Abstract
The production of odiferous metabolites, such as 2-methlyisoborneol (MIB), is a major concern for water utilities worldwide. Although MIB has no known biological function, the presence of the earthy/musty taste and odor attributed to this compound result in the reporting of numerous complaints by consumers, which undermines water utility performance and the safe and adequate provision of potable waters. Cyanobacteria are the major producers of MIB in natural waters, by mechanisms that have heretofore remained largely unstudied. To investigate the fundamental biological mechanism of MIB biosynthesis in cyanobacteria, the genome of a MIB-producing Pseudanabaena limnetica was sequenced using Next Generation Sequencing and the recombinant proteins derived from the putative MIB biosynthetic genes were biochemically characterized. We demonstrate that the biosynthesis of MIB in cyanobacteria is a result of 2 key reactions: 1) a S-adenosylmethionine -dependent methylation of the monoterpene precursor geranyl diphosphate (GPP) to 2-methyl-GPP catalyzed by geranyl diphosphate 2-methyltransferase (GPPMT), and 2) further cyclization of 2-methyl-GPP to MIB catalyzed by MIB synthase (MIBS) as part of a MIB operon. Based on a comparison of the component MIB biosynthetic genes in actinomycetes and cyanobacterial organisms, we hypothesize that there have been multiple rearrangements of the genes in this operon.
Keywords: methylisoborneol, MIB, geosmin, taste, odor, water
INTRODUCTION
The production of odorous compounds by organisms in water is a problem for water utilities worldwide. Both geosmin and 2-methylisoborneol (MIB), which each impart an earthy/musty taint to the water, are responsible for numerous customer complaints to water authorities regarding water quality (1,2). Aside from the unappealing taste and smell, consumers perceive the earthy/musty tasting water to be unsafe to drink, despite neither geosmin nor MIB having any known adverse biological or pathological effects. Currently, water remediation efforts focus on the removal of these compounds at the water treatment plant, primarily by applying activated carbon (3,4), at considerable expense and occupational health and safety risk to utility personnel, with no foreseeable changes in practice. The dosing of carbon is largely a reactive measure to a taste and odor episode that is firmly established, and the ability to predict the emergence of taste and odor compounds in waters will enable mitigating strategies to be deployed before there are customer complaints.
The majority of geosmin and MIB in waters is produced by cyanobacteria (5), and until recently the process by which this occurs has remained largely unstudied. Significant advances have been made, however, in the understanding of the fundamental biological mechanisms responsible for the production of geosmin in actinomycetes and more recently in cyanobacteria, where it was conclusively demonstrated that a single bi-functional terpene synthase, named geosmin synthase, is responsible for catalyzing the conversion of farnesyl diphosphate, the universal C15 sesquiterpene precursor, to geosmin (6–12). This information was used to identify several geosmin producing cyanobacterial species using real-time PCR, and resulted in the development of a sensitive and useful tool to correctly identify and provide advanced warning of geosmin-producing cyanobacteria in waters (8).
