests are required, but largely lacking. For example, as the focus of this study, one key factor to the assembly of NCPs is the effective inter-NCP potential that is usually computed following the Poisson-Boltzmann formulism for efficiency (11). However, such theoretical treatment has not been validated experimentally due to the lack of direct measurements of inter-NCP potentials. Comparisons with previously measured quantities are often difficult, e.g., the interpretation of sedimentation coefficient necessitates an ab initio hydrodynamic model.
Here, we report measurements of interactions between monomeric nucleosomes using solution small-angle x-ray scattering (SAXS) in conjunction with theories of polyelectrolytes. There are two outcomes we deem important: 1), Quantification of inter-NCP pair potentials by modeling full SAXS profiles; 2), Elucidation of the individual role of H3 and H4 tails. Although SAXS (and small-angle neutron scattering for neutron) has been used to study the structure and interaction of NCPs since the 1970s (20), no quantitative refinement of full SAXS profiles has been done to extract the inter-NCP interactions, which we obtain here by refining synchrotron SAXS data. One key advantage of full profile modeling is quantification of the inter-NCP potential as a function of inter-NCP distance (21), in contrast to single-parameter measurements such as second virial coefficients or osmotic pressures of NCP solutions. Bertin et al. (22) have compared full SAXS profiles with theoretical calculations. However, refinements were not performed, possibly due to the relatively dilute NCP concentrations (≤8 mg/ml, ∼0.04 mM) and limited angular range of the in-house SAXS data that lower SAXS sensitivity to inter-NCP interactions. In addition, using recombinant NCP constructs with either H3 or H4 tail deleted, to the best of our knowledge we report the first analysis of this kind on the respective role of H3 and H4 tails in mediating inter-NCP interactions.
Material and Methods
Natural-source monomeric NCPs (NS-NCP) were prepared from adult chicken erythrocytes as in (23). Gel electrophoresis was used to assess the DNA length (147 ± 5 bps) and the histone content (Fig. S1, a and b, in the Supporting Material). Recombinant nucleosomes (RC-NCP) were reconstituted by mixing 147-bp mouse mammary tumor virus DNA and histone octamers with sequences from Xenopus laevis following standard protocols (8) (Fig. S1, a and c). The globular H3 and H4 recombinant proteins were truncated at their N-terminal ends (referred to as gH3 and gH4 RC-NCPs), by deleting the coding sequences of amino acids (1–27 with +10 e of H3 and 1–10 with +3 e of H4) via site-directed mutagenesis. All NCP stocks were purified by size-exclusion chromatography (Sephacryl S300-HR medium) and then dialyzed against 10 mM KCl 10 mM Tris 0.1 mM EDTA pH 7.5 buffer. Salts were adjusted to studied conditions by adding concentrated salts. SAXS experiments were carried out at 20°C at the G1 station at the Cornell High Energy Synchrotron Source in Ithaca, New York. The incident beam had an energy of 9.97 keV and size of 250 × 250 μm. Samples of ∼30 μl were injected into an in-vacuum capillary flow cell to enable windowless data collection for background reduction. Radiation damage was mitigated by optimizing x-ray exposure time and oscillating the plug of sample solution. Six to eight two-second exposures of the same sample were taken and verified to be reproducible, indicative of the absence of radiation damage.
Results
Measured SAXS profile I(Q) is a product of the form factor P(Q) and the structure factor S(Q), i.e., I(Q) = P(Q) × S(Q), with S(Q) becoming significant under conditions such as high [NCP]s and strong inter-NCP interactions. I(Q) of a dilute NCP solution (i.e., S(Q) = 1) thus measures the structure of individual NCPs in terms of the form factor P(Q) = I(Q). This can reveal differences between molecular structures in solution and in crystal. Fig. 1 shows excellent agreement between the SAXS profile measured in solution and the profile calculated from a crystal structure in Q range of 0.01–0.28 Å−1, indicative of high sample purify and structural conformity. The same level of agreement was observed for all NCP constructs (i.e., NS-NCP, wild-type, gH3, and gH4 RC-NCPs). It is worth noting that the crystal structure P(Q) shows an extra dip around Q of 0.14 Å−1. This discrepancy has been observed in previous reports (22,24) and was attributed to the DNA unwrapping at the ends by a few basepairs. Tail deletions lead to small changes of the dip around Q = 0.14 Å−1 that can be attributed to increased DNA unwrapping (data not shown). Nonetheless, these dips appear in relatively high Q and do not affect our subsequent analysis. Guinier analysis of the low Q data (Fig. 1, inset) gives the radius of gyration Rg = 44.4 (3) Å, which is nearly identical to previous SAXS measurements of natural-source NCPs (25) and recombinant NCPs with α-satellite DNA (24). Note that NCPs reconstituted with the 601 sequence were reported to give slightly smaller Rg by 2–3 Å (22,24).
