Abstract
Alloimmune specificity and histocompatibility, driven by genetic polymorphism, are ancient determinants of self-/non-self-recognition. Recent molecular genetic evidence has revealed an allodeterminant in the cnidarian Hydractinia that consistently predicts histocompatibility reactions.
Early work on mammalian tissue transplantation has helped shape our modern concepts of immunological self-/non-self-recognition. The origins of allogeneic, as well as xenogeneic, immune-type recognition can be traced to the earliest metazoans. Most modern forms of these species spend at least a portion of their lives as sessile organisms in which they inhabit densely populated environments and aggressively compete for space [1]. The Mendelian inheritance of the recognition and rejection of ‘natural’ transplants is well established [2,3], and the biology and molecular determinants governing this process continue to be of considerable interest [4,5]. In this issue of Current Biology, Nicotra et al. [6] describe a specific, polymorphic allorecognition protein in Hydractinia symbiotongicarpus (Cnidaria: Hydrozoa) at the alr2 locus that predicts fusion/rejection responses and adds a new chapter in our understanding of the evolution of metazoan histocompatibility.
Despite the clarity of the findings reported by Nicotra et al. [6] and of other work in this area [7–9], defining allorecognition determinants has in and of itself had an interesting evolution. For well over two decades, various investigators unsuccessfully sought to explore genetic links between products of the major histocompatibility complex (MHC) — the primary determinant of tissue transplantation outcomes in jawed vertebrates — and potentially homologous structures in invertebrates, using both molecular genetic screening and computational methods. As the function of various families of MHC receptors became clearer, the merits of these approaches became far less compelling. Products of the MHC loci function in a wide range of immunobiological phenomena; for example, MHC class I and class II molecules play an integral role in our immune system by presenting peptide antigen to somatically derived, highly specific receptors on T lymphocytes. The extraordinary polymorphism and high-density expression on the cell surface of these receptors, which are confined to jawed vertebrates, make MHC molecules natural allodeterminants — the role of MHC class I molecules in transplantation dynamics is secondary to its function in the adaptive immune response [10,11].
At the same time that searches were on for MHC orthologs outside of the vertebrates, several laboratories, including that of Leo Buss, one of the authors of the new paper [6], were questioning if any relationship exists between the phenomenon of allorecognition in species such as Hydractinia and the histocompatibility revealed by tissue grafting studies in mammals [12]. Although it was advances in molecular genetics and genomics that opened new avenues for identifying and characterizing histocompatibility determinants in invertebrates, the discovery of their genes had its origins in the interpretation of their patterns of inheritance in several key organisms [7,9,13,14]. The compelling biological and genetic observations made earlier in Hydractinia [7–9] are entirely consistent with the principal molecular conclusions reached by Nicotra et al. [6].
Asexually expanding polyps, in Hydractinia and other cnidarians, or zooids, in protochordates such as Botryllus, are joined by gastrodermal or vascular-type canals and enveloped in a common encrusting tissue. When the leading edge of two asexually expanding colonies comes into contact, projections of the colonial tissue, termed stolens in cnidarians or ampullae in Botryllus, interact. In some invertebrates, such as Botryllus and Hydractinia — but not others, such as Hydra [15]—allogeneic transplantation leads to rejection, while autografts fuse permanently. In certain colonial forms, however, partial (albeit rare) genetic compatibilities lead to intermediate or incomplete fusion/rejection reactions (Figure 1).
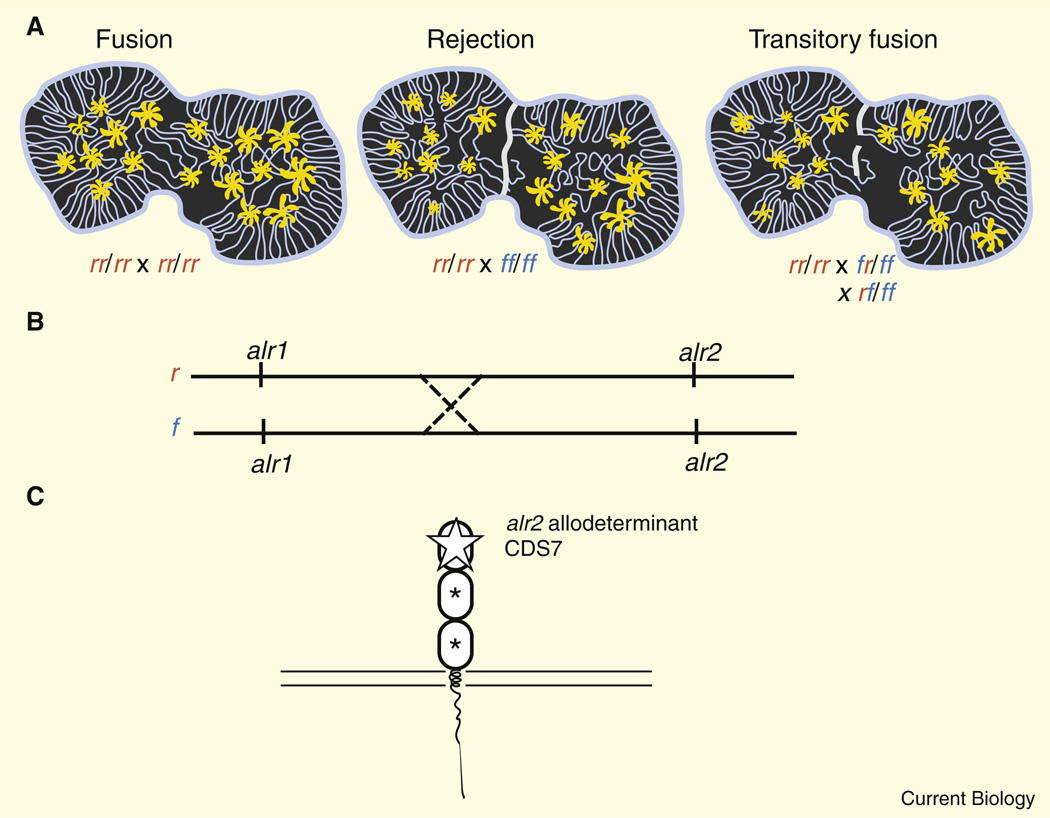
Figure 1. Allorecognition in a cnidarian.