The biosynthesis of MIB by actinomycetes has recently received much attention. In 2007, in the myxobacterium Nannocystis exedens, the incorporation of mevalonate isoprenoid precursors as well as methyl-labeled S-adenosylmethionine (SAM) into MIB was reported. Based on these in vivo results, the authors postulated a biosynthetic pathway in which geranyl diphosphate (GPP), the universal C10 monoterpene precursor, would undergo SAM-dependent methylation, resulting in the generation of a novel intermediate 2-methyl-GPP (13). This 2-methyl-GPP was proposed to then undergo direct cyclization to produce MIB. In 2008, the mechanistic details of MIB production in Streptomyces coelicolor A3(2) were unraveled. Wang and Cane (2008) discovered and characterized a two-gene MIB synthase operon in S. coelicolor A3(2) and showed that it was responsible for the conversion of GPP to MIB (14). The authors conclusively demonstrated that the previously uncharacterized SCO7701 protein of S. coelicolor is a SAM-dependent C-methyl transferas that catalyzes the methylation of GPP to produce 2-methyl-GPP, consistent with the results of Dickschat et al. (2007). They also demonstrated that the SCO7700 protein (a C11 homo-monoterpene synthase), encoded by the immediately upstream gene, catalyzes the cyclization of 2-methyl-GPP to MIB. The steady kinetic parameters (kcat and Km) for both reactions confirmed that incubation of GPP and SAM with both recombinant SCO7701 and SCO7700 proteins results directly in the formation of MIB. Simultaneously and independently, Komatsu and co-workers used bioinformatic analysis to identify the two candidate genes constituting the MIB synthase operon in seven microorganisms belonging to actinomycetes, including S. coelicolor A3(2) (15). This assignment was confirmed by PCR amplification and heterologous expression in S. avermitilis of the MIB biosynthetic operons from S. ambofaciens, S. lasaliensis, and Saccharopolyspora erythraea. Taken together, the results from both of these studies irrefutably demonstrated that only two structural genes and the derived protein products are required for MIB synthesis from a GPP precursor in actinomycetes. The monoterpene synthase involved was named MIB synthase (MIBS), while the SAM-dependent C-methyl transferase was named geranyl diphosphate 2-methyltransferase (GPPMT, also known as 2-methyl-GPP synthase [MGPPS]).
Previous work had suggested that geosmin and MIB are each synthesized from different, but closely related, isoprenoid precursors (16–18). With the confirmation that there is a similar biosynthetic mechanism for geosmin synthesis in both Streptomyces and cyanobacteria (8), it was hypothesized that a mechanism for MIB biosynthesis similar to that in actinomycetes and myxobacteria may also be present in cyanobacteria. Here we describe the identification, expression, and biochemical characterization of MIBS and GPPMT orthologs in a MIB-producing strain of Pseudanabaena limnetica (Castaic lake), as well as the discovery that the order of the genes in cyanobacterial MIB operons appears to be the reverse of that in the majority of actinomycetes.
MATERIALS AND METHODS
Cyanobacterial strains and DNA extraction
Pseudanabaena limnetica, isolated from Castaic lake (19), was a generous gift from George Izaguirre. DNA from Pseudanabaena limnetica (Lake Biwa, NIVA-CYA 111) and Oscillatoria limosa LBD 305b were provided by Sue Watson, National Water Research Institute, Burlington, Canada. DNA was extracted from P. limnetica (Castaic lake) by centrifuging 10 mL of high density culture at 4000 × g and subjecting the spun pellet to the tissue protocol procedure as outlined in the Qiagen DNA minikit (Qiagen), except that the pellet was digested with proteinase-K overnight, instead of 1–3 h. When necessary, DNA was concentrated using potassium acetate according to standard methods (20).
DNA sequencing and bioinformatics of Pseudanabaena limnetica data
Whole genome sequencing of Pseudanabaena limnetica (Castaic lake) was performed on an Illumina GAII next generation sequencer platform, with 75 bp paired end reads. Draft nucleotide sequence data was assembled using the VELVET algorithm (21). Protein matches retrieved from the 6 frames of amino acid sequence data of P. limnetica (Castaic lake) were achieved by utilizing the construction of a Pfam model for MIBS using actinomycete MIB synthases from the NCBI nr database (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/), and using hmmer-2.3.2 (ftp://selab.janelia.org/pub/software/hmmer/2.3.2/) as described previously (15).