Figure 1.
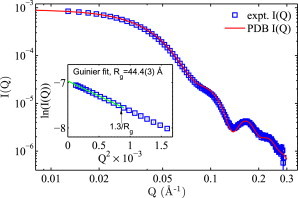
SAXS form factor P(Q) of NS-NCP from experiment (□) and crystal structure (red, PDB ID: 1KX5). Q = 4πsin(θ)/λ is the scattering vector, where 2θ is the scattering angle and λ is the x-ray wavelength. The experiment form factor was measured at a dilute NCP concentration of 0.003 mM (∼0.6 mg/ml) and an intermediate salt (40 mM KCl) to minimize the influence of inter-NCP interferences. Error bars of I(Q)s, in this and subsequent plots, are smaller than symbol sizes except at high Qs. Inset shows the Guinier fit (solid line) to obtain the radius of gyration (Rg). The linear region extends well over the empirical Qmax cutoff of 1.3/Rg for globular structures.
In addition to structure information, SAXS is capable of quantifying intermolecular interactions, via the structure factor S(Q) originating from spatial correlations between molecules. Qualitatively, repulsive interaction leads to a decrease of low Q intensities, i.e., a downturn, and attractive interaction gives rise to an increase at low Q, i.e., an upturn. Fig. 2 shows how SAXS profiles evolve with NCP and salt concentrations. As expected from strong inter-NCP repulsions at low salts (10 and 40 mM KCl, Fig. 2, a and b), low Q downturns are observed and deepen upon increasing [NCP]. In contrast, low Q upturns appear and heighten with [NCP] at high salts (100 and 200 mM KCl, Fig. 2, c and d), indicating inter-NCP attractions. These observations are consistent with reports of negative virial coefficients at >70 mM KCl (22). Divalent Mg2+, known to condense NCP at >2.5 mM [Mg2+] (4), was also studied and the onset of inter-NCP attraction was found to be between 1 and 2 mM [Mg2+] (Fig. S3).
Figure 2.
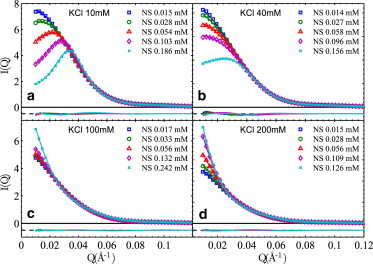
SAXS profiles, I(Q) = P(Q) × S(Q), of NS-NCPs at four KCl concentrations. I(Q)s are normalized by NCP concentrations to assist comparison. Each panel shows experimental I(Q)s (symbols) at a series of [NCP]s (0.1 mM gives ∼20 mg/ml) as indicated in the legends, together with their respective theoretical fits (lines). The residues are shown with an offset at the same scale. The pertinent pair-potential parameters are (discussed in detail in the main text), (a) Zeff = 22 (1) e, σ = 140 Å; (b) Zeff = 23 (1) e, σ = 110 Å; (c) Zattr =150 (5) e, σ = 100 Å; (d) Zattr = 200 (8) e, σ = 100 Å. Uncertainties of Z values originate from both curve fitting (via variance-covariance matrix, ∼1%) and small variations (∼3%) between values at different [NCP]s. The Zeff and Zattr values are poorly defined when [NCP] is small (<0.03 mM, or 6 mg/ml) and their values given here are from data at high [NCP]s.
To gain quantitative insights into the underlying inter-NCP interactions, we then applied the generalized one component method (GOCM) with the mean spherical approximation (21,26) to model the structure factors. As both repulsion and attraction exist, the general form of inter-NCP interaction potential as a function of inter-NCP distance is given by
where σ is the equivalent diameter of NCP, Zeff and Zattr are the effective charges to be fitted, ε = 80.4 is the dielectric constant of water at 20°C, and κ is the inverse Debye screening length calculated from the salt condition. Namely, the inter-NCP potential comprises a hard-sphere (HS) repulsive term, a Debye-Huckel (DH) electrostatic repulsive term, and a DH-like attractive term. Although it is possible to include both DH potentials in the fitting procedures, there exist significant correlations between the fitted repulsive and attractive DH terms despite better fits (see the Supporting Material for detailed results and discussions). Due to this complication, the attractive DH potential is turned off (i.e., Zattr = 0) under conditions where a net inter-NCP repulsion is observed. Likewise, Zeff is set to 0 under conditions where a net attraction is observed. As the physical origin of the attraction is poorly understood, we chose a short-range attraction of effective charge Zattr and 4.8 Å decay length for all data. The effect of NCP’s disk-like shape (axis ratio ∼2) on S(Q) only becomes significant at Q > 0.036 Å−1 (22) where S(Q) ∼1 as observed in the experiments; it is thus not considered. Such GOCM fits are shown in Fig. 2 as solid lines, noting that the form factor for each NCP construct is calculated based on the crystal structure (shown in Fig. S2). It is evident that the GOCM fits are in good agreement with experimental data over the entire Q range. Inter-NCP interactions are thus quantified in the forms of HS and DH potentials.
