Abstract
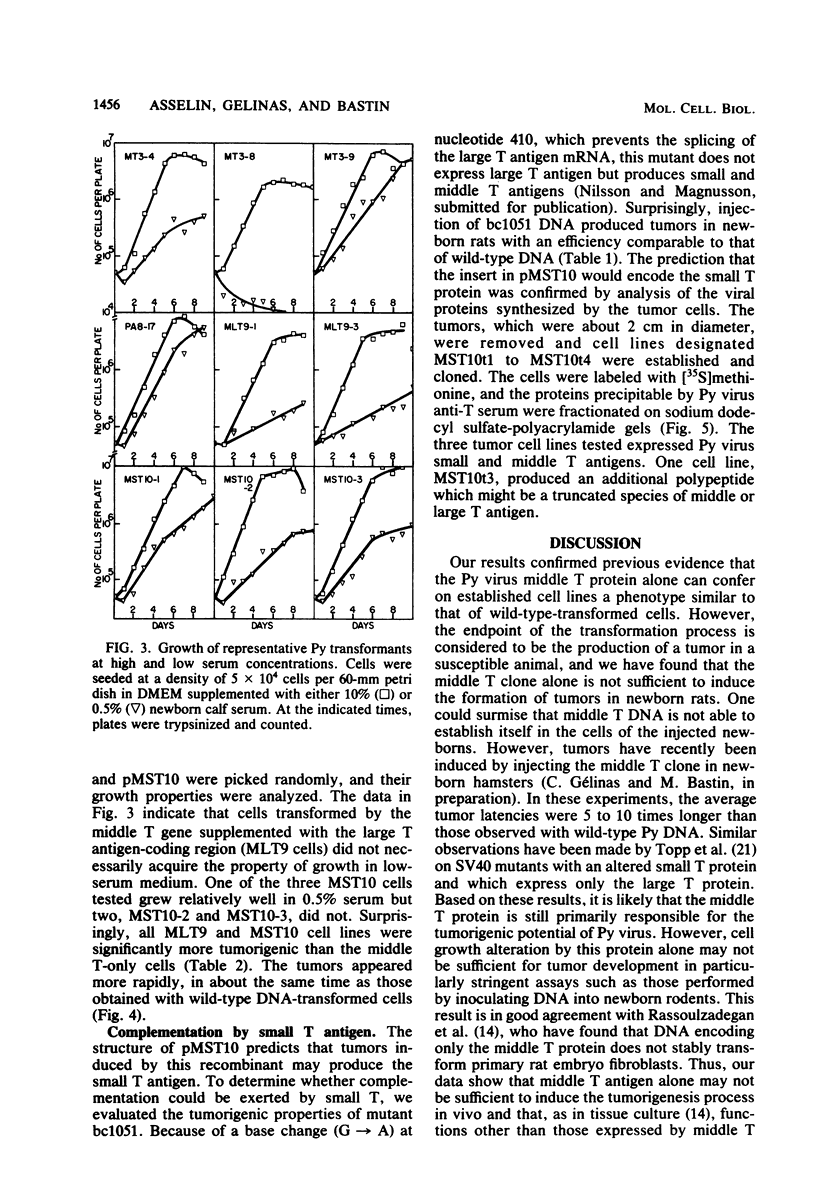
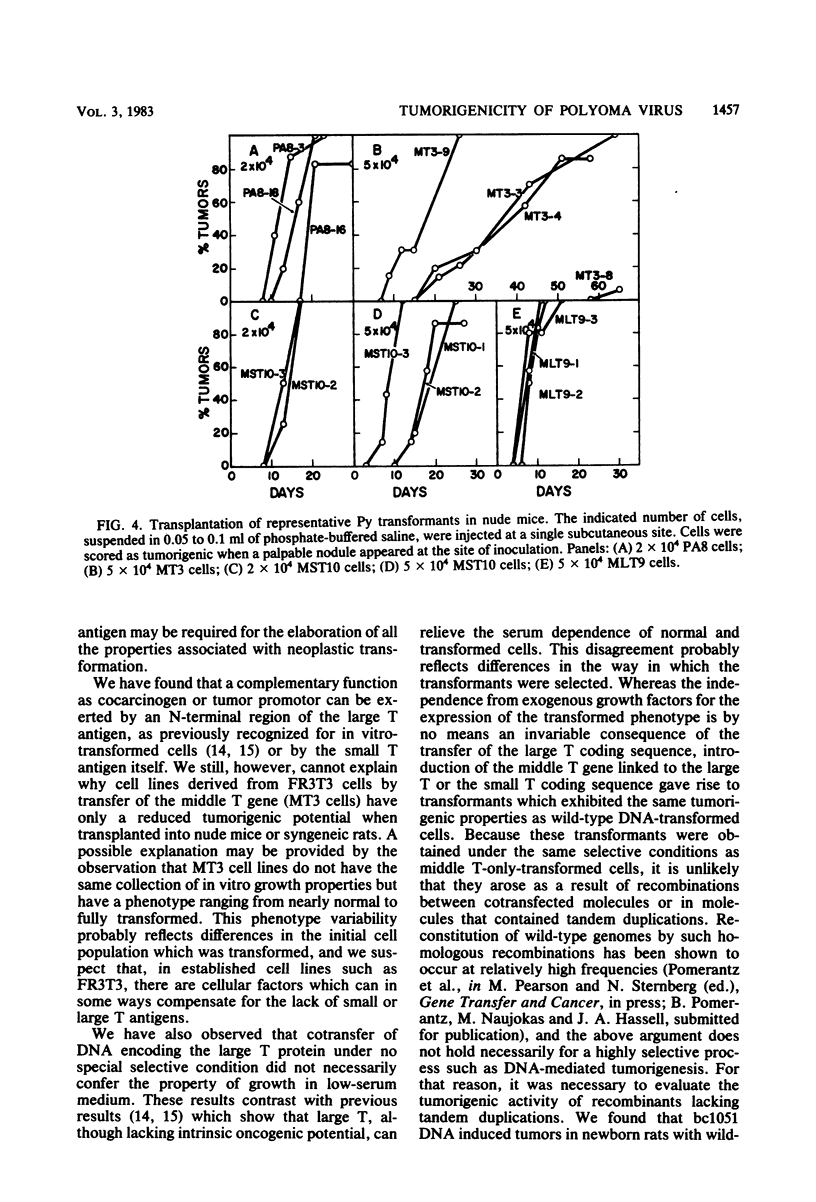
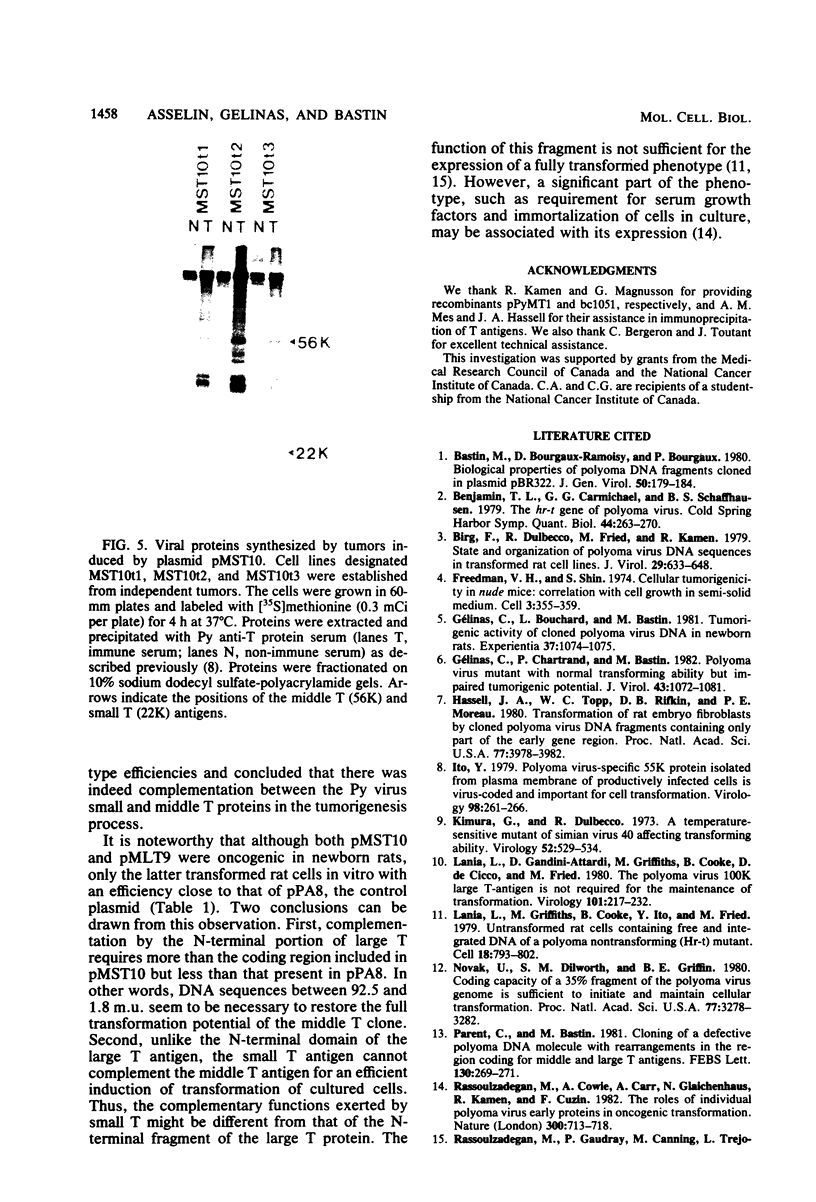
A modified polyoma virus genome which can encode the middle T protein but not the large or small T proteins transforms rat cells in culture with an efficiency about 20% that of the wild-type genome. Although middle T-transformed cells grow as tumors when transplanted into nude mice or syngeneic rats, the middle T gene alone is totally inactive when used in a more stringent and rigorous assay for tumorigenicity such as the injection of DNA into newborn rats. Thus, functions other than those expressed by middle T antigen are required for the elaboration of all the properties associated with tumorigenesis. To assess whether a complementary function could be exerted by the large or the small T antigen, we constructed plasmids containing two modified early regions which independently encoded middle T and one of the two other proteins. Both recombinants were tumorigenic in newborn rats. Cell lines derived by transfer of these plasmids under no special selective conditions did not acquire the property of growth in low-serum medium but exhibited the same tumorigenic properties as wild-type polyoma DNA-transformed cells. Furthermore, a recombinant which encoded the middle and small T antigens, but not the large T antigen, was tumorigenic in newborn rats. Although the small T antigen provides a complementary function for tumorigenicity, it cannot complement the middle T antigen for an efficient induction of transformation of cultured cells. This suggests that the complementary function exerted by the small T antigen is different from that of the N-terminal fragment of the large T protein.