Cloning, expression, and purification of putative MIBS and GPPMT proteins, incubation with GPP, and GC-MS analysis
Synthetic gene constructs of putative MIBS and GPPMT were synthesized by DNA2.0 in separate pJExpress vectors optimized for E. coli expression. The synthetic genes were each flanked by unique 5' NdeI and 3' XhoI restriction sites. Both the MIBS and GPPMT constructs were sub-cloned into pET-28a expression vectors and transformed into E. coli BL21(DE3) for over-expression of the corresponding N-terminal His6-tagged proteins, as previously described (14). The transformed E. coli cells were used to inoculate 5 mL of Terrific Broth medium, which was incubated overnight at 225 rpm and 37 °C. An aliquot of this culture was transferred to 500 mL of TB medium and incubated at 37 °C and 225 rpm until an OD600 of 0.6 was obtained. The culture was then induced with 0.2 mM of isopropylthio-D-galactopyranoside (IPTG) and incubated at 28 °C for 12 h with shaking at 225 rpm. The cells were harvested by centrifugation at 4200 g for 20 min and stored at −80 °C until required. For protein purification, the cell pellets were thawed and lysed by 2 passes through a French pressure cell and the cell debris cleared by centrifugation at 20,200 g for 1 h. The recombinant proteins were then purified using Ni-NTA affinity chromatography and visualized using SDS-PAGE by standard methods, generating approximately 40 mg of GPPMT and 20 mg of MIBS protein. Incubations of GPP and SAM with a mixture of GPPMT and MIBS, as well as GPP plus SAM with GPPMT and synthetic 2-methyl-GPP with MIBS, were carried out as previously described (Wang and Cane, 2008). Analysis by GC-MS and comparison with an authentic 2-MIB standard was performed as previously described (14).
PCR methods and DNA sequencing of PCR products
To confirm the orientation of the GPPMT and MIBS genes in Pseudanabaena limnetica (Castaic lake), primer combinations GPPF1/MIBR1 and MIBF1/GPPR1 (Table S1) were used in the following PCR reaction: approximately 10 ng of Pseudanabaena limnetica (Castaic lake) genomic DNA was added to a PCR reaction mixture containing 1 × PCR buffer (Invitrogen), 2.5 mM MgCl2, 200 nM dNTPs (Promega), 300 nM of each forward or reverse primer combination, and 1 U of Platinum Taq DNA polymerase (Invitrogen). Gradient PCRs were performed in a Corbett Palm Cycler (Qiagen), with an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 94 °C for 30 s (annealing at 45–60 °C for 30 s), and extension at 72 °C for 45 s. PCR products were visualized using 1% gel agarose electrophoresis supplemented with 1X SYBR Safe (Invitrogen). Several additional confirmatory assays for P. limnetica (Castaic lake) targets were similarly performed utilizing combinations of MIBF2/MIBR2, GPPF2/GPPR2, PSANROS1F/PSANDDR1, PSANROS1F/PSANNSER1, and PSAN562F/PSAN1140R as outlined in Table 1. For these confirmatory assays, PCR conditions similar to GPPR1/MIBR1 and MIBF1/GPPR1 PCRs were employed, except that annealing temperatures were at 50 and 60 °C only. PCR screening of possible homologous targets within P. limnetica (Lake Biwa) and Oscillatoria limosa utilized all of the PCR assays described in Table 1, except that PSANROS1F/PSANNSER1 PCR was omitted. When required, DNA from bands of the expected size was excised from the agarose gel and purified using QiaQuick Gel extraction kit (Qiagen). Purified amplification products were sequenced utilizing Big Dye Terminator 3 labeling and an AB3730xl 96-capillary sequencer (performed by the Australian Genome Research Facility, Adelaide). Data were analyzed and assembled using Lasergene SeqMan Pro (v. 8.0.2, DNASTAR). Bioiformatic analysis of sequence data was performed using MEGA v 4.0 (22), and ClustalX v2.0 algorithms (23).