Despite being quantitative, directly usable, and comparable with theoretical studies, it should be noted that the sum of HS and DH potentials describes an apparent inter-NCP interaction that reproduces the measured structure factor. In the case of inter-NCP repulsion, the effective charge Zeff, as the only fitting parameter, differs from the bare charge Zbare due to the nonlinear nature of ion screening and the validity of DH potentials in linear regime only. For the same reason, the equivalent diameter σ encloses the vicinity with a large electrostatic field (e.g., >3.8 kBT/e was used for dsDNA by the authors (27)), which is in the order of Debye length. Especially at low salt, σ is significantly larger than the 100 Å computed from the NCP crystal structure (21). We determined the value of σ at different salts based on cell model numerical calculations (27,28), and the same σ value was then fixed for all data at the same ionic condition for consistency. We show these relevant interaction potential parameters in the figure captions and in Table S1.
Fig. 3 shows data from recombinant gH3 and gH4 RC-NCPs at two salt conditions (10 and 100 mM KCl) together with GOCM fits (data at 20 and 40 mM KCl are shown in Fig. S4). We primarily focused on gH3 and gH4 RC-NCPs because recombinant samples were relatively limited in volume and previous studies have reported the second virial coefficients (though no inter-NCP potentials) of intact RC-NCPs and gH3gH4 RC-NCPs, i.e., with none or both tails deleted (22). To enable cross-comparisons, intact RC-NCPs were measured at a few selected conditions (Fig. S5). At low salt of 10 mM KCl, essentially no difference in the effective charge Zeff (∼22 e) was observed among all NCP constructs (i.e., NS-NCP, gH3, gH4, and intact RC-NCPs), noting that the negativity of Zeff is omitted for brevity. We further calculated the second virial coefficient from the inter-NCP potential using
and obtained a value of 2.5(1) × 10−4 ml·mol/g2. This value is in excellent agreement with the osmometry measurement by Mangenot et al. (29). Bertin et al. (22) used theoretical calculations of Zeff between 70 and 125 e and obtained much larger A2 values close to 5.0 × 10−4 ml·mol/g2. We attribute the difference to the discrepancy between theoretical and experimental Zeff values (discussed below) used by Bertin et al. and us, respectively. At high salt of 100 mM KCl, qualitative differences were observed: NS-NCP, gH3 and intact RC-NCPs show comparable levels of attraction; whereas gH4 and gH3gH4 RC-NCPs show no attraction (including observations in (22).). This suggests the major role of H4 tail in mediating inter-NCP attractions.
Figure 3.
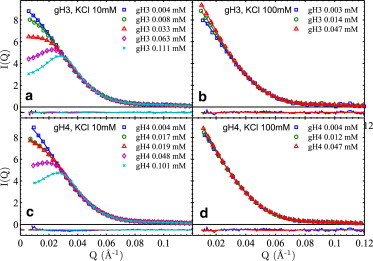
SAXS profiles, I(Q) = P(Q) × S(Q), of gH3 and gH4 RC-NCPs at 10 and 100 mM KCl. Annotations follow that of Fig. 2. Inter-NCP potential parameters are, (a) Zeff = 21 (1) e, σ = 140 Å; (b) Zattr = 123 (4) e, σ = 100 Å; (c) Zeff = 20 (1) e, σ = 140 Å; (d) Zattr = 100 (4) e, σ = 100 Å. It is worth noting that, although gH4 RC-NCP shows no interaction at 100 mM KCl, a significant Zattr exists so as to negate the hard sphere repulsion with σ = 100 Å, which makes Zattr values artificially large. Zattr would be zero if σ is zero for gH4 RC-NCP in 100 mM KCl. It is thus more appropriate to interpret Zattr-100 as contributed by the inter-NCP short-range attraction.