(A) Historecognition phenotypes in Hydractinia. Polyps are shown joined by a gastrovascular system (lines running through the colonial tissue; see text for details). Fusions, rejections or transitory fusions, which do not form stable chimeras, are determined by a pair of loci (r and f alleles in this example). (B) Transitory fusion is associated with polymorphism and haplotype recombination at two loci of the alr gene complex (illustrated by a reciprocal exchange), making chimeric fusions very rare. (C) Analysis of transitory fusions contributed to the identification of CDS7, a membrane-anchored protein possessing three extracellular domains in which the amino-terminal domain is hyperpolymorphic, as a determinant of allorecognition. Asterisks denote limited polymorphism; open star denotes extensive polymorphism.
Allorecognition in Hydractinia is associated with at least two histocompatibility loci, alr1 and alr2. Individual Hydractinia that share one allele at both loci (rr/rr × rr/rf) will undergo fusion during which colonies become chimeric and share vasculature. In contrast, if no alleles are shared in common (rr/rr × ff/ff), a rejection response occurs, which is marked by several characteristic responses: a failure of ectodermal cells to adhere; nematocyst discharge (in cnidarians); extension of hyperplastic tissues; and necrosis at the contact zones. Thus, transplantations can lead to fusion or rejection, and in some cases partial fusion in which colonies fuse and shortly thereafter separate (Type I transitory fusions) or partial rejections in which the animal cycles between fusion and rejection (Type II transitory fusions) [8]. Transitory fusions appear to be associated with multi-loci allorecognition [9,14] in which allele frequencies can be modified by crossover events [8]. Multi-locus histocompatibility systems are not limited to Hydractinia, as a diversity of allogeneic rejection responses also has been noted in corals [16,17]. By contrast, only fusions or rejections are seen in Botryllus, in which histocompatibility appears to be controlled by a single highly polymorphic locus, termed FuHC [18].
Analysis of the region of the Hydractinia genome corresponding to the alr2 locus led Nicotra et al. [6] to a coding sequence CDS7, which seemed a good candidate for being the gene encoding the major allodeterminant associated with histocompatibility reactions in both laboratory-derived and field-collected colonies. CDS7 encodes a putative transmembrane receptor with three extracellular domains, exhibiting some similarity with immunoglobulin superfamily-like molecules. The amino-terminal structural domain of CDS7 is longer than an immunoglobulin I-type domain and exhibits extensive polymorphism (Figure 1). Furthermore, certain canonical residues are missing. The qualified interpretation offered by the authors for this predicted structure is that it represents a divergent V-type gene. Such assignments are not insignificant as immunoglobulin-like domains, as with leucine-rich repeats and lectin-like domains, are commonly mobilized as units of immune recognition, providing highly efficient scaffolds for genome innovation in the host-pathogen battleground and like MHC may relate to historecognition indirectly (see [19]).
Notwithstanding its actual physical character, which will require definitive analysis at the structural level, Nicotra et al. [6] found that polymorphism in the first domain of CDS7 is an obligatory element of histocompatibility, as evidenced by screening of field-collected colonies and characterizing their ability to fuse to laboratory inbred lines. Of over 1,000 crosses examined, only two failed to reject. One of these colonies underwent a type II transitory fusion with an f tester colony, which is consistent with it possessing an f-like allele at the alr1 locus. The other colony (LH82) underwent a type I transitory fusion with the f tester colony, which is consistent with an f-like allele at alr2. PCR data confirmed that this colony was identical to the f tester allele over the first (recognition) domain. LH82 was backcrossed and subsequent results collectively indicated that CDS7 predicted fusion/rejection. Collection of additional data is warranted to further explore how polymorphism in the first domain of CDS7 relates to allorecognition.