RESULTS AND DISCUSSION
Pseudanabaena limnetica (Castaic lake) sequence analysis
A draft genome sequence of Pseudanabaena limnetica (Castaic lake) was assembled using VELVET to generate 11,258 contigs. The nucleotide sequence of each contig was translated to 6 frames of amino acid sequences. Initial attempts to identify the putative terpene synthases using a profile hidden Markov model to search against PF03936 (terpene synthase family, metal binding domain) were unsuccessful, despite using a relaxed E-value of approximately 10−2 for the hidden Markov model search. A new profile of hidden Markov model using alignment data of actinomycete MIB synthases and putative terpene synthases was generated and the P. limnetica (Castaic lake) data were scrutinized again for orthologous MIBS and terpene synthases. A single open reading frame (ORF) of 397 aa within the contig NODE_9452 was revealed (E-value 3.8 × 10−48). A BLAST homology search of this ORF (GenBank Accession XXXXX) demonstrated that this predicted terpene synthase was similar to several MIB synthases, the highest similarity being to ZP_05479642 of Streptomyces sp. AA4 (370 aa, 58% identity and 70% positive matches) at the amino acid sequence level (see Table S2 for additional sequence information for closely related bacterial orthologs). Immediately upstream of this putative MIBS ORF, a putative methyltransferase (277 aa, GenBank Accession XXXXX) was located, with a high similarity to GPPMT found in ZP_04609213 of Micromonospora sp. ATCC 39149 (289 aa, 73% identity and 84% positive matches) at the amino acid sequence level (see Table S3 for additional sequence information for closely related bacterial orthologs). Additionally, a putative nucleotide binding protein (GenBank Accession XXXXX) was found downstream of the putative MIBS ORF. This ORF had a high similarity to NP_824183 of Streptomyces avermitilis (468 aa, 60% identity and 78% positive matches) at the amino acid sequence level (see Table S4 for for additional sequence information for closely related bacterial orthologs).
Confirmation of Pseudanabaena limnetica (Castaic lake) putative MIB operon gene order
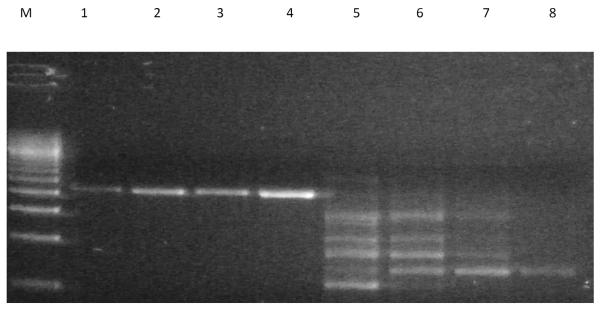
The combined demonstration of a methyl transferase, a terpene synthase, and a nucleotide binding protein (in this order) strongly suggested that the region located within contig NODE_9452 corresponded to the MIB operon. Interestingly, the order of the genes within this operon was the reverse of that observed in the majority of actinomycetes in which the gene order is nucleotide binding protein, followed downstream by MIBS, and then GPPMT (Figure 1). To confirm the unexpected gene order of the MIB operon within Pseudanabaena limnetica (Castaic lake), confirmatory PCRs were carried out using the GPPF1/MIBR1 and MIBF1/GPPR1 primer pairs. The predicted result, based on the order of the genes from the sequencing data, was that the PCR with GPPF1 (forward primer, anchored on start codon of the putative GPPMT) and MIBR1 (reverse primer, anchored to stop codon of the putative MIBS) would generate an amplicon of 2087 bp. If, however, the sequencing data were incorrect, and the order of the genes were instead MIBS followed immediately downstream by GPPMT, then a specific PCR amplification product would not be seen (since the forward and reverse primers would be extending away from each other). Figure 2 shows that a specific product of the expected size was indeed obtained for GPPF1/MIBR1 PCR, while PCR with the MIBF1/GPPR1 pair resulted in the production of only non-target PCR products.
Figure 1.
MIB operon gene arrangement in actinomycetes and Pseudanabaena limnetica (Castaic lake). Apart from Streptomyces sp. AA4, Catenulipsora acidiphila, and Pseudanabaena limnetica (Castaic lake), the gene order in this operon is nucleotide binding protein, followed downstream by 2-methylisoborneol synthase and GPP 2-methyl transferase.
Figure 2.
Agarose gel electrophoresis of gene order confirmatory PCRs for Pseudanabaena limnetica (Castaic lake). Lanes 1–4 display specific 2078 bp products for GPPF1/MIBR1 PCR at 45, 50, 55, and 60°C respectively; lanes 5–8 do not have any specific products of expected size for MIBF1/GPPR1 PCRs at 45, 50, 55, and 60°C respectively. M. 500 bp marker.