Discussion
We consider the quantification of inter-NCP pair potentials from SAXS structure factors as the most interesting result of our study. Compared with the traditional analysis of second virial coefficients that relies on I(Q = 0) only, the GOCM (and methods alike) uses the full SAXS profiles and obtains self-consistent pair potentials allowing quantitative comparisons with theoretical predictions. At low salts such as 10 mM KCl, NCP-NCP interactions are dominated by repulsion as expected. Essentially the same Zeff is obtained for all four NCP constructs. The minimal effect of tail deletions on inter-NCP repulsion is not surprising given that tail deletions change NCP bare charges by <8% and that cations can effectively fill in for the deleted monocharged amino acids. Quantitatively, it is instructive to compare the measured Zeff with the theoretical renormalized charge Zren that can be calculated following the charge renormalization theorem (28). For dsDNA with negatively charged groups only, our previous work showed that the measured Zeff is fairly consistent (within 10–25%) with its renormalized charge Zren in monovalent salts (27). However, for the nucleosome, its Zeff (∼22 e) is far smaller than theoretical Zren (>100 e for all experimental conditions), noting that Zbare is ∼150 e and Zren weakly depends on NCP and salt concentrations (28). Given that the Zren is calculated by treating NCP as a uniformly charged sphere, such drastic and physically significant discrepancy is likely due to the charge nonuniformity of NCP in terms of spatial distribution and sign, resulting in highly anisotropic behaviors where the uniformly charged surface consideration breaks down.
Inter-NCP interactions at short range are further complicated by the flexible histone tails (11,16). At elevated salts where electrostatic repulsion is greatly weakened, histone tail deletions can significantly affect NCP-NCP interactions. At 100 mM KCl (Fig. 2 c, Fig. 3, b and d), NS-NCP shows significant attraction (i.e., low Q upturn), gH3 RC-NCP shows attraction of comparable magnitude, and gH4 RC-NCP shows nearly no attraction. The difference between gH3 and gH4 RC-NCPs, which have not been studied by SAXS previously, suggests that H4 tail is more important in mediating NCP-NCP attractions than H3 tail. This corresponds well with the observed most important role of H4 tail in assembling the 30-nm chromatin fiber (7). Such trend is also consistent with the repulsion between gH3gH4 RC-NCPs up to 200 mM KCl (22). The more important role of the H4 tail in mediating internucleosome interactions has been suggested by Arya and Schlick (30) analyzing a coarse-grained mesoscale structure model with a tailored configurational-bias Monte Carlo method. As H4 tail deletion removes fewer charges than H3 tail deletion (+3 vs. +10 e) but shows a larger effect, this alludes to specific interactions of H4 tails with adjacent NCPs in close approach. To probe the likely H4-tail-DNA interactions, Korolev and Nordenskiöld (31) performed all-atomic molecular dynamics simulations of DNA and H4 tails and suggested possible lysine-mediated bridging and minor groove dynamic binding. Another possible explanation is H4-tail binding with the H2A/H2B anionic patch on neighboring NCPs, as suggested by crystallography studies (32,33). This is in agreement with recent experimental studies of the effects of H4 tail lysine acetylation on nucleosome array folding that proposed site-specific lysine binding to the H2B histone (9). To fully establish the underlying molecular mechanisms, experimental approaches probing the local structure would be needed to resolve the physical origins of inter-NCP short range attraction.
Conclusion
In conclusion, we have measured repulsive and attractive NCP-NCP interactions under various salt conditions with distinct NCP constructs. Quantitative knowledge of inter-NCP pair potentials should aid theoretical studies of nucleosome and chromatin high-order structure and dynamics. The physical understanding to be gained will shed light on the broad class of multiphasic macromolecules with two or more different interacting groups. Work is underway to relate the inter-NCP interactions to the conformation and energetics of nucleosome arrays through measurement and modeling.
Acknowledgments
The authors thank Arthur Woll, Richard Gillian, Soren Nielsen, and Qi Xia for assistance with experiment, and thank Yun Liu for discussions on data modeling. C.Y. acknowledges Prof. Timothy Richmond (ETH, Zurich) for generously providing us the plasmids of histone proteins and MMTV DNA.
This work was supported by George Washington University (to X.Q.), Research Corporation for Science Advancement and Gettysburg College (to K.A.), and the Purdue University (to C.Y.).