Full text
PDF

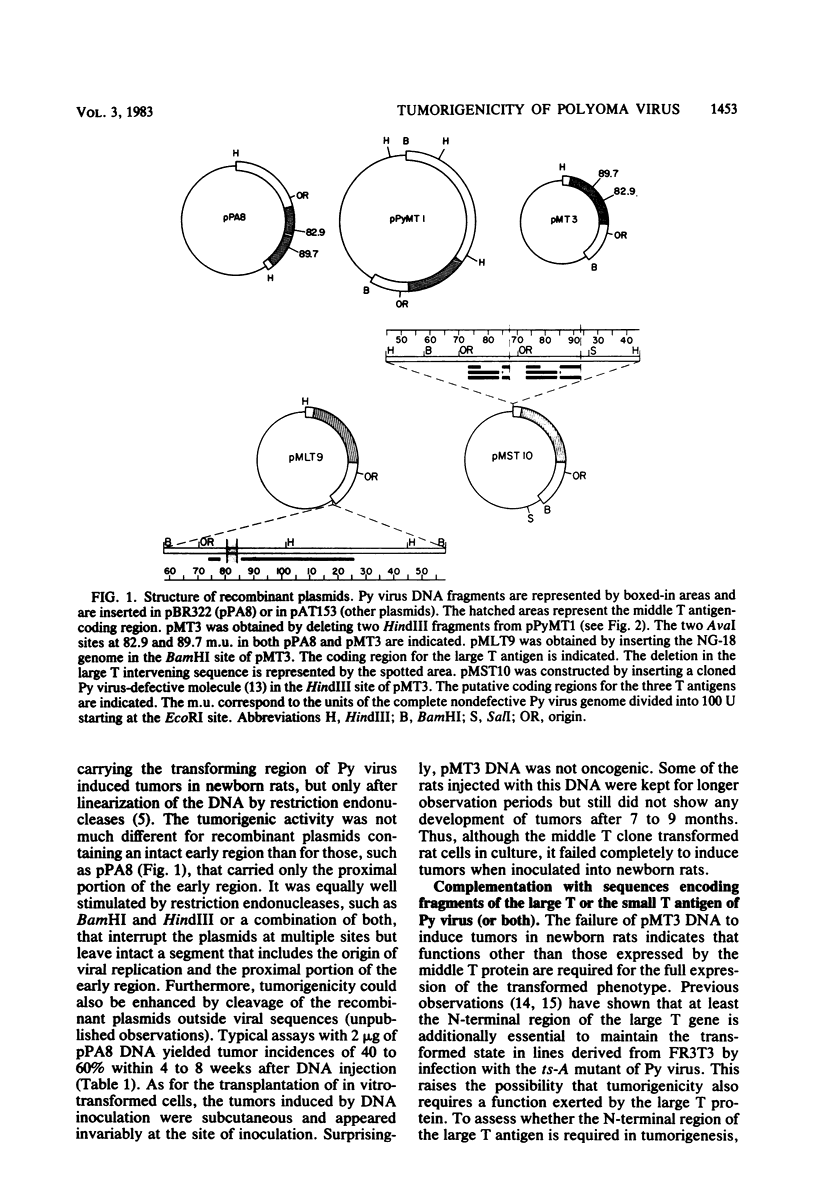
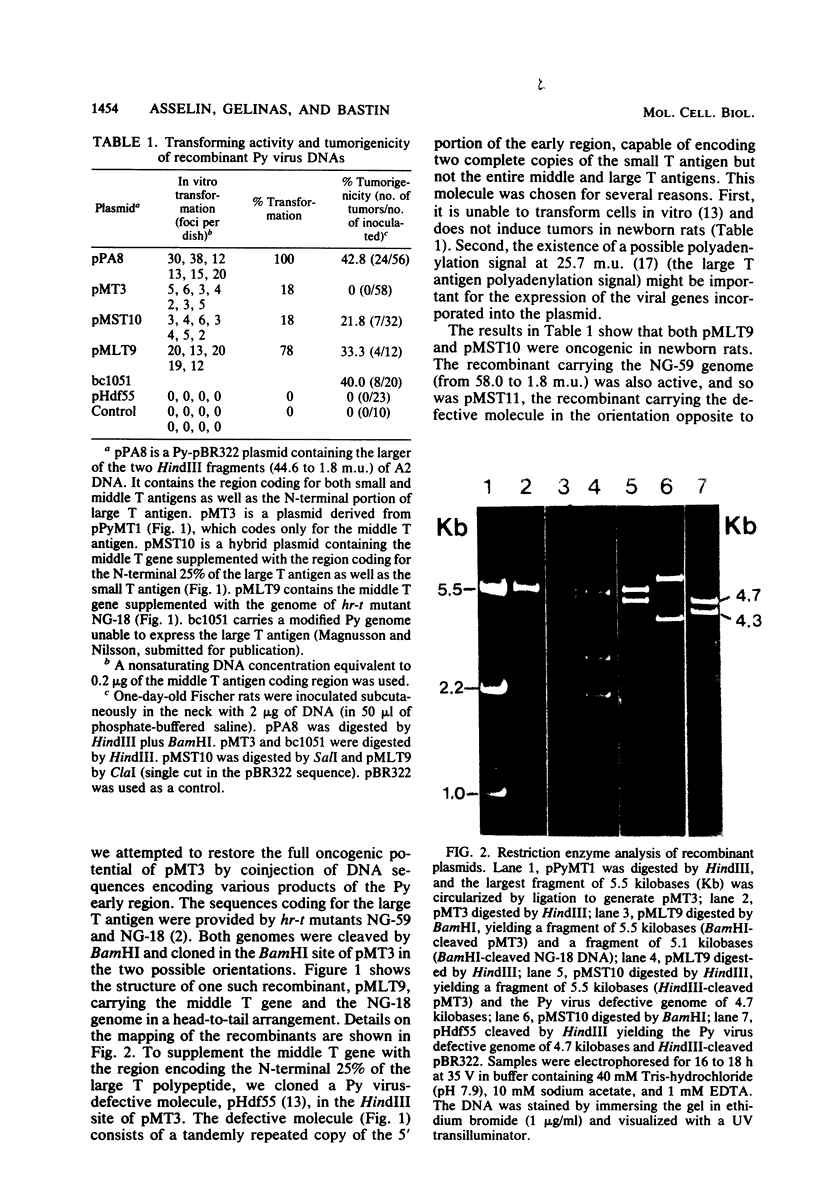


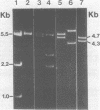
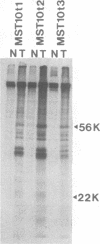
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bastin M., Bourgaux-Ramoisy D., Bourgaux P. Biological properties of polyoma DNA fragments cloned in plasmid pBR322. J Gen Virol. 1980 Sep;50(1):179–184. doi: 10.1099/0022-1317-50-1-179. [DOI] [PubMed] [Google Scholar]
- Benjamin T. L., Carmichael G. G., Schaffhausen B. S. The hr-t gene of polyoma virus. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):263–270. doi: 10.1101/sqb.1980.044.01.030. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman V. H., Shin S. I. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. 1974 Dec;3(4):355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- Gelinas C., Chartrand P., Bastin M. Polyoma virus mutant with normal transforming ability but impaired tumorigenic potential. J Virol. 1982 Sep;43(3):1072–1081. doi: 10.1128/jvi.43.3.1072-1081.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gélinas C., Bouchard L., Bastin M. Tumorigenic activity of cloned polyoma virus DNA in newborn rats. Experientia. 1981 Oct 15;37(10):1074–1075. doi: 10.1007/BF02085017. [DOI] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Kimura G., Dulbecco R. A temperature-sensitive mutant of simian virus 40 affecting transforming ability. Virology. 1973 Apr;52(2):529–534. doi: 10.1016/0042-6822(73)90348-6. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lania L., Griffiths M., Cooke B., Ito Y., Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (Hr-t) mutant. Cell. 1979 Nov;18(3):793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C., Bastin M. Cloning of a defective polyoma DNA molecule with rearrangements in the region coding for middle and large T antigens. FEBS Lett. 1981 Aug 3;130(2):269–271. doi: 10.1016/0014-5793(81)81136-2. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M., Cowie A., Carr A., Glaichenhaus N., Kamen R., Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982 Dec 23;300(5894):713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M., Gaudray P., Canning M., Trejo-Avila L., Cuzin F. Two polyoma virus gene functions involved in the expression of the transformed phenotype in FR 3T3 rat cells. I. Localization of a transformation maintenance function in the proximal half of the large T coding region. Virology. 1981 Oct 30;114(2):489–500. doi: 10.1016/0042-6822(81)90228-2. [DOI] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Stow N. D., Wilkie N. M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976 Dec;33(3):447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972 Oct;10(4):591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp W. C., Rifkin D. B., Sleigh M. J. SV40 mutants with an altered small-t protein are tumorigenic in newborn hamsters. Virology. 1981 Jun;111(2):341–350. doi: 10.1016/0042-6822(81)90338-x. [DOI] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]