Several additional confirmatory PCRs (MIBF2/MIBR2, GPPF2/GPPR2, PSANROS1F/PSANDDR1, PSANROS1F/PSANNSER1, and PSAN562F/PSAN1140R, detailed in Table S1) were performed, targeting either the GPPMT or MIBS gene, or regions spanning both these genes, with primers anchored on conserved regions, in particular, the Rossmann-fold motif of the methyl transferase, and the two magnesium binding domains (the aspartate-rich –xDDxx[D/E] or –xDDxxx[D/E]- and the “NSE” triad, -xNxxxSxxE-) that are universally conserved within monoterpene and sesquiterpene synthases. The DNA sequences within these conserved regions were not particularly favorable for the design of optimal PCRs, with only PSAN562F/PSAN1140R PCR primers specifically designed and optimized for the region spanning the GPPMT and MIBS in Pseudanabaena limnetica (Castaic lake). Consequently, GPPF2/GPPR2, PSANROS1F/PSANDDR1, and PSANROS1F/PSANNSER1 PCRs yielded several PCR products, including a predominant band of the expected size, while PCR with MIBF2/MIBR2 and PSAN562F/PSAN1140R produced relatively pure amplicons of the expected sizes (data not shown). Combined, these PCR data confirm that the order of the MIB genes originally deduced from the genomic sequencing data was indeed correct, and that the experimentally determined gene order with the 3-gene MIB operon in Pseudanabaena limnetica (Castaic lake) was in fact the reverse of that found in the overwhelming majority of actinomycetes.
Demonstration of GPPMT and MIBS activity from Pseudanabaena limnetica (Castaic lake)
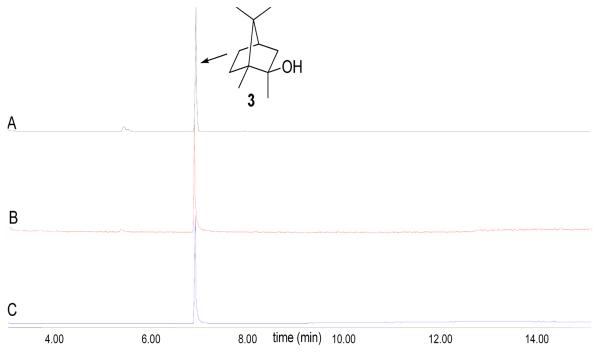
GC-MS analysis of the pentane soluble extract from the incubation of both recombinant GPPMT and MIBS protein with GPP and SAM revealed the formation of MIB as the major organic product as depicted in Scheme 1. This major component was identical by both GC retention time (Figure 3) and EI mass fragmentation pattern with an authentic sample of MIB (Figure S2, Supporting Information). Consistent with these results, incubation of GPPMT with GPP and SAM was shown to yield the expected product, (E)-2-methyl-GPP, while incubation of synthetic (E)-2-methyl-GPP with MIBS gave MIB as the major reaction production. The kcat/Km for the MIBS reaction was 1.45±0.04 × 103 M−1 s−1, comparable to the value of 0.9±0.1 × 103 M−1s−1 previously determined for the corresponding S. coelicolor MIBS (24). Neither MIBS protein exhibited saturation within the concentration limits of the 2-methyl-GPP substrate. Detailed information on the incubation of GPP with GPPMT and 2-methyl-GPP with MIBS can be found in the Supporting Information file.
Scheme 1.
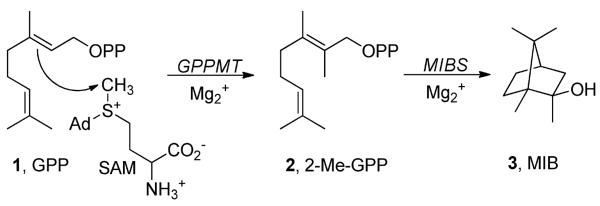
Conversion of GPP to MIB catalyzed by GPPMT and MIBS.
Figure 3.
GC-MS analysis of the in vitro production of 2-MIB. (A) Pentane extract from the incubation of MIBS and 2-Me-GPP (60 μM). (B) Pentane extract from the coupled incubation of GPP (60 μM) and SAM (120 μM) with GPPMT and MIBS. (C) MIB standard.