Supporting Material
References
- 1.Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 2.Clark D.J., Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 3.Korolev N., Allahverdi A., Nordenskiöld L. Electrostatic origin of salt-induced nucleosome array compaction. Biophys. J. 2010;99:1896–1905. doi: 10.1016/j.bpj.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Frutos M., Raspaud E., Livolant F. Aggregation of nucleosomes by divalent cations. Biophys. J. 2001;81:1127–1132. doi: 10.1016/S0006-3495(01)75769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J., Zhang Q., Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc. Natl. Acad. Sci. USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 7.Dorigo B., Schalch T., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 8.Gordon F., Luger K., Hansen J.C. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 9.Allahverdi A., Yang R., Nordenskiöld L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlick T., Hayes J., Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korolev N., Fan Y., Nordenskiöld L. Modelling chromatin structure and dynamics: status and prospects. Curr. Opin. Struct. Biol. 2012;22:151–159. doi: 10.1016/j.sbi.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 13.Gan H.H., Schlick T. Chromatin ionic atmosphere analyzed by a mesoscale electrostatic approach. Biophys. J. 2010;99:2587–2596. doi: 10.1016/j.bpj.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Materese C.K., Savelyev A., Papoian G.A. Counterion atmosphere and hydration patterns near a nucleosome core particle. J. Am. Chem. Soc. 2009;131:15005–15013. doi: 10.1021/ja905376q. [DOI] [PubMed] [Google Scholar]
- 15.Potoyan D.A., Papoian G.A. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J. Am. Chem. Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 16.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mühlbacher F., Schiessel H., Holm C. Tail-induced attraction between nucleosome core particles. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:031919. doi: 10.1103/PhysRevE.74.031919. [DOI] [PubMed] [Google Scholar]
- 18.Diesinger P.M., Heermann D.W. Depletion effects massively change chromatin properties and influence genome folding. Biophys. J. 2009;97:2146–2153. doi: 10.1016/j.bpj.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulić I.M., Schiessel H. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys. Rev. Lett. 2003;91:148103. doi: 10.1103/PhysRevLett.91.148103. [DOI] [PubMed] [Google Scholar]
- 20.Hjelm R.P., Kneale G.G., Ibel K. Small angle neutron scattering studies of chromatin subunits in solution. Cell. 1977;10:139–151. doi: 10.1016/0092-8674(77)90148-9. [DOI] [PubMed] [Google Scholar]
- 21.Chen S.H., Sheu E.Y., Hoffmann H. Small-angle neutron-scattering investigation of correlations in charged macromolecular and supramolecular solutions. J. Appl. Cryst. 1988;21:751–769. [Google Scholar]
- 22.Bertin A., Renouard M., Durand D. H3 and H4 histone tails play a central role in the interactions of recombinant NCPs. Biophys. J. 2007;92:2633–2645. doi: 10.1529/biophysj.106.093815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yager T.D., van Holde K.E. Dynamics and equilibria of nucleosomes at elevated ionic strength. J. Biol. Chem. 1984;259:4212–4222. [PubMed] [Google Scholar]
- 24.Yang C., van der Woerd M.J., Luger K. Biophysical analysis and small-angle X-ray scattering-derived structures of MeCP2-nucleosome complexes. Nucleic Acids Res. 2011;39:4122–4135. doi: 10.1093/nar/gkr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangenot S., Leforestier A., Livolant F. Salt-induced conformation and interaction changes of nucleosome core particles. Biophys. J. 2002;82:345–356. doi: 10.1016/S0006-3495(02)75399-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Fratini E., Chen S.-H. Effective long-range attraction between protein molecules in solutions studied by small angle neutron scattering. Phys. Rev. Lett. 2005;95:118102. doi: 10.1103/PhysRevLett.95.118102. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X., Kwok L.W., Pollack L. Measuring inter-DNA potentials in solution. Phys. Rev. Lett. 2006;96:138101. doi: 10.1103/PhysRevLett.96.138101. [DOI] [PubMed] [Google Scholar]
- 28.Trizac E., Bocquet L., von Grunberg H.H. Alexander’s prescription for colloidal charge renormalization. Langmuir. 2003;19:4027–4033. [Google Scholar]
- 29.Mangenot S., Raspaud E., Livolant F. Interactions between isolated nucleosome core particles: a tail-bridging effect? Eur. Phys. J. E. 2002;7:221–231. [Google Scholar]
- 30.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korolev N., Nordenskiöld L. H4 histone tail mediated DNA-DNA interaction and effects on DNA structure, flexibility, and counterion binding: a molecular dynamics study. Biopolymers. 2007;86:409–423. doi: 10.1002/bip.20749. [DOI] [PubMed] [Google Scholar]
- 32.Dorigo B., Schalch T., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 33.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