A compelling biological advantage for the polymorphisms that govern histocompatibility has been invoked for colonial invertebrates. Specifically, fusion of two or more conspecifics can lead to chimerism, which in turn can result in somatic (and stem cell) parasitism in which population diversity can be rapidly eroded. It is highly likely that allogeneic recognition may have evolved as a means to protect the genetic integrity of self, thereby maintaining population diversity [5,10,12].
With this new report [6], three major animal groups are now seen to exhibit different modes of histocompatibility. The answer to the question as to whether or not homology exists in disparate histocompatibility systems appears to be no. It also is equally unlikely that these are the only mechanisms of allorecognition yet to be defined. Other molecules have been implicated in protochordate allorecognition, some of which suggest interconnectivity to innate immune defenses [4]. Notably, not all invertebrates are responsive to allogeneic differences [16]. In the solitary freshwater hydrozoan, Hydra, allorecognition does not lead to rejection [15]; presumably it lacks allorecognition determinants of the types recognized above. Immunity to pathogenic threat in this species has been demonstrated directly at the ectodermal layer, involving specific and regulated induction of innate immune responses [20]. In this case, a solitary lifestyle (no encrusting colonial tissue) may have allowed for the secondary loss of a histocompatibility effector system. The interplay between diversity of histocompatibility systems, recognition specificity, and innate immunity is becoming increasingly clearer and provides an important chapter in our understanding of self-/non-self-recognition systems.
The lineage-specific innovations in allorecognition exemplified in cnidarians, protochordates, and vertebrates are a harbinger of the extraordinary diversity of form that effects a similar general function. The mechanisms whereby the genes governing histocompatibility are diversified and stabilized in the population represent fascinating models of evolutionary adaptation and selection. Their study is likely to yield insights far beyond histocompatibility as a natural process.
References
- 1.Grosberg RK. The evolution of allorecognition specificity in colonial invertebrates. Quart. Rev. Biol. 1988;63:377–412. [Google Scholar]
- 2.Hildemann WH. Immunocompetence and allogeneic polymorphism among invertebrates. Transplantation. 1979;27:1–3. [PubMed] [Google Scholar]
- 3.Burnet FM. ‘Self-recognition’ in colonial marine forms and flowering plants in relation to the evolution of immunity. Nature. 1971;232:230–235. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 4.Khalturin K, Bosch TC. Self/nonself discrimination at the basis of chordate evolution: limits on molecular conservation. Curr. Opin. Immunol. 2007;19:4–9. doi: 10.1016/j.coi.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Boehm T. Quality control in self/nonself discrimination. Cell. 2006;125:845–858. doi: 10.1016/j.cell.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Nicotra ML, Powell AE, Rosengarten RD, Moreno M, Grimwood J, Lakkis FG, Dellaporta SL, Buss LW. A hypervariable invertebrate allodterminant. Curr. Biol. 2009;19:583–589. doi: 10.1016/j.cub.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokady O, Buss LW. Transmission genetics of allorecognition in Hydractinia symbiolongicarpus (Cnidaria:Hydrozoa) Genetics. 1996;143:823–827. doi: 10.1093/genetics/143.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell AE, Nicotra ML, Moreno MA, Lakkis FG, Dellaporta SL, Buss LW. Differential effect of allorecognition loci on phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa) Genetics. 2007;177:2101–2107. doi: 10.1534/genetics.107.075689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadavid LF, Powell AE, Nicotra ML, Moreno M, Buss LW. An invertebrate histocompatibility complex. Genetics. 2004;167:357–365. doi: 10.1534/genetics.167.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borghans JA, Beltman JB, De Boer RJ. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- 11.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 12.Buss LW. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. USA. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scofield VL, Schlumpberger JM, West LA, Weissman IL. Protochordate allorecognition is controlled by a MHC-like gene system. Nature. 1982;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 14.Grosberg RK, Levitan DR, Cameron BB. Evolutionary genetics of allorecognition in the colonial hydroid Hydractinia symbiolongicarpus. Evolution. 1996;50:2221–2240. doi: 10.1111/j.1558-5646.1996.tb03612.x. [DOI] [PubMed] [Google Scholar]
- 15.Kuznetsov SG, Bosch TC. Self/nonself recognition in Cnidaria: contact to allogeneic tissue does not result in elimination of nonself cells in Hydra vulgaris. Zoology (Jena) 2003;106:109–116. doi: 10.1078/0944-2006-00105. [DOI] [PubMed] [Google Scholar]
- 16.Bigger CH. Historecognition and immunocompetence in selected marine invertebrates. In: Grosberg RK, Hedgecock D, Nelson K, editors. Invertebrate Historecognition. New York: Plenum Press; 1988. pp. 55–65. [Google Scholar]
- 17.Rinkevich B. Allorecognition and xenorecognition in reef corals: a decade of interactions. Hydrobiologia. 2004;530/531:443–450. [Google Scholar]
- 18.De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat. Rev. Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]