Detection of GPPMT and MIBS genes in additional cyanobacterial strains
MIBF2/MIBR2, GPPF2/GPPR2, PSANROS1F/PSANDDR1, and PSAN562F/PSAN1140R PCRs were used to screen Pseudanabaena limnetica (Lake Biwa) and Oscillatoria limosa for homologous MIB operon genes. At 50 °C, MIBF2/MIBR2 and PSAN562F/PSAN1140 PCRs yielded single specific PCR products of expected size for P. limnetica (Lake Biwa), while for O. limosa major bands of expected size were detected amongst some non-target amplicons. For both P. limnetica (Lake Biwa) and Oscillatoria limosa, GPPF2/GPPR2 and PSANROS1F/PSANDDR1 PCRs produced major bands of expected size, again amongst non-target PCR amplicons (data not shown). The generation of non-target amplicons for the majority of these PCRs was not unexpected. Firstly, the primers were designed specifically for P. limnetica (Castaic lake) based on genomic sequence data from the draft assembly; secondly, the placement of primers on target motif sequences (magnesium binding domains, and Rossmann fold domain) are in regions that do not allow the design of optimal PCR primers, thereby compromising the efficiency of the PCRs; and thirdly, these target motifs are also likely to be found in other DNA sequences encoding for methyltransferases and magnesium binding domains scattered throughout target organisms, which may result in aberrant amplification of non-target DNA sequences. The PCRs in this manuscript are not intended to be diagnostic for all MIB-producing organisms, and caution must therefore be exercised if using these specific primer combinations and PCR assays to screen for potential MIB-producing organisms.
DNA bands of the expected size from agarose gels were excised, purified, sequenced, and assembled. A Blastx homology search of translated sequence data from the predicted MIBS of O. limosa (359 aa, GenBank Accession XXXXX) revealed closest similarity to a MIBS found in ZP_05479642 of Streptomyces sp. AA4 (370 aa, 61% identity and 71% positive matches) at the amino acid sequence level. A Blastx of the predicted MIBS from Pseudanabaena limnetica (Lake Biwa, 379 aa, GenBank Accession XXXXX) yielded similar results, with similar identities and positive matches (see Table S5 for additional sequence information for closely related bacterial orthologs). A similar search of translated sequence data from the predicted GPPMT of O. limosa (257 aa, GenBank Accession XXXXX) revealed the highest similarity to GPPMT found in ZP_04609213 of Micromonospora sp. ATCC 39149 (289 aa, 74% identity and 84% positive matches) . A Blastx of the predicted GPPMT from Pseudanabaena limnetica (Lake Biwa, 257 aa, GenBank Accession XXXXX) also yielded similar results, with similar identities and positive matches (see Table S6 for additional sequence information for closely related bacterial orthologs).
The above similarity searches clearly identify the presence of both putative GPPMT and MIBS in O. limosa and P. limnetica (Lake Biwa), with a high amino acid conservation when compared to P. limnetica (Castaic Lake) and the clostes related bacterial orthologs (See Table S7 and S8). Moreover, the detection of specific PCR products spanning these genes, using PSAN562F/PSAN1140 and PSANROS1F/PSANDDR1 PCRs, demonstrate that the gene order is similar to P. limnetica (Castaic lake), Streptomyces sp. AA4, and C. acidiphila, in which the MIBS is immediately downstream of the GPPMT.
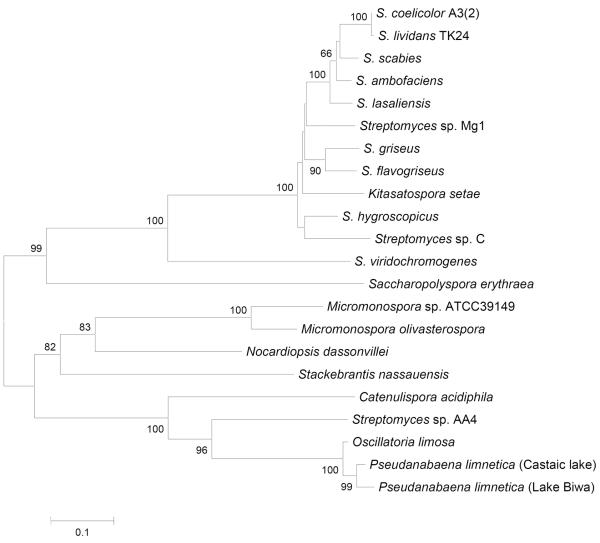
Upon closer scrutiny of the deduced MIBS amino acid sequence data, a number of differences within the conserved magnesium binding domains were observed. The upstream, aspartate-rich magnesium binding motif of monoterpene synthases (usually –xVDDxx[D/E]- or –xVDDxxx[D/E]-) appears to have 2 predominant signatures in MIB synthases; in several of the Streptomyces species, DDCYCED is present, whereas a DDYYAD variant is found in cyanobacteria (and the closely related proteins in Streptomyces sp. AA4 and C. acidiphila). Similarly, the “NSE” triad of residues (usually –xxNxxxSxxxE-) has a slight C-terminal acid variant in cyanobacteria and Streptomyces sp. AA4 of –xxNxxxSxxxD (Figure 4). It is interesting to note that the primary differences in both of these motifs is in the last of the conserved amino acids, where an aspartic acid residue (D) is used in preference to a glutamic acid residue (E) as found predominantly within the Streptomyces group. The organisms in which these variants occur also form a distinct cluster when examining the phylogeny of MIB synthases (Figure 5), where those organisms with variants in these key motifs also have possible recombination events altering the MIB operon gene order. Considering the sequence similarity amongst the group with the variant motifs, it is likely that the change in gene order predates the acquisition of the operon by cyanobacteria. The gene order and the tree branching order in this group suggests that multiple rearrangements have occurred; initially the placement of GPPMT between MIBS and the nucleotide binding protein (as in C. acidiphila), followed by placement of the nucleotide binding protein downstream of MIBS (as in AA4 and P. limnetica, Castaic lake). The role of the conserved nucleotide binding protein as a possible regulatory element in MIB biosynthesis has not yet been examined in any system.
Figure 4.
Alignment of amino acid sequences of MIB synthases with predicted MIB synthases. Shadow boxes indicate metal (Mg2+)-binding motif of terpene synthase.
Figure 5.
Phylogenetic analysis of MIB synthases from bacterial databases. Phylogenetic analysis of aligned sequences was performed using the bootstrap method (bootstrap number; 1,000, seed number; 111)
The significance of the reversal in gene order for the 3-gene MIB operon between cyanobacteria and the majority of other MIB-producing organisms is unclear. There are some interesting peculiarities in S. scabies 87.22 and the gram negative bacterium Pseudomonas fluorescens, where putative MIBS (SCAB82161 and Rcas_0622 respectively) are found in the absence of accompanying GPPMTs. In S. avermitilis MA-4680, there is also an apparent truncation of a putative MIBS (1,245,680 to 1,246,412 nt of the S. avermitilis genome, gene not annotated), where the second “NSE” motif is absent. In S. avermitilis MA-4680, however, the truncated MIBS is flanked downstream by a putative GPPMT, and the loss of the second magnesium binding domain in the truncated MIBS presumably does not allow for the full conversion of 2-methyl-GPP to MIB (15).
The identification, expression, and biochemical characterization of MIBS and GPPMT orthologs in P. limnetica (Castaic lake) confirm the assignment of isoprenoid-derived MIB in cyanobacteria. This information will provide an opportunity to further explore the occurrence of the MIB operon and its gene organization in other cyanobacteria. The resultant data may facilitate the development of a PCR screening assay for MIB-producing microorganisms in waters. As already demonstrated for the detection of geosmin synthases in cyanobacteria, direct detection of MIB operon genes may similarly provide an early detection tool for water utilities, and enable further research to investigate the production of this odiferous compound within cyanobacteria and associated water storages.
Supplementary Material
ACKNOWLEDGMENTS
We thank the South Australian Water Corporation, United Water International and the Water Research Foundation (WRF) for financial, technical, and administrative assistance in funding the project through which this information was discovered. Mention of trade names or commercial products does not constitute WRF endorsement or recommendations for use. Similarly, omission of products or trade names indicates nothing concerning WRF's position regarding product effectiveness or applicability. The comments and views detailed herein may not necessarily reflect the views of the WRF, its officers, directors, affiliates or agents. Bioinformatics advice from Jun Ishikawa, National Institute of Infectious Diseases, Tokyo, Japan, is gratefully acknowledged. D.E.C was supported by a grant from the US National Institutes of Health, GM30301. SG thanks Sue Watson and George Izaguirre for support and belief in this area of research.
Footnotes
Supporting Information available. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- (1).Izaguirre G, Hwang CJ, Krasner SW, McGuire MJ. Production of 2-methlyisoborneol by two benthic cyanophyta. Wat Sci Tech. 1983;15:211–220. [Google Scholar]
- (2).Izaguirre G, Taylor WD. A guide to geosmin- and MIB-producing cyanobacteria in the United States. Water Sci Technol. 2004;49:19–24. [PubMed] [Google Scholar]
- (3).Cook D, Newcombe G, Sztajnbok P. The application of powdered activated carbon for MIB and geosmin removal: predicting PAC doses in four raw waters. Water Res. 2001;35:1325–1333. doi: 10.1016/s0043-1354(00)00363-8. [DOI] [PubMed] [Google Scholar]
- (4).Bruce D, Westerhoff P, Brawley-Chesworth A. Removal of 2-methylisoborneol and geosmin in surface water treatment plants in Arizona. Journal of Water Supply Research and Technology-Aqua. 2002;51:183–197. [Google Scholar]
- (5).Juttner F, Watson SB. Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Appl Environ Microbiol. 2007;73:4395–4406. doi: 10.1128/AEM.02250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cane DE, He X, Kobayashi S, Omura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J Antibiot (Tokyo) 2006;59:471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- (7).Cane DE, Watt RM. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc Natl Acad Sci U S A. 2003;100:1547–1551. doi: 10.1073/pnas.0337625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Giglio S, Jiang J, Saint CP, Cane DE, Monis PT. Isolation and characterization of the gene associated with geosmin production in cyanobacteria. Environ Sci Technol. 2008;42:8027–8032. doi: 10.1021/es801465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jiang J, Cane DE. Geosmin biosynthesis. Mechanism of the fragmentation-rearrangement in the conversion of germacradienol to geosmin. J Am Chem Soc. 2008;130:428–429. doi: 10.1021/ja077792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jiang J, He X, Cane DE. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J Am Chem Soc. 2006;128:8128–8129. doi: 10.1021/ja062669x. [DOI] [PubMed] [Google Scholar]
- (12).Jiang J, He X, Cane DE. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat Chem Biol. 2007;3:711–715. doi: 10.1038/nchembio.2007.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dickschat JS, Nawrath T, Thiel V, Kunze B, Muller R, Schulz S. Biosynthesis of the off-flavor 2-methylisoborneol by the myxobacterium Nannocystis exedens. Angew Chem Int Ed Engl. 2007;46:8287–8290. doi: 10.1002/anie.200702496. [DOI] [PubMed] [Google Scholar]
- (14).Wang CM, Cane DE. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor. J Am Chem Soc. 2008;130:8908–8909. doi: 10.1021/ja803639g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci U S A. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm. 2008;5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- (17).Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bentley R, Meganathan R. Geosmin and methylisoborneol biosynthesis in streptomycetes. Evidence for an isoprenoid pathway and its absence in non-differentiating isolates. FEBS Lett. 1981;125:220–222. doi: 10.1016/0014-5793(81)80723-5. [DOI] [PubMed] [Google Scholar]
- (19).Izaguirre G, Taylor WD. A Pseudanabaena species from Castaic Lake, California, that produces 2-methylisoborneol. Water Research. 1998;32:1673–1677. [Google Scholar]
- (20).Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual. Second ed. Cold Spring Harbor Laboratory Press; NY: 1989. [Google Scholar]
- (21).Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- (23).Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang CM, Cane DE. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor. J Am Chem Soc. 2010;132:9509. doi: 10.1021/ja803639g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.